Introduction
The Encyclopedia of DNA Elements (ENCODE) Project (http://www.genome.gov/10005107) is an international collaboration of research groups funded by the National Human Genome Research Institute, with the goal of delineating all functional elements encoded in the human genome (1). This project began in 2003 with a targeted analysis of a selected 1% of the human genome. The results from the pilot project were published in 2007 (2), and a second phase of funding was then provided to scale the project to the entire human genome. Genome-scale projects in ENCODE involve the identification and quantification of RNA species in whole cells and subcellular compartments, mapping of noncoding and protein-coding genes by manual review and experimental methods, delineation of chromatin and DNA accessibility, mapping of histone modifications and transcription factor-binding sites by ChIP, and measurement of DNA methylation. More recently, ENCODE has adopted additional approaches that have not yet resulted in extensive data sets, including the examination of long-range chromatin interactions, analysis of RNA-binding proteins, and validation of transcriptional enhancers and silencers. To date, >2000 data sets have been deposited for public use by the ENCODE Project at the University of California Santa Cruz (UCSC) Genome Browser (3); to encourage public use of the data sets, a “user's guide” to the ENCODE data sets has been published (4). As the second phase of the ENCODE Project nears completion, the ENCODE Consortium has prepared a large integrative manuscript that includes analyses of experiments from 147 cell types and provides a summary of their functional annotation of the human genome (5). Additionally, other more narrowly focused studies on subsets of ENCODE data have been or will soon be published; for a list of ENCODE publications, see the ENCODE tab at the UCSC Genome Bioinformatics site.
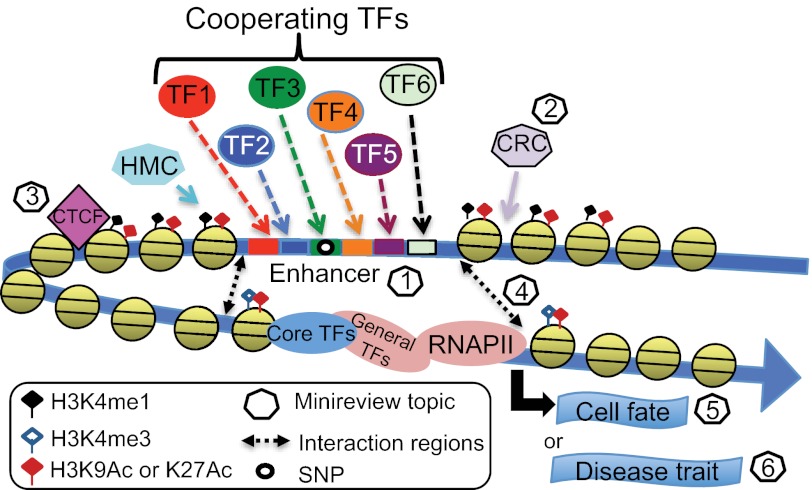
Many new insights concerning the organization and function of genomic elements have come from the ENCODE Project, including the findings that most transcription factors have many thousands of binding sites in the human genome and that these binding sites are distributed non-randomly, with only approximately one-third being located near a transcription start site (5). Many of these distally located regions of transcription factor-binding sites are thought to be transcriptional enhancers. Because enhancers are far from genes, can work in either orientation, and can sometimes skip over the nearest gene, in the past, they have been difficult to characterize. However, the study of enhancers has gained enormous momentum from high throughput methods such as ChIP-seq and comprehensive analyses from genomic projects such as ENCODE and the Roadmap Epigenomics Project. Based on current estimates of up to 50,000 enhancers in any given cell type and the fact that enhancers tend to be cell type-specific, it has been estimated that there are perhaps 105–106 enhancers in the human genome. Studies indicate that the majority of enhancers are composed of transcription factor-binding sites residing within nucleosome-free regions flanked by specific patterns of histone modifications (Fig. 1). The minireview entitled “Chromatin Fingerprint of Gene Enhancer Elements” by Gabriel E. Zentner and Peter C. Scacheri reviews the types of variant and modified histones and histone-modifying complexes found at enhancers and describes subclasses of active and poised enhancers.
FIGURE 1.
Genome-wide characterizations of regulatory regions. Recent genome-wide ChIP-seq studies have revealed 100,00–200,000 regions of open chromatin per cell type, tens of thousands of which are marked by specifically modified histones (e.g. H3K4me1 and H3K27ac) and bound by many different site-specific transcription factors (TFs; see “Chromatin Fingerprint of Gene Enhancer Elements” by Gabriel E. Zentner and Peter C. Scacheri). Creation of these open chromatin regions requires the actions of histone-modifying complexes (HMC), chromatin-remodeling complexes (CRC), and boundary proteins such as CTCF (see “SWI/SNF Chromatin-remodeling Factors: Multiscale Analyses and Diverse Functions” by Ghia Euskirchen, Raymond K. Auerbach, and Michael Snyder and “Genome-wide Studies of CCCTC-binding Factor (CTCF) and Cohesin Provide Insight into Chromatin Structure and Regulation” by Bum-Kyu Lee and Vishwanath R. Iyer). Distal regulatory regions are thought to function by interaction of bound site-specific factors with other transcription factors bound to core promoters via looping of the intervening DNA. For a review of recent experimental and computational methods used to identify intra- and interchromosomal interactions, see “Uncovering Transcription Factor Modules Using One-dimensional and Three-dimensional Analyses” by Xun Lan, Peggy J. Farnham, and Victor X. Jin. Finally, it is becoming increasingly clear that distal regions are critically important in specifying cell fate via regulation of specific cohorts of genes (see “Transcription Factor-mediated Epigenetic Reprogramming” by Camille Sindhu, Payman Samavarchi-Tehrani, and Alexander Meissner) and that SNPs located within distal regulatory regions contribute to the development of many human diseases (see “Genome-wide Epigenetic Data Facilitate Understanding of Disease Susceptibility Association Studies” by Ross C. Hardison). RNAPII, RNA polymerase II.
An emerging concept is that transcription factors bound to distal enhancer elements regulate genes by looping out the intervening DNA and interacting with other factors bound at promoter regions that can be tens to hundreds of kilobases away. It is becoming clear that not only are chromatin-remodeling complexes required to achieve and maintain the nucleosome-free regions of enhancers that are bound by the site-specific factors but that they are also involved in the formation of chromosomal loops. One such chromatin-remodeling complex is SWI/SNF, a DNA-dependent ATPase. Human SWI/SNF complexes contain 10–12 subunits, many of which have alternative forms encoded by different members of gene families, resulting in many different possible SWI/SNF complexes. Components of SWI/SNF have specific protein domains that can recognize the acetylated or methylated histones that are found at enhancer regions, thus providing anchoring to nucleosomes. SWI/SNF can also interact with a variety of site-specific DNA-binding transcription factors. The ability of the SWI/SNF complex to interact with both DNA-bound factors and nucleosomes may contribute to its ability to form or stabilize chromosomal loops. As described in the minireview by Ghia Euskirchen, Raymond K. Auerbach, and Michael Snyder entitled “SWI/SNF Chromatin-remodeling Factors: Multiscale Analyses and Diverse Functions,” changes in abundance, structure, or activity of different components can alter the function of SWI/SNF in different types of normal or diseased cells.
In a recent genome-wide study of SWI/SNF components, it was found that many of the binding sites are also bound by the CCCTC-binding factor (CTCF)2 (6). CTCF is a sequence-specific transcription factor that is thought to serve as a chromatin organizer by acting as a barrier to spreading of epigenomic marks and by associating with the nuclear matrix to form distinct chromatin domains. It can also co-localize with both enhancers and repressors and can prevent communication between distal regions and promoters. Accumulating evidence suggests that CTCF mediates many of its functions by regulating DNA looping. CTCF genomic binding patterns have recently been defined in many different cell types. Most CTCF-binding sites (perhaps those involved in setting up generalized chromosomal domains) are invariant across many cell types. However, some CTCF sites do show cell-type specificity (perhaps those most involved in gene regulation). At a subset of sites, CTCF co-localizes with cohesin, a protein involved in keeping sister chromatids together until the anaphase stage of mitosis. The intriguing idea that cohesin may also be involved in the CTCF-mediated looping of enhancer regions by keeping the two ends of distally located DNA regions together is discussed in the minireview by Bum-Kyu Lee and Vishwanath R. Iyer entitled “Genome-wide Studies of CCCTC-binding Factor (CTCF) and Cohesin Provide Insight into Chromatin Structure and Regulation.”
Methods to identify the sites of intra- and interchromosomal interactions mediated by transcription factors interacting with other transcription factors or chromatin-modifying complexes are rapidly evolving. Current methods used to detect chromosomal loops are described in the minireview by Xun Lan, Peggy J. Farnham, and Victor X. Jin entitled “Uncovering Transcription Factor Modules Using One-dimensional and Three-dimensional Analyses.” Such methods include protein-directed analyses such as ChIA-PET (chromatin interaction analysis with paired-end tag sequencing), which is similar to ChIP-seq except that distal regions brought into close proximity by the factor under analysis are ligated prior to the immunoprecipitation step. Other methods such as 3C (chromosome conformation capture), 4C (circularized chromosome conformation capture), and 5C (carbon-copy chromosome conformation capture) can also detect pairs of genomic loci that are far apart on the genome but close in three-dimensional space. One of the newer methods, Hi-C, provides an unbiased identification of chromosomal interactions across the genome but does not provide information as to which factors mediate the looping. At the present time, both the experimental and analytical steps of these chromosomal interaction methods are difficult and as such are being performed only in a few laboratories. However, it is anticipated that improvements in the protocols and analysis programs will eventually move methods such as ChIA-PET and Hi-C into the toolkit of more research groups.
Understanding how genomic sequence elements regulate normal development and differentiation and how variants in the genome contribute to human diseases are the leading challenges of 21st century medicine. The minireview entitled “Transcription Factor-mediated Epigenetic Reprogramming” by Camille Sindhu, Payman Samavarchi-Tehrani, and Alexander Meissner highlights an important use of genomic and epigenomic data in translational medicine. Studies showing that transcription factors play a pivotal role in regulating and maintaining cellular states suggest that it may be possible to reprogram any cell to another cell type, which could be of enormous importance for treatment of diseases that result in loss of cell function or viability. However, such studies require that we have a deep understanding of the relationships between sets of lineage-specific transcription factors and epigenomic regulators. Sindhu et al. provide a list of interactions between transcription factors and chromatin remodelers that have been identified by genomic profiling and compare the results of different types of molecular profiling of embryonic stem cells and induced pluripotent stem cells, including analysis of coding and noncoding RNAs, histone modifications, and DNA methylation.
A genome-wide association study (GWAS) attempts to define SNPs that are significantly more prevalent in a disease-affected group than in a non-affected group. The National Human Genome Research Institute maintains a catalogue of GWAS results (www.genome.gov/gwastudies/). An important recent finding from the integrative analysis of the ENCODE data sets is that a majority of SNPs associated with human disease lie in or near ENCODE-defined regions that are outside of protein-coding genes (5). As reviewed in “Genome-wide Epigenetic Data Facilitate Understanding of Disease Susceptibility Association Studies” by Ross C. Hardison, the integrative analysis of ENCODE data has shown that the phenotype-associated SNPs in the GWAS catalogue are enriched in nucleosome-free regions bound by transcription factors, i.e. putative enhancer regions. Thus, high throughput genomic assays are providing significant aid to our understanding of how SNPs identified by GWAS can contribute to human disease. Of course, simply determining that a SNP falls within a region having the hallmarks of an enhancer does not identify the gene whose regulation is affected by that SNP. However, it is clear that as the field of genomics moves further into the 21st century, the combination of GWAS, ChIP-seq of transcription factors and modified histones, and application of techniques to globally map chromosomal interactions will provide important new insights into human diseases.
Footnotes
- CTCF
- CCCTC-binding factor
- GWAS
- genome-wide association study.
REFERENCES
- 1. ENCODE Project Consortium (2004) The ENCODE (ENCyclopedia Of DNA Elements) Project. Science 306, 636–640 [DOI] [PubMed] [Google Scholar]
- 2. ENCODE Project Consortium (2007) Identification and analysis of functional elements in 1% of the human genome by the ENCODE Pilot Project. Nature 447, 799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Rosenbloom K. R., Dreszer T. R., Long J. C., Malladi V. S., Sloan C. A., Raney B. J., Cline M. S., Karolchik D., Barber G. P., Clawson H., Diekhans M., Fujita P. A., Goldman M., Gravell R. C., Harte R. A., Hinrichs A. S., Kirkup V. M., Kuhn R. M., Learned K., Maddren M., Meyer L. R., Pohl A., Rhead B., Wong M. C., Zweig A. S., Haussler D., Kent W. J. (2012) ENCODE whole-genome data in the UCSC Genome Browser: update 2012. Nucleic Acids Res. 40, D912–D917 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. ENCODE Project Consortium (2011) A user's guide to the Encyclopedia of DNA Elements (ENCODE). PLoS Biol. 9, e1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. ENCODE Consortium (2012) An integrated encyclopedia of DNA elements in the human genome. Nature, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Euskirchen G. M., Auerbach R. K., Davidov E., Gianoulis T. A., Zhong G., Rozowsky J., Bhardwaj N., Gerstein M. B., Snyder M. (2011) Diverse roles and interactions of the SWI/SNF chromatin remodeling complex revealed using global approaches. PLoS Genet. 7, e1002008. [DOI] [PMC free article] [PubMed] [Google Scholar]