Abstract
Accounting for target flexibility and selecting “hot spots” most likely to be able to bind an inhibitor continue to be challenges in the field of structure-based drug design, especially in the case of protein-protein interactions. Computational fragment-based approaches employing molecular dynamics (MD) simulations are a promising emerging technology having the potential to address both of these challenges. However, the optimal MD conditions permitting sufficient target flexibility while also avoiding fragment-induced target denaturation remain ambiguous. Using one such technology (SILCS: Site Identification by Ligand Competitive Saturation), conditions were identified to either prevent denaturation or identify and exclude trajectories in which subtle but important denaturation was occurring. The target system employed was the well-characterized protein cytokine IL-2, which is involved in a protein-protein interface and, in its un-liganded crystallographic form, lacks surface pockets that can serve as small-molecule binding sites. Nonetheless, small-molecule inhibitors have previously been discovered that bind to two “cryptic” binding sites that emerge only in the presence of ligand binding, highlighting the important role of IL-2 flexibility. Using the above conditions, SILCS with hydrophobic fragments was able to identify both sites based on favorable fragment binding while avoiding IL-2 denaturation. An important additional finding was that acetonitrile, a water-miscible fragment, fails to identify either site yet can induce target denaturation, highlighting the importance of fragment choice.
Keywords: interleukin-2, immunosuppression, computer-aided drug design, lead optimization, molecular dynamics simulations, protein-protein interactions
Introduction
Two important problems in structure-based drug design are identification of a binding site and the need to account for target flexibility. These two problems are in fact coupled, in that many targets undergo conformational rearrangements in the presence of a bound ligand, with these rearrangements directly altering the binding site relative to the unbound form of the target.1 One means to simultaneously address both problems is to use a target:ligand complex for the target atomic coordinates, since the bound ligand both identifies the binding site and captures or induces the relevant conformational rearrangements.2 However, experimental atomic coordinates of the complex, typically from a high-resolution x-ray crystal structure, are not always available, and the apo structure must therefore be used as the starting point.
With regard to identification of a binding site, technology to detect surface pockets on a target using geometric considerations was developed early in the history of structure-based computer-aided drug design, enabling identification and directed computational screening of a binding site.3 A variety of different approaches have since been developed and reviewed,4–7 and research applying geometric, energetic, and physicochemical criteria for detecting pockets and also determining whether such pockets would in fact be good small-molecule binding sites continues to the present day.8–17 The Site Identification by Ligand Competitive Saturation (SILCS) approach18 for fragment-based computer-aided drug design is a recently-developed methodology that can be put to such use. In SILCS, the target is computationally soaked in an aqueous solution of compounds corresponding to chemical moieties commonly found in drug-like small molecules (“fragments”), analogous to experimental approaches for fragment-based drug design.19 The system is then simulated using molecular dynamics (MD), and snapshots from the trajectory are used to compute three-dimensional residence probability maps (FragMaps) for fragment binding to the target surface. Importantly, SILCS not only identifies pockets, but can differentiate which pockets are most likely to bind fragments of drug-like compounds (“hot spots”) as judged by FragMap probabilities.20 That is, the high-probability regions in SILCS FragMaps can be used not only for pocket identification, but also the selection of pockets most likely to be capable of binding a specific chemical moiety or class of small molecules, thereby determining whether a pocket is also a potential small-molecule binding site.
Accounting for flexibility in a target continues to remain a challenging problem in the field of computer-aided drug design.21–25 An important advance in accounting for target flexibility has been to dock the same ligand library to multiple different static conformations of the same target.26,27 Approaches to generating target conformational ensembles have included using multiple different experimental atomic-resolution structures of the same protein28,29 or multiple different conformations generated by in silico methods such as MD or Monte Carlo simulations.30–32 Recently, Eyrisch and Helms demonstrated that snapshots from explicit-solvent MD simulations, started from apo crystal structures, contained a subset of conformations revealing “cryptic” pockets either only partly detectable or completely absent in the apo crystal structures but known to “appear” and serve as binding sites in complexes containing small molecule inhibitors.33 Raunest and Kandt have taken this approach to its logical conclusion by collating MD snapshots into three-dimensional residence probability maps and surveying the protein and solvent residence probabilities to detect cavities, tunnels, and clefts.17 Such results provide support for the ability of MD to generate different, relevant conformations for drug discovery.
Because MD simulations are used to generate SILCS FragMaps, in principle full target flexibility can be accounted for by doing unrestrained simulations. In practice, SILCS was originally developed using harmonic positional restraints on protein backbone Cα atoms so as to allow for use of a rectangular simulation box proportional in dimensions to an oblong protein without danger of the protein undergoing rotation and colliding into a periodic image of itself.18 In the presence of such restraints, it was shown that important sidechain conformational changes associated with ligand binding to protein targets were indeed accounted for.18,20 With regard to unrestrained simulations, using a similar protocol in the context of the canonical model system hen egg-white lysozyme, Lexa and Carlson concluded that full protein flexibility was essential for proper hot-spot mapping.34 They further emphasized the importance of short (10 ns) trajectories to allow solvent equilibration and convergence while avoiding possible target denaturation, though they did not note signs of any denaturation in their study. Additionally, the target used – hen egg-white lysozyme – has four disulfide bonds, which, as they are not subject to breaking or rearrangement in the molecular mechanics framework, are stabilizing of the folded state.
For systems with few or no stabilizing disulfide bonds, the possibility of fragment-induced denaturation of the target is a potential issue with SILCS and related methods.34,35 This issue points to the inter-relatedness of binding site or “hot spot” identification and target flexibility, where flexibility may be required for binding site identification, but allowing too much flexibility can leave the target prone to denaturation. Additionally, it is not apparent a priori what the necessary timescale is either to achieve relevant conformational sampling of a particular target or for target denaturation to occur. Indeed, it may be the case that denaturation occurs on the shorter timescale. A basic assumption of the present study is that protein denaturation, which involves the loss of a protein’s secondary, tertiary, and quarternary structures, is undesirable in that it is associated with large-scale structural perturbations that are not relevant to conformational changes associated with the identification of cryptic pockets or binding sites.
The protein IL-2, which is one of the three targets studied by Eyrisch and Helms using MD,33 lacks deep pockets for structure-based drug design,36–38 a not altogether surprising fact given that its natural “ligand” is the trimeric IL-2 receptor (IL-2R),39,40 a protein complex much larger than IL-2 itself. Despite this, IL-2 has been shown to be capable of binding small molecules. From small-molecule discovery efforts toward immunosuppressive therapeutics, two distinct binding sites not apparent in apo IL-236–38 have been identified in atomic-resolution crystal structures of IL-2:small-molecule complexes,41–43 with the first of these binding sites (cryptic binding site 1) corresponding to the single cryptic pocket identified in the above MD study.33 Here, the SILCS methodology is applied to determine if MD conditions can be found that detect both of the IL-2 cryptic pockets while also either identifying or preventing denaturation in the MD trajectories. Importantly, in the present approach, the cryptic pockets are detected by their ability to preferentially bind fragments, thus identifying them as binding sites, a fact not revealed by pocket-detection approaches based solely on geometric considerations.
Results and Discussion
The SILCS methodology entails immersing the protein in a high-concentration (~1 M) aqueous solution of fragments, running multiple nanosecond-length MD simulations of the composite protein+fragment+water system, and computing residence probability maps for fragment and water binding around the protein for each simulation. The probability maps of the same type from all simulations are then combined to generate a single probability map (FragMap) of each fragment type (please see the “Methods” section for additional details on system construction, MD protocols, and analysis). As SILCS FragMaps are probability histograms generated by binning fragment non-hydrogen-atom positions from each MD snapshot into voxels in a Cartesian grid, high-probability regions correspond to those regions of the target surface where there are favorable target:fragment interactions. Importantly, in addition to the standard force field terms, a repulsive potential is applied to fragment:fragment pairs to prevent hydrophobic fragment aggregation during the MD simulations to maintain a near-ideal solution of fragments at the high concentrations used in SILCS.18 In the present case, as well as in prior work,18,20 the fragment identities were propane and benzene; that is, IL-2 SILCS simulations are done using a ternary solution that is ~1 M in propane and benzene. That said, it is trivial to use fragments of a different identity.
The remainder of the Results and Discussion is organized as follows. First, target denaturation is analyzed in four different protocols having: (1) no positional restraints; (2) weak positional restraints on Cα positions (the original SILCS protocol18) (3) positional restraints only on non-hydrogen atoms near the core of the target protein; (4) no positional restraints and no repulsive potential preventing fragment aggregation. Subsequently, each of these protocols is evaluated with regard to its ability to identify the two known cryptic binding sites in IL-2. For comparison, the SILCS-like protocol used on hen egg-white lysozyme in which the fragment was acetonitrile (Acn)34 is also evaluated (Table 1).
Table 1.
SILCS protocol parameters
| Protocol # | Restrained atoms | Restraining potential ka | Inter-fragment repulsive potential? |
|---|---|---|---|
| Control | None | n/a | No fragments in simulated system |
| 1 | None | n/a | Yes |
| 2b | Cα | 0.12 | Yes |
| 3 | Non-hydrogen atoms within 7 Å of protein center of mass | 0.1 | Yes |
| 4 | None | n/a | No |
| Acn | Same as Protocol 4, but using a 50/50 w/w solution of acetonitrile in water |
The restraining potential on atomic positions is of the form k(Δr)2, where Δr is the displacement in Å from the crystallographic position and k has units of kcal*mol−1*Å−2.
Same protocol as that originally described for SILCS.18 n.b.: The restraining potential k value was originally incorrectly stated to be “0.01 kcal*mol−1*Å−2”18 whereas it should have been described as “0.01 kcal*mol−1*Å−2 per atomic mass unit,” i.e. 0.12 kcal*mol−1*Å−2 for Cα atoms, as stated here and in Reference 20.
Target denaturation
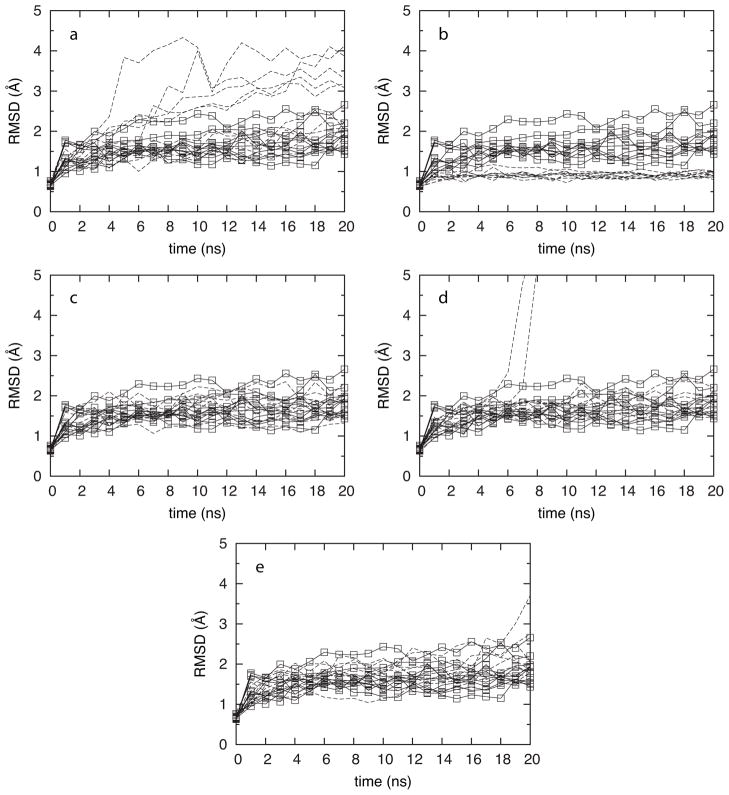
The extent of target denaturation was evaluated based on root mean squared deviation (RMSD) in atomic coordinates from the starting crystal structure as a function of simulation times (Figure 1). Results from control simulations that lacked both fragments and restraints (Table 1: “control”) are included in each panel of Figure 1. What is immediately apparent from the MD time courses of IL-2 RMSD is that maintenance of the crystallographic structure depends very strongly on the parameters used in the MD simulations. In the case where full flexibility is included (Table 1: “Protocol 1”), half of the ten SILCS trajectories have RMSD time courses that are visually different from the ten control trajectories, whereas the other half are in line with the “control” protocol (Figure 1a). Application of Cα positional restraints, per the “original” SILCS protocol (Table 1: “Protocol 2”), leads to sampled conformations that are less variable and more similar to the starting crystal structure than seen in the “control” data (Figure 1b). In contrast, replacing positional restraints on all Cα with positional restraints on only non-hydrogen atoms near the core of the target (Table 1: “Protocol 3”) leads to RMSD behavior indistinguishable from the “control” data (Figure 1c).
Figure 1.
IL-2 RMSD for ten independent trajectories (dashed lines) using Protocols 1 (a), 2 (b), 3 (c), 4 (d), and Acn (e). Data from the control protocol (solid lines with boxes) are duplicated in each frame.
As stated previously, a key ingredient of SILCS simulations is the addition of a repulsive potential to prevent fragment aggregation. Without this repulsive potential, significant observable aggregation occurs on the MD timescale of nanoseconds, whereas inclusion of the potential guarantees a uniform distribution of fragments in aqueous solution.18 As aggregation lowers the effective fragment concentration, it is anticipated that inclusion of the repulsive potential will lead to better sampling, an issue discussed in the next section. Additionally, aggregation of hydrophobic molecules is thought to contribute to non-specific false positives in inhibition assays by denaturing the target and mimicking results anticipated in the presence of a true inhibitor.44 Such target denaturation is directly observed here in the absence of the repulsive potential (i.e., Protocol 4). Two of the ten trajectories employing Protocol 4 undergo catastrophic changes in structure prior to 10 ns, with RMSD values exceeding 5 Å at 7 ns and 8 ns, respectively (Figure 1d). Interestingly, the other eight trajectories are indistinguishable from the “control” data for the length of the 20-ns simulations. This kinetic difference in behavior highlights the utility of running multiple simulations.
For comparison, IL-2 was simulated without restraints in a 50/50 w/w solution of water and acetonitrile (i.e., Protocol Acn) as per the prior hen egg-white lysozyme work. Up until the 18 ns time point, all ten trajectories had RMSD data indistinguishable from the “control” simulations. At the 18 ns time point, one of the ten trajectories underwent a rapid increase in RMSD from 2.5 Å up to 3.7 Å at 20 ns, which is a value inconsistent with IL-2 behavior in “control” simulations without fragments (Figure 1e). Thus, it appears that Protocol Acn, along with Protocol 1 and Protocol 4 run the risk of denaturing the target protein during the course of the simulation.
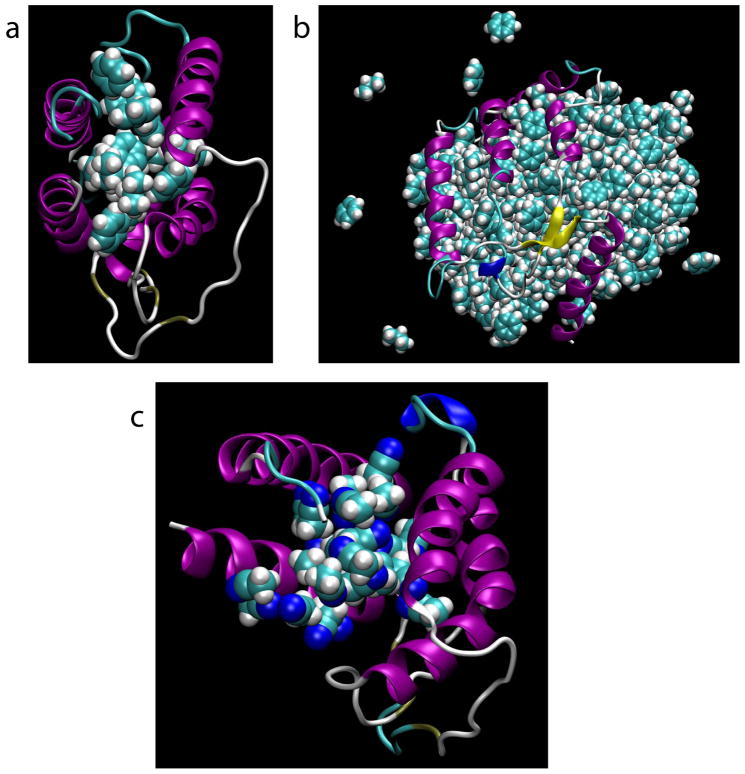
Visualization of the final snapshots with the highest RMSD values from Protocols 1, 4, and Acn confirms that indeed in all three cases, target denaturation is occurring. In the case of Protocol 1, benzene and propane molecules are observed to infiltrate the center of the protein and to thus disrupt tertiary structure contacts between spatially adjacent helices (Figure 2a). However, the inter-fragment repulsive potential prevents further recruitment of denaturing fragments by the infiltrating fragments and additional disruption of the protein structure. This is not the case for Protocol 4, in which, owing to the lack of inter-fragment repulsion, the fragments form a compact globule that phase-separates from the aqueous compartment and leads to catastrophic disruption of hydrophobic tertiary structure contacts (Figure 2b), thereby leading to the very large RMSD of 12 Å. Finally, in the case of Protocol Acn, acetonitrile molecules infiltrate the core of the protein, resulting in a situation similar to that observed for Protocol 1 (Figure 2c).
Figure 2.
Final (20 ns) snapshots from denaturing trajectories. Snapshots are from Protocols 1 (a; RMSD = 4.1 Å), 4 (b; RMSD = 12.3 Å), and Acn (c; RMSD = 3.7 Å).
While the RMSD data for Protocols 1, 4, and Acn suggested target denaturation, it was only in the case of Protocol 4 that the data were unambiguous. In the case of the other two protocols, it is possible to envision a situation where the observed RMSD values of 3 to 4 Å arise not because of denaturation due to fragment infiltration of the protein hydrophobic core, but because of relevant conformational changes such as large loop rearrangements or switching from one state to another involving the sliding of spatially-adjacent helices. To help rule out this possibility, a simple metric to detect fragment infiltration of the protein hydrophobic core is proposed. As apparent from pictures (Figures 2a and 2c), fragment infiltration leads to swelling of the protein, thereby suggesting the protein radius of gyration (Rgyr) as a useful measurement, especially given its prior application in all-atom explicit-solvent protein folding studies.45–47
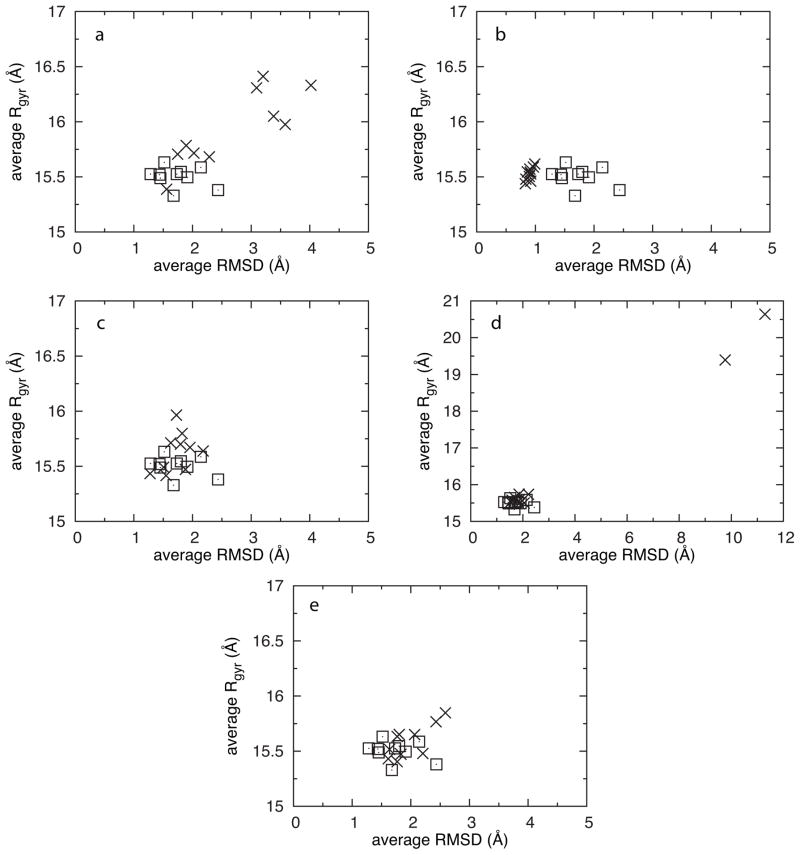
Previous work demonstrated that FragMap convergence improved with simulation time, leading to the use of the final 5-ns block from 20-ns trajectories to compute FragMaps.20 As such, this time block from each trajectory was used to compute average RMSD and Rgyr values for each simulation in each protocol (Figure 3). Based on this analysis, the five Protocol 1 trajectories with high RMSD (Figure 1a) also are swelling and therefore being infiltrated by fragments (Figure 3a), recommending that these blocks of data be excluded from the tabulation of FragMaps. The situation is less clear in the case of Protocol Acn, where it is not obvious from the Rgyr vs. RMSD data which trajectory is showing denaturation (Figure 3e). This is due to the fact that swelling does not start occurring until the 18 ns timepoint, whereas the average reflects behavior across the full 15–20 ns interval. That said, the final conformation in this trajectory (Figure 2c) has an RMSD of 3.7 Å and an Rgyr of 16.2 Å, placing it squarely in the cluster of data from Protocol 1 associated with fragment infiltration (Figure 3a). Thus, the proposed Rgyr vs. RMSD metric is a simple way to detect target denaturation due to fragment infiltration of the target core.
Figure 3.
Average radius of gyration (Rgyr) vs. average RMSD. Averages were taken over the 15–20-ns interval of each trajectory. Data in X’s are for Protocols 1 (a), 2 (b), 3 (c), 4 (d), and Acn (e). Control protocol data are in squares and are duplicated in each frame; panel d is on a different scale than the other panels.
Cryptic binding site identification
Based on the above, fragment-induced target denaturation is a real risk when full protein flexibility is included, regardless of whether the fragments are hydrophobic (e.g. benzene and propane) or miscible with water (e.g. acetonitrile). The two options presented for dealing with target denaturation are (1) use of restraining potentials to maintain the target structure, or (2) identification of denaturing trajectories and exclusion of data from these trajectories. The next question is whether using either of these approaches is compatible with proper sampling of protein flexibility as required to identify fragment binding sites, which in the case of IL-2 are cryptic binding sites (CBS) 1 and 2. In a sense, protein flexibility with conformational sampling away from a static atomic-resolution experimental conformation and protein denaturation are points on a continuum. Thus, there is the danger that by abrogating denaturation using either of the above two options, relevant flexibility is also excluded.
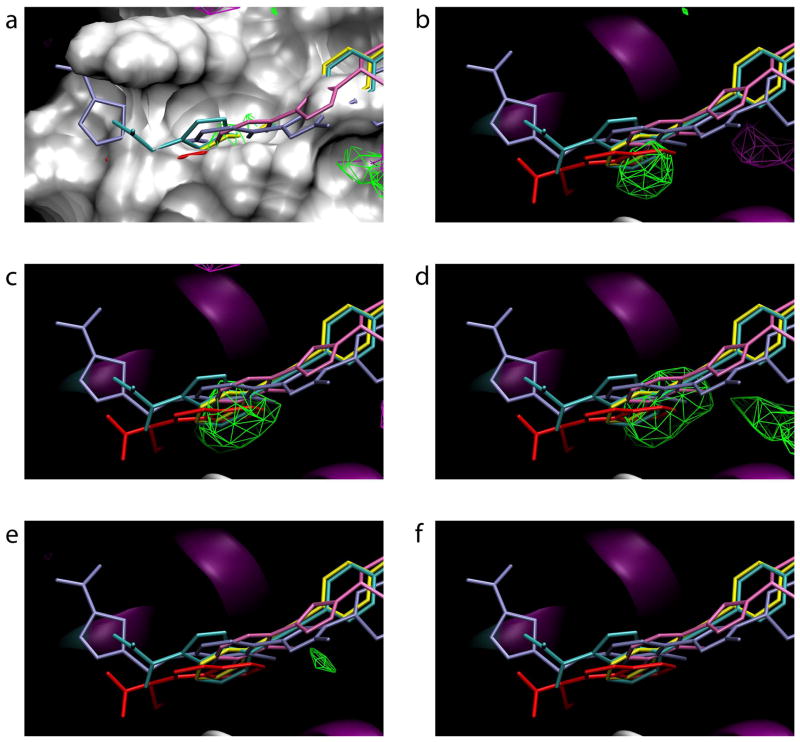
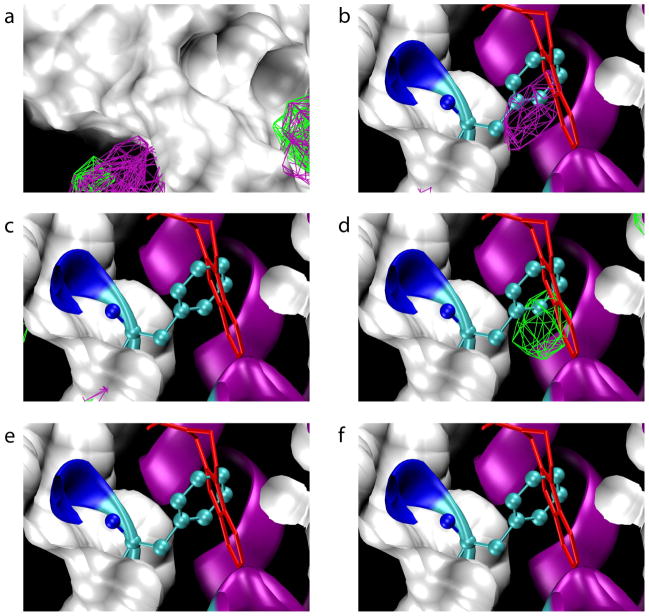
Based on FragMaps from the various protocols, it is apparent that both approaches are viable options for avoiding denaturation while maintaining relevant flexibility. Comparison of the crystal structure of unbound IL-236–38 with co-crystal crystal structures of IL-2 containing non-covalent inhibitors41,42 reveals conformational change is required to accommodate inhibitor binding. After alignment of IL-2 in all these structures, in all cases a significant subset of inhibitor atoms are buried below the molecular surface of apo IL-242,48 (Figure 4a). Removing the molecular surface reveals that FragMaps from several protocols have high-probability areas, located underneath the molecular surface of the starting conformation, that correspond to CBS1. In particular, the relevant FragMaps were generated from Protocols 1, 2, and 3 (Figure 4b, c, d). In the case of Protocol 1, which included full flexibility, the FragMaps were generated using only the five trajectories that did not denature based on the RMSD vs. Rgyr analysis. This addresses the important question of whether exclusion of data will compromise sampling; in the case of IL-2, the answer is quite clearly “no,” though a more general conclusion requires additional experience with a number of other systems. In contrast to Protocol 1, Protocols 2 and 3 both had positional restraints on IL-2 atomic positions. Interestingly, the results for CBS1 are not sensitive to the particular nature of the restraints. Regardless of whether Cα atoms per the original SILCS protocol (Protocol 2) or non-hydrogen atoms near the center of the protein (Protocol 3) are used (Table 1), sampling of CBS1 is achieved (Figure 4c, d). Thus, IL-2 denaturation can be handled either through restraining potentials to avoid denaturation itself, or by identifying and excluding denaturing trajectories, with both approaches still capable of detecting CBS1.
Figure 4.
Small-molecule compounds bound to cryptic binding site 1 and FragMaps for all protocols. Green contours are for propane, purple for benzene, and blue for acetonitrile. (a) The molecular surface of the starting x-ray crystallographic conformation of apo IL-2 is shown along with inhibitor conformations from IL-2:inhibitor complexes aligned with unbound IL-2. (b-f) FragMaps from Protocols 1 through 4 and Acn respectively, along with IL-2 inhibitors and the crystallographic secondary structure of apo IL-2.
Protocols 1, 2, and 3 each identify only one region, approximately the size of a six-member ring, of the broad inhibitor-binding groove that encompasses CBS1. Additionally, all three protocols identify the same region (Figure 4b, c, d) as CBS1. In the orientation depicted, one of the crystallographically-identified ligands extends only to the left of CBS1, two extend only to the right, and two span outward on both sides; however, regardless of how they are positioned in the inhibitor-binding groove, all ligands have a functional group bound in CBS1, highlighting its importance.
Functionally, IL-2 forms a complex with IL-2R, which is composed of three subunits α,β, and γ. Although CBS1 is located at the IL-2:IL-2Rα interface, CBS1 does not make contact with any IL-2R residues in structures of IL-2 complexed either with just the IL-2Rα chain49 or with complete IL-2R.39,40 In the binary complex with the IL-2Rα chain, CBS1 is not revealed and that region of the IL-2 surface looks similar to the apo IL-2 structure. In both quaternary complexes with the complete IL-2R, CBS1 is revealed, as it is revealed in IL-2:inhibitor complexes, yet remains unoccupied. Thus, starting with the apo IL-2 structure, the IL-2:IL-2Rα complex, or the complete IL-2:IL-2R complex, traditional structure-based discovery approaches would have been unproductive, either because no pocket exists or because the pocket, while present, is empty, thereby suggesting it is not a potential binding site. This is in contrast to the SILCS Protocols 1, 2, and 3, which all identify this important cryptic binding site and furthermore point to what appears to be a “hot spot” within the binding site, as identified by the fact that that region is occupied by all of the various small-molecule inhibitors in available crystallographic complexes.
In contrast to the above favorable results, neither Protocol 4 nor Protocol Acn is able to identify CBS1 (Figure 4e, f). In the case of Protocol 4, the two denaturing trajectories were excluded when computing FragMaps (as per Protocol 1). The poor sampling, despite full target flexibility, is most likely due to the fact that the effective fragment concentration is greatly reduced below the computed ~1 M concentration owing to fragment aggregation caused by the lack of the SILCS inter-fragment repulsive potential. The introduction of this repulsive potential is a key differentiator of SILCS from other similar computational protocols, and allows for the use of relevant fragments that would normally phase separate from water. For the same reason, the repulsive potential greatly extends the palette of testable fragments relative to that of experimental protocols, which allows SILCS to probe the utility of fragments that would be difficult or impossible to study experimentally.
The case of Protocol 4 presents one extreme, where sampling is compromised because the fragments are too hydrophobic and phase separate from water. Water-miscible fragments fall on the other side of the spectrum in that they may fail to identify hydrophobic binding sites because they prefer to stay solvated in the aqueous phase. This is apparently the case for Protocol Acn where, on visual inspection, the acetonitrile molecules remain uniformly distributed in the aqueous phase, but based on the FragMaps, fail to sample CBS1 to any appreciable degree (Figure 4f). This again highlights the utility of the SILCS approach, which is not limited by considerations of fragment hydrophilicity or hydrophobicity because of the use of the inter-fragment repulsive potential.
The structure of CBS2 is less characterized than CBS1 because there is only one available example of a small-molecule:IL-2 crystal structure. Moreover, in this lone example the inhibitor is covalently tethered to the protein at a Tyr31Cys mutation.43 Another difference between CBS1 and 2 is that CBS1 lies at the IL-2:IL-2Rα interface whereas CBS2 does not. Additionally, while it was initially suggested that the location of the covalently tethered inhibitor in CBS1 lay near the IL-2:IL-2Rβ interface,43 subsequent crystal structures that included all three chains of IL-2R show that this is clearly not the case.39,40 Thus, even though CBS2 is an inducible pocket, it is not a binding site for native ligands, the α, β, and γ chains of IL-2R. What is interesting about CBS2 in the context of co-crystals with complete IL-2R is that this region is occupied by the sidechain of IL-2 residue Leu80 while in the apo IL-2 crystal it is occupied by Phe78. Thus, CBS2 may not have biological relevance from the standpoint of binding directly to a component of IL-2R, but rather acts as a slot to allow conformational switching of IL-2 to better accommodate IL-2R binding. Indeed, the Tyr31Cys-tethered small-molecule binding to CBS2 has a cooperative effect on the small-molecule binding at CBS1,43 suggesting that conformational changes at CBS2 are communicated to the IL-2Rα binding interface of IL-2.
Similar results are seen for CBS2 with regard to the relative effectiveness of the different protocols, with one notable exception (Figure 5). As with CBS1, Protocol 1 and 3 identify CBS2 and, as with CBS1, Protocols 4 and Acn fail to identify CBS2 (Figure 5). The one difference with regard to the effectiveness of the protocols is the failure of Protocol 2 to identify CBS2 even though it does identify CBS1. CBS2 is even deeper than CBS1 (Figure 5), and this may be one reason why the presence of the Cα atom positional restraints diminishes protein flexibility to a degree as to be incompatible with robust sampling of CBS2. This argues that, for sampling of particularly deep cryptic binding sites, the preferable approach is to use SILCS either with full protein flexibility and post-run determination and exclusion of denaturing trajectories (Protocol 1) or with “core” restraints on the protein similar to those in Protocol 3. Nevertheless, it should be emphasized that CBS1 was successfully sampled using the original SILCS protocol having Cα atom positional restraints, indicating that full protein flexibility is not required in all cases as previously suggested.34 In addition to CBS1 and CBS2, there are other sites that demonstrate benzene and propane binding at the free-energy threshold used for CBS visualization. However, it is unclear whether these sites are true or false positives due to the lack of available experimental data. That said, the same balance of forces between the protein, fragment, and water atoms that resulted in detection of CBS1 and CBS2 is at play throughout the system, suggesting that these additional sites would have some experimental affinity for the fragments.
Figure 5.
Small-molecule compound binding to cryptic binding site 2 and FragMaps for all protocols. Green contours are for propane, purple for benzene, and blue for acetonitrile. (a) The molecular surface of the starting x-ray crystallographic conformation of unbound IL-2 is shown along with FragMaps from all protocols. (b-f) FragMaps from Protocols 1 through 4 and Acn, respectively, along with the inhibitor conformation from the single, covalent IL-2:inhibitor complex aligned with unbound IL-2, as well as the secondary structure of unbound crystallographic IL-2 (ribbons) and Phe78 (cyan balls-and-sticks) in that structure. For visibility, the molecular surface has been cut away in panels b-f. The inhibitor is not visible in panel a as it is underneath the molecular surface.
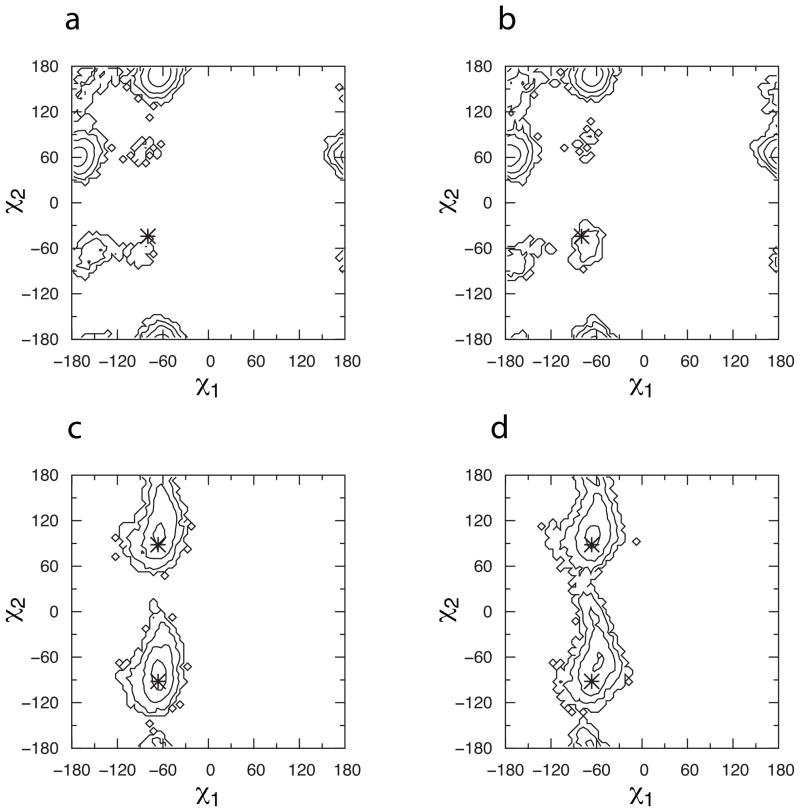
When superimposed on the starting crystallographic conformation, the high-density region located in CBS1 overlaps with the Leu72 sidechain and the high-density region located in CBS2 overlaps with the Phe78 sidechain. Analysis of sidechain dihedral distributions and contact distances was done to elucidate conformational changes that might enable fragment sampling of CBS1 and CBS2. Comparing Protocol 3 with the control protocol lacking fragments, there is no difference in the conformational properties of either of these two sidechains as judged by the sampling of the χ1 and χ2 sidechain dihedrals (Figure 6). In the case of Phe78, the sidechain samples free-energy minima consistent with the crystallographic conformation. For Leu72, while there is no sampling difference between the two protocols, the two deep free-energy minima sampled are located in a different region of conformational space than the starting crystallographic conformation of χ1/χ2 = −80°/−44° (it should be noted that all the other IL-2 crystal structures considered in this study – 1M4A, 1M4B, 1M48, 1M49, 1NBP, 1PW6, 1PY2, 1Z92, 2B5I, and 2ERJ – have their Leu72 sidechain in a conformation consistent with the sampled free-energy minimum at χ1/χ2 = −60°/180°). However, neither of the sampled sidechain minima is consistent with a conformational change sufficient to displace the Leu72 sidechain from CBS1. Therefore, Leu72 and Phe78 sidechain conformational sampling is not affected by the presence of fragments and does not account for the opening of either CBS1 or CBS2.
Figure 6.
Cryptic binding site sidechain dihedral distributions. (a) CBS1 Leu72, Protocol 3; (b) CBS1 Leu72, control protocol; (c) CBS2 Phe78, Protocol 3; (d) CBS2 Phe78, control protocol. Data are shown as Boltzmann-inverted probabilities aggregated across the final 5 ns of all simulations in a given protocol. Contours are every 1 kcal/mol, and *’s indicate starting crystallographic values, with two *’s for Phe78 due to rotational symmetry in χ2. Leu72 χ2 is defined using atom CD1.
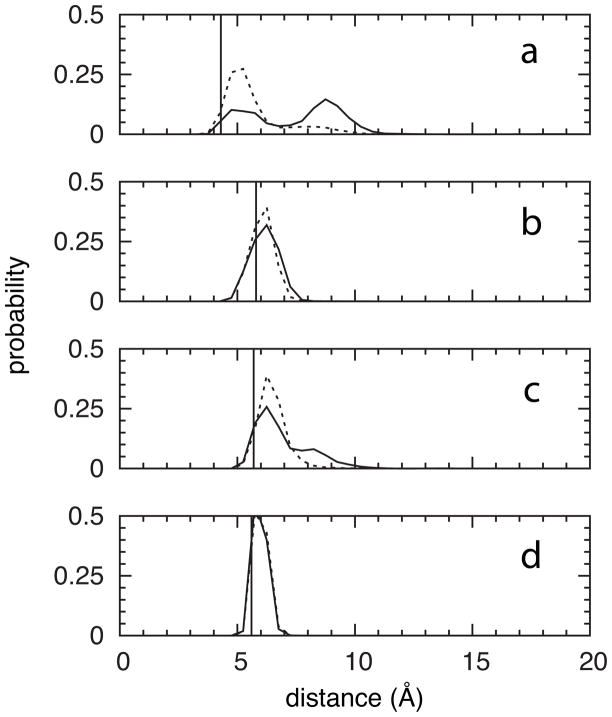
In contrast to this finding, there is a clear fragment effect on the contacts that the Leu72 and Phe78 sidechains make with other residues. All crystallographic atoms within 6 Å of Leu72 CG were initially selected, and from these a subset of atoms was defined such that at most one atom was included from each residue’s backbone and from each residue’s sidechain, yielding a list of 11 nearby non-hydrogen atoms. Analysis of the distance distributions for these atoms revealed that fragments induce opening of the cleft between residues 35–42 on one side and residues 69–72 on the other (Figure 7a), whereas the relative position of Leu72 in the residue 69–72 stretch is unperturbed (Figure 7b). A similar procedure using Phe78 CG – resulting in a list of 18 nearby non-hydrogen atoms – showed separation from residues 23–24 relative to the crystallographic and control protocol distances (Figure 7c), as well as separation from residues 70–74, 80–81, and 85, no change with respect to residues 27–28 and 75–79 (Figure 7d), and, interestingly, compaction with respect to residue 82. In summary, local changes in inter-residue contacts relative to the crystallographic and control protocol distances accommodate fragment binding, as opposed to motion of the Leu72 and Phe78 sidechains that occlude CBS1 and CBS2, respectively, in the crystallographic conformation of the apo protein.
Figure 7.
Cryptic binding site contact distances. Probabilities are shown for the (a) CBS1 Leu72 CG – Met39 CG, (b) CBS1 Leu72 CG – Val69 CB, (c) CBS2 Phe78 CG – Ile24 CA, and (d) CBS2 Phe78 CG – Lys76 C distances. Data have been aggregated across the final 5 ns of all simulations in a given protocol. Solid line = Protocol 3, dotted line = control protocol, and vertical line = starting crystallographic value.
It is worth emphasizing that the present results demonstrate that the identity, and hence physical properties, of the fragments play a critical role in detecting hot spots, and that no amount of protein flexibility can mitigate for mismatch between the type of binding sites (e.g. hydrophobic) and the physical properties (e.g. hydrophilic) of the fragments employed to probe for such binding sites. The SILCS inter-fragment repulsive potential permits the inclusion of fragments that would normally phase-separate from water, allowing such hydrophobic fragments to be included in the palette of fragments used to probe a target’s surface, and, in the case of IL-2, to identify hydrophobic cryptic binding sites.
Conclusions
One key finding from the present work is that, either through judicious selection of restraining potentials or by identifying and excluding trajectories in which target denaturation occurs, it is possible to incorporate target flexibility in SILCS and related calculations while avoiding fragment-induced target denaturation. The results on the example system, IL-2, are particularly promising because using either approach it was possible to identify both of the “cryptic” binding sites that were absent from the apo crystal structure used to seed the simulations but that are seen in IL-2:small-molecule complexes. Another key finding from the present work is that the physical properties of the fragments used for the simulations have important implications with regard to the ability to detect binding sites. In particular, hydrophobic fragments were able to detect both IL-2 cryptic binding sites, while the water-miscible fragment acetonitrile detected neither site. A key innovation of the SILCS approach is the use of a repulsive potential to prevent aggregation of hydrophobic fragments, thereby allowing such fragments to be used.18 This is in contrast to other computational approaches and to experimental approaches, in which fragment aggregation would limit the types of fragments that can be used. Finally, the results show that even water-miscible fragments can cause target denaturation on the timescale of the MD simulations used to probe for fragment binding, as should be anticipated from MD studies probing the atomic details of, for example, urea- or guanidinium-induced protein50 and nucleic acid denaturation.51 Thus, the methodologies described for preventing target denaturation and for identifying and excluding denaturing trajectories from analysis are anticipated to be useful for both hydrophobic and hydrophilic fragment-based computational approaches to inhibitor discovery.
Methods
Force field and software
The protein52–54 and small molecule55 components of the CHARMM all-atom pairwise-additive biomolecular force field were used for simulations, along with a modified version56 of the rigid TIP3P model57 to represent water molecules. In addition to the standard CHARMM force field bonded and nonbonded potential terms, a nonbonded intermolecular repulsive potential was introduced to prevent aggregation of propane and benzene molecules in the aqueous milieu depending on the simulation protocol (Table 1). This was accomplished through the addition of a massless interaction site to the geometric center of each propane and benzene molecule that resulted in propane:propane, propane:benzene, and benzene:benzene intermolecular repulsion, but which was “invisible” to all other parts of the system, thereby leaving all other propane:system and benzene:system intermolecular interactions unperturbed.18 The CHARMM software58,59 was used for system construction, minimization, simulations, and simulation analyses and the VMD software60 was used for molecular visualization.
System construction
The un-liganded crystal structure of IL-2 from the Protein Data Bank61 (PDB ID: 3INK,36–38 chain C) was used as the starting conformation for all simulations. The Reduce algorithm62 was applied to add hydrogen atoms and optimize adjustable groups (OH, SH, NH3+, Met-CH3, and Asn, Gln, and His sidechain orientation). To account for the missing residues 1–5 and 99–104 in the 133-residue sequence, atomic coordinates were spliced from a solution NMR structure of the IL-2 F52A mutant63 (PDB ID: 1IRL) after alignment of the two structures. This entailed first applying Reduce to 1IRL followed by performing a root-mean-squared (RMS) alignment of the backbone atoms of residues 7–8, copying the NMR residue 1–6 coordinates into the crystal structure, performing an RMS alignment of the backbone atoms of residues 97, 98, 105, and 106, and copying the NMR residue 99–104 coordinates into the crystal structure. The non-spliced coordinates were constrained to their crystallographic positions and, using only the bonded and Lennard-Jones terms of the force field, the spliced coordinates were minimized for 10000 steps using the steepest-descent algorithm64 in order to relieve any bonded strain or steric clashes.
Ten aqueous fragment solutions with ~1 M propane + ~1 M benzene were created by randomly placing either a propane or benzene molecule on each grid point of a grid of the appropriate density overlaid on a box of TIP3P water at the experimental density of water. To create ten different systems, each fragment solution box was combined with the prepared IL-2 coordinates, and solution molecules overlapping protein atoms were deleted. The systems were trimmed to dimensions of 62 Å × 62 Å × 62 Å, which allowed for at least two layers of water separating the protein measured along its longest principal axis from the nearest box face and at least six layers of water when measured along the shortest principal axis. No counterions were used as the protein is net neutral. A protein+water system lacking propane and benzene was similarly prepared for control simulations.
Ten different aqueous fragment solutions consisting of 50% w/w water:acetonitrile (H3CCN) were created by randomly replacing water molecules with acetonitrile molecules in a box of water having the experimental density of water until in each solution the ratio of watermolecules to acetonitrile molecules equaled the ratio of MWacetonitrile to MWwater (41.05 amu /18.02 amu). From these ten water:acetonitrile solutions, ten different protein:water:acetonitrile systems were created using the procedure detailed for the generation of protein:water:benzene:propane systems.
Molecular dynamics
Protein atoms were held in place with mass-weighted harmonic restraining potentials on atomic positions and the entire system was minimized using steepest descents.64 With the restraints still in place, the system was gradually heated to 298 K over 10 ps by periodic reassignment of velocities from a Gaussian distribution and then equilibrated for 10 ps at 298 K. Subsequently, the restraints were removed and replaced according to the particular protocol (Table 1), and the system was simulated at 298 K and 1 atm for 20 ns. Conditions of constant temperature and constant pressure were maintained using the Nosé-Hoover thermostat65,66 and the Langevin barostat.67 The equations of motion were integrated using the “leapfrog” integrator68 and a 2 fs timestep. The SHAKE algorithm69 was applied to constrain bonds to hydrogen atoms to their equilibrium lengths and to maintain rigid water geometries through the addition of a virtual bond connecting the hydrogen atoms in molecular water. Periodic boundary conditions70 were employed, Lennard-Jones interactions were smoothly truncated to zero at the cutoff distance of 8 Å by applying a switching function to the energy in the range of 5 Å to 8 Å,71 and electrostatic interactions beyond the cutoff distance were accounted for with the particle-mesh Ewald method72 with a grid spacing of ~1 Å. An isotropic correction to the virial was included to account for Lennard-Jones contributions to the pressure beyond the cutoff distance.70 To simplify post-simulation analysis of MD snapshots, a weak harmonic restraining potential of the form 1.0*(Δr)2 kcal*mol−1*Å−2, where Δr is the displacement in Å, was applied to the center of mass of the protein to keep it centered in the periodic box.
Visualization and analysis
Molecular graphics were generated using VMD v. 1.9,60 and molecular surfaces were generated using SURF.73 PDB structures of IL-2, namely 3INK (chain C),37,38 1M48 (chain A),42 1M4A,42 1M4B42, 1M49 (chain A),42 1NBP,43 1PW6 (chain A),41 1PY2 (chain A),41 1Z92 (chain A),49 2B5I (chain A),39 and 2ERJ (chain D),40 were aligned using the STAMP structural alignment program74 as implemented in the MultiSeq VMD plugin.75 All reported RMSD values are the Cα RMSD values relative to the 3INK crystallographic coordinates, and exclude residues 1–6 and 99–104 to account for the splicing required to construct missing coordinates. FragMaps were prepared from each simulation by binning fragment non-hydrogen atom positions from MD snapshots taken every 2 ps during the 15 ns-20 ns interval. In a given snapshot, only those atoms within 5 Å of the protein were binned, and bins consisted of 1 Å × 1 Å × 1 Å cubic volume elements of a grid spanning the entire system. For each type of FragMap, the respective FragMaps from each of the selected simulations were added together to create a single FragMap. Bin occupancies were converted to Grid Free Energies (GFEs) as detailed previously,20 with average bulk densities computed from simulations of just the respective fragment solutions (propane+benzene+water or acetonitrile+water) at 1 atm and 298 K. Isocontours were −1.2 kcal/mol for benzene and propane maps, and a more permissive −0.5 kcal/mol for acetonitrile maps.
Acknowledgments
This work was supported by funding from the Samuel Waxman Cancer Foundation (A. D. M.), University of New England College of Pharmacy start-up (O. G.), and NIH grants CA107331 (A. D. M.), CA120215 (A. D. M.), and R15GM099022 (O. G.). O. G. is grateful to Professor Ronald D. Hills, Jr. for helpful discussions.
References
- 1.Spyrakis F, BidonChanal A, Barril X, Luque F. J Curr Top Med Chem. 2011;11(2):192–210. doi: 10.2174/156802611794863571. [DOI] [PubMed] [Google Scholar]
- 2.Jorgensen WL. Acc Chem Res. 2009;42(6):724–733. doi: 10.1021/ar800236t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kuntz ID, Blaney JM, Oatley SJ, Langridge R, Ferrin TE. J Mol Biol. 1982;161(2):269–288. doi: 10.1016/0022-2836(82)90153-x. [DOI] [PubMed] [Google Scholar]
- 4.Sotriffer C, Klebe G. Farmaco. 2002;57(3):243–251. doi: 10.1016/s0014-827x(02)01211-9. [DOI] [PubMed] [Google Scholar]
- 5.Campbell SJ, Gold ND, Jackson RM, Westhead DR. Curr Opin Struct Biol. 2003;13(3):389–395. doi: 10.1016/s0959-440x(03)00075-7. [DOI] [PubMed] [Google Scholar]
- 6.Laurie AT, Jackson RM. Curr Protein Pept Sci. 2006;7(5):395–406. doi: 10.2174/138920306778559386. [DOI] [PubMed] [Google Scholar]
- 7.Leis S, Schneider S, Zacharias M. Curr Med Chem. 2010;17(15):1550–1562. doi: 10.2174/092986710790979944. [DOI] [PubMed] [Google Scholar]
- 8.Zhang X, Bajaj C. Comput Syst Bioinformatics Conf. 2007;6:275–286. [PubMed] [Google Scholar]
- 9.Zhong S, MacKerell AD., Jr J Chem Inf Model. 2007;47(6):2303–2315. doi: 10.1021/ci700149k. [DOI] [PubMed] [Google Scholar]
- 10.Kawabata T, Go N. Proteins. 2007;68(2):516–529. doi: 10.1002/prot.21283. [DOI] [PubMed] [Google Scholar]
- 11.Kim D, Cho CH, Cho Y, Ryu J, Bhak J, Kim DS. J Mol Graph Model. 2008;26(7):1104–1112. doi: 10.1016/j.jmgm.2007.10.002. [DOI] [PubMed] [Google Scholar]
- 12.Kupas K, Ultsch A, Klebe G. Proteins. 2008;71(3):1288–1306. doi: 10.1002/prot.21823. [DOI] [PubMed] [Google Scholar]
- 13.Le Guilloux V, Schmidtke P, Tuffery P. BMC Bioinformatics. 2009;10:168. doi: 10.1186/1471-2105-10-168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tripathi A, Kellogg GE. Proteins. 2010;78(4):825–842. doi: 10.1002/prot.22608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Coleman RG, Sharp KA. J Chem Inf Model. 2010;50(4):589–603. doi: 10.1021/ci900397t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Till MS, Ullmann GM. J Mol Model. 2010;16(3):419–429. doi: 10.1007/s00894-009-0541-y. [DOI] [PubMed] [Google Scholar]
- 17.Raunest M, Kandt C. J Mol Graph Model. 2011;29(7):895–905. doi: 10.1016/j.jmgm.2011.02.003. [DOI] [PubMed] [Google Scholar]
- 18.Guvench O, MacKerell AD., Jr PLoS Comp Bio. 2009;5(7):e1000435. doi: 10.1371/journal.pcbi.1000435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Murray CW, Blundell TL. Curr Opin Struct Biol. 2010;20(4):497–507. doi: 10.1016/j.sbi.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 20.Raman EP, Yu W, Guvench O, MacKerell AD., Jr J Chem Inf Model. 2011;51(4):877–896. doi: 10.1021/ci100462t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cozzini P, Kellogg GE, Spyrakis F, Abraham DJ, Costantino G, Emerson A, Fanelli F, Gohlke H, Kuhn LA, Morris GM, Orozco M, Pertinhez TA, Rizzi M, Sotriffer CA. J Med Chem. 2008;51(20):6237–6255. doi: 10.1021/jm800562d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rao B-C, Subramanian J, Sharma SD. Drug Discov Today. 2009;14(7–8):394–400. doi: 10.1016/j.drudis.2009.01.003. [DOI] [PubMed] [Google Scholar]
- 23.Guvench O, MacKerell AD., Jr Curr Opin Struct Biol. 2009;19(1):56–61. doi: 10.1016/j.sbi.2008.11.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bastard K, Saladin A, Prévost C. Int J Mol Sci. 2011;12(2):1316–1333. doi: 10.3390/ijms12021316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ivetac A, McCammon JA. Curr Pharm Des. 2011;17(17):1663–1671. doi: 10.2174/138161211796355056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Totrov M, Abagyan R. Curr Opin Struct Biol. 2008;18(2):178–184. doi: 10.1016/j.sbi.2008.01.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhong SJ, Macias AT, MacKerell AD., Jr Curr Top Med Chem. 2007;7(1):63–82. doi: 10.2174/156802607779318334. [DOI] [PubMed] [Google Scholar]
- 28.Barril X, Morley SD. J Med Chem. 2005;48(13):4432–4443. doi: 10.1021/jm048972v. [DOI] [PubMed] [Google Scholar]
- 29.Damm KL, Carlson HA. J Am Chem Soc. 2007;129(26):8225–8235. doi: 10.1021/ja0709728. [DOI] [PubMed] [Google Scholar]
- 30.Wong CF, Kua J, Zhang Y, Straatsma TP, McCammon JA. Proteins. 2005;61(4):850–858. doi: 10.1002/prot.20688. [DOI] [PubMed] [Google Scholar]
- 31.Bisson WH, Cheltsov AV, Bruey-Sedano N, Lin B, Chen J, Goldberger N, May LT, Christopoulos A, Dalton JT, Sexton PM, Zhang XK, Abagyan R. Proc Natl Acad Sci U S A. 2007;104(29):11927–11932. doi: 10.1073/pnas.0609752104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yu W-M, Guvench O, MacKerell AD, Jr, Qu C-K. J Med Chem. 2008;51(23):7396–7404. doi: 10.1021/jm800229d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eyrisch S, Helms V. J Med Chem. 2007;50(15):3457–3464. doi: 10.1021/jm070095g. [DOI] [PubMed] [Google Scholar]
- 34.Lexa KW, Carlson HA. J Am Chem Soc. 2011;133(2):200–202. doi: 10.1021/ja1079332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Seco J, Luque FJ, Barril X. J Med Chem. 2009;52(8):2363–2371. doi: 10.1021/jm801385d. [DOI] [PubMed] [Google Scholar]
- 36.Brandhuber BJ, Boone T, Kenney WC, McKay DB. J Biol Chem. 1987;262(25):12306–12308. [PubMed] [Google Scholar]
- 37.Brandhuber BJ, Boone T, Kenney WC, McKay DB. Science. 1987;238(4834):1707–1709. doi: 10.1126/science.3500515. [DOI] [PubMed] [Google Scholar]
- 38.McKay DB. Science. 1992;257(5068):412–413. doi: 10.1126/science.257.5068.412. [DOI] [PubMed] [Google Scholar]
- 39.Wang X, Rickert M, Garcia KC. Science. 2005;310(5751):1159–1163. doi: 10.1126/science.1117893. [DOI] [PubMed] [Google Scholar]
- 40.Stauber DJ, Debler EW, Horton PA, Smith KA, Wilson IA. Proc Natl Acad Sci U S A. 2006;103(8):2788–2793. doi: 10.1073/pnas.0511161103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thanos CD, Randal M, Wells JA. J Am Chem Soc. 2003;125(50):15280–15281. doi: 10.1021/ja0382617. [DOI] [PubMed] [Google Scholar]
- 42.Arkin MR, Randal M, DeLano WL, Hyde J, Luong TN, Oslob JD, Raphael DR, Taylor L, Wang J, McDowell RS, Wells JA, Braisted AC. Proc Natl Acad Sci U S A. 2003;100(4):1603–1608. doi: 10.1073/pnas.252756299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Hyde J, Braisted AC, Randal M, Arkin MR. Biochemistry. 2003;42(21):6475–6483. doi: 10.1021/bi034138g. [DOI] [PubMed] [Google Scholar]
- 44.Coan KE, Maltby DA, Burlingame AL, Shoichet BK. J Med Chem. 2009;52(7):2067–2075. doi: 10.1021/jm801605r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boczko EM, Brooks CL., III Science. 1995;269(5222):393–396. doi: 10.1126/science.7618103. [DOI] [PubMed] [Google Scholar]
- 46.Sheinerman FB, Brooks CL., III J Mol Biol. 1998;278(2):439–456. doi: 10.1006/jmbi.1998.1688. [DOI] [PubMed] [Google Scholar]
- 47.Shea JE, Brooks CL., III Annu Rev Phys Chem. 2001;52:499–535. doi: 10.1146/annurev.physchem.52.1.499. [DOI] [PubMed] [Google Scholar]
- 48.Arkin MR, Wells JA. Nat Rev Drug Discovery. 2004;3(4):301–317. doi: 10.1038/nrd1343. [DOI] [PubMed] [Google Scholar]
- 49.Rickert M, Wang X, Boulanger MJ, Goriatcheva N, Garcia KC. Science. 2005;308(5727):1477–1480. doi: 10.1126/science.1109745. [DOI] [PubMed] [Google Scholar]
- 50.Bennion BJ, Daggett V. Proc Natl Acad Sci U S A. 2003;100(9):5142–5147. doi: 10.1073/pnas.0930122100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Priyakumar UD, Hyeon C, Thirumalai D, Mackerell AD., Jr J Am Chem Soc. 2009;131(49):17759–17761. doi: 10.1021/ja905795v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.MacKerell AD, Jr, Bashford D, Bellott M, Dunbrack RL, Evanseck JD, Field MJ, Fischer S, Gao J, Guo H, Ha S, Joseph-McCarthy D, Kuchnir L, Kuczera K, Lau FTK, Mattos C, Michnick S, Ngo T, Nguyen DT, Prodhom B, Reiher WE, Roux B, Schlenkrich M, Smith JC, Stote R, Straub J, Watanabe M, Wiórkiewicz-Kuczera J, Yin D, Karplus M. J Phys Chem B. 1998;102(18):3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 53.MacKerell AD, Jr, Feig M, Brooks CL., III J Comput Chem. 2004;25(11):1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 54.MacKerell AD, Jr, Feig M, Brooks CL., III J Am Chem Soc. 2004;126(3):698–699. doi: 10.1021/ja036959e. [DOI] [PubMed] [Google Scholar]
- 55.Vanommeslaeghe K, Hatcher E, Acharya C, Kundu S, Zhong S, Shim J, Darian E, Guvench O, Lopes P, Vorobyov I, Mackerell AD., Jr J Comput Chem. 2010;31(4):671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Durell SR, Brooks BR, Ben-Naim A. J Phys Chem. 1994;98(8):2198–2202. [Google Scholar]
- 57.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. J Chem Phys. 1983;79(2):926–935. [Google Scholar]
- 58.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. J Comput Chem. 1983;4(2):187–217. [Google Scholar]
- 59.Brooks BR, Brooks CL, III, MacKerell AD, Jr, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M. J Comput Chem. 2009;30(10):1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Humphrey W, Dalke A, Schulten K. J Mol Graph. 1996;14(1):33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 61.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. Nucleic Acids Res. 2000;28(1):235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Word JM, Lovell SC, Richardson JS, Richardson DC. J Mol Biol. 1999;285(4):1735–1747. doi: 10.1006/jmbi.1998.2401. [DOI] [PubMed] [Google Scholar]
- 63.Mott HR, Baines BS, Hall RM, Cooke RM, Driscoll PC, Weir MP, Campbell ID. J Mol Biol. 1995;247(5):979–994. doi: 10.1006/jmbi.1994.0194. [DOI] [PubMed] [Google Scholar]
- 64.Levitt M, Lifson S. J Mol Biol. 1969;46(2):269–279. doi: 10.1016/0022-2836(69)90421-5. [DOI] [PubMed] [Google Scholar]
- 65.Nosé S. Mol Phys. 1984;52(2):255–268. [Google Scholar]
- 66.Hoover WG. Phys Rev A. 1985;31(3):1695–1697. doi: 10.1103/physreva.31.1695. [DOI] [PubMed] [Google Scholar]
- 67.Feller SE, Zhang YH, Pastor RW, Brooks BR. J Chem Phys. 1995;103(11):4613–4621. [Google Scholar]
- 68.Hockney RW. In: Methods in Computational Physics. Alder B, Fernbach S, Rotenberg M, editors. Academic Press; New York: 1970. pp. 136–211. [Google Scholar]
- 69.Ryckaert JP, Ciccotti G, Berendsen HJC. J Comput Phys. 1977;23(3):327–341. [Google Scholar]
- 70.Allen MP, Tildesley DJ. Computer Simulation of Liquids. Oxford University Press; Oxford: 1987. [Google Scholar]
- 71.Steinbach PJ, Brooks BR. J Comput Chem. 1994;15(7):667–683. [Google Scholar]
- 72.Darden T, York D, Pedersen L. J Chem Phys. 1993;98(12):10089–10092. [Google Scholar]
- 73.Varshney A, Brooks FP, Jr, Wright AF. IEEE Computer Graphics and Applications. 1994;14:19–25. [Google Scholar]
- 74.Russell RB, Barton GJ. Proteins. 1992;14(2):309–323. doi: 10.1002/prot.340140216. [DOI] [PubMed] [Google Scholar]
- 75.Roberts E, Eargle J, Wright D, Luthey-Schulten Z. BMC Bioinformatics. 2006;7:382. doi: 10.1186/1471-2105-7-382. [DOI] [PMC free article] [PubMed] [Google Scholar]