Abstract
The transcription factor Sox2 is necessary for foregut morphogenesis. Sox2 is also required for the normal development of the trachea and lung. Recently, Sox2 amplifications were investigated using large-scale single nucleotide polymorphism arrays in esophageal and lung cancer. We hypothesized that Sox2 overexpression might be correlated with clinicopathological features of lung cancers. The increased copy number of the Sox2 gene was analyzed by real-time polymerase chain reaction amplifications in 127 surgically treated non-small cell lung cancer cases from Nagoya City University Hospital, Japan. A total of 87 squamous cell carcinoma (SCC) cases were involved. An increased Sox2 gene copy number was found in 42 (33.1%) lung cancer patients. Increased Sox2 copy number status was significantly correlated with gender (females, 9.5% vs. males, 34.1%; p=0.0026), smoking status (never smoker, 4.8% vs. smoker, 32.9%; p=0.0003) and pathological subtypes (squamous cell carcinoma, 44.8% vs. non-squamous cell carcinoma, 7.5%; p<0.0001). However, among the SCCs, the Sox2 copy number status was not significantly correlated with gender, smoking status, pathological stage or differentiation status. An increased Sox2 copy number is common within SCC.
Keywords: Sox2, lung cancer, squamous cell carcinoma, copy number, differentiation
Introduction
Lung cancer is a major cause of mortality among malignant diseases due to its high incidence and malignant behavior as well as a lack of major advancements in treatment strategy (1). Lung cancer was the leading indication for respiratory surgery (46.7%) in 2007 in Japan (2), and more than 25,000 patients underwent surgery for lung cancer at Japanese institutions in the same year (2). The clinical behavior of lung cancer is largely associated with its stage. The cure of the disease by surgery is only achieved in cases representing an early stage of lung cancer (3).
Sox2 belongs to a family of evolutionarily conserved transcription factors containing a Sry-related high mobility group (HMG) box (4,5). Sox proteins are recognized as key players in the regulation of embryonic development and determination of cell fate and maintenance (6–9). Later in development, Sox2 is expressed in the endodermal epithelium of the tongue, esophagus, trachea and lung, and plays crucial roles in the differentiation and morphogenesis of these organ systems (5,10–12). Recently, Sox2 amplifications were investigated using large-scale single nucleotide polymorphism (SNP) arrays in esophageal and lung cancer (13). Sox2 is highly expressed in squamous cell carcinoma (SCC) of the gastrointestinal tract (14). Although Sox2 expression was also investigated in small cell lung cancers (15) and lung SCC (mRNA) (16), the correlation between Sox2 gene status and lung cancer in Japan has not been previously reported.
To determine the Sox2 copy number status in Japanese lung carcinoma patients for screening purposes, we investigated the Sox2 copy number using real-time polymerase chain reaction (real-time PCR) amplifications. The findings were compared against the clinicopathological features of lung cancer.
Patients and methods
Patients
The study group included 127 lung cancer patients who had undergone surgery at the Department of Surgery II, Nagoya City University Medical School, Japan, between 2001 and 2008. All tumor samples were immediately frozen and stored at −80°C until assayed. The clinical and pathological characteristics of the 127 lung cancer patients were as follows: 78 cases at stage I, 24 at stage II, and 25 at stages III–IV. The mean age was 66.0 years (range, 29–86). Among the patients, 30 (23.6%) were non-smokers, 94 (74.0%) were male and 87 (68.5%) were diagnosed as having SCC (this study focused mainly on SCC). The samples from these patients had previously been sequenced for EGFR (17–20).
PCR assays for Sox2
Genomic DNA was extracted from lung cancer tissues using the Wizard SV Genomic DNA Purification System (Promega, Madison, WI, USA) according to the manufacturer’s instructions. The DNA concentration was determined using a NanoDrop spectrophotometer (NanoDrop Technologies, Inc., Rockland, DE, USA) and adjusted to a concentration of 2.5 ng/ml.
We then used 5 μl of each DNA for the PCR assays. The Sox2 copy number was analyzed by quantitative real-time PCR and performed on the 7500 Real-Time PCR System (Applied Biosystems, Foster City, CA, USA) using a QuantiTect SYBR-Green kit (Qiagen, Inc., Valencia, CA, USA) (21,22). The PCR run was performed in triplicate for each patient. The Sox2 primers used for amplification were 5′-CAAAGAAAAA CGAGGGAAAT-3′ and 5′-ATGGGATTGGTGTTCTCTTT-3′. Total DNA content was estimated by assaying LINE-1 elements for each sample using the primers 5′-AAAGCCGCTCAA CTACATGG-3′ and 5′-TGCTTTGAATGCGTCCCAGAG-3′.
The cycling conditions were as follows: initial denaturation at 95°C for 15 min, followed by 40 cycles at 94°C for 15 sec, 56°C for 30 sec, and 72°C for 34 sec.
Immunohistochemistry
Sox2 and p63 protein expression were evaluated by immunohistochemistry (IHC) using an anti-Sox2 antibody (rabbit polyclonal; Thermo Scientific, Rockford, IL, USA) and a p63 ready-to-use antibody (mouse, clone 4A4; Dako, Carpinteria, CA, USA). We used a standard protocol for the immunostaining of the samples. Sections (4 μm) were cut from paraffin tissue blocks from the non-small cell lung cancer tumors. The slides were treated with xylene then dehydrated in alcohol. For epitope retrieval, specimens were exposed to 10 mM citrate buffer (pH 6.0) and heated to approximately 10 min in a microwave. Endogenous peroxidase activity was blocked with H2O2 in methanol. Sections were incubated with blocking solution (10% Block Ace) and then reacted with anti-Sox2 (x250) or p63 antibody overnight at 4°C. After washing out the excess antibody with phosphate-buffered saline (PBS), samples were incubated with a peroxidase-conjugated anti-mouse antibody (Mouse HRP EnVision™+; Dako) for 45 min. After washing out the excess antibody with PBS, 3,3′-diaminobenzidine (DAB) substrate (10 min) was used to visualize the antibody binding, and the sections were counterstained with hematoxylin. Sox2 and p63 staining was evaluated under a light microscope at x400 magnification. Tumor nuclear staining intensity was graded on a scale of 1–4. The percentage of positive tumor nuclei was evaluated and a proportion score was attributed (1, <5%; 2, 5–25%; 3, 25–50% and 4, >50%), as previously described (23,24).
Statistical analysis
Statistical analyses were performed using the Mann-Whitney U-test for unpaired samples and Wilcoxon’s signed rank test for paired samples. Linear relationships between variables were determined by means of simple linear regression. Correlation coefficients were determined by rank correlation using the Spearman’s test and the χ2 test. The overall survival of lung cancer patients was evaluated by the Kaplan-Meier method, and differences were determined by the log-rank test. All analyses were performed using the StatView software package (Abacus Concepts Inc., Berkeley, CA, USA), and p<0.05 was considered significant.
Results
Sox2 gene status in Japanese lung cancer patients
Using the primer sets for the Sox2 gene, 42 of the 127 lung cancer patients were found to have more than 4 copies of the Sox2 gene. The clinicopathological background of the patients is shown in Table I. The Sox2 gene copy status was significantly correlated with gender (male, 40.4% vs. female, 12.1%; p=0.0026), tobacco smoking (non-smokers, 6.6% vs. smokers, 41.2%; p=0.0003) and pathological subtype (squamous cell carcinoma, 44.8% vs. non-squamous cell carcinoma, 7.5%; p<0.0001), but not with pathological stage (stage I vs. II–IV, p=0.3878) or age (≤65 vs. >65; p=0.8052). The increased Sox2 gene copy number was observed in 2/13 (15.4%) of the adenosquamous cell carcinomas and 1/26 (3.8%) of the adenocarcinomas. There was a tendency towards a higher degree of differentiation within the normal Sox2 copy number group when compared with the increased Sox2 copy number group (p=0.1046). The overall survival of the 127 lung cancer patients from Nagoya City University, who were followed-up until December 31 2009, was studied in reference to the Sox2 gene status. The survival rate of patients with an increased Sox2 gene copy number (n=42, 13 were deceased) and patients with a normal Sox2 copy number (n=85, 27 were deceased) was not significantly different (log-rank test, p=0.9299; Fig. 1).
Table I.
Clinicopathological data of 127 lung cancer patients.
| Factors |
Sox2 gene status
|
p-value | ||
|---|---|---|---|---|
| Amplified Patients (n=42) | Normal Patients (n=85) | |||
| Mean age (years) | 66.0±10.2 | 67.1±8.7 | 65.5±10.8 | 0.4134 |
| Stage | ||||
| I | 26 (61.9%) | 52 (61.2%) | 0.9999 | |
| II–IV | 16 (38.1%) | 33 (38.8%) | ||
| Lymph node metastasis | ||||
| N0 | 29 (55.8%) | 56 (65.9%) | 0.2779 | |
| N(+) | 23 (44.2%) | 29 (34.1%) | ||
| Smoking status | ||||
| Never smoker | 2 (4.8%) | 28 (32.9%) | 0.0003 | |
| Smoker | 40 (95.2%) | 57 (67.1%) | ||
| Differentiation | ||||
| Well-differentiated | 6 (15.4%) | 21 (31.3%) | 0.1046 | |
| Moderate/poor | 33 (64.6%) | 46 (68.7%) | ||
| Pathological subtype | ||||
| Squamous | 39 (92.9%) | 48 (56.5%) | ||
| Non-squamous | 3 (7.1%) | 37 (43.5%) | <0.0001 | |
| Age | ||||
| ≤65 | 18 (42.9%) | 39 (45.9%) | 0.8501 | |
| >65 | 24 (57.1%) | 46 (54.1%) | ||
| Gender | ||||
| Male | 38 (90.5%) | 56 (65.9%) | 0.0026 | |
| Female | 4 (9.5%) | 29 (34.1%) | ||
N0, lymph node metastasis negative; N(+), lymph node metastasis positive; Squamous, squamous cell carcinoma.
Figure 1.
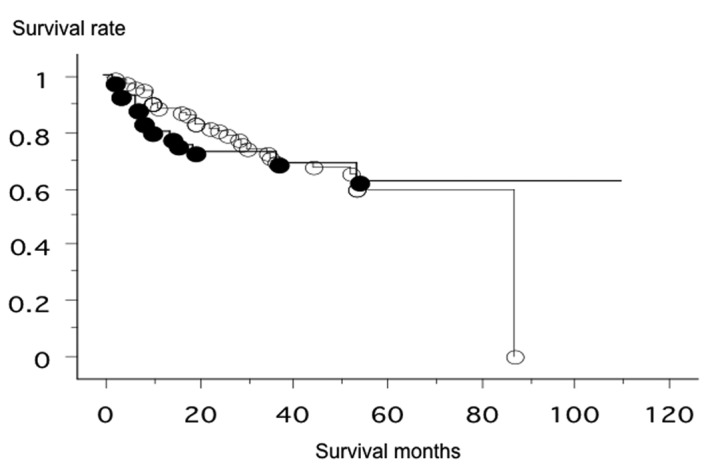
The prognosis of patients with an increased Sox2 gene copy number (•, n=42, 13 were deceased; mean survival, 40.8 months) and patients with a normal Sox2 copy number (○, n=85, 27 were deceased; mean survival, 62.8 months) was not significantly different (log-rank test, p=0.9299).
Immunohistochemistry
The immunohistochemical evaluation was performed according to the scoring system described in Materials and methods. IHC was performed for 72 patients, since the tissue blocks were not available for the other patients. The p63-positive (≥4) ratio (Fig. 2) was more closely correlated with squamous histology (adenosquamous + SCC, 95.2% vs. adenocarcinoma, 60%; p=0.0004) than the Sox2-positive ratio (adenosquamous + SCC, 85.4% vs. adenocarcinoma, 45.5%; p=0.001) (Fig. 3). The p63 IHC was correlated with an increased Sox2 copy number (p=0.0226). The Sox2-positive IHC cases had a tendency towards an increased Sox2 copy number (p=0.1035). However, the p63 IHC status (p=0.8188) and Sox2 IHC status (p=0.3394) did not correlate with the prognosis of lung cancers.
Figure 2.
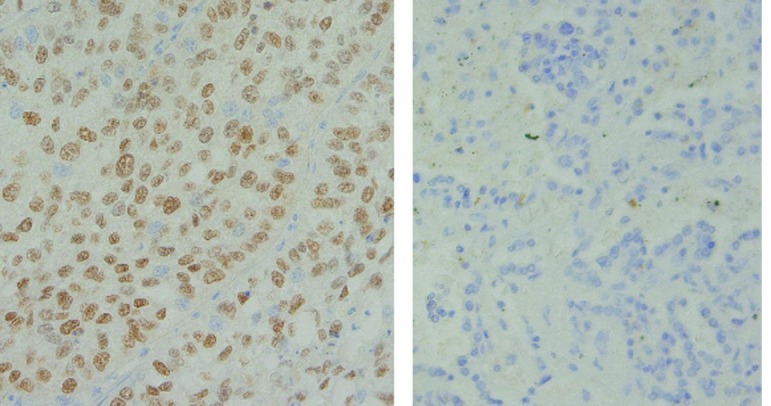
Immunohistochemistry for p63. Left, p63-positive (≥4) section. Right, p63-negative section.
Figure 3.
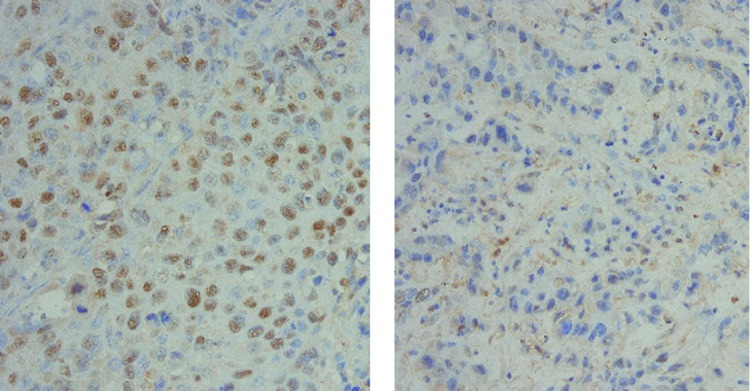
Immunohistochemistry for Sox2. Left, Sox2-positive (≥4) section. Right, Sox2-negative section.
Discussion
In this analysis, we found an increased Sox2 gene copy number in 33% of the Japanese lung cancer patients. The Sox2 gene statuses were mainly correlated with squamous histology.
The HMG domain is a DNA-binding motif that was originally discovered in abundant nonhistone components of chromatin. The proteins containing this motif are considered as architectural components in the assembly of nucleoprotein complexes, and they regulate transcription by interacting with the minor groove of the DNA helix and modulating DNA structure by bending the DNA helix (25). At least 20 Sox genes have so far been identified in mammals and more in other vertebrates (26–28), and they have been implicated in the regulation of a variety of developmental processes. Sox2 has a recognized role in lung branching morphogenesis and differentiation in embryonic development. Mice engineered to overexpress Sox2 in the developing lung epithelium have a marked reduction in the number of airways (5). Sox2 is modulated in concert during the course of tracheal and esophageal development (12).
Sox2 is highly expressed in SCC of the gastrointestinal tract (14). Sox2 was expressed in 81% of esophageal SCC cases and 91% of anal canal SCC cases, compared with 13% and 17% of esophageal and rectal adenocarcinoma cases, respectively (14). In the upper gastrointestinal tract, Sox2 is expressed in the developing foregut endoderm and is thought to play a role in establishing the boundary between the keratinized, squamous-lined esophagus and the glandular hindstomach (12). The high correlation between p63 and Sox2 expression was reported in esophageal cancer and raised the issue of a possible contribution of Sox2 to squamous cell carcinogenesis, perhaps via disturbed differentiation and patterning in the context of other procarcinogenic events.
Copy number increase in the 3q26-qter region is observed in various squamous cell cancers including lung, esophagus, head and neck and cervix (13). Two studies have reported the identification of Sox2 as a novel oncogene in lung and esophageal SCC (13,29). The knockdown experiments demonstrated that Sox2 was necessary for lung squamous cell viability by protecting cells from apoptosis (13,29). The suppression of Sox2 with shRNA constructs reduced proliferation and colony formation in the 3q26.33-amplified cell lines but not in the controls (13). Although Sox2 itself was not transforming, the combination of Sox2 with the FOXE1 or FGFR2 isoforms promoted anchorage-independent growth in cancers (13).
Sholl et al demonstrated that Sox2 IHC expression was more common in poorly differentiated adenocarcinoma of the lung (23), and this study also demonstrated that there was a tendency towards a higher degree of differentiation within the normal Sox2 copy number group compared to the increased Sox2 copy number group (p=0.1046). However, Hussenet et al addressed the relationship between the Sox2 expression level and histological differentiation status in lung SCC and found no significant changes (29). They found that Sox2 is highly expressed in both aggressive and non-aggressive tumors with no statistically significant differences between the two groups (29). This result is consistent with the very high expression levels of Sox2 protein observed in the majority of primary SCC cases (13,23,29).
Acknowledgments
The authors would like to thank Mrs. Akiha Kuramoto for her excellent technical assistance. This study was supported by Grants-in-Aid for scientific research from the Japan Society for the Promotion of Science (JSPS) (nos. 23659674, 21390394 and 21591820) and a grant for cancer research of the Program for Developing the Supporting System for Upgrading Education and Research (2009) from the Ministry of Education, Culture, Sports, Science and Technology of Japan.
References
- 1.Ginsberg RJ, Kris MK, Armstrong JG. Principles and Practice of Oncology. 4th edition. Lippincott; Philadelphia: 1993. Cancer of the lung; pp. 673–682. [Google Scholar]
- 2.Ueda Y, Fujii Y, Kuwano H. Thoracic and cardiovascular surgery in Japan during 2007. Annual report by the Japanese association for thoracic surgery. Gen Thorac Cardiothorac Surg. 2007;57:488. doi: 10.1007/s11748-009-0460-y. [DOI] [PubMed] [Google Scholar]
- 3.Postmus PE. Chemotherapy for non-small cell lung cancer: the experience of the Lung Cancer Cooperative Group of the European Organization for Research and Treatment of Cancer. Chest. 1998;113:28S–31S. doi: 10.1378/chest.113.1_supplement.28s. [DOI] [PubMed] [Google Scholar]
- 4.Que J, Luo X, Schwartz RJ, et al. Multiple roles for Sox2 in the developing and adult mouse trachea. Development. 2009;136:1899–1907. doi: 10.1242/dev.034629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gontan C, de Munck A, Vermeij M, et al. Sox2 is important for two crucial processes in lung development: Branching morphogenesis and epithelial cell differentiation. Dev Biol. 2008;317:296–309. doi: 10.1016/j.ydbio.2008.02.035. [DOI] [PubMed] [Google Scholar]
- 6.Kamachi Y, Uchikawa M, Kondoh H. Pairing SOX off: with partners in the regulation of embryonic development. Trends Genet. 2000;16:182–187. doi: 10.1016/s0168-9525(99)01955-1. [DOI] [PubMed] [Google Scholar]
- 7.Lefebvre V, Dumitriu B, Penzo-Mendez A, et al. Control of cell fate and differentiation by Sry-related high-mobility group box (Sox) transcription factors. Int J Biochem Cell Biol. 2007;39:2195–2214. doi: 10.1016/j.biocel.2007.05.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schepers G, Teasdale R, Koopman P. Twenty pairs of sox, extent, homology, and nomenclature of the mouse and human sox transcription factor gene families. Dev Cell. 2002;3:167. doi: 10.1016/s1534-5807(02)00223-x. [DOI] [PubMed] [Google Scholar]
- 9.Wilson M, Koopman P. Matching SOX: partner proteins and co-factors of the SOX family of transcriptional regulators. Curr Opin Genet Dev. 2002;12:441–446. doi: 10.1016/s0959-437x(02)00323-4. [DOI] [PubMed] [Google Scholar]
- 10.Ishii Y, Rex M, Scotting PJ, et al. Region-specific expression of chicken Sox2 in the developing gut and lung epithelium: regulation by epithelial-mesenchymal interations. Dev Dyn. 1998;213:464–475. doi: 10.1002/(SICI)1097-0177(199812)213:4<464::AID-AJA11>3.0.CO;2-Z. [DOI] [PubMed] [Google Scholar]
- 11.Okubo T, Pevny LH, Hogan BL. Sox2 is required for development of taste bud sensory cells. Genes Dev. 2006;20:2654–2659. doi: 10.1101/gad.1457106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Que J, Okubo T, Goldenring JR, et al. Multiple dose-dependent roles for Sox2 in the patterning and differentiation of anterior foregut endoderm. Development. 2007;134:2521–2531. doi: 10.1242/dev.003855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bass AJ, Watanabe H, Mermel CH, et al. SOX2 is an amplified lineage-survival oncogene in lung and esophageal squamous cell carcinomas. Nature Genet. 2009;41:1238–1242. doi: 10.1038/ng.465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Long KB, Hornick JL. SOX2 is highly expressed in squamous cell carcinomas of the gastrointestinal tract. Hum Pathol. 2009;40:1768–1773. doi: 10.1016/j.humpath.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 15.Titulaer MJ, Klooster R, Portman M, et al. SOX antibodies in small-cell lung cancer and Lambert-Eaton myasthenic syndrome: frequency and relation with survival. J Clin Oncol. 2009;27:4260–4267. doi: 10.1200/JCO.2008.20.6169. [DOI] [PubMed] [Google Scholar]
- 16.Yuan P, Kadara H, Behrens C, et al. Sex determining region Y-box 2 is a potential cell-lineage gene highly expressed in the pathogenesis of squamous cell carcinomas of the lung. PLoS One. 2010;5:e9112. doi: 10.1371/journal.pone.0009112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Paez JG, Janne PA, Lee LC, et al. EGFR mutations in lung cancer: correlation with clinical response to gefitinib therapy. Science. 2004;304:1497–1500. doi: 10.1126/science.1099314. [DOI] [PubMed] [Google Scholar]
- 18.Sasaki H, Shimizu S, Endo K, et al. EGFR and erbB2 mutation status in Japanese lung cancer patients. Int J Cancer. 2006;118:180–184. doi: 10.1002/ijc.21301. [DOI] [PubMed] [Google Scholar]
- 19.Endo K, Konishi A, Sasaki H, et al. Epidermal growth factor receptor gene mutation in non-small cell lung cancer using highly sensitive and fast TaqMan PCR assay. Lung Cancer. 2005;50:375–384. doi: 10.1016/j.lungcan.2005.08.009. [DOI] [PubMed] [Google Scholar]
- 20.Sasaki H, Endo K, Konishi A, et al. EGFR mutation status in Japanese lung cancer patients: genotyping analysis using LightCycler. Clin Cancer Res. 2005;11:2924–2929. doi: 10.1158/1078-0432.CCR-04-1904. [DOI] [PubMed] [Google Scholar]
- 21.Endo K, Sasaki H, Yano M, et al. Evaluation of the epidermal growth factor receptor gene mutation and copy number in non-small cell lung cancer with gefitinib therapy. Oncol Rep. 2006;16:533–541. [PubMed] [Google Scholar]
- 22.Wang TL, Maierhofer C, Speicher MR, et al. Digital karyotyping. Proc Natl Acad Sci USA. 2002;99:16156–16161. doi: 10.1073/pnas.202610899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sholl LM, Long KB, Hornick JL. Sox2 expression in pulmonary non-small cell and neuroendocrine carcinomas. Appl Immunohistochem Mol Morphol. 2010;18:55–61. doi: 10.1097/PAI.0b013e3181b16b88. [DOI] [PubMed] [Google Scholar]
- 24.Nonaka D. Differentiatial expression of SOX2 and SOX17 in testicular germ cell tumors. Am J Clin Pathol. 2009;131:731–736. doi: 10.1309/AJCP7MNCNBCRN8NO. [DOI] [PubMed] [Google Scholar]
- 25.Grosschedl R, Giese K, Pagel J. HMG domain proteins: Architectual elements in the assembly of nucleoprotein structures. Trends Genet. 1994;10:94–99. doi: 10.1016/0168-9525(94)90232-1. [DOI] [PubMed] [Google Scholar]
- 26.Denny P, Swift S, Brand N, et al. A conserved family of genes related to the testis determining gene, SRY. Nucleic Acids Res. 1992;20:2887. doi: 10.1093/nar/20.11.2887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hudson C, Clements D, Friday RV, et al. Xsox17-α and -β mediate endoderm formation in Xenopus. Cell. 1997;91:397–405. doi: 10.1016/s0092-8674(00)80423-7. [DOI] [PubMed] [Google Scholar]
- 28.Russell SRH, Sanchez-Soriano N, Wright CR, Ashburner M. The dechaete gene of Drosophila melanogaster encodes a SOX-domain protein required for embryonic segmentation. Development. 1996;122:3669–3676. doi: 10.1242/dev.122.11.3669. [DOI] [PubMed] [Google Scholar]
- 29.Hussenet T, Dali S, Exinger J, et al. SOX2 is an oncogene activated by recurrent 3q26.3 amplification in human lung squamous cell carcinomas. PLoS One. 2010;5:8960. doi: 10.1371/journal.pone.0008960. [DOI] [PMC free article] [PubMed] [Google Scholar]


