Abstract
While several mutagenesis methods have been successfully applied in zebrafish, these mutations do not allowed tissue- or temporal-specific functional analysis. We have developed a strategy that will allow tissue- or temporal-specific disruption of genes in zebrafish. This strategy combines gene trap mutagenesis and FlEx modules containing target sites for site specific recombinases. The gene trap cassette is highly mutagenic in one orientation and non-mutagenic in the opposite orientation, with different fluorescent proteins as indicators of the orientation. The inclusion of the FlEx modules allows two rounds of stable inversion mediated by the Cre and Flp recombinases. This gene trap cassette can be easily delivered via transposons. Through large-scale community-wide efforts, broad genome coverage can be obtained. This should allow investigation of cell/tissue specific gene function of a wide-range of genes.
Introduction
Zebrafish has proven to be a powerful model for forward genetic analysis of vertebrate development. It can be argued that the advantages of zebrafish, including high fecundity, low cost maintenance, and feasibility of live imaging, also make it a good model for reverse genetic analysis. Several strategies have been developed to generate and identify mutations in a gene of interest after chemical mutagenesis or insertional mutagenesis (Jao et al., 2008; Moens et al., 2008; Sood et al., 2006; Wienholds et al., 2003). Furthermore, targeted mutagenesis using preselected zinc finger nucleases is also feasible (Doyon et al., 2008; Foley et al., 2009; Meng et al., 2008). Despite their proven power, one limitation of these approaches is that the genes are mutated in all cells and at all times. This often complicates determining the function of the mutated genes in a specific tissue or a specific process. Conditional mutations are more desirable because they allow inactivation of genes in somatic cells in a temporal or tissue specific manner.
In the mouse conditional mutations generally refer to alleles containing two target sites of a site-specific recombinase configured to delete either an essential part of or the entire gene. These mutations are constructed by modifying a cloned fragment of the gene of interest, which is then used to replace the endogenous locus via homologous recombination in ES cells (Feil, 2007; Gu et al., 1993). Recombination between two target sites mediated by the site-specific recombinase leads to the deletion of the intervening sequence. The Cre recombinase and its target loxP sites are most frequently used in mouse, although the Flp recombinase and its target FRT are also used, but to a lesser extent. Both of these recombinases recognize a 34 bp target site that consists of two 13bp inverted repeats and an 8 bp spacer (Branda and Dymecki, 2004). Each repeat provides a binding site for one recombinase molecule and the spacer dictates the configuration of the heteroduplex intermediate and the outcome of recombination. If the targets are in the same orientation, as used in conditional alleles in mouse, recombination results in the deletion of the intervening sequence. If the targets are in opposite orientation, recombination will result in the inversion of the intervening sequence. With tissue-specific expression of Cre and ligand-dependent Cre (e.g. CreER), conditional alleles allow functional determination of genes in different tissues/organs and at different stages during development as well as in adult animals.
Rationale for conditional mutations
Most genes have a dynamic expression patterns and likely function in multiple cells types and multiple stages. Traditional mutations abolish gene function in all cells from the beginning of embryogenesis, often revealing only the earliest or most conspicuous functions and concealing later or less pronounced functions. This is particularly true for mutations that lead to embryonic lethality, a common feature for most of the available zebrafish mutations. Conditional mutations of these essential genes can reveal additional functional insights. A good example comes from mutation of the insulin receptor in mouse. Global mutation of insulin receptor leads to growth retardation and perinatal death due to diabetic ketoacidosis. But tissue-specific inactivation has revealed that insulin signaling may play distinct roles in different tissues (Kitamura et al., 2003). In skeletal muscle, insulin resistance caused by the lack of insulin receptor has a key role in the development of metabolic syndrome. In the liver, insulin resistance is involved in the development of diabetes; whereas in fat, insulin signaling has a role in the regulation of life span (Bluher et al., 2003; Fisher and Kahn, 2003). Insulin resistance in the CNS regulates both appetite and reproduction (Kahn, 2003). By specifically inhibiting the insulin signaling in these individual tissues, a better overall understanding of the role insulin signaling in multiple physiological processes has been gained. Undoubtedly an approach for tissue specific gene inactivation in zebrafish may reveal a more precise functional understanding of genes, particularly the essential genes. The Ekker lab has developed a “gene-breaking” transposon contains a gene trap cassette that can be deleted (Petzold et al., 2009). However, it only allows temporal or tissue specific rescue but not temporal or tissue specific inactivation of the mutated genes.
Rationale for gene trap mutagenesis to generate conditional mutations
While gene targeting is not available for zebrafish because of the lack of ES cells, gene trapping using the Tol2 transposon system (Kawakami, 2004) is very efficient. Because the mutagenicity of an intronic gene trap insertion is orientation dependent, it is possible to make such alleles conditional if the orientation of the gene trap can be stably switched (Schnutgen et al., 2005; Xin et al., 2005). The orientation switch utilizes site-specific recombinases and a configuration of heterodimers of heterotypic (incompatible) recombinase target sites for Flip and Excision (FlEx) (Floss and Schnutgen, 2008; Schnutgen et al., 2003). With a FlEx approach incorporating targets sites for both Cre and Flp, the intervening sequence can be inverted in two successive rounds. Coupling the FlEx strategy to the efficient Tol2 transgenesis in zebrafish will allow conditional gene inactivation without the need for ES cells.
Rationale for vector design
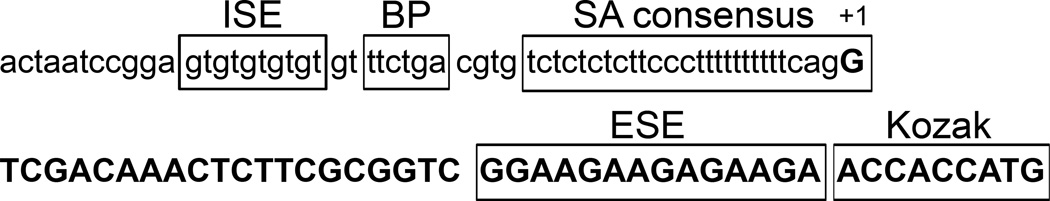
There are three keys to generating an ideal gene-trap vector. First, for a conditional gene trap to be successful the cassette needs to be highly mutagenic in one orientation and non-mutagenic in the other orientation. The mutagenicity is determined by how efficiently the gene trap is recognized as a terminal exon; intercepting and prematurely terminating the transcription of the trapped genes. Because recognition of a terminal exon requires both a splice acceptor and a polyadenylation signal (Levitt et al., 1989; Niwa et al., 1992), inclusion of a strong splice acceptor and a strong polyadenylation signal in the cassette would ensure a highly mutagenic gene trap that is efficiently recognized as a terminal exon. For the splice acceptor, we used the consensus splice acceptor from zebrafish (Yeo et al., 2004) along with intronic and exonic splice enhancers (Figure 1). We also used the tandem repeats of the well characterized bovine growth hormone gene polyadenylation and transcription termination sequence as the polyadenylation signal. Tandem repeat of polyadenylation signal is known to stop transcription more effectively (Maxwell et al., 1989; Soriano, 1999). Conversely, a weak splice acceptor coupled with a weak polyadenylation site will likely produce a nonmutagenic gene trap. For the non mutagenic gene trap, we chose a truncated zebrafish splice acceptor consensus without intronic or exonic splicing enhancers and a truncated polyadenylation signal of the SV40 genome. Second, a mechanism is needed to allow two rounds of stable inversion of the gene trap. Two rounds of stable inversion are necessary to achieve tissue specific gene inactivation for insertions in an initially mutagenic orientation. For this we included FlEx modules for both Cre and Flp to allow two rounds of stable inversion. Third, the vector used to deliver the gene trap needs to be non mutagenic. A mutagenic vector will negate the conditionality of the gene trap. Although retrovirus is the most efficient insertional mutagen, it is not a suitable to for delivering the conditional gene trap since the virus itself is highly mutagenic (Jao et al., 2008). Transposons are suitable for generating conditional gene trap insertions because they require only minimal inverted terminal repeats for transposition and are easy to work with. Furthermore, multiple transposons such as Tol2, piggyBac, and Ac/Ds are available (Emelyanov et al., 2006; Ivics et al., 2009), each potentially with different integration site preferences. The combination of these transposons should make the entire genome more accessible for mutagenesis. Fourth, although not essential, fluorescent reporter genes are useful to indicate the expression pattern of the mutated genes, as well as the orientation of the insertion. The small size of zebrafish makes it difficult to isolate a specific tissue for verification of the mutational status of the trapped gene using DNA-based methods. A fluorescent marker for each orientation would allow visualization of the genotype of individual cells. However, only a fraction of insertions are in frame, and not all will result in levels high enough for detection.
Figure 1.
Methods
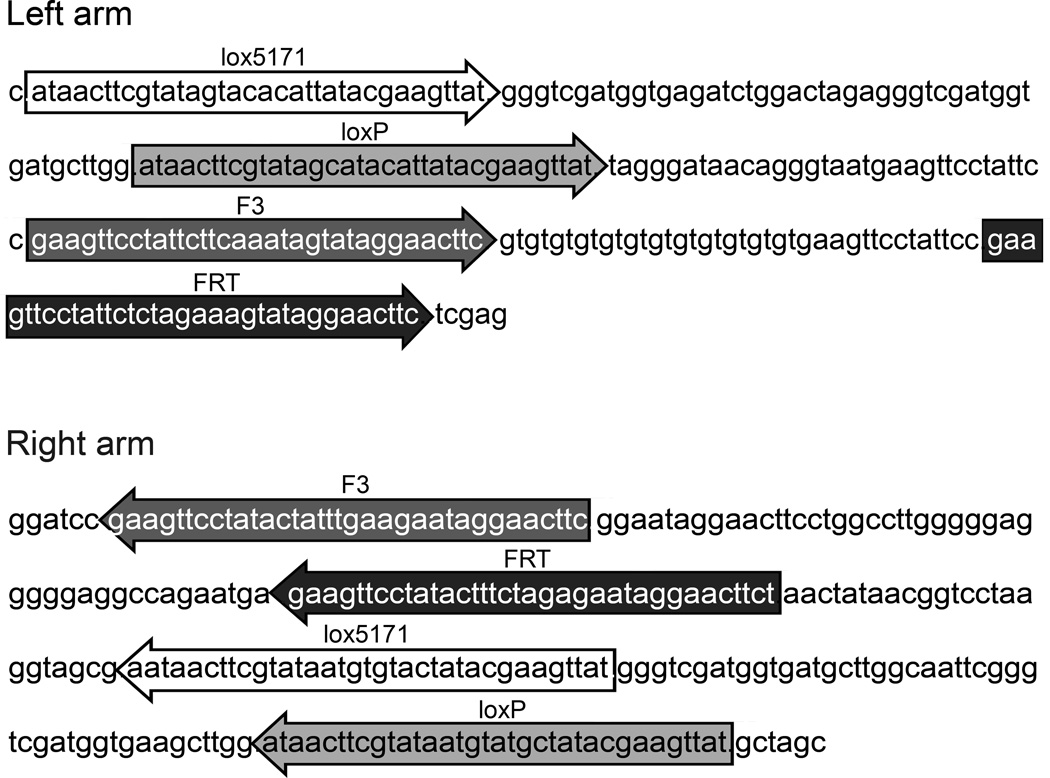
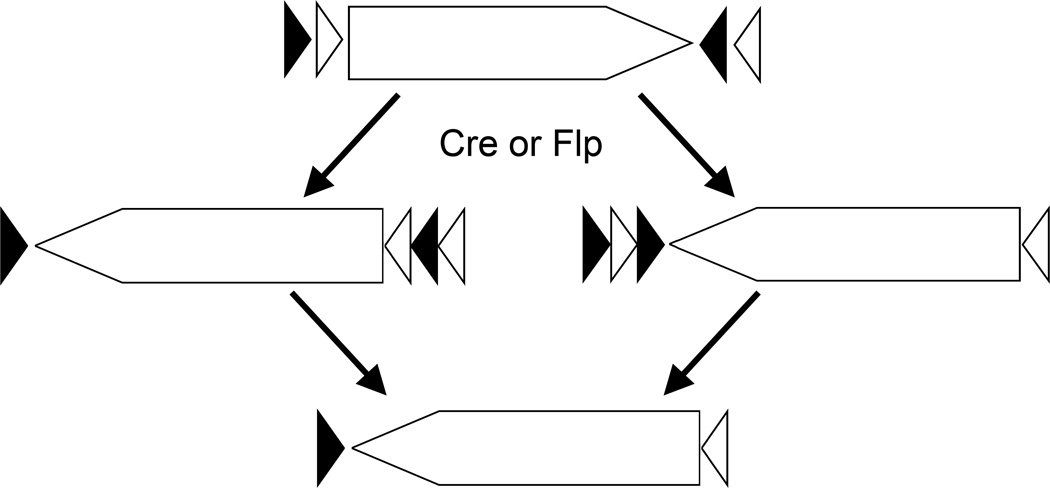
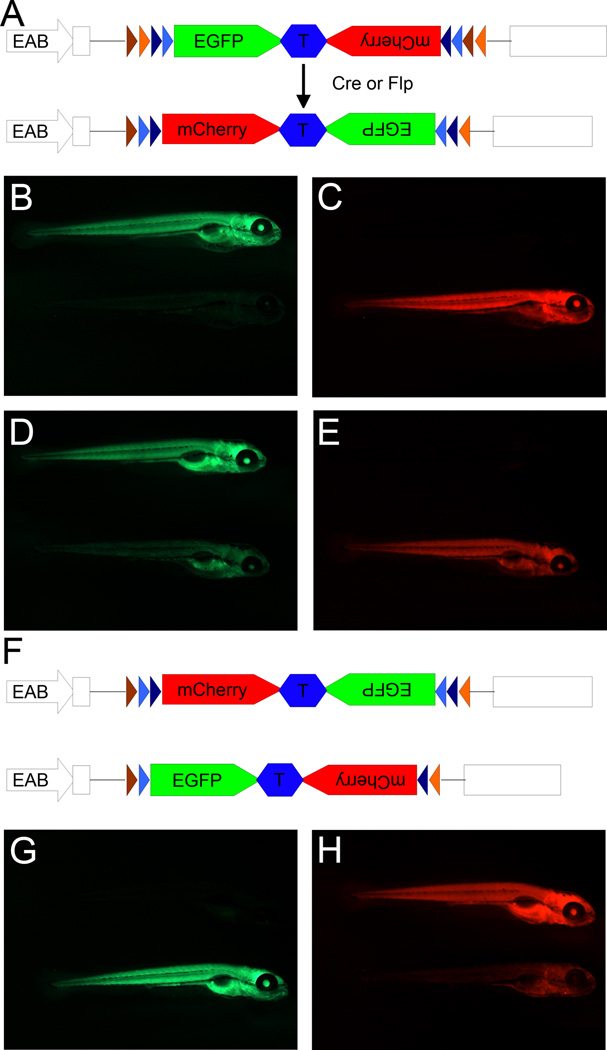
We have constructed a FlEx module that allows two rounds of inversion, incorporated it in a reporter gene for Cre and Flp, and demonstrated its function in zebrafish (Boniface et al., 2009). The FlEx module contains two heterotypic Cre recognition sites, loxP and lox5171, and two heterotypic Flp sites, FRT and F3. Orientation and organization of the module is critical to allow FlEx (Figure 2). For simplicity, Figure 3 outlines the flip and excision process using recognition sites for a single recombinase. Following the initial flipping of the cassette between one set of homotypic recognition sites, the other set of homotypic recognition sites are in an orientation that will result in excision. However, the excision event does not impact the cassette and the end result is an inverted cassette with heterotypic recognition sites on either side of the cassette, which is resistant to further recombination by the same recombinase. But the recognition sites may be used for recombination mediated cassette exchange. Figure 4 demonstrates inversion induced by Tg(hsp70I:Cre) or Tg(hsp70I:Flp) as indicated by switching from EGFP expression to mCherry expression in the reporter. Furthermore, in fish with the reporter stably inverted by Cre recombinase, Tg(hsp70I:Flp) can reinvert the reporter to EGFP following heat shock.
Figure 2.
Figure 3.
Figure 4.
We have been testing the feasibility of FlEx based gene trap mutagenesis in zebrafish and a manuscript detailing this project is forthcoming. For the purposes here, we outline the approach to generate conditional alleles with full details of a pilot screen to be presented elsewhere.
Construction of gene trap cassettes
The mutagenic gene trap consists of a strong splice acceptor, mCherry as the reporter gene, and a strong polyadenylation signal. The strong splice acceptor was generated by annealing and repairing primers SAF and SAR (Table 1). The strong polyadenylation signal is 5× tandem repeats of the bgh polyadenylation site that was produced by conventional molecular cloning. The nonmutagenic gene trap was generated by introducing a truncated zebrafish splice acceptor consensus to the 5’ end of mCitrine by PCR and removing sequence downstream of the HpaI site of the SV40 polyadenylation signal in pmCitrine-N1. The two gene traps were combined tail to tail and inserted between the two FlEx arms in a Tol2 vector and injected into 1-cell stage embryos. An in-frame gene trap resulted in detectable expression of one of the fluorescent reporter genes dependant on the orientation. The mCherry expressing gene trap is much more mutagenic than the mCitrine expressing gene trap using RT-PCR analysis of the trapped gene (data not shown).
Table 1.
primers used for constructing synthetic splice acceptor
| Primer name | Primer sequence 5’ – 3’ |
|---|---|
| SAF | CTCGAGGACTATCCGGAGTGTGTGTGTGTTTCTGACGTGTCTCTCTCTTCCCTTTTTTTTTTCAGGTCGACAAACTCTTCG |
| SAR | CCATGGTGGTTCTTCTCTTCTTCCGACCGCGAAGAGTTTGTCGACCTG |
| LM-PCR Long | CTAGGATTAGCTGCTGGAGTACACGATCGCTTAATAGAGGCACGTGGAACGCGGGC |
| LM-PCR Short CG | PCGGCCCGCGTTCCACGTGZZOG |
| LM-PCR long CATG | CTAGGATTAGCTGCTGGAGTACACGATCGCTTAATAGAGGCACGTGGAACGCGGGCCGCATG |
| LM-PCR UP1 | CTAGGATTAGCTGCTGGAGTACACG |
| LM-PCR NUP2 | ATCGCTTAATAGAGGCACGTGGAAC |
| 5’ TIR1 | CCAAAGGACCAATGAACATGTCTGAC |
| 5’ TIR2 | AACTGGGCATCAGCGCAATTCA |
| 5’ TIR3 | TTGTACTCAAGTAAAGTAAAAATCCC |
| 3’ TIR1 | TCAGCCCCAAAAGAGCTAGGCTTG |
| 3’ TIR2 | GCGTGTACTGGCATTAGATTGTCTGTC |
| 3’ TIR3 | TCAAGTAAGATTCTAGCCAGATAC |
| GT1 | CTTCCGACCGCGAAGAGTTTGTC |
| GT2 | CTTCTAACTATAACGGTCCTAAGGTAGCG |
Generating insertions
Tol2 transgenesis
-
DNA and RNA preparation
-
Prepare the Tol2 gene trap plasmid DNA using standard plasmid preparation protocols.
Purify the gene-trap plasmid using GeneClean (Bio101) according to manufacturer’s instructions.
-
Prepare transposase RNA using mMESSAGE mMACHINE (Ambion) according to manufacturer’s instructions.
Dilute the RNA to 300ng/µl and store in 2µl aliquots at −80°C until use.
-
-
Injection
-
Prepare injection solution
For 5µl injection solution
150 ng purified gene-trap DNA
150 ng transposase RNA
0.5µl 0.5% phenol red
RNAse, DNAse free water to 5 µl
Inject 1nl solution into the cell of 1-cell stage wild-type embryos avoiding injection into the yolk.
Place injected embryos in 0.3× Danieau solution with 1% penicillin and streptomycin stock solution (Invitrogen) and incubate at 28°C
-
-
Recovery
-
4–6 hours after injection remove unfertilized and dead embryos
keep no more than 70 embryos per dish
24 hours after injection remove any dead embryos
once the embryos hatch at 3 days after injection, change to fresh 0.3× Danieau solution without antibiotics, removing the chorions and any abnormal embryos
5 days after injection begin feeding the larvae and raise to maturity
-
Screening for gene-trap events
-
Embryo production
Cross individual gene-trap injected fish to a wild-type fish
-
Collect eggs from successful matings
Hold the gene-trap fish in a separate small tank
-
Screening for fluorescence
-
At 24hpf, 48hpf and 5 dpf
Check for fluorescence on both red and yellow channels
Keep embryos with fluorescent protein expression and raise them to maturity. Genomic DNA can be isolated from tailfin biopsies at 6 weeks of age for insert identification.
if more than 10 embryos with identical expression pattern are available from a single clutch, 1 to 3 embryos can be used for insert identification and the rest raised to maturity.
-
Insert identification via linker-mediated PCR
All oligonucleotide sequences are in Table 1
-
DNA isolation
-
From tailfin
Anaesthetize the fish in 0.05% MESAB
Cut the tail with a scalpel
Transfer the tail into a tube containing 250 µL of 20 mM NaOH
Place fish in individual plastic cups with 350 ml of system water
-
From embryos
Place individual embryos in a tube containing 100 µL of 20 mM NaOH
Incubate the samples at 95 °C for 20 minutes
-
Add 1/5 volume of 1M Tris.HCl to neutralize the samples
50 µl for tailfin, 20 µl for embryos
-
-
Template preparation
-
Genomic DNA digestion
-
In a 50 µl reaction, digest 20 µl of tailfin or embryo lysate (about 1 µg DNA) with one of the following for more than 2 hrs
1µl TaqI at 65°C
1µl NlaIII at 37°C
Heat inactivate the enzyme at 80°C for 20 min
-
-
Linker preparation
-
Determine appropriate linker pair
For Taq1 digestion – CG linker (LM-PCR long + LM-PCR Short CG)
For NlaIII digestion – CATG linker (LM-PCR long CATG + LM-PCR Short CG)
- Anneal linkers
4.5 µl 20mM long linker 4.5 µl 20mM short linker 1.0 µl 10× NEB3 buffer 95°C 2 min 45°C 10 min 4°C 5 min
-
- Ligate linkers to genomic DNA fragments at 16°C for more than 2 hours
50.0 µl Digested genomic DNA 6.0 µl 10× T4 DNA ligase buffer 2.0 µl 10uM annealed linkers 1.0 µl T4 DNA ligase Purify DNA using Qiagen PCR purification columns and elute DNA in 40 µl 10mM Tris pH 8.0
-
-
Amplification of 5’ side flanking sequence
-
First round PCR
- Mix the following
5.0 µl 5× GoTaq buffer (Promega) 3.0 µl 25 mM MgCl2 5.0 µl DNA with linker 0.5 µl 10mM dNTPs 0.5 µl 10 µM UP1 0.5 µl 10 µM 5’TIR1 10 µl H20 0.2 µl Taq DNA polymerase (5U/µl) - Perform PCR using the following thermal profile
1 cycle 94°C 2 minutes 25 cycles 94°C 20 seconds 65°C 3 minutes 1 cycle 72°C 5 min Dilute the PCR 1:25
-
Second round PCR
- Mix the following
10.0 µl 5× GoTaq buffer (Promega) 6.0 µl 25 mM MgCl2 1.0 µl Diluted 1st round product 1.0 µl 10mM dNTPs 1.0 µl 10 µM NUP2 1.0 µl 10 µM 5’TIR2 31.0 µl H20 0.2 µl Taq DNA polymerase (5U/µl) - Perform PCR using the following thermal profile
1 cycle 94°C 2 minutes 35 cycles 94°C 20 seconds 65°C 3 minutes 1 cycle 72°C 5 min
Run the products on a 1.0% agarose gel
Isolate bands, purify fragments and directly sequence using 5’ TIR3
-
-
Amplification of 3’ side flanking sequence
-
First round PCR
- Mix the following
5.0 µl 5× GoTaq buffer (Promega) 3.0 µl 25 mM MgCl2 5.0 µl DNA with linker 0.5 µl 10mM dNTPs 0.5 µl 10 µM UP1 0.5 µl 10 µM 3’TIR1 10 µl H20 0.2 µl Taq DNA polymerase (5U/µl) - Perform PCR using the following thermal profile
1 cycle 94°C 2 minutes 25 cycles 94°C 20 seconds 65°C 3 minutes 1 cycle 72°C 5 min Dilute the PCR 1:25
-
Second round PCR
- Mix the following
10.0 µl 5× GoTaq buffer (Promega) 6.0 µl 25 mM MgCl2 1.0 µl Diluted 1st round product 1.0 µl 10mM dNTPs 1.0 µl 10 µM NUP2 1.0 µl 10 µM 3’TIR2 31.0 µl H20 0.2 µl Taq DNA polymerase (5U/µl) - Perform PCR using the following thermal profile
1 cycle 94°C 2 minutes 35 cycles 94°C 20 seconds 65°C 3 minutes 1 cycle 72°C 5 min
Run the products on a 1.0% agarose gel
Isolate bands, purify fragments and directly sequence using 3’ TIR3
-
Genotyping
A 3-primer PCR genotyping protocol is used to determine the genotype of individual embryos. Two of the three primers target the genomic DNA flanking the insertion and will produce an amplicon from the wildtype allele. The third primer is specific to the insertion and will produce an amplicon with one of the other two primers when the insertion is present. We have used the TIR primers in LM-PCR protocol (5’ TIR1, 5’TIR2, 3’ TIR1 and 3’TIR2) as the gene-trap primers. However, these TIR-specific primers are not ideal to distinguish the orientation of the gene trap. To determine the orientation of the gene trap, we used either one of two primers that are internal of the FlEx modules (GT1 and GT2, Table 1). When designing the gene specific primers, it is important to make sure that the size of the amplicon from the wild-type allele is sufficiently different from the amplicon from the mutant allele to easily distinguish them in a conventional agarose gel.
-
DNA isolation
-
For tailfin from adult fish
Anaesthetize the fish in 0.05% MESAB
Cut the tail with a scalpel
Transfer the tail into a tube containing 250 µL of 20 mM NaOH
Place fish in individual plastic cups with 350 ml of system water
-
For embryos
Place individual embryos in a tube containing 100 µL of 20 mM NaOH
Incubate the samples at 95 °C for 20 minutes
-
Add 1/5 volume of 1M Tris.HCl to neutralize the samples
50 µl for tailfin, 20 µl for embryos
-
-
PCR
-
Prepare a PCR reaction cocktail sufficient for the number of samples
- For a single reaction
5× GoTaq Buffer (Promega) 5.0 µl Sterile Water 15.0 µl 25 mM MgCl2 2.5 µl 10 µM gene-specific primer 1 0.5 µl 10 µM gene-specific primer 2 0.5 µl 10 µM transposon-specific primer 0.5 µl Taq DNA Polymerase (5U/µl) 0.05 µl
Add 24 µl of PCR reaction cocktail to each PCR tube
Add 1 µl of the DNA sample to the corresponding PCR tube
- Perform PCR using the following thermal profile
1 cycle 94°C 2 minutes 35 cycles 94°C 15 seconds 65°C 15 seconds 72°C 1 minute/kb amplicon 1 cycle 72°C 5 min Run 15 µl of each sample on a 1 to 2% agarose gel depending on amplicon size.
-
Work flow for using conditional mutations
Once the genomic location and orientation of the insertions have been determined, one can manipulate the orientation of the gene trap either using Cre or Flp transgenic lines, or by RNA injection. Before utilizing a conditional mutation, one should determine whether the gene of interest is expressed in the target tissue as well as other tissues. One should also determine whether global inactivation of the gene of interest causes a discernable defect outside of the target tissues. Due to the greater availability of transgenic Cre lines in zebrafish, Flp recombinase is ideal for germline inversion. Although transgenic Flp lines can be used, we prefer to globally invert the gene trap by injecting Flp RNA into the 1-cell embryos, which can result in 100% germline inversion. By injecting the Flp RNA into progeny of a gene trap carrier and a Cre transgenic fish, one accomplishes both germline inversion and generation of Cre-expressing carriers of conditional alleles in one generation. We prefer to use a male Cre carrier in these cases. In our hands we have found that even when Cre expression is driven by a previously characterized tissue specific promoter with no maternal activity as indicated by a fluorescent reporter, germline inversion may still occur. This is potentially because the amount of Cre protein required for recombination is much less than the amount of fluorescent protein needed for visual detection. Therefore, using a male Cre carrier avoids the complication of germline inversion. The following outlines how conditional mutations can be manipulated depending on the initial orientation of the insertion.
Starting with a nonmutagenic insertion
-
Global gene inactivation
Cross the insertion carriers to wild-type fish.
Inject Flp RNA in one-cell stage progeny and raise to maturity.
-
Identify injected fish with high levels of germline inversion.
Cross the injected fish with wildtype.
Determine insert orientation in 48 embryos to determine frequency of inversion.
Incross fish with high levels (close to 100%) of germline inversion and observe progeny for phenotypes.
-
Tissue-specific gene inactivation.
Cross the insertion carriers to Cre transgenic fish.
Identify double carrier progeny.
Cross a double carrier to a Cre-negative, insertion carrier and observe progeny for a phenotype in the target tissue.
-
Tissue specific rescue
-
Cross insertion carriers with Cre transgenic fish.
Inject Flp RNA in one-cell stage embryos and raise to maturity.
-
Identify injected fish with high levels of germline inversion.
Cross the injected fish with wildtype.
Determine insert orientation in 48 embryos to determine frequency of inversion.
Genotype the fish with high levels of germline inversion for the Cre transgene.
Cross a germline inverted, Cre-positive fish to a germline inverted Cre-negative fish and observe progeny for rescue of phenotype presented in global inactivation mutants.
-
Starting with a mutagenic insertion
-
Global gene inactivation
Cross two insertion carriers and observe embryos for phenotypes
-
Tissue-specific gene inactivation.
-
Cross insertion carriers with Cre transgenic fish
Inject Flp RNA in one-cell stage embryos and raise to maturity
-
Identify injected fish with high levels of germline inversion
Cross the injected fish with wildtype
Determine insert orientation in 48 embryos to determine frequency of inversion.
Cross a double germline inverted, Cre-positive fish to a Cre-negative, germline inverted fish and observe progeny for a phenotype in the target tissue.
-
-
Tissue specific rescue
Cross the insertion carriers to Cre transgenic fish
Identify double carrier progeny
Cross a double carrier to a Cre-negative, insertion carrier and observe progeny for the rescue in the target tissue.
Injection of Cre or Flp RNA
-
Prepare RNA using mMESSAGE mMACHINE (Ambion) according to manufacturer’s instructions
Dilute the RNA to 300 ng/µl and store in 2µl aliquots at −80°C until use
-
Injection
Dilute RNA with RNase/DNase free water to 100 ng/µl.
Inject 1nl (100 pg RNA) into the cell of 1-cell stage embryos.
Keep injected embryos (no more than 70 per dish) in 0.3× Danieau solution plus antibiotics.
-
Recovery
4–6 hours after injection remove unfertilized and dead embryos.
24 hours after injection remove any dead embryos.
3 days after injection once the embryos hatch, change the solution to antibiotic-free 0.3× Danieau solution, removing the chorions and any abnormal embryos.
5 days after injection begin feeding the larvae and raise to maturity.
Discussion
This approach to generate and use conditional mutations should allow gene functions to be investigated not only in the entire organism but also in a tissue specific manner. Although this approach is not as efficient as tilling or retroviral mutagenesis and is not targeted as ZFN mutagenesis, these types of alleles will have a greater utility and are worth the effort. An ambitious goal is to achieve a scale large enough to approach genome saturation. With the availability of multiple transposon systems and the ease of generating insertions using them, this could be achieved. An attractive perspective is for many laboratories to perform conditional gene trap mutagenesis using different transposon vectors. This will insure the most efficient production of conditional mutations with minimal redundancy.
An essential component to achieve spatial and temporal control of gene inactivation is the availability of suitable Cre- and Flp-expressing lines. In mice, a large collection of such lines has been generated by many labs over the past 15 years [Cre transgenic database, http://www.mshri.on.ca/nagy/Cre-pub.html]. Comparatively, very few lines suitable for conditional gene inactivation exist in zebrafish. The majority of lines used in zebrafish have been based on the hsp70 promoter, although several labs have recently used specific promoters to express the Cre recombinase in a cell/tissue specific manner. For example Cre lines specific for cardiomyocytes (Boniface et al., 2009; Jopling et al.; Kikuchi et al.), oocytes (Liu et al., 2008), pancreatic beta-cells (Hesselson et al., 2009) among others. The predominantly used Cre transgenic line has been Tg(hsp70:Cre) which in theory allows spatial and temporal regulation of Cre expression(Halloran et al., 2000). However, the zebrafish hsp70l promoter has basal activity in the absence of heat shock (Blechinger et al., 2002; Le et al., 2007; Scott et al., 2007) and can be induced even at standard laboratory conditions (Feng et al., 2007) including in germ cells. Another way to achieve precise control of recombinase activity is to use a conditional variant of Cre. The most commonly used conditional recombinase is Cre fused to mutant versions of the ligand-binding domain of estrogen receptors that are insensitive to endogenous hormone but remain sensitive to tamoxifen, a synthetic anti-estrogen (Casanova et al., 2002; Feil et al., 1997; Hayashi and McMahon, 2002; Indra et al., 1999). The combination of a tissue-specific promoter and a tamoxifen-inducible Cre could provide a powerful means for gene functional analysis. However, for robust analysis of tissue specific inhibition, the majority of cells in the tissue would need to be affected. There is some question of the ligand induced efficiency of the Cre-ER and Cre-ERT2 fusion proteins and the ability to affect all cells.
Summary
We have presented here an approach to generating conditional mutations in zebrafish. The core of this approach rests with an asymmetrically mutagenic gene-trap vector and FlEx modules which allow two rounds of stable inversion. This allows the insertion to be flipped from a non-mutagenic to a mutagenic orientation in a tissue or temporal specific manner using an appropriate Cre or Flp transgenic line. Through large-scale, community-wide efforts incorporating different transposon systems, extensive genome coverage can be achieved. This will likely allow conditional mutagenesis of most genes of interest and further extend the usefulness of the zebrafish.
Acknowledgements
We thank members in the Chen laboratory for discussions. The work is supported a grant from National Eye Institute at NIH (EY016092).
References
- Blechinger SR, Evans TG, Tang PT, Kuwada JY, Warren JT, Jr, Krone PH. The heat-inducible zebrafish hsp70 gene is expressed during normal lens development under non-stress conditions. Mech Dev. 2002;112:213–215. doi: 10.1016/s0925-4773(01)00652-9. [DOI] [PubMed] [Google Scholar]
- Bluher M, Kahn BB, Kahn CR. Extended longevity in mice lacking the insulin receptor in adipose tissue. Science. 2003;299:572–574. doi: 10.1126/science.1078223. [DOI] [PubMed] [Google Scholar]
- Boniface EJ, Lu J, Victoroff T, Zhu M, Chen W. FlEx-based transgenic reporter lines for visualization of Cre and Flp activity in live zebrafish. Genesis. 2009;47:484–491. doi: 10.1002/dvg.20526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Branda CS, Dymecki SM. Talking about a revolution: The impact of site-specific recombinases on genetic analyses in mice. Dev Cell. 2004;6:7–28. doi: 10.1016/s1534-5807(03)00399-x. [DOI] [PubMed] [Google Scholar]
- Casanova E, Fehsenfeld S, Lemberger T, Shimshek DR, Sprengel R, Mantamadiotis T. ER-based double iCre fusion protein allows partial recombination in forebrain. Genesis. 2002;34:208–214. doi: 10.1002/gene.10153. [DOI] [PubMed] [Google Scholar]
- Doyon Y, McCammon JM, Miller JC, Faraji F, Ngo C, Katibah GE, Amora R, Hocking TD, Zhang L, Rebar EJ, Gregory PD, Urnov FD, Amacher SL. Heritable targeted gene disruption in zebrafish using designed zinc-finger nucleases. Nat Biotechnol. 2008;26:702–708. doi: 10.1038/nbt1409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emelyanov A, Gao Y, Naqvi NI, Parinov S. Trans-kingdom transposition of the maize dissociation element. Genetics. 2006;174:1095–1104. doi: 10.1534/genetics.106.061184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feil R. Conditional somatic mutagenesis in the mouse using site-specific recombinases. Handb Exp Pharmacol. 2007:3–28. doi: 10.1007/978-3-540-35109-2_1. [DOI] [PubMed] [Google Scholar]
- Feil R, Wagner J, Metzger D, Chambon P. Regulation of Cre recombinase activity by mutated estrogen receptor ligand-binding domains. Biochem Biophys Res Commun. 1997;237:752–757. doi: 10.1006/bbrc.1997.7124. [DOI] [PubMed] [Google Scholar]
- Feng H, Langenau DM, Madge JA, Quinkertz A, Gutierrez A, Neuberg DS, Kanki JP, Look AT. Heat-shock induction of T-cell lymphoma/leukaemia in conditional Cre/lox-regulated transgenic zebrafish. Br J Haematol. 2007;138:169–175. doi: 10.1111/j.1365-2141.2007.06625.x. [DOI] [PubMed] [Google Scholar]
- Fisher SJ, Kahn CR. Insulin signaling is required for insulin's direct and indirect action on hepatic glucose production. J Clin Invest. 2003;111:463–468. doi: 10.1172/JCI16426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Floss T, Schnutgen F. Conditional gene trapping using the FLEx system. Methods Mol Biol. 2008;435:127–138. doi: 10.1007/978-1-59745-232-8_9. [DOI] [PubMed] [Google Scholar]
- Foley JE, Maeder ML, Pearlberg J, Joung JK, Peterson RT, Yeh JR. Targeted mutagenesis in zebrafish using customized zinc-finger nucleases. Nat Protoc. 2009;4:1855–1867. doi: 10.1038/nprot.2009.209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu H, Zou YR, Rajewsky K. Independent control of immunoglobulin switch recombination at individual switch regions evidenced through Cre-loxP-mediated gene targeting. Cell. 1993;73:1155–1164. doi: 10.1016/0092-8674(93)90644-6. [DOI] [PubMed] [Google Scholar]
- Halloran MC, Sato-Maeda M, Warren JT, Su F, Lele Z, Krone PH, Kuwada JY, Shoji W. Laser-induced gene expression in specific cells of transgenic zebrafish. Development. 2000;127:1953–1960. doi: 10.1242/dev.127.9.1953. [DOI] [PubMed] [Google Scholar]
- Hayashi S, McMahon AP. Efficient recombination in diverse tissues by a tamoxifeninducible form of Cre: a tool for temporally regulated gene activation/inactivation in the mouse. Dev Biol. 2002;244:305–318. doi: 10.1006/dbio.2002.0597. [DOI] [PubMed] [Google Scholar]
- Hesselson D, Anderson RM, Beinat M, Stainier DY. Distinct populations of quiescent and proliferative pancreatic beta-cells identified by HOTcre mediated labeling. Proc Natl Acad Sci U S A. 2009;106:14896–14901. doi: 10.1073/pnas.0906348106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Indra AK, Warot X, Brocard J, Bornert JM, Xiao JH, Chambon P, Metzger D. Temporally-controlled site-specific mutagenesis in the basal layer of the epidermis: comparison of the recombinase activity of the tamoxifen-inducible Cre-ERT) and Cre-ERT2) recombinases. Nucleic Acids Res. 1999;27:4324–4327. doi: 10.1093/nar/27.22.4324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivics Z, Li MA, Mates L, Boeke JD, Nagy A, Bradley A, Izsvak Z. Transposonmediated genome manipulation in vertebrates. Nat Methods. 2009;6:415–422. doi: 10.1038/nmeth.1332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jao LE, Maddison L, Chen W, Burgess SM. Using retroviruses as a mutagenesis tool to explore the zebrafish genome. Brief Funct Genomic Proteomic. 2008;7:427–443. doi: 10.1093/bfgp/eln038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jopling C, Sleep E, Raya M, Marti M, Raya A, Belmonte JC. Zebrafish heart regeneration occurs by cardiomyocyte dedifferentiation and proliferation. Nature. 2010;464:606–609. doi: 10.1038/nature08899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kahn CR. Knockout mice challenge our concepts of glucose homeostasis and the pathogenesis of diabetes. Exp Diabesity Res. 2003;4:169–182. doi: 10.1155/EDR.2003.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawakami K. Transgenesis and gene trap methods in zebrafish by using the Tol2 transposable element. Methods Cell Biol. 2004;77:201–222. doi: 10.1016/s0091-679x(04)77011-9. [DOI] [PubMed] [Google Scholar]
- Kikuchi K, Holdway JE, Werdich AA, Anderson RM, Fang Y, Egnaczyk GF, Evans T, Macrae CA, Stainier DY, Poss KD. Primary contribution to zebrafish heart regeneration by gata4(+) cardiomyocytes. Nature. 2010;464:601–605. doi: 10.1038/nature08804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kitamura T, Kahn CR, Accili D. Insulin receptor knockout mice. Annu Rev Physiol. 2003;65:313–332. doi: 10.1146/annurev.physiol.65.092101.142540. [DOI] [PubMed] [Google Scholar]
- Le X, Langenau DM, Keefe MD, Kutok JL, Neuberg DS, Zon LI. Heat shock-inducible Cre/Lox approaches to induce diverse types of tumors and hyperplasia in transgenic zebrafish. Proc Natl Acad Sci U S A. 2007;104:9410–9415. doi: 10.1073/pnas.0611302104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levitt N, Briggs D, Gil A, Proudfoot NJ. Definition of an efficient synthetic poly(A) site. Genes Dev. 1989;3:1019–1025. doi: 10.1101/gad.3.7.1019. [DOI] [PubMed] [Google Scholar]
- Liu X, Li Z, Emelyanov A, Parinov S, Gong Z. Generation of oocyte-specifically expressed cre transgenic zebrafish for female germline excision of loxP-flanked transgene. Dev Dyn. 2008;237:2955–2962. doi: 10.1002/dvdy.21701. [DOI] [PubMed] [Google Scholar]
- Maxwell IH, Harrison GS, Wood WM, Maxwell F. A DNA cassette containing a trimerized SV40 polyadenylation signal which efficiently blocks spurious plasmid-initiated transcription. Biotechniques. 1989;7:276–280. [PubMed] [Google Scholar]
- Meng X, Noyes MB, Zhu LJ, Lawson ND, Wolfe SA. Targeted gene inactivation in zebrafish using engineered zinc-finger nucleases. Nat Biotechnol. 2008;26:695–701. doi: 10.1038/nbt1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moens CB, Donn TM, Wolf-Saxon ER, Ma TP. Reverse genetics in zebrafish by TILLING. Brief Funct Genomic Proteomic. 2008;7:454–459. doi: 10.1093/bfgp/eln046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa M, MacDonald CC, Berget SM. Are vertebrate exons scanned during splicesite selection? Nature. 1992;360:277–280. doi: 10.1038/360277a0. [DOI] [PubMed] [Google Scholar]
- Petzold AM, Balciunas D, Sivasubbu S, Clark KJ, Bedell VM, Westcot SE, Myers SR, Moulder GL, Thomas MJ, Ekker SC. Nicotine response genetics in the zebrafish. Proc Natl Acad Sci U S A. 2009;106:18662–18667. doi: 10.1073/pnas.0908247106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnutgen F, De-Zolt S, Van Sloun P, Hollatz M, Floss T, Hansen J, Altschmied J, Seisenberger C, Ghyselinck NB, Ruiz P, Chambon P, Wurst W, von Melchner H. Genomewide production of multipurpose alleles for the functional analysis of the mouse genome. Proc Natl Acad Sci U S A. 2005;102:7221–7226. doi: 10.1073/pnas.0502273102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schnutgen F, Doerflinger N, Calleja C, Wendling O, Chambon P, Ghyselinck NB. A directional strategy for monitoring Cre-mediated recombination at the cellular level in the mouse. Nat Biotechnol. 2003;21:562–565. doi: 10.1038/nbt811. [DOI] [PubMed] [Google Scholar]
- Scott EK, Mason L, Arrenberg AB, Ziv L, Gosse NJ, Xiao T, Chi NC, Asakawa K, Kawakami K, Baier H. Targeting neural circuitry in zebrafish using GAL4 enhancer trapping. Nat Methods. 2007;4:323–326. doi: 10.1038/nmeth1033. [DOI] [PubMed] [Google Scholar]
- Sood R, English MA, Jones M, Mullikin J, Wang DM, Anderson M, Wu D, Chandrasekharappa SC, Yu J, Zhang J, Paul Liu P. Methods for reverse genetic screening in zebrafish by resequencing and TILLING. Methods. 2006;39:220–227. doi: 10.1016/j.ymeth.2006.04.012. [DOI] [PubMed] [Google Scholar]
- Soriano P. Generalized lacZ expression with the ROSA26 Cre reporter strain. Nat Genet. 1999;21:70–71. doi: 10.1038/5007. [DOI] [PubMed] [Google Scholar]
- Wienholds E, van Eeden F, Kosters M, Mudde J, Plasterk RH, Cuppen E. Efficient target-selected mutagenesis in zebrafish. Genome Res. 2003;13:2700–2707. doi: 10.1101/gr.1725103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xin HB, Deng KY, Shui B, Qu S, Sun Q, Lee J, Greene KS, Wilson J, Yu Y, Feldman M, Kotlikoff MI. Gene trap and gene inversion methods for conditional gene inactivation in the mouse. Nucleic Acids Res. 2005;33:e14. doi: 10.1093/nar/gni016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yeo G, Hoon S, Venkatesh B, Burge CB. Variation in sequence and organization of splicing regulatory elements in vertebrate genes. Proc Natl Acad Sci U S A. 2004;101:15700–15705. doi: 10.1073/pnas.0404901101. [DOI] [PMC free article] [PubMed] [Google Scholar]