Abstract
Eukaryotic genomes are organized into higher order chromatin architectures by protein-mediated long-range interactions in the nucleus. CCCTC-binding factor (CTCF), a sequence-specific transcription factor, serves as a chromatin organizer in building this complex chromatin structure by linking chromosomal domains. Recent genome-wide studies mapping the binding sites of CTCF and its interacting partner, cohesin, using chromatin immunoprecipitation coupled with deep sequencing (ChIP-seq) revealded that CTCF globally co-localizes with cohesin. This partnership between CTCF and cohesin is emerging as a novel and perhaps pivotal aspect of gene regulatory mechanisms, in addition to playing a role in the organization of higher order chromatin architecture.
Keywords: Chromatin Immunoprecipitation (ChIP), Chromatin Regulation, Cohesin, Functional Genomics, Transcription Factors, CTCF
Introduction
Genetic information in the eukaryotic genome is highly organized. Higher order chromatin organization includes interactions between DNA elements on the same or different chromosomes (1–3). Transcription factors can bridge promoters and distal cis-regulatory elements such as enhancers, insulators, and silencers, with looping out of the intervening DNA. For example, estrogen receptor-α (ER-α)2 mediates intra- and interchromosomal interactions across the human genome, as observed by chromatin interaction analysis with paired-end tag sequencing (ChIA-PET) (4). Similar techniques designed to investigate chromatin-interacting loci suggest the existence of spatial chromatin compartments where genes are differentially regulated based on higher order chromatin structure (5–7). Although some of these methods cannot distinguish between true interactions and mere proximity, they demonstrate that the chromosome does not exist in linear form. Significant progress has been made toward understanding local chromatin structure in terms of histone modifications, but the higher order architecture of chromosomes remains less well understood.
Recent genome-wide assays have shown that the transcription factor CTCF can link chromatin domains through long-range interactions between distal genomic regions, suggesting a crucial role for it as an architect of chromatin conformation (8). Cohesin, the protein complex that holds two sister chromatids together until they partition into daughter cells during mitosis (9), is also seen to interact with CTCF. Genome-wide analysis of cohesin binding using chromatin immunoprecipitation followed by microarrays (ChIP-chip) or deep sequencing (ChIP-seq) indicates that cohesin can also regulate gene expression in conjunction with CTCF (10). Thus, together with cohesin, CTCF is becoming recognized as an important factor involved in transcriptional gene regulation and chromatin architecture organization. Groups like the ENCODE Project Consortium are continuing to generate systematic data sets of CTCF and cohesin binding along with other assays of chromatin structure in a variety of human cells, promising to yield further insights (11).
Here, we review the diverse roles of CTCF and the possible mechanisms behind them and discuss advances in our understanding of the pivotal roles of CTCF and its partner, cohesin, in higher order chromatin architecture and transcription regulation.
Versatile Roles of CTCF
CTCF, first identified as a transcriptional repressor of c-MYC (12), is a zinc-finger protein that is ubiquitously expressed and highly conserved in vertebrates (13). A paralog of CTCF called BORIS is expressed only in testes and may be involved in resetting epigenetic marks in the male germ line (14). In conjunction with hormone receptors, CTCF mediates synergistic transcriptional repression of the chicken lysozyme gene (15) and activates the amyloid β-protein precursor gene (16). CTCF binds to insulators and prevents communication between promoters and enhancers. Its enhancer-blocking activity is seen at many insulators such as the 5′-end of the chicken β-globin locus (17), the imprinting control region (ICR) of the mouse Igf2/H19 gene (18), and the DM1 locus underlying myotonic dystrophy (19). By flanking the 3′- and 5′-boundaries of the β-globin locus in human and mouse, CTCF isolates the β-globin gene from strong neighboring enhancers, suggesting another role for it as a barrier element (17, 20).
CTCF is also implicated in genomic imprinting, namely differential expression of maternally and paternally inherited genes. In somatic cells, the Igf2/H19 ICR shows differential DNA methylation according to parental origin (18, 21, 22). Hypermethylation of the paternally inherited ICR inhibits CTCF binding to it, which silences expression of the H19 gene and also, in conjunction with its distal enhancers, indirectly activates transcription of Igf2. In contrast, CTCF binding to the hypomethylated maternal ICR leads to activation of H19 while repressing Igf2 by isolating its promoter from its distal enhancer through looping interactions.
In mammals, CTCF is a trans-acting factor in X chromosome inactivation, which silences one X chromosome in females. This process involves counting of X chromosome number, mutually exclusive choice, and silencing by noncoding RNAs Xist and Tsix (23, 24). Overexpression of CTCF can cause cell cycle arrest and apoptosis, whereas its knockdown induces proliferation and inhibits differentiation (25, 26), suggesting a role for it as a tumor suppressor.
Genome-wide Mapping of CTCF-binding Sites
Many genome-wide studies have revealed tens of thousands of CTCF-binding sites and shed light on its functions (27–29). An initial study in mouse fetal liver using ChIP-chip found >200 methylation-free CTCF-binding sites, many of which functioned as insulators (30). DNase I hypersensitivity mapping showed that 10% of 3904 DNase I sites were also bound by CTCF, suggesting that a subset of CTCF sites are functional cis-regulatory elements (31). The first genome-wide analysis of CTCF binding using ChIP-chip identified 13,804 sites in human fibroblasts (32), mostly outside promoters. More than 75% of these binding sites contained the consensus CTCF-binding motif (CCGCGNGGNGGCAG) (18). A landmark ChIP-seq study by Barski et al. (33) identified 20,262 CTCF-binding sites in human CD4+ T cells. Analyzed in conjunction with a variety of histone modifications, many CTCF sites were seen to be located between active and repressive chromatin domains, suggesting that CTCF functions as a barrier preventing the spread of these domains. An analysis of 13 pluripotency-related transcription factors and CTCF in mouse embryonic stem cells (ESCs) showed that a subset of CTCF sites coincided with multiple transcription factor-binding loci, in particular with c-MYC and n-MYC, implying that CTCF may cooperate with other transcription factors at some target promoters (34).
Another genome-wide study in human CD4+ T, HeLa, and Jurkat cells showed that CTCF-binding sites were largely invariant across these cell types, but a subset showed cell-type specificity (35). This study provided additional evidence that CTCF acts as a chromatin domain barrier because it was seen to occupy the boundaries between active chromatin regions marked by H2AK5ac and repressive regions marked by H3K27me3. The ENCODE Project Consortium has now identified tens of thousands of CTCF-binding sites in a large number of human cell types, confirming on a genomic scale that CTCF binding is associated with both gene activation and repression (27). However, it is still unclear what proportion of CTCF sites function as insulators involved in enhancer blocking or function as barrier elements.
How Can CTCF Perform These Diverse Functions?
It has been proposed that CTCF plays diverse roles through the ability of its 11 zinc-finger domains to recognize various sequences (36). However, recent genome-wide studies of its binding found only one consensus motif to be overrepresented in most CTCF-binding sites (27, 32, 35). Analysis of cohesin binding (see below) revealed many CTCF-independent cohesin sites, most of which (84%) also contained the consensus CTCF motif (37). Therefore, the CTCF motif may not be the only determinant of its binding to DNA. Given that ChIP does not detect only direct interactions between a protein and its target sites on DNA, it is possible that CTCF plays diverse roles through its binding partners, which it recognizes using its zinc-finger domains. Because many transcription factors can co-localize with CTCF, it is also possible that CTCF acts as a landmark to recruit other transcription factors by constitutively binding on the genome.
Post-transcriptional modification can also contribute to the diverse roles of CTCF. So far, phosphorylation (38, 39), poly(ADP-ribosylation) (40, 41), and sumoylation (42, 43) have been reported. Several lines of evidence support a role for these modifications in CTCF function. A point mutation of CTCF phosphorylation sites affected the expression of its target, c-MYC (38). An insulator assay coupled with 3-aminobenzamide, an inhibitor of poly(ADP-ribose) polymerase, demonstrated that poly(ADP-ribosylation) is required for the chromatin insulator activity of CTCF (40). Although not influencing the DNA binding of CTCF, sumoylation nevertheless represses the c-MYC P2 promoter through its co-localization with the repressive Polycomb protein Pc2 (43). These examples indicate that post-translational modifications of CTCF contribute to its versatile roles.
CTCF binds to the linker regions between nucleosomes at the ICR of the imprinted Igf2/H19 and DM1 loci (19, 44), and these sites are enriched in the H2A.Z histone variant. DNase I hypersensitivity studies have revealed that enhancer blocking is correlated with nucleosome depletion at insulators (45), suggesting that nucleosome positioning is an important aspect of CTCF-mediated insulator function. A genome-wide analysis of nucleosomes and DNase I-hypersensitive sites in relation to CTCF binding identified positioned nucleosomes flanking CTCF sites (46), suggesting that CTCF acts as a landmark to position nucleosomes around its binding sites.
CTCF-mediated Higher Order Chromatin Architecture
An accumulating body of evidence supports that CTCF mediates long-range chromosomal interactions by looping. Chromatin conformation capture assays and fluorescence in situ hybridization indicate that CTCF mediates interchromosomal interactions by promoting the co-localization of the Igf2/H19 locus with the Wsb/Nf1 locus on a different chromosome into a “transcription factory.” Only one allele of each locus is included in such interactions, and it is possible that such interchromosomal interactions mediated by CTCF are a common feature of higher order chromatin structure (47).
An integrative analysis combining data from Hi-C assays, which use proximity ligation to identify long-range chromosomal interactions (7), with genome-wide data on CTCF binding supports the idea that long-range intra- and interchromosomal interactions could be mediated by CTCF (48). Further support comes from the mapping of CTCF-mediated chromatin interaction sites using ChIA-PET, combining ChIP, proximity ligation, and deep sequencing. This study identified 1480 cis- and 336 trans-interacting sites in mouse ESCs (8) and suggested that CTCF organizes the genome into discrete regions enriched with distinct epigenetic marks.
CTCF-mediated interactions may be particularly important in immune cells. CTCF has a crucial role in regulating SATB1 (special AT-rich binding protein 1)-dependent Th2 cytokine expression (49). SATB1 is also a chromatin organizer in thymocytes (50) and organizes loop structures in the MHC class I locus by attaching chromatin to the nuclear matrix, leading to alteration of gene expression (51). There is as yet no evidence for a direct interaction between CTCF and SATB1, but because CTCF can also interact with the nuclear matrix, it is possible that CTCF interacts with SATB1 either directly or indirectly to construct the appropriate higher order chromatin structure (52, 53). Genome-wide comparison between CTCF- and SATB1-binding sites may illuminate the relationship between these chromatin organizers. Recent work has shown that CTCF, possibly in conjunction with cohesin, mediates looping interactions important for Ig locus contraction and V(D)J recombination (54–56).
How Can CTCF Manage Chromatin Organization?
The association of CTCF with matrix-associated regions in the genome allows chromatin to attach to the nuclear matrix and form functionally and structurally distinct domains (52). Such topologically independent loops may result in gene silencing, but most insulator elements may not necessarily be attached to the nuclear matrix. Attachment of chromosomal domains to the nuclear lamina (NL) can also contribute to the higher order structure of chromatin (57, 58). In human fibroblasts, lamina-associated domains (LADs) separate the genome into large discrete domains that interact with the NL. These LADs contain repressive chromatin modifications, and their borders are demarcated by CTCF (52, 59). It is conceivable that CTCF prevents chromatin from anchoring onto the NL at the boundary of LADs by masking interacting sites of chromatin and NL components.
Many CTCF binding partners have been reported, which could aid in the diverse functions of CTCF. They include DNA-binding proteins (YB1, YY1, RFX, CIITA, and Kaiso), chromatin proteins (H2A.Z, SIN3A, CHD8, TAF-1/Set, Suz12, and cohesin), and others (lamin, RNA polymerase II, importins, CP190, topoisomerase II, and nucleophosmin) (60, 61). CTCF interacts with a nuclear matrix phosphoprotein, nucleophosmin, which is localized in the periphery of the nucleolus (62). Co-localization of CTCF and nucleophosmin onto the β-globin insulator resulted in localization of the insulator to the peripheral region of the nucleolus (62). Based on such observations, it is plausible that some CTCF binding partners may be important for organizing chromatin structure by localizing chromatin to specific areas in the nucleus in combination with CTCF.
Cohesin Binds in Association with CTCF and Is Involved in Insulator Function
A spate of genome-wide binding studies have shown co-localization of CTCF and cohesin at their chromosomal sites, suggesting a functional connection (63–66). The cohesin complex includes SMC1, SMC3, SCC3, and the α-kleisin SCC1, also known as MCD1 or RAD21. SMC1 and SMC3 dimerize at one end of their linear, folded-back, coiled-coil structures. SCC1 links the nucleotide-binding domains at the other end of SMC1/3 to form a closed V-shaped ring and also recruits SCC3 to the complex (9). The two sister chromatids are encircled by cohesin rings from S phase to G2, preventing their premature segregation until anaphase of mitosis (67, 68). Cohesin expression is not limited to dividing cells, suggesting additional cellular roles for it. Furthermore, mutations in cohesin underlie diseases such as Cornelia de Lange syndrome (69, 70) and developmental defects (71). Because most of these disease-linked cohesin mutations do not completely abrogate cell division, these disorders may be triggered by malfunction of non-canonical cohesin roles such as altered gene expression rather than loss of chromatid cohesion (66).
In yeast, cohesin is localized to centromeres, pericentric domains, intergenic regions, and ORFs, without a consensus sequence (72). Unlike yeast, mammalian cohesin sites contain consensus sequences similar to the CTCF motif (63, 66). SCC3 associates with CTCF at the c-MYC insulator and also interacts with the ICR of the imprinted Igf2/H19 locus in an allele-specific manner (64). Another genome-wide ChIP-chip analysis identified 8811 cohesin (SCC1) sites and 13,894 CTCF sites in the human genome (66). Like CTCF, the largest proportion of cohesin sites (49%) were located in intergenic regions, and 89% of SCC1 sites overlapped with those of CTCF. An insulator assay revealed that cohesin is essential for CTCF insulator function at the Igf2/H19 imprinting locus, although it is dispensable for CTCF binding on the genome (66). In agreement with these studies, an independent mapping of SCC1/RAD21 sites in the human and mouse genomes also revealed overlap between their binding sites and demonstrated a dependence on CTCF for the recruitment of the cohesin subunits SCC1 and SMC3 to specific CTCF sites (63). However, CTCF was not required for the sister chromatid cohesion functions of cohesin. Other studies mapping different cohesin subunits such as SMC1A and SMC3 in mouse ESCs showed similar results (73). These studies also revealed many CTCF/cohesin sites at promoters, suggesting a joint regulatory role. Although most cohesin sites overlap with CTCF, a significant proportion of each factor's sites are independent of the other, implying CTCF-independent functions of cohesin as well as cohesin-independent CTCF roles.
Several studies have demonstrated cohesin-mediated long-range looping (37, 74–78). Although CTCF-binding sites are highly conserved across cell types (27, 35), loop formation at these CTCF sites occurs in a cell type-specific manner (79). If such loops are mediated by cohesin, it remains unclear how cell type-specific looping is achieved. Moreover, CTCF functions as an enhancer blocker as well as a barrier preventing the spread of heterochromatin. The enhancer-blocking function depends on looping, and cohesin plays an important role in CTCF enhancer-blocking activity. However, it is not clear whether cohesin functions with CTCF to construct chromatin barriers because barrier activity at the CTCF sites of the human and mouse homeobox gene A (HOXA) loci appears to be independent of cohesin (80), suggesting that distinct functions of CTCF may or may not depend on cohesin.
Cohesin Regulates Gene Expression with and without CTCF
A CTCF-independent role for cohesin in gene regulation has been shown in multiple cell types (78). Without any CTCF co-binding, cohesin was functionally co-localized with ER-α in human MCF-7 breast cancer cells and with liver-specific transcription factors in hepatic cells. An integrative analysis of cohesin binding in MCF-7 cells with chromosomal interaction data from ChIA-PET (4) indicated that cohesin was enriched in ER-α-bound regions that were involved in intrachromosomal loops. Cohesin could thus mediate long-range looping in a CTCF-independent manner. SCC1 co-localizes with pluripotency-related factors, including KLF4, OCT4, SOX2, ESSRB, and NANOG, in a CTCF-independent manner, suggesting that cohesin also plays a role in ESC identity (37).
Direct regulation of oncogenic c-MYC by cohesin has been reported in human cells and model organisms (64, 65, 81–84). Expression profiling of disease/mutant cohesin cell lines identified several hundred dysregulated genes (82). Many cohesin sites were enriched in the promoters of dysregulated genes, suggesting direct regulation by cohesin. Some studies have shown significant overlap between genes dysregulated in cohesin mutant cell lines and genes dysregulated in mutants of NIPBL (SCC2), a cohesin-loading factor (81, 82). Accordingly, depletion of cohesin and NIPBL results in similar dysregulation of cohesin target genes (73). These mutants do not show significant defects in chromosome segregation. Thus, it is possible that NIPBL affects gene expression by regulating cohesin loading at regulatory sites.
In one recent example of cohesin-mediated looping in gene regulation, cohesin interacted with the mediator complex, which is a transcriptional coactivator. NIPBL binds to the cohesin-mediator complex and loads cohesin onto promoters, resulting in DNA looping between enhancers and promoters of key pluripotency genes such as OCT4 and NANOG (73). In another example, TAF3, a core promoter factor, localized to a subset of CTCF/cohesin sites and regulated genes by long-range looping, which is indispensible for endoderm lineage differentiation and prevention of premature differentiation of neuroectoderm and mesoderm in mouse ESCs (85). These examples suggest that looping often involves cohesin, but the positioning of the loops and the location of cohesin depend on other factors such as mediator or CTCF.
CTCF and RNA Polymerase II
CTCF can interact with the initiation and elongation forms of RNA polymerase II (RNAPII) in vitro, but in vivo, it displays a preference for the initiation form (86). It has been observed that RNAPII tends to stall at CTCF/cohesin-binding sites (87). It is possible that a CTCF-cohesin complex constrains RNAPII from proceeding along its DNA template. In contrast, a single CTCF-binding site cloned into a luciferase assay vector exhibits transcriptional activity (60). Thus, CTCF may help recruit RNAPII to promoters for initiation but could also mediate promoter-proximal pausing of RNAPII, which poises it for swift expression of developmental or stress-responsive genes (88).
Because splicing occurs co-transcriptionally, pausing or slowing down of RNAPII can affect splicing in addition to elongation and overall transcript levels. CTCF also appears to be involved in RNAPII pausing-dependent splicing. At the CD45 gene, a model for alternative splicing, as well as other genes genome-wide, CTCF binding causes RNAPII pausing and promotes the incorporation of “weak” upstream exons into spliced transcripts. Just as in the case of the Igf2/H19 ICR, CTCF binding to these pause sites is inhibited by DNA methylation, raising the possibility that this epigenetic mark affects splicing by a CTCF-mediated process (89). Interestingly, an independent study showed that cohesin is also recruited to genes where RNAPII pausing occurs (90). Although pausing was shown to be independent of cohesin binding, it is not clear to what extent cohesin and CTCF work together and how in RNAPII pausing-related functions.
Model for the Role of CTCF, Cohesin, and Other Factors in Chromatin Architecture
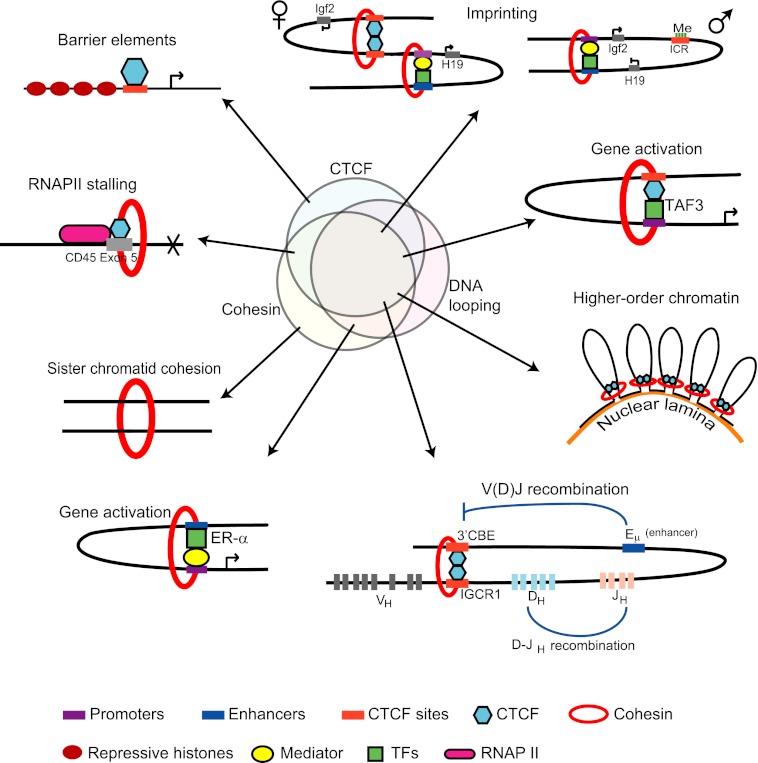
The diverse chromosomal and cellular contexts in which CTCF, cohesin, and other factors function, particularly with regard to long-range looping interactions, are unlikely to have a single underlying mechanism. However, some unifying principles suggest themselves (Fig. 1).
FIGURE 1.
Overview of CTCF and cohesin function. The realms of function of CTCF and cohesin partially overlap, and some of these functions also involve DNA looping. Distinct combinations of these factors and DNA looping are observed at different locations and conditions in cells. Maternally and paternally inherited chromosomes are indicated by appropriate symbols under Imprinting. Although this view is partly speculative, experimental evidence exists for many aspects of CTCF and cohesin function. 3′CBE, 3′-CTCF-binding element; TFs, transcription factors.
First, an attractive explanation for the role of cohesin is that a cohesin ring could encircle two DNA segments from distant loci on the same chromosome to form an intramolecular loop. Although this notion has been proposed before (9, 68, 91), there is as yet no direct molecular evidence for such structures. Second, in this view of looping interactions, the important mechanical stabilizing influence that maintains loops is provided by cohesin rather than other factors like CTCF that are also part of this structure. However, because the cohesin ring lacks DNA-binding specificity and could slide along DNA (92), factors like CTCF, SATB1, or others could serve critical roles as positioning factors, specifying where cohesin locates on the chromosome to generate loops. These positioning factors interact with specific DNA loci, as well as with cohesin, directly or indirectly, to target the placement of loops and lock them in place once formed.
Third, different combinations of factors could be required under different circumstances (Fig. 1). Loops in stable structural features such as chromosome scaffolding interactions, matrix-associated regions, or distinct heterochromatin or euchromatin domains could be somewhat invariant across cell types, and such “permanent” loops are more likely to require CTCF as a positioning factor and cohesin as the clasp. Other situations in which the long-term structural integrity and topology of the loops are important, such as in imprinting and Ig locus contraction, are also likely to require cohesin in conjunction with CTCF as the preferred positioning partner. However, some functions of CTCF, such as its barrier function preventing the spread of heterochromatin could conceivably be achieved just by its strong binding, without the requirement for topological loops, and therefore do not have a requirement for cohesin (78). Interactions between proteins binding at distant loci, such as enhancers and promoters, could form transient loops depending on cell type and physiological state, and they may require cohesin, although perhaps at a subset of these “temporary” regulatory loops. Such regulatory loops could use the ubiquitous CTCF as a positioning factor, but they are more likely to involve alternative positioning factors, such as in the case of ER and mediator in cell type-specific regulation (73, 78).
Finally, cohesin-loading factors like NIPBL/SCC2 could be important not just for the act of cohesin loading but also to ensure that it occurs at proper locations, in conjunction with the appropriate positioning factors. For sister chromatid cohesion, cohesin loading along chromosomes is undoubtedly critical, but perhaps not its location. In contrast, intramolecular looping depends on proper positioning, and it is possible that Nipbl and other mutations seen in Cornelia de Lange syndrome disrupt the location of the intramolecular cohesin clasps and thus have pathological effects in a cell- and tissue-specific manner while largely sparing the chromatid cohesion function.
Although the ideas discussed above can account for many of the current observations, many questions arise. During cell division, cohesin is deposited on individual DNA chromatids before S phase, and DNA replication creates the two catenated chromatids within a cohesin ring. Is it possible for a cohesin ring to be loaded across two non-homologous chromosomal segments to form an intramolecular loop? In yeast, intramolecular cohesin rings can occur at pericentric heterochromatin, so this may not be an insurmountable problem (93). Although much evidence supports the so-called single-ring model for cohesin in chromatid cohesion, alternative models involving two rings are also possible (9, 67). Is it possible that intramolecular loops involve an alternative form of the ring? Once formed, are cohesin rings at the base of intramolecular loops permanent, or can they be removed when necessary? If the latter, how might this occur? Dissociation of cohesin rings around sister chromatids occurs due to proteolytic cleavage of SCC1/RAD21 by separase at the start of anaphase, so this is a highly cell cycle-regulated process. It is unclear whether this mode of cohesin ring dissolution or an alternative mechanism involving ATP hydrolysis or removal by sliding occurs at intramolecular loops.
Conclusions
Eukaryotic genomes are compartmentalized into functional domains that can dynamically reorganize depending on cellular demands. The spatiotemporal architecture of chromatin can establish and maintain active, poised, or repressive chromatin states with regard to gene expression. Genome-wide mapping of CTCF- and cohesin-binding sites has advanced our understanding of the roles of these key proteins in establishing and maintaining chromatin architecture and gene regulation. These studies indicate that CTCF- and cohesin-mediated chromatin organization is prevalent in the genome and is pivotal for malleability in pluripotency, development, and cell identity.
More work remains, however, to determine the molecular details of how cohesin works with CTCF and other factors to organize topological looping and establish regulatory interactions between distant regions of the genome. It also remains to be seen how long-range chromatin-organizing factors such as CTCF and cohesin might cooperate with locally acting chromatin-remodeling factors and chromatin-modifying enzymes to generate distinct transcriptional environments. How this long-range and higher order epigenetic architecture of chromosomes is stably maintained or altered during cell division and development is largely unknown. Continued investigation of the genomic binding locations of these chromatin factors in different cellular contexts coupled with mechanistic studies will be required to shed light on these questions.
Acknowledgment
We thank M. Jayaram for discussions.
This work was supported, in whole or in part, by National Institutes of Health Grants CA130075, CA095548, and HG004563. This work was also supported by Cancer Prevention and Research Institute of Texas Grant RP120194. This is the third article in the Thematic Minireview Series on Results from the ENCODE Project: Integrative Global Analyses of Regulatory Regions in the Human Genome.
- ER-α
- estrogen receptor-α
- CTCF
- CCCTC-binding factor
- ICR
- imprinting control region
- ESC
- embryonic stem cell
- NL
- nuclear lamina
- LAD
- lamina-associated domain
- RNAPII
- RNA polymerase II.
REFERENCES
- 1. Dekker J., Rippe K., Dekker M., Kleckner N. (2002) Capturing chromosome conformation. Science 295, 1306–1311 [DOI] [PubMed] [Google Scholar]
- 2. Simonis M., Klous P., Splinter E., Moshkin Y., Willemsen R., de Wit E., van Steensel B., de Laat W. (2006) Nuclear organization of active and inactive chromatin domains uncovered by chromosome conformation capture-on-chip (4C). Nat. Genet. 38, 1348–1354 [DOI] [PubMed] [Google Scholar]
- 3. Dostie J., Richmond T. A., Arnaout R. A., Selzer R. R., Lee W. L., Honan T. A., Rubio E. D., Krumm A., Lamb J., Nusbaum C., Green R. D., Dekker J. (2006) Chromosome conformation capture carbon copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 16, 1299–1309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Fullwood M. J., Liu M. H., Pan Y. F., Liu J., Xu H., Mohamed Y. B., Orlov Y. L., Velkov S., Ho A., Mei P. H., Chew E. G., Huang P. Y., Welboren W. J., Han Y., Ooi H. S., Ariyaratne P. N., Vega V. B., Luo Y., Tan P. Y., Choy P. Y., Wansa K. D., Zhao B., Lim K. S., Leow S. C., Yow J. S., Joseph R., Li H., Desai K. V., Thomsen J. S., Lee Y. K., Karuturi R. K., Herve T., Bourque G., Stunnenberg H. G., Ruan X., Cacheux-Rataboul V., Sung W. K., Liu E. T., Wei C. L., Cheung E., Ruan Y. (2009) An estrogen receptor-α-bound human chromatin interactome. Nature 462, 58–64 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Duan Z., Andronescu M., Schutz K., McIlwain S., Kim Y. J., Lee C., Shendure J., Fields S., Blau C. A., Noble W. S. (2010) A three-dimensional model of the yeast genome. Nature 465, 363–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Dixon J. R., Selvaraj S., Yue F., Kim A., Li Y., Shen Y., Hu M., Liu J. S., Ren B. (2012) Topological domains in mammalian genomes identified by analysis of chromatin interactions. Nature 485, 376–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lieberman-Aiden E., van Berkum N. L., Williams L., Imakaev M., Ragoczy T., Telling A., Amit I., Lajoie B. R., Sabo P. J., Dorschner M. O., Sandstrom R., Bernstein B., Bender M. A., Groudine M., Gnirke A., Stamatoyannopoulos J., Mirny L. A., Lander E. S., Dekker J. (2009) Comprehensive mapping of long-range interactions reveals folding principles of the human genome. Science 326, 289–293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Handoko L., Xu H., Li G., Ngan C. Y., Chew E., Schnapp M., Lee C. W., Ye C., Ping J. L., Mulawadi F., Wong E., Sheng J., Zhang Y., Poh T., Chan C. S., Kunarso G., Shahab A., Bourque G., Cacheux-Rataboul V., Sung W. K., Ruan Y., Wei C. L. (2011) CTCF-mediated functional chromatin interactome in pluripotent cells. Nat. Genet. 43, 630–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Nasmyth K., Haering C. H. (2009) Cohesin: its roles and mechanisms. Annu. Rev. Genet. 43, 525–558 [DOI] [PubMed] [Google Scholar]
- 10. Dorsett D. (2011) Cohesin: genomic insights into controlling gene transcription and development. Curr. Opin. Genet. Dev. 21, 199–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. ENCODE Project Consortium (2011) A user's guide to the Encyclopedia of DNA Elements (ENCODE). PLoS Biol. 9, e1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Klenova E. M., Nicolas R. H., Paterson H. F., Carne A. F., Heath C. M., Goodwin G. H., Neiman P. E., Lobanenkov V. V. (1993) CTCF, a conserved nuclear factor required for optimal transcriptional activity of the chicken c-myc gene, is an 11-zinc-finger protein differentially expressed in multiple forms. Mol. Cell. Biol. 13, 7612–7624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Filippova G. N., Fagerlie S., Klenova E. M., Myers C., Dehner Y., Goodwin G., Neiman P. E., Collins S. J., Lobanenkov V. V. (1996) An exceptionally conserved transcriptional repressor, CTCF, employs different combinations of zinc fingers to bind diverged promoter sequences of avian and mammalian c-myc oncogenes. Mol. Cell. Biol. 16, 2802–2813 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Loukinov D. I., Pugacheva E., Vatolin S., Pack S. D., Moon H., Chernukhin I., Mannan P., Larsson E., Kanduri C., Vostrov A. A., Cui H., Niemitz E. L., Rasko J. E., Docquier F. M., Kistler M., Breen J. J., Zhuang Z., Quitschke W. W., Renkawitz R., Klenova E. M., Feinberg A. P., Ohlsson R., Morse H. C., 3rd, Lobanenkov V. V. (2002) BORIS, a novel male germ line-specific protein associated with epigenetic reprogramming events, shares the same 11-zinc-finger domain with CTCF, the insulator protein involved in reading imprinting marks in the soma. Proc. Natl. Acad. Sci. U.S.A. 99, 6806–6811 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Lutz M., Baniahmad A., Renkawitz R. (2000) Modulation of thyroid hormone receptor silencing function by co-repressors and a synergizing transcription factor. Biochem. Soc. Trans. 28, 386–389 [PubMed] [Google Scholar]
- 16. Vostrov A. A., Quitschke W. W. (1997) The zinc-finger protein CTCF binds to the APBβ domain of the amyloid β-protein precursor promoter. Evidence for a role in transcriptional activation. J. Biol. Chem. 272, 33353–33359 [DOI] [PubMed] [Google Scholar]
- 17. Bell A. C., West A. G., Felsenfeld G. (1999) The protein CTCF is required for the enhancer-blocking activity of vertebrate insulators. Cell 98, 387–396 [DOI] [PubMed] [Google Scholar]
- 18. Bell A. C., Felsenfeld G. (2000) Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405, 482–485 [DOI] [PubMed] [Google Scholar]
- 19. Filippova G. N., Thienes C. P., Penn B. H., Cho D. H., Hu Y. J., Moore J. M., Klesert T. R., Lobanenkov V. V., Tapscott S. J. (2001) CTCF-binding sites flank CTG/CAG repeats and form a methylation-sensitive insulator at the DM1 locus. Nat. Genet. 28, 335–343 [DOI] [PubMed] [Google Scholar]
- 20. Ristimäki A., Ylikorkala O., Pesonen K., Perheentupa J., Viinikka L. (1991) Human milk stimulates prostacyclin production by cultured human vascular endothelial cells. J. Clin. Endocrinol. Metab. 72, 623–627 [DOI] [PubMed] [Google Scholar]
- 21. Szabó P., Tang S. H., Rentsendorj A., Pfeifer G. P., Mann J. R. (2000) Maternal-specific footprints at putative CTCF sites in the H19 imprinting control region give evidence for insulator function. Curr. Biol. 10, 607–610 [DOI] [PubMed] [Google Scholar]
- 22. Kanduri C., Pant V., Loukinov D., Pugacheva E., Qi C. F., Wolffe A., Ohlsson R., Lobanenkov V. V. (2000) Functional association of CTCF with the insulator upstream of the H19 gene is parent of origin-specific and methylation-sensitive. Curr. Biol. 10, 853–856 [DOI] [PubMed] [Google Scholar]
- 23. Augui S., Nora E. P., Heard E. (2011) Regulation of X chromosome inactivation by the X-inactivation center. Nature Rev. Genet. 12, 429–442 [DOI] [PubMed] [Google Scholar]
- 24. Chao W., Huynh K. D., Spencer R. J., Davidow L. S., Lee J. T. (2002) CTCF, a candidate trans-acting factor for X-inactivation choice. Science 295, 345–347 [DOI] [PubMed] [Google Scholar]
- 25. Torrano V., Chernukhin I., Docquier F., D'Arcy V., León J., Klenova E., Delgado M. D. (2005) CTCF regulates growth and erythroid differentiation of human myeloid leukemia cells. J. Biol. Chem. 280, 28152–28161 [DOI] [PubMed] [Google Scholar]
- 26. Qi C. F., Martensson A., Mattioli M., Dalla-Favera R., Lobanenkov V. V., Morse H. C., 3rd. (2003) CTCF functions as a critical regulator of cell cycle arrest and death after ligation of the B cell receptor on immature B cells. Proc. Natl. Acad. Sci. U.S.A. 100, 633–638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lee B. K., Bhinge A. A., Battenhouse A., McDaniell R. M., Liu Z., Song L., Ni Y., Birney E., Lieb J. D., Furey T. S., Crawford G. E., Iyer V. R. (2012) Cell type-specific and combinatorial usage of diverse transcription factors revealed by genome-wide binding studies in multiple human cells. Genome Res. 22, 9–24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Schmidt D., Schwalie P. C., Wilson M. D., Ballester B., Gonçalves A., Kutter C., Brown G. D., Marshall A., Flicek P., Odom D. T. (2012) Waves of retrotransposon expansion remodel genome organization and CTCF binding in multiple mammalian lineages. Cell 148, 335–348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Maurano M. T., Wang H., Kutyavin T., Stamatoyannopoulos J. A. (2012) Widespread site-dependent buffering of human regulatory polymorphism. PLoS Genet. 8, e1002599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Mukhopadhyay R., Yu W., Whitehead J., Xu J., Lezcano M., Pack S., Kanduri C., Kanduri M., Ginjala V., Vostrov A., Quitschke W., Chernukhin I., Klenova E., Lobanenkov V., Ohlsson R. (2004) The binding sites for the chromatin insulator protein CTCF map to DNA methylation-free domains genome-wide. Genome Res. 14, 1594–1602 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Xi H., Shulha H. P., Lin J. M., Vales T. R., Fu Y., Bodine D. M., McKay R. D., Chenoweth J. G., Tesar P. J., Furey T. S., Ren B., Weng Z., Crawford G. E. (2007) Identification and characterization of cell type-specific and ubiquitous chromatin regulatory structures in the human genome. PLoS Genet. 3, e136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Kim T. H., Abdullaev Z. K., Smith A. D., Ching K. A., Loukinov D. I., Green R. D., Zhang M. Q., Lobanenkov V. V., Ren B. (2007) Analysis of the vertebrate insulator protein CTCF-binding sites in the human genome. Cell 128, 1231–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Barski A., Cuddapah S., Cui K., Roh T. Y., Schones D. E., Wang Z., Wei G., Chepelev I., Zhao K. (2007) High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837 [DOI] [PubMed] [Google Scholar]
- 34. Chen X., Xu H., Yuan P., Fang F., Huss M., Vega V. B., Wong E., Orlov Y. L., Zhang W., Jiang J., Loh Y. H., Yeo H. C., Yeo Z. X., Narang V., Govindarajan K. R., Leong B., Shahab A., Ruan Y., Bourque G., Sung W. K., Clarke N. D., Wei C. L., Ng H. H. (2008) Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133, 1106–1117 [DOI] [PubMed] [Google Scholar]
- 35. Cuddapah S., Jothi R., Schones D. E., Roh T. Y., Cui K., Zhao K. (2009) Global analysis of the insulator-binding protein CTCF in chromatin barrier regions reveals demarcation of active and repressive domains. Genome Res. 19, 24–32 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Filippova G. N., Qi C. F., Ulmer J. E., Moore J. M., Ward M. D., Hu Y. J., Loukinov D. I., Pugacheva E. M., Klenova E. M., Grundy P. E., Feinberg A. P., Cleton-Jansen A. M., Moerland E. W., Cornelisse C. J., Suzuki H., Komiya A., Lindblom A., Dorion-Bonnet F., Neiman P. E., Morse H. C., 3rd, Collins S. J., Lobanenkov V. V. (2002) Tumor-associated zinc finger mutations in the CTCF transcription factor selectively alter its DNA-binding specificity. Cancer Res. 62, 48–52 [PubMed] [Google Scholar]
- 37. Nitzsche A., Paszkowski-Rogacz M., Matarese F., Janssen-Megens E. M., Hubner N. C., Schulz H., de Vries I., Ding L., Huebner N., Mann M., Stunnenberg H. G., Buchholz F. (2011) RAD21 cooperates with pluripotency transcription factors in the maintenance of embryonic stem cell identity. PLoS ONE 6, e19470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Klenova E. M., Chernukhin I. V., El-Kady A., Lee R. E., Pugacheva E. M., Loukinov D. I., Goodwin G. H., Delgado D., Filippova G. N., León J., Morse H. C., 3rd, Neiman P. E., Lobanenkov V. V. (2001) Functional phosphorylation sites in the C-terminal region of the multivalent multifunctional transcriptional factor CTCF. Mol. Cell. Biol. 21, 2221–2234 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. El-Kady A., Klenova E. (2005) Regulation of the transcription factor CTCF by phosphorylation with protein kinase CK2. FEBS Lett. 579, 1424–1434 [DOI] [PubMed] [Google Scholar]
- 40. Yu W., Ginjala V., Pant V., Chernukhin I., Whitehead J., Docquier F., Farrar D., Tavoosidana G., Mukhopadhyay R., Kanduri C., Oshimura M., Feinberg A. P., Lobanenkov V., Klenova E., Ohlsson R. (2004) Poly(ADP-ribosyl)ation regulates CTCF-dependent chromatin insulation. Nat. Genet. 36, 1105–1110 [DOI] [PubMed] [Google Scholar]
- 41. Guerrero P. A., Maggert K. A. (2011) The CCCTC-binding factor (CTCF) of Drosophila contributes to the regulation of the ribosomal DNA and nucleolar stability. PLoS ONE 6, e16401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Kitchen N. S., Schoenherr C. J. (2010) Sumoylation modulates a domain in CTCF that activates transcription and decondenses chromatin. J. Cell. Biochem. 111, 665–675 [DOI] [PubMed] [Google Scholar]
- 43. MacPherson M. J., Beatty L. G., Zhou W., Du M., Sadowski P. D. (2009) The CTCF insulator protein is post-translationally modified by SUMO. Mol. Cell. Biol. 29, 714–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Kanduri M., Kanduri C., Mariano P., Vostrov A. A., Quitschke W., Lobanenkov V., Ohlsson R. (2002) Multiple nucleosome positioning sites regulate the CTCF-mediated insulator function of the H19 imprinting control region. Mol. Cell. Biol. 22, 3339–3344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Zhao H., Kim A., Song S. H., Dean A. (2006) Enhancer blocking by chicken β-globin 5′-HS4: role of enhancer strength and insulator nucleosome depletion. J. Biol. Chem. 281, 30573–30580 [DOI] [PubMed] [Google Scholar]
- 46. Fu Y., Sinha M., Peterson C. L., Weng Z. (2008) The insulator-binding protein CTCF positions 20 nucleosomes around its binding sites across the human genome. PLoS Genet. 4, e1000138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Ling J. Q., Li T., Hu J. F., Vu T. H., Chen H. L., Qiu X. W., Cherry A. M., Hoffman A. R. (2006) CTCF mediates interchromosomal colocalization between Igf2/H19 and Wsb1/Nf1. Science 312, 269–272 [DOI] [PubMed] [Google Scholar]
- 48. Botta M., Haider S., Leung I. X., Lio P., Mozziconacci J. (2010) Intra- and interchromosomal interactions correlate with CTCF binding genome-wide. Mol. Syst. Biol. 6, 426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Ribeiro de Almeida C., Heath H., Krpic S., Dingjan G. M., van Hamburg J. P., Bergen I., van de Nobelen S., Sleutels F., Grosveld F., Galjart N., Hendriks R. W. (2009) Critical role for the transcription regulator CCCTC-binding factor in the control of Th2 cytokine expression. J. Immunol. 182, 999–1010 [DOI] [PubMed] [Google Scholar]
- 50. Yasui D., Miyano M., Cai S., Varga-Weisz P., Kohwi-Shigematsu T. (2002) SATB1 targets chromatin remodeling to regulate genes over long distances. Nature 419, 641–645 [DOI] [PubMed] [Google Scholar]
- 51. Kumar P. P., Bischof O., Purbey P. K., Notani D., Urlaub H., Dejean A., Galande S. (2007) Functional interaction between PML and SATB1 regulates chromatin-loop architecture and transcription of the MHC class I locus. Nat. Cell Biol. 9, 45–56 [DOI] [PubMed] [Google Scholar]
- 52. Dunn K. L., Zhao H., Davie J. R. (2003) The insulator-binding protein CTCF associates with the nuclear matrix. Exp. Cell Res. 288, 218–223 [DOI] [PubMed] [Google Scholar]
- 53. Ohlsson R., Lobanenkov V., Klenova E. (2010) Does CTCF mediate between nuclear organization and gene expression? BioEssays 32, 37–50 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Degner S. C., Verma-Gaur J., Wong T. P., Bossen C., Iverson G. M., Torkamani A., Vettermann C., Lin Y. C., Ju Z., Schulz D., Murre C. S., Birshtein B. K., Schork N. J., Schlissel M. S., Riblet R., Murre C., Feeney A. J. (2011) CCCTC-binding factor (CTCF) and cohesin influence the genomic architecture of the Igh locus and antisense transcription in pro-B cells. Proc. Natl. Acad. Sci. U.S.A. 108, 9566–9571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Guo C., Gerasimova T., Hao H., Ivanova I., Chakraborty T., Selimyan R., Oltz E. M., Sen R. (2011) Two forms of loops generate the chromatin conformation of the immunoglobulin heavy chain gene locus. Cell 147, 332–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Guo C., Yoon H. S., Franklin A., Jain S., Ebert A., Cheng H. L., Hansen E., Despo O., Bossen C., Vettermann C., Bates J. G., Richards N., Myers D., Patel H., Gallagher M., Schlissel M. S., Murre C., Busslinger M., Giallourakis C. C., Alt F. W. (2011) CTCF-binding elements mediate control of V(D)J recombination. Nature 477, 424–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Goldman R. D., Gruenbaum Y., Moir R. D., Shumaker D. K., Spann T. P. (2002) Nuclear lamins: building blocks of nuclear architecture. Genes Dev. 16, 533–547 [DOI] [PubMed] [Google Scholar]
- 58. Schermelleh L., Carlton P. M., Haase S., Shao L., Winoto L., Kner P., Burke B., Cardoso M. C., Agard D. A., Gustafsson M. G., Leonhardt H., Sedat J. W. (2008) Subdiffraction multicolor imaging of the nuclear periphery with three-dimensional structured illumination microscopy. Science 320, 1332–1336 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Guelen L., Pagie L., Brasset E., Meuleman W., Faza M. B., Talhout W., Eussen B. H., de Klein A., Wessels L., de Laat W., van Steensel B. (2008) Domain organization of human chromosomes revealed by mapping of nuclear lamina interactions. Nature 453, 948–951 [DOI] [PubMed] [Google Scholar]
- 60. Zlatanova J., Caiafa P. (2009) CTCF and its protein partners: divide and rule? J. Cell Sci. 122, 1275–1284 [DOI] [PubMed] [Google Scholar]
- 61. Wallace J. A., Felsenfeld G. (2007) We gather together: insulators and genome organization. Curr. Opin. Genet. Dev. 17, 400–407 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Yusufzai T. M., Tagami H., Nakatani Y., Felsenfeld G. (2004) CTCF tethers an insulator to subnuclear sites, suggesting shared insulator mechanisms across species. Mol. Cell 13, 291–298 [DOI] [PubMed] [Google Scholar]
- 63. Parelho V., Hadjur S., Spivakov M., Leleu M., Sauer S., Gregson H. C., Jarmuz A., Canzonetta C., Webster Z., Nesterova T., Cobb B. S., Yokomori K., Dillon N., Aragon L., Fisher A. G., Merkenschlager M. (2008) Cohesins functionally associate with CTCF on mammalian chromosome arms. Cell 132, 422–433 [DOI] [PubMed] [Google Scholar]
- 64. Rubio E. D., Reiss D. J., Welcsh P. L., Disteche C. M., Filippova G. N., Baliga N. S., Aebersold R., Ranish J. A., Krumm A. (2008) CTCF physically links cohesin to chromatin. Proc. Natl. Acad. Sci. U.S.A. 105, 8309–8314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Stedman W., Kang H., Lin S., Kissil J. L., Bartolomei M. S., Lieberman P. M. (2008) Cohesins localize with CTCF at the KSHV latency control region and at cellular c-myc and H19/Igf2 insulators. EMBO J. 27, 654–666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Wendt K. S., Yoshida K., Itoh T., Bando M., Koch B., Schirghuber E., Tsutsumi S., Nagae G., Ishihara K., Mishiro T., Yahata K., Imamoto F., Aburatani H., Nakao M., Imamoto N., Maeshima K., Shirahige K., Peters J. M. (2008) Cohesin mediates transcriptional insulation by CCCTC-binding factor. Nature 451, 796–801 [DOI] [PubMed] [Google Scholar]
- 67. Haering C. H., Farcas A. M., Arumugam P., Metson J., Nasmyth K. (2008) The cohesin ring concatenates sister DNA molecules. Nature 454, 297–301 [DOI] [PubMed] [Google Scholar]
- 68. Wood A. J., Severson A. F., Meyer B. J. (2010) Condensin and cohesin complexity: the expanding repertoire of functions. Nat. Rev. Genet. 11, 391–404 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Krantz I. D., McCallum J., DeScipio C., Kaur M., Gillis L. A., Yaeger D., Jukofsky L., Wasserman N., Bottani A., Morris C. A., Nowaczyk M. J., Toriello H., Bamshad M. J., Carey J. C., Rappaport E., Kawauchi S., Lander A. D., Calof A. L., Li H. H., Devoto M., Jackson L. G. (2004) Cornelia de Lange syndrome is caused by mutations in NIPBL, the human homolog of Drosophila melanogaster Nipped-B. Nat. Genet. 36, 631–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Musio A., Selicorni A., Focarelli M. L., Gervasini C., Milani D., Russo S., Vezzoni P., Larizza L. (2006) X-linked Cornelia de Lange syndrome owing to SMC1L1 mutations. Nat. Genet. 38, 528–530 [DOI] [PubMed] [Google Scholar]
- 71. Liu J., Krantz I. D. (2008) Cohesin and human disease. Annu. Rev. Genomics Hum. Genet. 9, 303–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Glynn E. F., Megee P. C., Yu H. G., Mistrot C., Unal E., Koshland D. E., DeRisi J. L., Gerton J. L. (2004) Genome-wide mapping of the cohesin complex in the yeast Saccharomyces cerevisiae. PLoS Biol. 2, E259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Kagey M. H., Newman J. J., Bilodeau S., Zhan Y., Orlando D. A., van Berkum N. L., Ebmeier C. C., Goossens J., Rahl P. B., Levine S. S., Taatjes D. J., Dekker J., Young R. A. (2010) Mediator and cohesin connect gene expression and chromatin architecture. Nature 467, 430–435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Gause M., Schaaf C. A., Dorsett D. (2008) Cohesin and CTCF: cooperating to control chromosome conformation? BioEssays 30, 715–718 [DOI] [PubMed] [Google Scholar]
- 75. Hadjur S., Williams L. M., Ryan N. K., Cobb B. S., Sexton T., Fraser P., Fisher A. G., Merkenschlager M. (2009) Cohesins form chromosomal cis-interactions at the developmentally regulated IFNG locus. Nature 460, 410–413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Mishiro T., Ishihara K., Hino S., Tsutsumi S., Aburatani H., Shirahige K., Kinoshita Y., Nakao M. (2009) Architectural roles of multiple chromatin insulators at the human apolipoprotein gene cluster. EMBO J. 28, 1234–1245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Nativio R., Wendt K. S., Ito Y., Huddleston J. E., Uribe-Lewis S., Woodfine K., Krueger C., Reik W., Peters J. M., Murrell A. (2009) Cohesin is required for higher order chromatin conformation at the imprinted IGF2-H19 locus. PLoS Genet. 5, e1000739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Schmidt D., Schwalie P. C., Ross-Innes C. S., Hurtado A., Brown G. D., Carroll J. S., Flicek P., Odom D. T. (2010) A CTCF-independent role for cohesin in tissue-specific transcription. Genome Res. 20, 578–588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Hou C., Dale R., Dean A. (2010) Cell-type specificity of chromatin organization mediated by CTCF and cohesin. Proc. Natl. Acad. Sci. U.S.A. 107, 3651–3656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Kim Y. J., Cecchini K. R., Kim T. H. (2011) Conserved, developmentally regulated mechanism couples chromosomal looping and heterochromatin barrier activity at the homeobox gene A locus. Proc. Natl. Acad. Sci. U.S.A. 108, 7391–7396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Kawauchi S., Calof A. L., Santos R., Lopez-Burks M. E., Young C. M., Hoang M. P., Chua A., Lao T., Lechner M. S., Daniel J. A., Nussenzweig A., Kitzes L., Yokomori K., Hallgrimsson B., Lander A. D. (2009) Multiple organ system defects and transcriptional dysregulation in the Nipbl+/− mouse, a model of Cornelia de Lange syndrome. PLoS Genet. 5, e1000650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Liu J., Zhang Z., Bando M., Itoh T., Deardorff M. A., Clark D., Kaur M., Tandy S., Kondoh T., Rappaport E., Spinner N. B., Vega H., Jackson L. G., Shirahige K., Krantz I. D. (2009) Transcriptional dysregulation in NIPBL and cohesin mutant human cells. PLoS Biol. 7, e1000119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Schaaf C. A., Misulovin Z., Sahota G., Siddiqui A. M., Schwartz Y. B., Kahn T. G., Pirrotta V., Gause M., Dorsett D. (2009) Regulation of the Drosophila Enhancer of split and invected-engrailed gene complexes by sister chromatid cohesion proteins. PLoS ONE 4, e6202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Rhodes J. M., Bentley F. K., Print C. G., Dorsett D., Misulovin Z., Dickinson E. J., Crosier K. E., Crosier P. S., Horsfield J. A. (2010) Positive regulation of c-myc by cohesin is direct and evolutionarily conserved. Dev. Biol. 344, 637–649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Liu Z., Scannell D. R., Eisen M. B., Tjian R. (2011) Control of embryonic stem cell lineage commitment by core promoter factor TAF3. Cell 146, 720–731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Chernukhin I., Shamsuddin S., Kang S. Y., Bergström R., Kwon Y. W., Yu W., Whitehead J., Mukhopadhyay R., Docquier F., Farrar D., Morrison I., Vigneron M., Wu S. Y., Chiang C. M., Loukinov D., Lobanenkov V., Ohlsson R., Klenova E. (2007) CTCF interacts with and recruits the largest subunit of RNA polymerase II to CTCF target sites genome-wide. Mol. Cell. Biol. 27, 1631–1648 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Wada Y., Ohta Y., Xu M., Tsutsumi S., Minami T., Inoue K., Komura D., Kitakami J., Oshida N., Papantonis A., Izumi A., Kobayashi M., Meguro H., Kanki Y., Mimura I., Yamamoto K., Mataki C., Hamakubo T., Shirahige K., Aburatani H., Kimura H., Kodama T., Cook P. R., Ihara S. (2009) A wave of nascent transcription on activated human genes. Proc. Natl. Acad. Sci. U.S.A. 106, 18357–18361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Zeitlinger J., Stark A., Kellis M., Hong J. W., Nechaev S., Adelman K., Levine M., Young R. A. (2007) RNA polymerase stalling at developmental control genes in the Drosophila melanogaster embryo. Nat. Genet. 39, 1512–1516 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Shukla S., Kavak E., Gregory M., Imashimizu M., Shutinoski B., Kashlev M., Oberdoerffer P., Sandberg R., Oberdoerffer S. (2011) CTCF-promoted RNA polymerase II pausing links DNA methylation to splicing. Nature 479, 74–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Fay A., Misulovin Z., Li J., Schaaf C. A., Gause M., Gilmour D. S., Dorsett D. (2011) Cohesin selectively binds and regulates genes with paused RNA polymerase. Curr. Biol. 21, 1624–1634 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Phillips J. E., Corces V. G. (2009) CTCF: master weaver of the genome. Cell 137, 1194–1211 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Lengronne A., Katou Y., Mori S., Yokobayashi S., Kelly G. P., Itoh T., Watanabe Y., Shirahige K., Uhlmann F. (2004) Cohesin relocation from sites of chromosomal loading to places of convergent transcription. Nature 430, 573–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Yeh E., Haase J., Paliulis L. V., Joglekar A., Bond L., Bouck D., Salmon E. D., Bloom K. S. (2008) Pericentric chromatin is organized into an intramolecular loop in mitosis. Curr. Biol. 18, 81–90 [DOI] [PMC free article] [PubMed] [Google Scholar]