Abstract
The aim of this study was to investigate whether par-3 partitioning defective 3 homolog (C. elegans) (PARD3) single nucleotide polymorphisms (SNPs) are associated with schizophrenia. A total of 204 Korean schizophrenic patients [117 male, 41.1±9.6 years (mean age ± SD); 87 female, 42.6±11.5] and 351 control subjects (170 male, 43.8±6.6 years; 181 female, 44.2±5.8) were enrolled. We genotyped nine SNPs of the PARD3 gene [rs7075263 (intron), rs10827392 (intron), rs773970 (intron), rs2252655 (intron), rs10763984 (intron), rs3781128 (Ser889Ser), rs1936429 (intron), rs671228 (intron) and rs16935163 (intron)]. Genotypes of PARD3 polymorphisms were evaluated by direct sequencing. We used SNPStats, SPSS 18.0 and Haploview 4.2 software for analysis of genetic data. Multiple logistic regression models were used to calculate the odds ratio (OR), 95% confidence interval (CI), and corresponding p-values (p), controlling for age and gender as covariables. Allele frequencies of the PARD3 SNPs were significantly associated with schizophrenia (rs3781128, p=0.041; rs1936429, p=0.030; rs671228, p=0.028). Certain genotype frequencies of the PARD3 SNPs also showed significant associations with schizophrenia (p<0.05, rs7075263, rs773970, rs2252655, rs10763984, rs3781128, rs1936429, rs16935163). To the best of our knowledge, this is the first report showing that PARD3 is associated with susceptibility to schizophrenia in a Korean population. In conclusion, our findings suggest that PARD3 may contribute to genetic susceptibility to schizophrenia.
Keywords: PARD3, single nucleotide polymorphism, schizophrenia, association study
Introduction
Schizophrenia is a severe, debilitating, psychiatric disorder. Although the exact etiology of schizophrenia is unknown, twin, family and adoption studies have provided consistent evidence that genetic factors play a major role in the pathogenesis of schizophrenia.
In a recent study, the data showed that polymorphisms of several genes affect gene expression or the function of the encoded protein in the human brain (1). Clinical, epidemiological, neuroimaging and postmortem data suggest that schizophrenia is a neurodevelopmental disorder, and that synaptic disturbances may play a critical role in the development of the disease (2). In addition, it is hypothesized that a loss of function of glial gap junctions may cause severe cognitive impairment in schizophrenia. Thus, several gap junction proteins may be capable of differentiating between the operation qualities of the cognate synapses defined by the neurotransmitter types (3). The major component for barrier-forming tight junctions may play a crucial role in response to the changing natural, physiological and pathological conditions of schizophrenia (4).
In this study, we investigated whether par-3 partitioning defective 3 homolog (C. elegans) (PARD3) is associated with schizophrenia in a Korean population.
Subjects and methods
Subjects
All subjects were ethnically Korean and unrelated to each other. A total of 204 Korean schizophrenic patients [117 male, 41.1±9.6 years (mean age ± SD); 87 female, 42.6±11.5] and 351 control subjects (170 male, 43.8±6.6 years; 181 female, 44.2±5.8) were enrolled in the study. All patients were diagnosed with schizophrenia by three psychiatrists according to the Diagnostic and Statistical Manual of Mental Disorders (4th edition) using all available information from interviews and clinical records. All patients were assessed using the Operational Criteria Checklist for Psychotic Illness (OPCRIT). The OPCRIT was designed as a best estimate diagnostic procedure, in which symptom and course features are coded by an experienced clinician, allowing integration of the relative prominence of clinical features over the entire course of the illness. The control subjects were recruited after they had been assessed as mentally healthy in a general health examination program. Written informed consent was obtained from all subjects involved in the study, and the study was approved by the Ethics Review Committee of the Medical Research Institute, Kyung Hee University Medical Center, Seoul, Republic of Korea. All studies were carried out according to the guidelines of the Declaration of Helsinki.
SNP selection and genotyping
We searched the SNPs of the PARD3 gene in the SNP database of the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/ SNP, BUILD 132). We selected nine SNPs [rs7075263 (intron), rs10827392 (intron), rs773970 (intron), rs2252655 (intron), rs10763984 (intron), rs3781128 (Ser889Ser), rs1936429 (intron), rs671228 (intron) and rs16935163 (intron)] in the PARD3 gene. SNP genotyping was conducted using direct sequencing. For direct sequencing, genomic DNA was amplified using polymerase chain reaction (PCR). The PCR products were sequenced by an ABI PRISM 3730XL analyzer (PE Applied Biosystems, Foster City, CA, USA), and sequence data were analyzed using SeqManII software (DNASTAR Inc., Madison, WI, USA).
Statistical analysis
The Chi-square test was used to assess Hardy-Weinberg equilibrium (HWE). Genetic data were analyzed using SNPStats (http://bioinfo.iconcologia.net/index.php?module=Snpstats), Helixtree (Golden Helix Inc., Bozeman, MT, USA) and SPSS 18.0 (SPSS Inc., Chicago, IL, USA). The analysis of SNPs was performed using multiple logistic models (codominant 1, codominant 2, dominant, recessive and log-additive models). The Haploview version 4.2 (5) was used for linkage disequilibrium (LD) and haplo-type analysis between the nine SNPs.
Results
We selected nine SNPs of PARD3. The position of the PARD3 SNPs is illustrated in Fig. 1A. Table I documents the allele frequencies of the nine examined SNPs in schizophrenia patients and control subjects. The HWEs of nine SNPs revealed no deviation in the control groups, respectively (p>0.05, data not shown). Multiple logistic regression analysis with adjustment for age and gender was performed. Three SNPs were associated with schizophrenia (rs3781128, OR=0.76, 95% CI=0.59–0.99, p=0.041; rs1936429, OR=0.75, 95% CI=0.58–0.97, p=0.030; rs16935163, OR=1.69, 95% CI=1.06–2.68, p=0.028, respectively, Table I). These results suggest that the C allele of rs3781128, the A allele of rs1936429 and the C allele of rs16935163 may be risk factors for schizophrenia.
Figure 1.
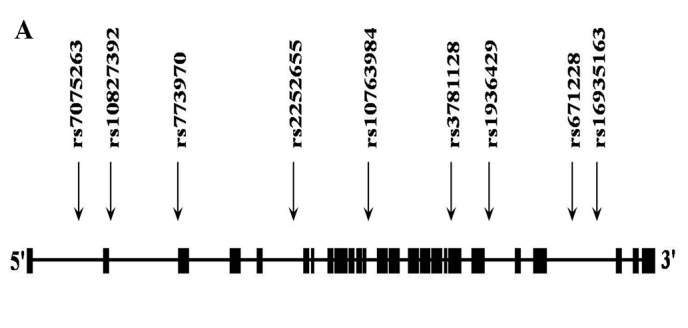
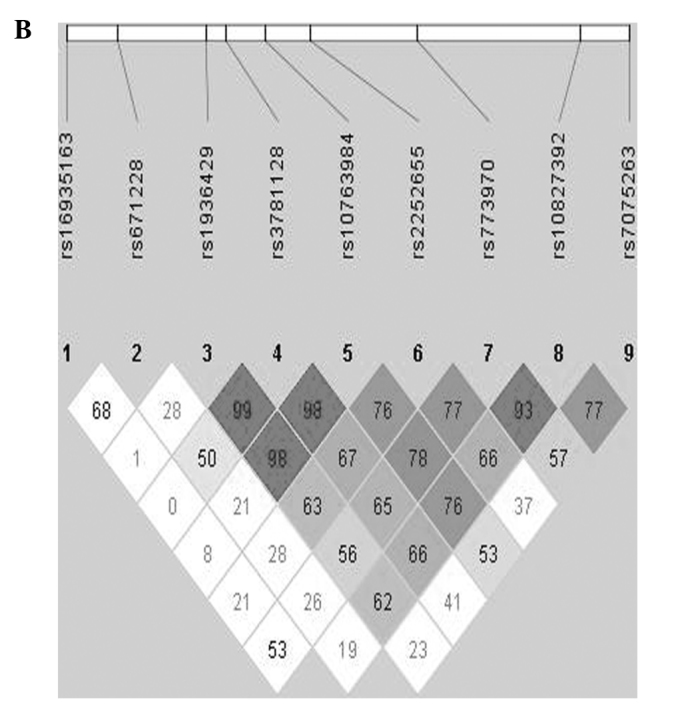
Gene map and linkage disequilibrium (LD) in the PARD3 gene. (A) Gene map and single nucleotide polymorphisms (SNPs) in the PARD3 gene on chromosome 10. Exons are marked with a box. The coding regions are black boxes. Arrow indicates the location of each SNP. (B) LD blocks among PARD3 SNPs.
Table I.
Allele frequencies of polymorphisms of the PARD3 gene in schizophrenia patients and control subjects.
| Control
|
Schizophrenia
|
||||||
|---|---|---|---|---|---|---|---|
| SNP | Allele | n | % | n | % | OR (95% CI) | P-value |
| rs7075263 | T | 623 | 88.7 | 354 | 86.8 | 1 | |
| Intron | C | 79 | 11.3 | 54 | 13.2 | 1.20 (0.83–1.74) | 0.33 |
| rs10827392 | T | 631 | 89.9 | 358 | 87.7 | 1 | |
| Intron | C | 71 | 10.1 | 50 | 12.3 | 1.24 (0.85–1.82) | 0.27 |
| rs773970 | G | 385 | 57.3 | 227 | 59.1 | 1 | |
| Intron | A | 287 | 42.7 | 157 | 40.9 | 0.93 (0.72–1.20) | 0.56 |
| rs2252655 | A | 343 | 50.7 | 216 | 53.5 | 1 | |
| Intron | G | 333 | 49.3 | 188 | 46.5 | 0.90 (0.70–1.15) | 0.38 |
| rs10763984 | C | 348 | 49.6 | 221 | 54.4 | 1 | |
| Intron | T | 354 | 50.4 | 185 | 45.6 | 0.82 (0.64–1.05) | 0.12 |
| rs3781128 | C | 435 | 62.5 | 272 | 68.7 | 1 | |
| Ser889Ser | T | 261 | 37.5 | 124 | 31.3 | 0.76 (0.59–0.99) | 0.041 |
| rs1936429 | A | 394 | 57.9 | 256 | 64.6 | 1 | |
| Intron | G | 286 | 42.1 | 140 | 35.4 | 0.75 (0.58–0.97) | 0.030 |
| rs671228 | C | 565 | 80.5 | 330 | 81.7 | 1 | |
| Intron | T | 137 | 19.5 | 74 | 18.3 | 0.93 (0.68–1.27) | 0.63 |
| rs16935163 | C | 658 | 94.3 | 361 | 90.7 | 1 | |
| Intron | A | 40 | 5.7 | 37 | 9.3 | 1.69 (1.06–2.68) | 0.028 |
P-values were calculated from logistic regression analyses. Bold numbers indicate significant associations. SNP, singe nucleotide polymorphism; OR, odds ratio; CI, confidence interval; n, number of subjects.
Table II shows genotype frequencies of the nine examined SNPs in schizophrenia patients and control subjects. The seven SNPs among the PARD3 polymorphisms showed significant associations with schizophrenia [rs7075263, OR=6.00, 95% CI=1.21–26.98, p=0.028 in codominant 2 (T/T versus T/T), OR=6.06, 95% CI=1.23–29.87, p=0.014 in recessive (T/T+C/T versus C/C); rs773970, OR=0.52, 95% CI=0.30–0.89, p=0.014 in recessive (G/G+A/G versus A/A); rs2252655, OR=0.58, 95% CI=0.37–0.91, p=0.016 in recessive (A/A+A/G versus G/G); rs10763984, OR=0.53, 95% CI=0.34–0.84, p=0.005 in recessive (C/C+C/T versus T/T); rs3781128, OR=0.65, 95% CI=0.45–0.95, p=0.026 in codominant 1 (C/C versus C/T), OR=0.66, 95% CI=0.46–0.94, p=0.021 in dominant (C/C versus C/T+T/T); rs1936429, OR=0.57, 95% CI=0.32–0.99, p=0.047 in codominant 2 (A/A versus G/G), OR=0.69, 95% CI=0.47–0.99, p=0.045 in dominant (A/A versus A/G+G/G), OR=0.75, 95% CI=0.57–0.97, p=0.028 in log-additive (A/A versus A/G versus G/G); and rs16935163, OR=1.68, 95% CI=1.02–2.76, p=0.044 in dominant (C/C versus A/C+A/A), OR=1.74, 95% CI=1.08–2.81, p=0.024 in log-additive (C/C versus A/C versus A/A) models, respectively]. The rs10827392 and rs671228 SNPs had no differences between the schizophrenia and control groups.
Table II.
Genotype frequencies of polymorphisms of the PARD3 gene in schizophrenia patients and control subjects.
| Control
|
Schizophrenia
|
|||||||
|---|---|---|---|---|---|---|---|---|
| SNP | Genotype | n | % | n | % | Model | OR (95% CI) | P-value |
| rs7075263 | T/T | 274 | 78.1 | 156 | 76.8 | Codominant 1 | 0.96 (0.62–1.48) | 0.85 |
| intron | C/T | 75 | 21.4 | 40 | 19.7 | Codominant 2 | 6.00 (1.21–29.68) | 0.028 |
| C/C | 2 | 0.6 | 7 | 3.5 | Dominant | 1.09 (0.72–1.66) | 0.68 | |
| Recessive | 6.06 (1.23–29.87) | 0.014 | ||||||
| Log-additive | 1.22 (0.84–1.77) | 0.30 | ||||||
| rs10827392 | T/T | 281 | 80.1 | 157 | 77.3 | Codominant 1 | 1.11 (0.72–1.72) | 0.63 |
| intron | C/T | 69 | 19.7 | 42 | 20.7 | Codominant 2 | 7.50 (0.82–68.89) | 0.08 |
| C/C | 1 | 0.3 | 4 | 2.0 | Dominant | 1.20 (0.78–1.84) | 0.40 | |
| Recessive | 7.32 (0.80–67.16) | 0.044 | ||||||
| Log-additive | 1.28 (0.86–1.91) | 0.22 | ||||||
| rs773970 | G/G | 108 | 32.1 | 55 | 28.8 | Codominant 1 | 1.36 (0.91–2.04) | 0.14 |
| intron | A/G | 169 | 50.3 | 116 | 60.7 | Codominant 2 | 0.63 (0.34–1.16) | 0.14 |
| A/A | 59 | 17.6 | 20 | 10.5 | Dominant | 1.16 (0.79–1.72) | 0.44 | |
| Recessive | 0.52 (0.30–0.89) | 0.014 | ||||||
| Log-additive | 0.90 (0.68–1.18) | 0.44 | ||||||
| rs2252655 | A/A | 88 | 26.0 | 47 | 23.4 | Codominant 1 | 1.34 (0.87–2.07) | 0.18 |
| intron | A/G | 167 | 49.4 | 121 | 60.2 | Codominant 2 | 0.71 (0.41–1.22) | 0.22 |
| G/G | 83 | 24.6 | 33 | 16.4 | Dominant | 1.13 (0.75–1.71) | 0.56 | |
| Recessive | 0.58 (0.37–0.91) | 0.016 | ||||||
| Log-additive | 0.87 (0.67–1.13) | 0.29 | ||||||
| rs10763984 | C/C | 87 | 24.8 | 49 | 24.3 | Codominant 1 | 1.21 (0.79–1.86) | 0.37 |
| intron | C/T | 174 | 49.6 | 122 | 60.4 | Codominant 2 | 0.61 (0.35–1.04) | 0.07 |
| T/T | 90 | 25.6 | 31 | 15.3 | Dominant | 1.01 (0.67–1.52) | 0.97 | |
| Recessive | 0.53 (0.34–0.84) | 0.005 | ||||||
| Log-additive | 0.80 (0.62–1.04) | 0.10 | ||||||
| rs3781128 | C/C | 130 | 37.4 | 94 | 47.7 | Codominant 1 | 0.65 (0.45–0.95) | 0.026 |
| Ser889Ser | C/T | 175 | 50.3 | 82 | 41.6 | Codominant 2 | 0.68 (0.37–1.22) | 0.19 |
| T/T | 43 | 12.4 | 21 | 10.7 | Dominant | 0.66 (0.46–0.94) | 0.021 | |
| Recessive | 0.84 (0.48–1.48) | 0.55 | ||||||
| Log-additive | 0.76 (0.58–1.00) | 0.045 | ||||||
| rs1936429 | A/A | 113 | 33.2 | 81 | 41.1 | Codominant 1 | 0.73 (0.49–1.07) | 0.11 |
| intron | A/G | 168 | 49.4 | 92 | 46.7 | Codominant 2 | 0.57 (0.32–0.99) | 0.047 |
| G/G | 59 | 17.4 | 24 | 12.2 | Dominant | 0.69 (0.47–0.99) | 0.045 | |
| Recessive | 0.68 (0.41–1.14) | 0.14 | ||||||
| Log-additive | 0.75 (0.57–0.97) | 0.028 | ||||||
| rs671228 | C/C | 225 | 64.1 | 135 | 67.2 | Codominant 1 | 0.83 (0.56–1.22) | 0.35 |
| intron | C/T | 115 | 32.8 | 58 | 28.9 | Codominant 2 | 1.17 (0.45–3.00) | 0.75 |
| T/T | 11 | 3.1 | 8 | 4.0 | Dominant | 0.86 (0.59–1.25) | 0.43 | |
| Recessive | 1.24 (0.48–3.16) | 0.66 | ||||||
| Log-additive | 0.92 (0.67–1.26) | 0.59 | ||||||
| rs16935163 | C/C | 309 | 88.5 | 163 | 82.3 | Codominant 1 | 1.58 (0.95–2.62) | 0.08 |
| intron | A/C | 40 | 11.5 | 33 | 16.7 | Codominant 2 | NA (0.00-NA) | NA |
| A/A | 0 | 0.0 | 2 | 1.0 | Dominant | 1.68 (1.02–2.76) | 0.044 | |
| Recessive | NA (0.00-NA) | 0.040 | ||||||
| Log-additive | 1.74 (1.08–2.81) | 0.024 | ||||||
P-values were calculated from logistic regression analyses. Bold numbers indicated significant associations. SNP, singe nucleotide polymorphism; OR, odds ratio; CI, confidence interval; n, number of subjects; NA, not applicable.
The LD block was estimated using Haploview version 4.2. The LD block did not be made among SNPs of PARD3 (Fig. 1B, data not shown).
Discussion
In this study, we found that PARD3 gene polymorphisms were associated with schizophrenia. The allele distributions of three SNPs (rs3781128, rs1936429 and rs16935163) and genotype frequencies of seven SNPs (rs7075263, rs773970, rs2252655, rs10763984, rs3781128, rs1936429 and rs16935163) were significantly different between schizophrenia patients and control subjects.
PAR-3 is a scaffold-like PDZ-containing protein. PAR-3 forms a complex with PAR-6 and atypical protein kinase C (PAR-3-atypical protein kinase C-PAR-6 complex) and is associated with the establishment of cell polarization (6,7). Cell polarity has an important role in various cellular processes such as cell migration, asymmetric cell division, and axon and dendrite specification.
Chan et al reported that PAR-3 is directly associated and recruits the p75 neurotrophin receptor to the axon glial junction, forming a complex necessary for myelination (8). It was found that depletion of PAR-3 in mammalian epithelial cells profoundly disrupts tight junction assembly (9). Synapses of the junctions between nerve cells, through which the cells communicate, are formed by the coordinated assembly and tight attachment of presynaptic and postsynaptic specializations. Therefore, tight junction protein-related studies suggest that junctional protein polymorphisms are sufficient for a functional postsynaptic response change (10–12).
Tight junctions play a major role in cell polarity in vertebrate endothelial and epithelial cells. A protein complex consisting of the cell polarity proteins PAR-3, PAR-6 and the atypical protein kinase C localizes at tight junctions. It is crucial for tight junction formation. It has been shown that PAR-3 associates with the junctional adhesion molecule (JAM). This suggests that the ternary complex is targeted to tight junctions of epithelial cells through PAR-3 binding to JAM (13).
In conclusion, this is the first report showing that PARD3 is associated with the susceptibility to schizophrenia in a Korean population. Our results suggest that PARD3 may contribute to a genetic susceptibility to schizophrenia.
Acknowledgments
This research was supported by a grant from Kyung Hee University (KHU-20090641).
References
- 1.Kleinman JE, Law AJ, Lipska BK, et al. Genetic neuropathology of schizophrenia: new approaches to an old question and new uses for postmortem human brains. Biol Psychiatry. 2010;69:140–145. doi: 10.1016/j.biopsych.2010.10.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Faludi G, Mirnics K. Synaptic changes in the brain of subjects with schizophrenia. Int J Dev Neurosci. 2011;29:305–309. doi: 10.1016/j.ijdevneu.2011.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mitterauer B. Loss of function of glial gap junctions may cause severe cognitive impairments in schizophrenia. Med Hypotheses. 2009;73:393–397. doi: 10.1016/j.mehy.2009.04.003. [DOI] [PubMed] [Google Scholar]
- 4.Sun ZY, Wei J, Xie L, et al. The CLDN5 locus may be involved in the vulnerability to schizophrenia. Eur Psychiatry. 2004;19:354–357. doi: 10.1016/j.eurpsy.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 5.Barrett JC, Fry B, Maller J, et al. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- 6.Mizuno K, Suzuki A, Hirose T, et al. Self-association of PAR-3-mediated by the conserved N-terminal domain contributes to the development of epithelial tight junctions. J Biol Chem. 2003;278:31240–31250. doi: 10.1074/jbc.M303593200. [DOI] [PubMed] [Google Scholar]
- 7.Zen K, Yasui K, Gen Y, et al. Defective expression of polarity protein PAR-3 gene (PARD3) in esophageal squamous cell carcinoma. Oncogene. 2009;28:2910–2918. doi: 10.1038/onc.2009.148. [DOI] [PubMed] [Google Scholar]
- 8.Chan JR, Jolicoeur C, Yamauchi J, et al. The polarity protein Par-3 directly interacts with p75NTR to regulate myelination. Science. 2006;314:832–836. doi: 10.1126/science.1134069. [DOI] [PubMed] [Google Scholar]
- 9.Chen X, Macara IG. Par-3 controls tight junction assembly through the Rac exchange factor Tiam1. Nat Cell Biol. 2005;7:262–269. doi: 10.1038/ncb1226. [DOI] [PubMed] [Google Scholar]
- 10.Willott E, Balda MS, Fanning AS, et al. The tight junction protein ZO-1 is homologous to the Drosophila discs – large tumor suppressor protein of septate junctions. Proc Natl Acad Sci USA. 1993;90:7834–7838. doi: 10.1073/pnas.90.16.7834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mino A, Ohtsuka T, Inoue E, et al. Membrane-associated guanylate kinase with inverted orientation (MAGI)-1/brain angiogenesis inhibitor 1-associated protein (BAP1) as a scaffolding molecule for Rap small G protein GDP/GTP exchange protein at tight junctions. Genes Cells. 2000;5:1009–1016. doi: 10.1046/j.1365-2443.2000.00385.x. [DOI] [PubMed] [Google Scholar]
- 12.Biederer T, Sara Y, Mozhayeva M, et al. SynCAM, a synaptic adhesion molecule that drives synapse assembly. Science. 2002;297:1525–1531. doi: 10.1126/science.1072356. [DOI] [PubMed] [Google Scholar]
- 13.Ebnet K, Aurrand-Lions M, Kuhn A, et al. The junctional adhesion molecule (JAM) family members JAM-2 and JAM-3 associate with the cell polarity protein PAR-3: a possible role for JAMs in endothelial cell polarity. J Cell Sci. 2003;116:3879–3891. doi: 10.1242/jcs.00704. [DOI] [PubMed] [Google Scholar]


