Abstract
Input from various signaling pathways in conjunction with specific transcription factors (TFs), noncoding RNAs, and epigenetic modifiers governs the maintenance of cellular identity. Endogenous or exogenous TFs operate within certain boundaries, which are set, in part, by the cell type-specific epigenetic landscape. Ectopic expression of selected TFs can override the cellular identity and induce reprogramming to alternative fates. In this minireview, we summarize many of the classic examples and a large number of recent studies that have taken advantage of TF-mediated reprogramming to produce cell types of biomedical relevance.
Keywords: Epigenetics, Regenerative Medicine, Transcription, Transcription Regulation, Transcription/Developmental Factors
Introduction
For many years, it was unclear whether differentiation involves irreversible changes to the genome that would restrict a cell's developmental potential. Early work by Briggs and King (1) and later Gurdon (2) addressed this subject using somatic cell nuclear transfer (SCNT)2 from various donor nuclei into enucleated frog oocytes. Successful generation of viable organisms by SCNT demonstrated that the genome of a differentiated cell does retain all genetic information necessary for normal development. Later experiments were able to expand these seminal findings to mammals (3). SCNT using donor nuclei from genetically marked lymphoid cells (4) and olfactory receptor neurons (5) demonstrated further that even terminally differentiated and post-mitotic genomes could be reprogrammed. More recently, human somatic cell nuclei were also shown to be amenable to reprogramming by SCNT. However, in contrast to other species, this could be achieved only without prior removal of the oocyte nucleus (6).
Other cell types besides oocytes have been shown to possess factors capable of activating silenced loci in a somatic genome. For instance, introduction of a chicken erythrocyte nucleus into the cytoplasm of a HeLa cell results in chromatin decondensation and initiation of RNA synthesis from the previously inactive erythrocyte genome (7). Later studies in mice and humans demonstrated that fusion of somatic cells with embryonic stem cells (ESCs) or embryonic germ cells reactivates pluripotency-related genes from the somatic genome and creates pluripotent, albeit tetraploid (4N), cells (8–10). Although these have limited clinical value, their genesis has provided a useful experimental platform for studying cellular plasticity and reprogramming (11). Moreover, the fusion experiments offered evidence that ESCs, like oocytes, zygotes, and early blastomeres, contain factors that are sufficient to reprogram a somatic cell.
Initial evidence for the capacity of selected transcription factors (TFs) to direct cellular reprogramming came from the classic myoD experiments and subsequent lineage conversions in the hematopoietic system (reviewed in Ref. 12). However, a major advancement in the field occurred in 2006 with the landmark study by Takahashi and Yamanaka (13), who reported the generation of induced pluripotent stem cells (iPSCs) through ectopic expression of only four TFs.
Converting Cell States
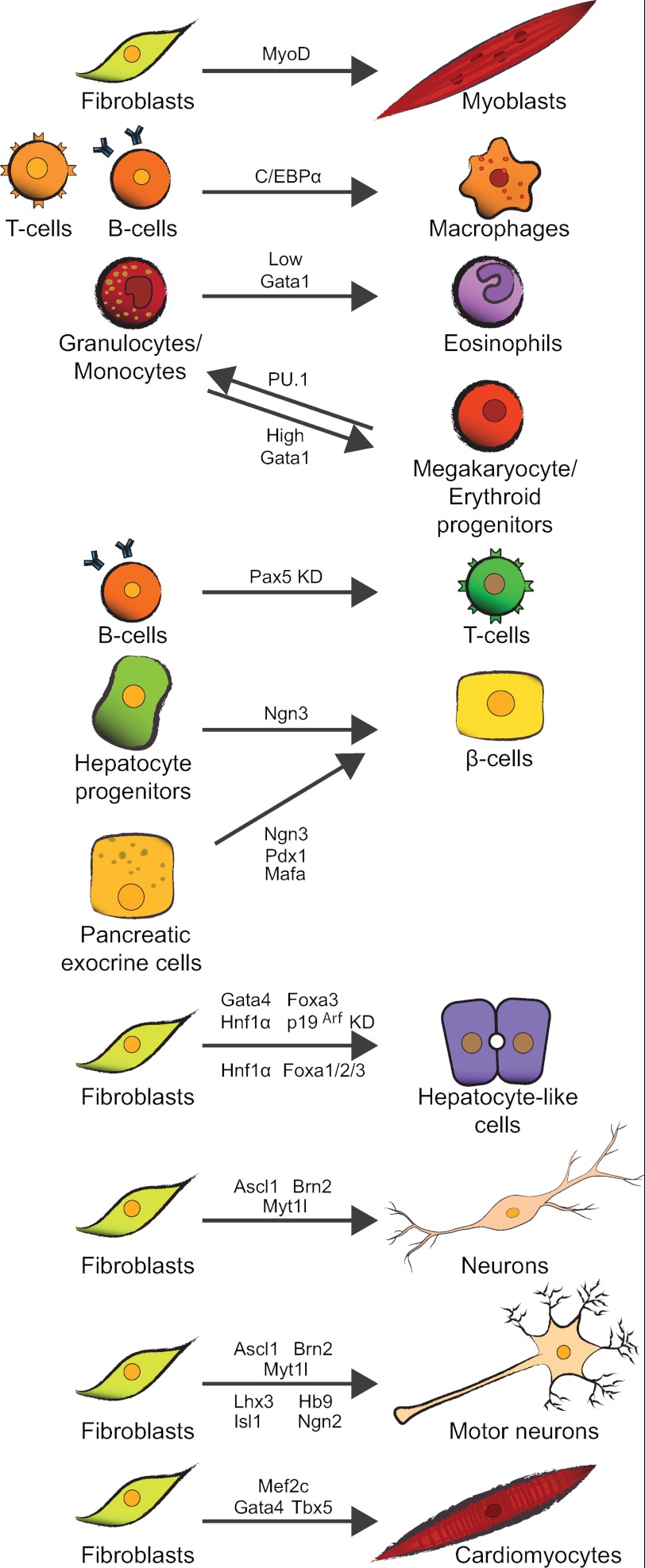
In contrast to the limited plasticity that exists in vivo, there are many examples of ectopic TF-mediated reprogramming in vitro (Fig. 1). Early unrelated studies led to the discovery of the basic helix-loop-helix (bHLH) TF myoD, whose expression in mouse embryonic fibroblasts could induce their conversion to myoblasts (14). MyoD appears to be a master regulator of muscle-specific transcriptional programs, and its ectopic expression can induce activation of muscle-specific genes in a variety of non-muscle cells (15, 16). Interestingly, although MyoD could induce complete phenotypic conversion in mesodermal cell types, ectodermal and endodermal cells responded less effectively (16).
FIGURE 1.
TF-mediated cell fate conversions in a variety of starting cell types. Shown are examples of published murine cell state conversions using modulation of TF expression. The majority utilize ectopic expression of one or more lineage-specific TFs, whereas others apply targeted depletion of TFs. Factors are shown next to the arrows. KD, knockdown.
Valuable insights into TF-mediated reprogramming were gained through fate conversions within the hematopoietic lineage, where developmental hierarchies are well understood. Common lymphoid progenitors (CLPs) can be induced to give rise to myeloid lineage cells such as granulocytes and monocytes by ectopic expression of the IL-2 receptor, which in turn leads to expression of GM-CSF receptors and the associated responsiveness to myeloid commitment-inducing signals (17). Although both CLPs and pro-T-cells could undergo this form of lineage redirection, the further downstream and lineage-committed pre-T- and pro-B-cells could not, suggesting a possible link between differentiation state and amenability to cell fate conversion. In another example, ectopic expression of the zinc finger TF GATA-1, a key regulator of megakaryocyte-erythrocyte precursor lineage commitment, was found to elicit conversion of CLP and GM progenitors into megakaryocytes and erythrocytes (18, 19). GATA-1 activates megakaryocyte-erythrocyte precursor-specific genes while concomitantly down-regulating markers of the other lineages. Alternatively, when GATA-1 is expressed at low levels in GM progenitors, a fate decision is forced whereby eosinophils or mast cells are generated (20), suggesting a critical role for factor stoichiometry in fate determination.
The transcription factor PU.1 (Spi-1) belongs to the Ets family and is a key regulator of myeloid and B-lymphoid lineage specification, functioning through interaction with other TFs such as GATA-1/2 and CCAAT enhancer-binding protein (C/EBP) α/β (21–23). Interestingly, lineage choice between these two fates is determined in part by graded expression of PU.1 (24). Through its interaction with GATA proteins, PU.1 transactivation of myeloid target genes is suppressed (25). Conversely, PU.1 inhibits the erythroid program by binding GATA-1 and inducing the formation of repressive chromatin structure at GATA-1 target loci (26, 27). Consistently, ectopic expression of PU.1 in multipotent progenitor cell lines leads to suppression of GATA-1 and concomitant conversion into myeloid cells (28). The antagonism between GATA-1 and PU.1 dictates one of two fates and highlights the key role for TF interactions in the process.
Another classic example involves the basic leucine zipper transcription factor C/EBPα, which is required for the common myeloid progenitor-to-GM progenitor transition in vivo (29). Ectopic expression of C/EBPα in primary bone marrow cells, lymphocytes (30, 31), or fibroblasts (32) can elicit myelomonocytic cell-type characteristics, a classification that includes both macrophages and granulocyte precursors. The function of C/EBPα in this context is dependent on synergism with PU.1, which is required for deposition of H3K4me1 at enhancer elements of target genes, suggesting that these factors cooperate to define cell type-specific binding patterns at regulatory elements (33, 34).
In addition to ectopic expression, loss of fate determinants can also induce cell fate changes. Differentiation of CLPs into the B-cell lineage depends on the TFs PU.1, E2A, and EBF1, which induce Pax5 to activate B-cell-specific genes while repressing genes associated with alternative lineages. In agreement with this model, Pax5−/− pro-B-cells fail to complete B-lymphopoiesis but are capable of differentiating into other hematopoietic cell types such as macrophages, dendritic cells, and granulocytes in response to specific signaling cascades (35–37). Additionally, Pax5 ablation in mature B-cells results in dedifferentiation into an uncommitted progenitor cell population, which can then undergo T-lymphopoiesis (38). Notably, the order in which factors are expressed can also impact the outcome. For example, altering the sequential expression of C/EBPα and GATA-2 in GM progenitors can instruct commitment to different hematopoietic cell types (39).
In summary, lessons from the hematopoietic system have provided strong evidence for the ability of TFs to redirect cell fate across related lineages derived from one germ layer or between specialized cell types of a particular lineage. Below, we will briefly review many of the more recent reprogramming studies.
Using a candidate gene approach, Melton and co-workers (40) identified three bHLH TFs, Ngn3 (or NeuroD1), Pdx1, and MafA, whose forced expression can convert exocrine pancreas tissue into insulin-secreting endocrine β-cells in vivo. Expressing only the endocrine progenitor-defining TF Ngn3 in hepatic progenitor cells generated physiologically responsive pancreatic endocrine cells. In contrast, expression of Ngn3 in mature hepatocytes induced only insulin expression (but not transdifferentiation) in islet cells (41). An additional example of reprogramming in vivo comes from the inner ear, where the TFs Atoh1 and Prox1, among others, were found to regulate the development of sensory hair cells and supporting cells from a common progenitor (42, 43). Ectopic expression of the bHLH TF Atoh1 (also known as Math1) results in conversion of non-sensory cochlear cells into functional sensory hair cells (44, 45). Conversely, expression of Prox1 in sensory hair cells leads to the repression of Gfi1 and Atoh1, factors required for sensory hair cell specification, resulting in cellular degeneration (43).
Recently, other groups have demonstrated direct reprogramming in vitro into additional endodermal cells types. The ectopic expression of GATA-4, Hnf1a, and Foxa3 in fibroblasts, along with inactivation of p19Arf, could give rise to induced hepatocyte-like cells (46). A separate study found that ectopic expression of Hnf4α and one of the three foxA genes in mouse embryonic or adult fibroblasts could also induce expression of multiple hepatocyte-specific features, converting the fibroblasts to hepatocyte-like cells (47).
Employing a similar candidate gene approach as Yamanaka and Melton, Wernig and co-workers (48) demonstrated that expression of three factors, Ascl1, Brn2 (also known as Pou3f2), and Myt1l, in mouse embryonic and postnatal fibroblasts induced conversion to neural cells, termed induced neuronal (iN) cells. Although the exact identity of these cells remains unclear, iN cells can form functional synapses and are physiologically responsive (48). These three factors could also induce neuronal differentiation of human ESCs, although reprogramming of human fetal fibroblasts to functional iN cells required additional coexpression of NeuroD1 (49). To definitively demonstrate the neural conversion of a differentiated non-ectodermal cell type, Marro et al. (50) recently reprogrammed mouse hepatocytes into iN cells. The hepatic transcriptome was largely repressed in the iN cells, and they retained only a limited epigenetic signature of their starting state. These initial findings demonstrate the ability of ectopic TFs to reprogram somatic cells across germ layers. More recently, others have shown that human and mouse fibroblasts can be reprogrammed to specialized functional neural subtypes such as dopaminergic neurons (51, 52) and spinal motor neurons (53). Interestingly, fibroblasts can be induced to express neural lineage markers and exhibit neuronal morphology solely by forced expression of microRNAs miR-9/9* and miR-124, which repress the Baf53a subunit of the BAF chromatin-remodeling complex (54). However, generating functional iN cells required expression of additional TFs (54, 55). Two of these recent iN studies also noted that reprogramming occurred in the absence of continued cell division, as the majority of reprogramming cells became post-mitotic within 24 h of factor induction (48, 50).
Along with the ability to generate a variety of neuronal cell types, effective production of cardiac muscle cells from ESCs or non-cardiac somatic cells is a compelling target of translational medicine. The first evidence of reprogramming non-cardiogenic tissues to a cardiac fate was provided by in vivo transfection of mouse embryos with core transcriptional regulators of cardiac development (56). More recently, Srivastava and co-workers (57) reported the TFs GATA-4, Mef2C, and Tbx5 could reprogram embryonic and adult fibroblasts directly into induced cardiomyocytes.
The diversity of cell types generated by TF-mediated reprogramming demonstrates the potential utility of this approach for therapeutic purposes. However, several key aspects of these converted cell types need further investigation. In particular, the stability and relative completion of functional, transcriptional, and epigenetic remodeling, as well as the in vivo equivalence of the generated cell types, remain unclear. A better understanding of potential risks posed by incomplete reprogramming or cellular memory is an important benchmark for the translational application of induced cell types.
Reprogramming Somatic Cells to Pluripotency
Perhaps the most striking example of factor-induced changes in cell state remains the reprogramming of somatic cells into pluripotent embryonic stem-like cells, termed iPSCs. The seminal work by Takahashi and Yamanaka (13) demonstrated that ectopic expression of Oct4, Sox2, Klf4, and c-Myc (OSKM) in embryonic and adult fibroblasts could convert these cells to iPSCs. Subsequently, numerous laboratories have improved the original protocol and generated iPSCs from a variety of species and cell types (58), highlighting the robustness and general conservation of the approach (Fig. 2).
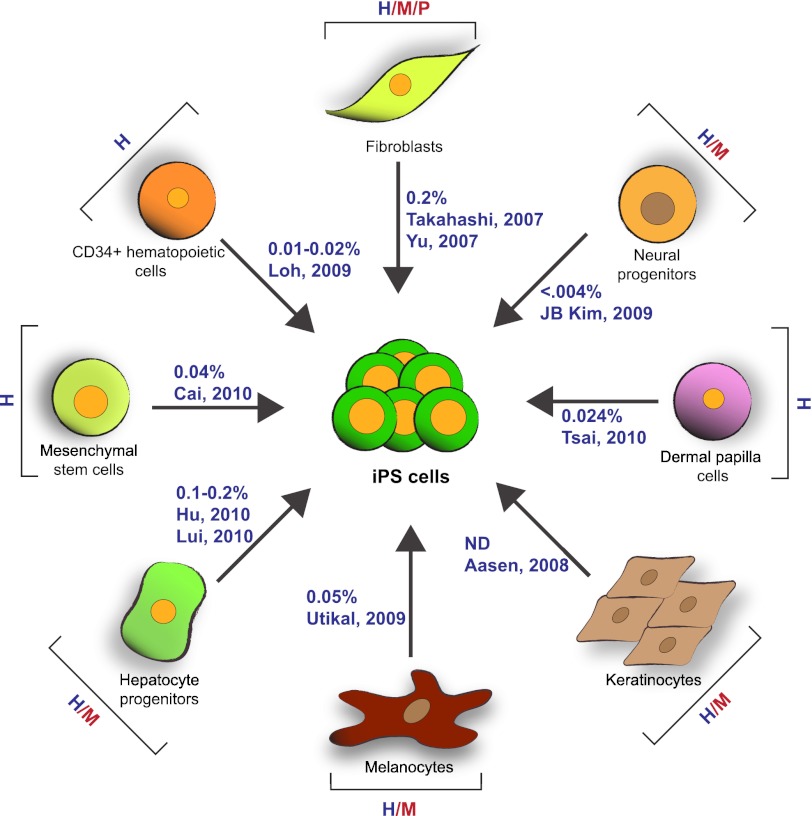
FIGURE 2.
Diverse somatic origin and efficiencies of iPSC generation. A wide variety of starting cell types originating from different species and representing all three embryonic germ layers can be reprogrammed to pluripotency with variable but consistently low efficiency. H, human; M, mouse; P, pig. Only human reprogramming efficiencies are indicated next to the arrows (highlighted in blue).
Forced expression of OSKM initiates dramatic phenotypic (59) and molecular (60–63) changes in the targeted somatic cell. Transcriptional and epigenetic changes eventually lead to the reactivation of endogenous pluripotency genes and acquisition of molecular and, in some cases, functional pluripotency. Commonly used markers of reprogramming include alkaline phosphatase and SSEA1 (stage-specific embryonic antigen-1) (62, 64). Although these markers appear earlier in the process, they are not indicative of complete reprogramming and are of relatively low stringency (65). Retroactive tracing of individual reprogramming fibroblasts has demonstrated that within the set of cells that undergo complete reprogramming, the initial response to reprogramming factor induction is characterized by a marked decrease in cell size and increased proliferation rate (59). The early response to the four TFs is marked by the transcriptional reversion from the somatic state, as evidenced by the down-regulation of the fibroblast-specific genes Snai1/2, Zeb1/2, and Cdh2 and the surface marker Thy1 (60, 63, 64). The loss of a mesenchymal phenotype coincides with the emerging expression of epithelium-associated markers such as E-cadherin and EpCAM, known as the mesenchymal-to-epithelial (MET) transition (63, 66). The importance of MET during reprogramming is highlighted by acceleration of reprogramming using bone morphogenetic protein ligand stimulation or expression of microRNAs from the miR-200 family (63), which have been implicated in MET (67). Notably, Klf4, the main driver of this transition, synergizes with Oct4 and Sox2 to direct reprogramming, and its absence can be replaced by stimulation with bone morphogenetic protein (68).
Establishment of the iPSC state in mice requires continuous expression of the OSKM transgenes for 8–10 days (64), leading to the eventual activation of endogenous pluripotency genes such as Oct4, Sox2, and Nanog after ∼2 weeks. The final progression of reprogramming occurs only in a small percentage of SSEA1-positive cells (62). The mechanism whereby the endogenous stem cell network is transcriptionally reactivated in the final transition to pluripotency remains poorly understood.
Although c-Myc is routinely included in the reprogramming factor mixture, and expression of this gene enhances the efficiency of somatic cell reprogramming, it is not a core member of the ESC pluripotency network. The network controlled by c-Myc is largely separate from the core ESC pluripotency network and primarily includes targets involved in metabolic, cell cycle, and proliferative processes (69–71). In ESCs, c-Myc target gene promoters are highly enriched for the active histone modification H3K4me3 and depleted of the repressive modification H3K27me3; this is a striking pattern that is not exhibited to the same extent by targets of the core pluripotency factors (69). The core factors reside at targets enriched for both activating and repressive histone modifications, reflecting their role in maintaining ESC identity by activating ESC-specific genes while repressing expression of lineage-specific transcriptional regulators (72). Conversely, c-Myc targets are more expansive in function, encompassing genes not unique to the pluripotent state (61, 69). The recent discovery that c-Myc plays a role in regulating pause release of polymerase II transcriptional elongation is in agreement with many of the early phenotypic and molecular responses to reprogramming factor induction (73). Notably, the latter suggests that c-Myc may not be directly involved in reactivating the pluripotency network but rather act more broadly to drive robust transcription of paused genes during the early stages of reprogramming. Genome-wide localization studies of pluripotency factors, along with affinity-based identification of their interacting partners, have revealed an interconnection between TFs (74, 75), chromatin-remodeling, and modifying complexes (Table 1). Although it is well established that ESC chromatin is predominantly euchromatic (76, 77) and possess characteristic bivalent enrichment of both active and repressive histone modifications at developmental genes (78), functional links between specific epigenetic modifiers and members of the ESC core transcriptional network have only recently come to light. Wdr5, a member of the mammalian Trithorax complex and an effector of activating H3K4 methylation, is an interacting partner of Oct4 (79). This provides a framework for conceptualizing how transcriptional regulators of pluripotency may cooperate with chromatin-associated factors to regulate the balance between self-renewal and differentiation.
TABLE 1.
Genome-wide localization studies of TFs and chromatin remodelers
Early Dynamics in TF-induced Reprogramming
Genome-wide chromatin mapping studies of selected epigenetic marks in populations of induced fibroblasts have provided many useful insights into early reprogramming dynamics (80). One of the earliest cellular responses to ectopic factor expression is de novo deposition or enhanced enrichment of euchromatin-associated H3K4me2 at a large number of promoters. Many of the target genes include developmental regulators and pluripotency genes (80). Importantly, these changes appear to precede transcriptional activation, which, in the case of the pluripotency-associated genes, will not occur until the late stages of reprogramming. Many of these dynamics can be observed in the absence of cell division. The fact that these regions are enriched for both targets of Oct4 and Sox2 suggests that one of the early steps in somatic cell reprogramming involves global coordinated epigenetic “priming.” The notable enrichment for H3K4 methylation at Oct4 target sites accords well with the aforementioned study by Ang et al. (79) connecting Oct4, Wdr5, and histone methylation of Oct4 targets. Importantly, the reestablishment and erasure of repressive marks such as H3K27me3 occur at a notably slower rate (80).
Interestingly, the majority of expression changes that are detectable during the first few cell divisions occur almost solely within regions of H3K4me3-enriched accessible chromatin environments (60, 80). This suggests that the action of the reprogramming factors is restricted, at least initially, to transcriptionally permissive genomic loci. Moreover, at these early stages, up-regulated (but not down-regulated) genes are strongly enriched for targets of c-Myc. Unlike Oct4 and Sox2, c-Myc is already endogenously expressed in somatic fibroblasts (13) and primarily regulates targets involved in cellular metabolism and proliferation. The observed transcriptional response to increased ectopic c-Myc is consistent with its proposed role in stabilizing transcriptional elongation, thereby enabling productive transcription at many shared (somatic and pluripotent) target genes (73). Consistent with these data and in accordance with a distinct binding pattern for c-Myc in ESCs (69), Plath and co-workers (61) showed that the most dramatic early changes in fibroblast gene levels were largely attributable to c-Myc expression.
Subsequent to the first few cell divisions, molecular events leading to up-regulation of endogenous pluripotency genes and transition to the iPSC state remain hard to define due to population heterogeneity. Treatment of reprogramming cells with chromatin-modifying agents can positively affect efficiency. For instance, the Dnmt inhibitor 5-azacytidine (5-AZA), which decreases overall DNA methylation levels and promotes chromatin decondensation, can improve overall efficiency and facilitate the transition of partially reprogrammed cells to pluripotency (60). Similarly, inhibitors of histone deacetylases and methyltransferases can elicit a similar improvement (81), as can members of ATP-dependent chromatin-remodeling complexes. In a recent screen for ESC factors able to facilitate the reprogramming process, Schöler and co-workers (82) identified components of the BAF chromatin-remodeling complex that act in part by facilitating Oct4 binding to target loci during reprogramming, perhaps by improving TF access to target DNA.
Full transition to pluripotency can also be mediated by kinase inhibition in partially reprogrammed cells that lack the expression of true pluripotency markers. Treatment of these cells with inhibitors of GSK3β and MEK (dual inhibition) induces the up-regulation of endogenous Nanog and Oct4 and demethylates the Nanog promoter to ESC-comparable levels, eventually leading to stable pluripotency (83). It is worth noting that although both 5-AZA treatment and dual inhibition culture can enhance reprogramming, their effects may be attributable in part to the selection against somatic or non-pluripotent intermediates. Both the toxicity of 5-AZA and the lethality of kinase inhibition for any mitogen-dependent cells could cause an apparent increase in efficiency. The acquisition of an ESC-like proliferative state, which is uniquely resistant to the growth inhibitory effects of these small molecules, may provide a selective advantage to successfully reprogramming cells compared with those that retain somatic cell cycle characteristics.
Some iPSCs Are Equivalent to ESCs
The functional and molecular equivalence of ESCs and iPSCs has been a key question since the initial report of iPSC derivation (13). Developmental potential can be determined with low stringency by in vitro differentiation and teratoma formation and with higher stringency in non-human species by chimera formation, germ-line contribution (84), or the production of entirely iPSC/ESC-derived animals by tetraploid complementation (85, 86). Multiple molecular screening strategies have been employed to compare ESC and iPSC lines quantitatively, including profiling transcriptional and epigenetic signatures on a genome scale. At the morphological, molecular, and functional levels, ESCs and iPSCs show a high degree of similarity (87), although the complete equivalence of these cell types remains controversial. Numerous studies have described molecular differences between iPSCs and ESCs (88–90). We have recently used three genomic assays to profile 20 human ESC and 12 iPSC lines and generated a reference map of DNA methylation, gene expression, and in vitro differentiation potential for the tested lines (91). Surprisingly, much of the variation observed within iPSC lines could be explained by the general variation between the different pluripotent lines tested, including the ESC lines. These findings accord with functional studies in the mouse, which have established that some (but not all) iPSC lines are both functionally and molecularly equivalent to ESCs (86, 89, 92).
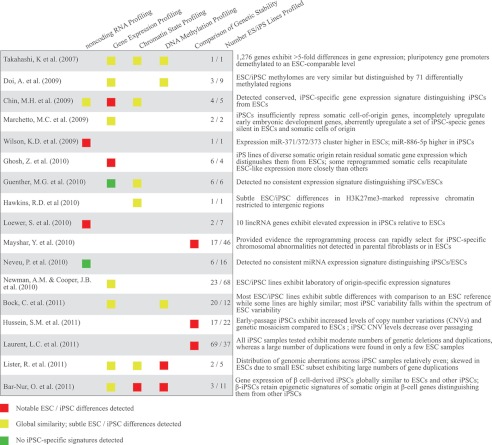
Experimentally determined variations in gene expression levels and epigenetic profiles have yet to provide a molecular signature specific to iPSCs that can consistently discriminate between the various pluripotent populations (91, 93). Establishing the criteria for such a signature has proven challenging, given that variability exists among individual ESC and iPSC lines. To avoid confounding effects of differences in genetic background, a recent study derived and compared the molecular and functional characteristics of genetically matched mouse ESCs and iPSCs. The findings suggest that the expression patterns of ESCs and iPSCs are remarkably similar, with the exception of a few genes of the imprinted Dlk1-Dio3 locus, which appeared to be aberrantly silenced in the majority of iPSC clones examined (89). A more recent study has shown this disparity to be greatly affected by the stoichiometry of the TFs (OSKM), suggesting the inactivation of this locus may be linked to the specific strategy or conditions in which the cells are generated and is not necessarily inherent to reprogramming (94). An additional caveat to the generality of the observations summarized here is the relatively small number of ESC and iPSC lines that are used in many studies (Table 2). In humans, another concern is the limited number of ESC lines, such as H1 and H9, which have been used as references or controls in many studies. For instance, a recent study reported aberrant hypomethylation of promoter regions in iPSC lines compared with the H1 and H9 ESC lines (95). However, comparison with a panel of 32 pluripotent cell lines (91) demonstrated that hypomethylation of the reported regions is a general characteristic of both ESCs and iPSCs, with the exception of a few cell lines, including the H1 and H9 ESC lines used as controls in the original report. This suggests that rather than aberrant hypomethylation in the iPSCs, these loci exhibit possibly aberrant hypermethylation in the control ESC lines.
TABLE 2.
Summary of recent studies comparing molecular characteristics of various human ESC and iPSC lines
Conclusion
Coordinated activity of TFs and epigenetic modifiers, acting downstream or in concert with signaling molecules, establishes and maintains stable cellular identities by driving cell type-specific gene expression programs. Epigenetic modifications affect the ability of the transcriptional machinery to access the chromatin template, thereby influencing gene expression patterns that ultimately determine cellular identity. Although cellular differentiation appears to be largely unidirectional during development and fixed in adult organisms, elegant experiments over the recent decades have demonstrated the remarkable plasticity by which differentiated cells can be redirected to alternative fates. Cell state can be considered the phenotypic output of regulatory networks, which are in turn influenced by and reflected in cell-specific epigenetic landscapes. By manipulating transcriptional programs, it should be theoretically possible to reprogram a cell's identity to any desired alternative state.
Many important questions remain open in the field of cell fate conversion experiments. It is still unknown how ectopic factors access their binding sites in a foreign chromatin context and how their function is influenced by the starting cell's molecular ensemble. A mechanistic description for the relevance of epigenetic or transcriptional similarities between starting and intended cell types in cell fate reprogramming is also lacking. For example, the higher efficiency of conversion between closely related cell types may be partly attributable to higher transcriptional similarity and thus availability of essential cofactors between the starting and intended cell types. In addition to the transcriptional repertoire, however, a more closely related epigenetic landscape may also enable more efficient conversions. Ultimately, the knowledge gained from experimental manipulation of cell fates will have far-reaching implications not only for the basic understanding of the molecular mechanisms behind cellular identity but also for the eventual application to personalized regenerative medicine.
Acknowledgments
We thank members of the Meissner laboratory (in particular, Zachary D. Smith and Casey A. Gifford) for critical comments.
This work was supported, in whole or in part, by National Institutes of Health Grants P01GM099117 and U01ES017155 (to A. M.). This work was also supported by the Pew Charitable Trusts (to A. M.). This is the fifth article in the Thematic Minireview Series on Results from the ENCODE Project: Integrative Global Analyses of Regulatory Regions in the Human Genome.
- SCNT
- somatic cell nuclear transfer
- ESC
- embryonic stem cell
- TF
- transcription factor
- iPSC
- induced pluripotent stem cell
- bHLH
- basic helix-loop-helix
- CLP
- common lymphoid progenitor
- C/EBP
- CCAAT enhancer-binding protein
- iN
- induced neuronal
- OSKM
- Oct4, Sox2, Klf4, and c-Myc
- MET
- mesenchymal-to-epithelial
- 5-AZA
- 5-azacytidine.
REFERENCES
- 1. Briggs R., King T. J. (1952) Transplantation of living nuclei from blastula cells into enucleated frog eggs. Proc. Natl. Acad. Sci. U.S.A. 38, 455–463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Gurdon J. B. (1962) Adult frogs derived from the nuclei of single somatic cells. Dev. Biol. 4, 256–273 [DOI] [PubMed] [Google Scholar]
- 3. Wilmut I., Schnieke A. E., McWhir J., Kind A. J., Campbell K. H. (1997) Viable offspring derived from fetal and adult mammalian cells. Nature 385, 810–813 [DOI] [PubMed] [Google Scholar]
- 4. Hochedlinger K., Jaenisch R. (2002) Monoclonal mice generated by nuclear transfer from mature B and T donor cells. Nature 415, 1035–1038 [DOI] [PubMed] [Google Scholar]
- 5. Eggan K., Baldwin K., Tackett M., Osborne J., Gogos J., Chess A., Axel R., Jaenisch R. (2004) Mice cloned from olfactory sensory neurons. Nature 428, 44–49 [DOI] [PubMed] [Google Scholar]
- 6. Noggle S., Fung H. L., Gore A., Martinez H., Satriani K. C., Prosser R., Oum K., Paull D., Druckenmiller S., Freeby M., Greenberg E., Zhang K., Goland R., Sauer M. V., Leibel R. L., Egli D. (2011) Human oocytes reprogram somatic cells to a pluripotent state. Nature 478, 70–75 [DOI] [PubMed] [Google Scholar]
- 7. Bolund L., Ringertz N. R., Harris H. (1969) Changes in the cytochemical properties of erythrocyte nuclei reactivated by cell fusion. J. Cell Sci. 4, 71–87 [DOI] [PubMed] [Google Scholar]
- 8. Tada M., Tada T., Lefebvre L., Barton S. C., Surani M. A. (1997) Embryonic germ cells induce epigenetic reprogramming of somatic nucleus in hybrid cells. EMBO J. 16, 6510–6520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Cowan C. A., Atienza J., Melton D. A., Eggan K. (2005) Nuclear reprogramming of somatic cells after fusion with human embryonic stem cells. Science 309, 1369–1373 [DOI] [PubMed] [Google Scholar]
- 10. Tada M., Takahama Y., Abe K., Nakatsuji N., Tada T. (2001) Nuclear reprogramming of somatic cells by in vitro hybridization with ES cells. Curr. Biol. 11, 1553–1558 [DOI] [PubMed] [Google Scholar]
- 11. Pereira C. F., Terranova R., Ryan N. K., Santos J., Morris K. J., Cui W., Merkenschlager M., Fisher A. G. (2008) Heterokaryon-based reprogramming of human B-lymphocytes for pluripotency requires Oct4 but not Sox2. PLoS Genet. 4, e1000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Graf T. (2011) Historical origins of transdifferentiation and reprogramming. Cell Stem Cell 9, 504–516 [DOI] [PubMed] [Google Scholar]
- 13. Takahashi K., Yamanaka S. (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126, 663–676 [DOI] [PubMed] [Google Scholar]
- 14. Davis R. L., Weintraub H., Lassar A. B. (1987) Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 51, 987–1000 [DOI] [PubMed] [Google Scholar]
- 15. Choi J., Costa M. L., Mermelstein C. S., Chagas C., Holtzer S., Holtzer H. (1990) MyoD converts primary dermal fibroblasts, chondroblasts, smooth muscle, and retinal pigmented epithelial cells into striated mononucleated myoblasts and multinucleated myotubes. Proc. Natl. Acad. Sci. U.S.A. 87, 7988–7992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Weintraub H., Tapscott S. J., Davis R. L., Thayer M. J., Adam M. A., Lassar A. B., Miller A. D. (1989) Activation of muscle-specific genes in pigment, nerve, fat, liver, and fibroblast cell lines by forced expression of MyoD. Proc. Natl. Acad. Sci. U.S.A. 86, 5434–5438 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Kondo M., Scherer D. C., Miyamoto T., King A. G., Akashi K., Sugamura K., Weissman I. L. (2000) Cell fate conversion of lymphoid-committed progenitors by instructive actions of cytokines. Nature 407, 383–386 [DOI] [PubMed] [Google Scholar]
- 18. Heyworth C., Pearson S., May G., Enver T. (2002) Transcription factor-mediated lineage switching reveals plasticity in primary committed progenitor cells. EMBO J. 21, 3770–3781 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Iwasaki H., Mizuno S., Wells R. A., Cantor A. B., Watanabe S., Akashi K. (2003) GATA-1 converts lymphoid and myelomonocytic progenitors into the megakaryocyte/erythrocyte lineages. Immunity 19, 451–462 [DOI] [PubMed] [Google Scholar]
- 20. Kulessa H., Frampton J., Graf T. (1995) GATA-1 reprograms avian myelomonocytic cell lines into eosinophils, thromboblasts, and erythroblasts. Genes Dev. 9, 1250–1262 [DOI] [PubMed] [Google Scholar]
- 21. Klemsz M. J., McKercher S. R., Celada A., Van Beveren C., Maki R. A. (1990) The macrophage- and B-cell-specific transcription factor PU.1 is related to the ets oncogene. Cell 61, 113–124 [DOI] [PubMed] [Google Scholar]
- 22. Gupta P., Gurudutta G. U., Saluja D., Tripathi R. P. (2009) PU.1 and partners: regulation of hematopoietic stem cell fate in normal and malignant hematopoiesis. J. Cell. Mol. Med. 13, 4349–4363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Scott E. W., Fisher R. C., Olson M. C., Kehrli E. W., Simon M. C., Singh H. (1997) PU.1 functions in a cell-autonomous manner to control the differentiation of multipotential lymphoid-myeloid progenitors. Immunity 6, 437–447 [DOI] [PubMed] [Google Scholar]
- 24. DeKoter R. P., Singh H. (2000) Regulation of B-lymphocyte and macrophage development by graded expression of PU.1. Science 288, 1439–1441 [DOI] [PubMed] [Google Scholar]
- 25. Zhang P., Behre G., Pan J., Iwama A., Wara-Aswapati N., Radomska H. S., Auron P. E., Tenen D. G., Sun Z. (1999) Negative cross-talk between hematopoietic regulators: GATA proteins repress PU.1. Proc. Natl. Acad. Sci. U.S.A. 96, 8705–8710 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stopka T., Amanatullah D. F., Papetti M., Skoultchi A. I. (2005) PU.1 inhibits the erythroid program by binding to GATA-1 on DNA and creating a repressive chromatin structure. EMBO J. 24, 3712–3723 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Rekhtman N., Radparvar F., Evans T., Skoultchi A. I. (1999) Direct interaction of hematopoietic transcription factors PU.1 and GATA-1: functional antagonism in erythroid cells. Genes Dev. 13, 1398–1411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Nerlov C., Graf T. (1998) PU.1 induces myeloid lineage commitment in multipotent hematopoietic progenitors. Genes Dev. 12, 2403–2412 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Zhang P., Iwasaki-Arai J., Iwasaki H., Fenyus M. L., Dayaram T., Owens B. M., Shigematsu H., Levantini E., Huettner C. S., Lekstrom-Himes J. A., Akashi K., Tenen D. G. (2004) Enhancement of hematopoietic stem cell repopulating capacity and self-renewal in the absence of the transcription factor C/EBPα. Immunity 21, 853–863 [DOI] [PubMed] [Google Scholar]
- 30. Xie H., Ye M., Feng R., Graf T. (2004) Stepwise reprogramming of B-cells into macrophages. Cell 117, 663–676 [DOI] [PubMed] [Google Scholar]
- 31. Laiosa C. V., Stadtfeld M., Xie H., de Andres-Aguayo L., Graf T. (2006) Reprogramming of committed T-cell progenitors to macrophages and dendritic cells by C/EBPα and PU.1 transcription factors. Immunity 25, 731–744 [DOI] [PubMed] [Google Scholar]
- 32. Feng R., Desbordes S. C., Xie H., Tillo E. S., Pixley F., Stanley E. R., Graf T. (2008) PU.1 and C/EBPα/β convert fibroblasts into macrophage-like cells. Proc. Natl. Acad. Sci. U.S.A. 105, 6057–6062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ghisletti S., Barozzi I., Mietton F., Polletti S., De Santa F., Venturini E., Gregory L., Lonie L., Chew A., Wei C. L., Ragoussis J., Natoli G. (2010) Identification and characterization of enhancers controlling the inflammatory gene expression program in macrophages. Immunity 32, 317–328 [DOI] [PubMed] [Google Scholar]
- 34. Heinz S., Benner C., Spann N., Bertolino E., Lin Y. C., Laslo P., Cheng J. X., Murre C., Singh H., Glass C. K. (2010) Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B-cell identities. Mol. Cell 38, 576–589 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Nutt S. L., Heavey B., Rolink A. G., Busslinger M. (1999) Commitment to the B-lymphoid lineage depends on the transcription factor Pax5. Nature 401, 556–562 [DOI] [PubMed] [Google Scholar]
- 36. Rolink A. G., Nutt S. L., Melchers F., Busslinger M. (1999) Long-term in vivo reconstitution of T-cell development by Pax5-deficient B-cell progenitors. Nature 401, 603–606 [DOI] [PubMed] [Google Scholar]
- 37. Mikkola I., Heavey B., Horcher M., Busslinger M. (2002) Reversion of B-cell commitment upon loss of Pax5 expression. Science 297, 110–113 [DOI] [PubMed] [Google Scholar]
- 38. Cobaleda C., Jochum W., Busslinger M. (2007) Conversion of mature B-cells into T-cells by dedifferentiation to uncommitted progenitors. Nature 449, 473–477 [DOI] [PubMed] [Google Scholar]
- 39. Iwasaki H., Mizuno S., Arinobu Y., Ozawa H., Mori Y., Shigematsu H., Takatsu K., Tenen D. G., Akashi K. (2006) The order of expression of transcription factors directs hierarchical specification of hematopoietic lineages. Genes Dev. 20, 3010–3021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Zhou Q., Brown J., Kanarek A., Rajagopal J., Melton D. A. (2008) In vivo reprogramming of adult pancreatic exocrine cells to β-cells. Nature 455, 627–632 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Yechoor V., Liu V., Espiritu C., Paul A., Oka K., Kojima H., Chan L. (2009) Neurogenin-3 is sufficient for transdetermination of hepatic progenitor cells into neo-islets in vivo but not transdifferentiation of hepatocytes. Dev. Cell 16, 358–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Edge A. S., Chen Z. Y. (2008) Hair cell regeneration. Curr. Opin. Neurobiol. 18, 377–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kirjavainen A., Sulg M., Heyd F., Alitalo K., Ylä-Herttuala S., Möröy T., Petrova T. V., Pirvola U. (2008) Prox1 interacts with Atoh1 and Gfi1 and regulates cellular differentiation in the inner ear sensory epithelia. Dev. Biol. 322, 33–45 [DOI] [PubMed] [Google Scholar]
- 44. Zheng J. L., Gao W. Q. (2000) Overexpression of Math1 induces robust production of extra hair cells in postnatal rat inner ears. Nat. Neurosci. 3, 580–586 [DOI] [PubMed] [Google Scholar]
- 45. Woods C., Montcouquiol M., Kelley M. W. (2004) Math1 regulates development of the sensory epithelium in the mammalian cochlea. Nat. Neurosci. 7, 1310–1318 [DOI] [PubMed] [Google Scholar]
- 46. Huang P., He Z., Ji S., Sun H., Xiang D., Liu C., Hu Y., Wang X., Hui L. (2011) Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature 475, 386–389 [DOI] [PubMed] [Google Scholar]
- 47. Sekiya S., Suzuki A. (2011) Direct conversion of mouse fibroblasts to hepatocyte-like cells by defined factors. Nature 475, 390–393 [DOI] [PubMed] [Google Scholar]
- 48. Vierbuchen T., Ostermeier A., Pang Z. P., Kokubu Y., Südhof T. C., Wernig M. (2010) Direct conversion of fibroblasts to functional neurons by defined factors. Nature 463, 1035–1041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Pang Z. P., Yang N., Vierbuchen T., Ostermeier A., Fuentes D. R., Yang T. Q., Citri A., Sebastiano V., Marro S., Südhof T. C., Wernig M. (2011) Induction of human neuronal cells by defined transcription factors. Nature 476, 220–223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Marro S., Pang Z. P., Yang N., Tsai M. C., Qu K., Chang H. Y., Südhof T. C., Wernig M. (2011) Direct lineage conversion of terminally differentiated hepatocytes to functional neurons. Cell Stem Cell 9, 374–382 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Caiazzo M., Dell'Anno M. T., Dvoretskova E., Lazarevic D., Taverna S., Leo D., Sotnikova T. D., Menegon A., Roncaglia P., Colciago G., Russo G., Carninci P., Pezzoli G., Gainetdinov R. R., Gustincich S., Dityatev A., Broccoli V. (2011) Direct generation of functional dopaminergic neurons from mouse and human fibroblasts. Nature 476, 224–227 [DOI] [PubMed] [Google Scholar]
- 52. Pfisterer U., Kirkeby A., Torper O., Wood J., Nelander J., Dufour A., Björklund A., Lindvall O., Jakobsson J., Parmar M. (2011) Direct conversion of human fibroblasts to dopaminergic neurons. Proc. Natl. Acad. Sci. U.S.A. 108, 10343–10348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Son E. Y., Ichida J. K., Wainger B. J., Toma J. S., Rafuse V. F., Woolf C. J., Eggan K. (2011) Conversion of mouse and human fibroblasts into functional spinal motor neurons. Cell Stem Cell 9, 205–218 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Yoo A. S., Sun A. X., Li L., Shcheglovitov A., Portmann T., Li Y., Lee-Messer C., Dolmetsch R. E., Tsien R. W., Crabtree G. R. (2011) MicroRNA-mediated conversion of human fibroblasts to neurons. Nature 476, 228–231 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ambasudhan R., Talantova M., Coleman R., Yuan X., Zhu S., Lipton S. A., Ding S. (2011) Direct reprogramming of adult human fibroblasts to functional neurons under defined conditions. Cell Stem Cell 9, 113–118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Takeuchi J. K., Bruneau B. G. (2009) Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature 459, 708–711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Ieda M., Fu J. D., Delgado-Olguin P., Vedantham V., Hayashi Y., Bruneau B. G., Srivastava D. (2010) Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 142, 375–386 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Masip M., Veiga A., Izpisúa Belmonte J. C., Simón C. (2010) Reprogramming with defined factors: from induced pluripotency to induced transdifferentiation. Mol. Human Reprod. 16, 856–868 [DOI] [PubMed] [Google Scholar]
- 59. Smith Z. D., Nachman I., Regev A., Meissner A. (2010) Dynamic single-cell imaging of direct reprogramming reveals an early specifying event. Nat. Biotechnol. 28, 521–526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Mikkelsen T. S., Hanna J., Zhang X., Ku M., Wernig M., Schorderet P., Bernstein B. E., Jaenisch R., Lander E. S., Meissner A. (2008) Dissecting direct reprogramming through integrative genomic analysis. Nature 454, 49–55 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Sridharan R., Tchieu J., Mason M. J., Yachechko R., Kuoy E., Horvath S., Zhou Q., Plath K. (2009) Role of the murine reprogramming factors in the induction of pluripotency. Cell 136, 364–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Brambrink T., Foreman R., Welstead G. G., Lengner C. J., Wernig M., Suh H., Jaenisch R. (2008) Sequential expression of pluripotency markers during direct reprogramming of mouse somatic cells. Cell Stem Cell 2, 151–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Samavarchi-Tehrani P., Golipour A., David L., Sung H. K., Beyer T. A., Datti A., Woltjen K., Nagy A., Wrana J. L. (2010) Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell 7, 64–77 [DOI] [PubMed] [Google Scholar]
- 64. Stadtfeld M., Maherali N., Breault D. T., Hochedlinger K. (2008) Defining molecular cornerstones during fibroblast to iPS cell reprogramming in mouse. Cell Stem Cell 2, 230–240 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Meissner A. (2010) Epigenetic modifications in pluripotent and differentiated cells. Nat. Biotechnol. 28, 1079–1088 [DOI] [PubMed] [Google Scholar]
- 66. Li R., Liang J., Ni S., Zhou T., Qing X., Li H., He W., Chen J., Li F., Zhuang Q., Qin B., Xu J., Li W., Yang J., Gan Y., Qin D., Feng S., Song H., Yang D., Zhang B., Zeng L., Lai L., Esteban M. A., Pei D. (2010) A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell 7, 51–63 [DOI] [PubMed] [Google Scholar]
- 67. Gregory P. A., Bert A. G., Paterson E. L., Barry S. C., Tsykin A., Farshid G., Vadas M. A., Khew-Goodall Y., Goodall G. J. (2008) The miR-200 family and miR-205 regulate epithelial to mesenchymal transition by targeting ZEB1 and SIP1. Nat. Cell Biol. 10, 593–601 [DOI] [PubMed] [Google Scholar]
- 68. Chen J., Liu J., Yang J., Chen Y., Chen J., Ni S., Song H., Zeng L., Ding K., Pei D. (2011) BMPs functionally replace Klf4 and support efficient reprogramming of mouse fibroblasts by Oct4 alone. Cell Res. 21, 205–212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Kim J., Chu J., Shen X., Wang J., Orkin S. H. (2008) An extended transcriptional network for pluripotency of embryonic stem cells. Cell 132, 1049–1061 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Liu X., Huang J., Chen T., Wang Y., Xin S., Li J., Pei G., Kang J. (2008) Yamanaka factors critically regulate the developmental signaling network in mouse embryonic stem cells. Cell Res. 18, 1177–1189 [DOI] [PubMed] [Google Scholar]
- 71. Papp B., Plath K. (2011) Reprogramming to pluripotency: stepwise resetting of the epigenetic landscape. Cell Res. 21, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Jaenisch R., Young R. (2008) Stem cells, the molecular circuitry of pluripotency and nuclear reprogramming. Cell 132, 567–582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Rahl P. B., Lin C. Y., Seila A. C., Flynn R. A., McCuine S., Burge C. B., Sharp P. A., Young R. A. (2010) c-Myc regulates transcriptional pause release. Cell 141, 432–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Wang J., Rao S., Chu J., Shen X., Levasseur D. N., Theunissen T. W., Orkin S. H. (2006) A protein interaction network for pluripotency of embryonic stem cells. Nature 444, 364–368 [DOI] [PubMed] [Google Scholar]
- 75. van den Berg D. L., Snoek T., Mullin N. P., Yates A., Bezstarosti K., Demmers J., Chambers I., Poot R. A. (2010) An Oct4-centered protein interaction network in embryonic stem cells. Cell Stem Cell 6, 369–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Efroni S., Duttagupta R., Cheng J., Dehghani H., Hoeppner D. J., Dash C., Bazett-Jones D. P., Le Grice S., McKay R. D., Buetow K. H., Gingeras T. R., Misteli T., Meshorer E. (2008) Global transcription in pluripotent embryonic stem cells. Cell Stem Cell 2, 437–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Fisher C. L., Fisher A. G. (2011) Chromatin states in pluripotent, differentiated, and reprogrammed cells. Curr. Opin. Genet. Dev. 21, 140–146 [DOI] [PubMed] [Google Scholar]
- 78. Bernstein B. E., Mikkelsen T. S., Xie X., Kamal M., Huebert D. J., Cuff J., Fry B., Meissner A., Wernig M., Plath K., Jaenisch R., Wagschal A., Feil R., Schreiber S. L., Lander E. S. (2006) A bivalent chromatin structure marks key developmental genes in embryonic stem cells. Cell 125, 315–326 [DOI] [PubMed] [Google Scholar]
- 79. Ang Y. S., Tsai S. Y., Lee D. F., Monk J., Su J., Ratnakumar K., Ding J., Ge Y., Darr H., Chang B., Wang J., Rendl M., Bernstein E., Schaniel C., Lemischka I. R. (2011) Wdr5 mediates self-renewal and reprogramming via the embryonic stem cell core transcriptional network. Cell 145, 183–197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Koche R. P., Smith Z. D., Adli M., Gu H., Ku M., Gnirke A., Bernstein B. E., Meissner A. (2011) Reprogramming factor expression initiates widespread targeted chromatin remodeling. Cell Stem Cell 8, 96–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Huangfu D., Maehr R., Guo W., Eijkelenboom A., Snitow M., Chen A. E., Melton D. A. (2008) Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat. Biotechnol. 26, 795–797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Singhal N., Graumann J., Wu G., Araúzo-Bravo M. J., Han D. W., Greber B., Gentile L., Mann M., Schöler H. R. (2010) Chromatin-remodeling components of the BAF complex facilitate reprogramming. Cell 141, 943–955 [DOI] [PubMed] [Google Scholar]
- 83. Theunissen T. W., van Oosten A. L., Castelo-Branco G., Hall J., Smith A., Silva J. C. (2011) Nanog overcomes reprogramming barriers and induces pluripotency in minimal conditions. Curr.. Biol. 21, 65–71 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Okita K., Ichisaka T., Yamanaka S. (2007) Generation of germ line-competent induced pluripotent stem cells. Nature 448, 313–317 [DOI] [PubMed] [Google Scholar]
- 85. Boland M. J., Hazen J. L., Nazor K. L., Rodriguez A. R., Gifford W., Martin G., Kupriyanov S., Baldwin K. K. (2009) Adult mice generated from induced pluripotent stem cells. Nature 461, 91–94 [DOI] [PubMed] [Google Scholar]
- 86. Zhao X. Y., Li W., Lv Z., Liu L., Tong M., Hai T., Hao J., Guo C. L., Ma Q. W., Wang L., Zeng F., Zhou Q. (2009) iPS cells produce viable mice through tetraploid complementation. Nature 461, 86–90 [DOI] [PubMed] [Google Scholar]
- 87. Plath K., Lowry W. E. (2011) Progress in understanding reprogramming to the induced pluripotent state. Nat. Rev. Genet. 12, 253–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Chin M. H., Mason M. J., Xie W., Volinia S., Singer M., Peterson C., Ambartsumyan G., Aimiuwu O., Richter L., Zhang J., Khvorostov I., Ott V., Grunstein M., Lavon N., Benvenisty N., Croce C. M., Clark A. T., Baxter T., Pyle A. D., Teitell M. A., Pelegrini M., Plath K., Lowry W. E. (2009) Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell 5, 111–123 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Stadtfeld M., Apostolou E., Akutsu H., Fukuda A., Follett P., Natesan S., Kono T., Shioda T., Hochedlinger K. (2010) Aberrant silencing of imprinted genes on chromosome 12qF1 in mouse induced pluripotent stem cells. Nature 465, 175–181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Kim K., Doi A., Wen B., Ng K., Zhao R., Cahan P., Kim J., Aryee M. J., Ji H., Ehrlich L. I., Yabuuchi A., Takeuchi A., Cunniff K. C., Hongguang H., McKinney-Freeman S., Naveiras O., Yoon T. J., Irizarry R. A., Jung N., Seita J., Hanna J., Murakami P., Jaenisch R., Weissleder R., Orkin S. H., Weissman I. L., Feinberg A. P., Daley G. Q. (2010) Epigenetic memory in induced pluripotent stem cells. Nature 467, 285–290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Bock C., Kiskinis E., Verstappen G., Gu H., Boulting G., Smith Z. D., Ziller M., Croft G. F., Amoroso M. W., Oakley D. H., Gnirke A., Eggan K., Meissner A. (2011) Reference maps of human ES and iPS cell variation enable high-throughput characterization of pluripotent cell lines. Cell 144, 439–452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Kang L., Wang J., Zhang Y., Kou Z., Gao S. (2009) iPS cells can support full-term development of tetraploid blastocyst-complemented embryos. Cell Stem Cell 5, 135–138 [DOI] [PubMed] [Google Scholar]
- 93. Guenther M. G., Frampton G. M., Soldner F., Hockemeyer D., Mitalipova M., Jaenisch R., Young R. A. (2010) Chromatin structure and gene expression programs of human embryonic and induced pluripotent stem cells. Cell Stem Cell 7, 249–257 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Carey B. W., Markoulaki S., Hanna J. H., Faddah D. A., Buganim Y., Kim J., Ganz K., Steine E. J., Cassady J. P., Creyghton M. P., Welstead G. G., Gao Q., Jaenisch R. (2011) Reprogramming factor stoichiometry influences the epigenetic state and biological properties of induced pluripotent stem cells. Cell Stem Cell 9, 588–598 [DOI] [PubMed] [Google Scholar]
- 95. Ohi Y., Qin H., Hong C., Blouin L., Polo J. M., Guo T., Qi Z., Downey S. L., Manos P. D., Rossi D. J., Yu J., Hebrok M., Hochedlinger K., Costello J. F., Song J. S., Ramalho-Santos M. (2011) Incomplete DNA methylation underlies a transcriptional memory of somatic cells in human iPS cells. Nat. Cell Biol. 13, 541–549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96. Chen X., Xu H., Yuan P., Fang F., Huss M., Vega V. B., Wong E., Orlov Y. L., Zhang W., Jiang J., Loh Y. H., Yeo H. C., Yeo Z. X., Narang V., Govindarajan K. R., Leong B., Shahab A., Ruan Y., Bourque G., Sung W. K., Clarke N. D., Wei C. L., Ng H. H. (2008) Integration of external signaling pathways with the core transcriptional network in embryonic stem cells. Cell 133, 1106–1117 [DOI] [PubMed] [Google Scholar]
- 97. Boyer L. A., Plath K., Zeitlinger J., Brambrink T., Medeiros L. A., Lee T. I., Levine S. S., Wernig M., Tajonar A., Ray M. K., Bell G. W., Otte A. P., Vidal M., Gifford D. K., Young R. A., Jaenisch R. (2006) Polycomb complexes repress developmental regulators in murine embryonic stem cells. Nature 441, 349–353 [DOI] [PubMed] [Google Scholar]
- 98. Lee T. I., Jenner R. G., Boyer L. A., Guenther M. G., Levine S. S., Kumar R. M., Chevalier B., Johnstone S. E., Cole M. F., Isono K., Koseki H., Fuchikami T., Abe K., Murray H. L., Zucker J. P., Yuan B., Bell G. W., Herbolsheimer E., Hannett N. M., Sun K., Odom D. T., Otte A. P., Volkert T. L., Bartel D. P., Melton D. A., Gifford D. K., Jaenisch R., Young R. A. (2006) Control of developmental regulators by Polycomb in human embryonic stem cells. Cell 125, 301–313 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Kidder B. L., Palmer S., Knott J. G. (2009) SWI/SNF-Brg1 regulates self-renewal and occupies core pluripotency-related genes in embryonic stem cells. Stem cells 27, 317–328 [DOI] [PubMed] [Google Scholar]
- 100. Ku M., Koche R. P., Rheinbay E., Mendenhall E. M., Endoh M., Mikkelsen T. S., Presser A., Nusbaum C., Xie X., Chi A. S., Adli M., Kasif S., Ptaszek L. M., Cowan C. A., Lander E. S., Koseki H., Bernstein B. E. (2008) Genome-wide analysis of PRC1 and PRC2 occupancy identifies two classes of bivalent domains. PLoS Genet. 4, e1000242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Li G., Margueron R., Ku M., Chambon P., Bernstein B. E., Reinberg D. (2010) Jarid2 and PRC2, partners in regulating gene expression. Genes Dev. 24, 368–380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Ho L., Jothi R., Ronan J. L., Cui K., Zhao K., Crabtree G. R. (2009) An embryonic stem cell chromatin-remodeling complex, esBAF, is an essential component of the core pluripotency transcriptional network. Proc. Natl. Acad. Sci. U.S.A. 106, 5187–5191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Xie L., Pelz C., Wang W., Bashar A., Varlamova O., Shadle S., Impey S. (2011) KDM5B regulates embryonic stem cell self-renewal and represses cryptic intragenic transcription. EMBO J. 30, 1473–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Boyer L. A., Lee T. I., Cole M. F., Johnstone S. E., Levine S. S., Zucker J. P., Guenther M. G., Kumar R. M., Murray H. L., Jenner R. G., Gifford D. K., Melton D. A., Jaenisch R., Young R. A. (2005) Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122, 947–956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Yang J., Chai L., Fowles T. C., Alipio Z., Xu D., Fink L. M., Ward D. C., Ma Y. (2008) Genome-wide analysis reveals Sall4 to be a major regulator of pluripotency in murine embryonic stem cells. Proc. Natl. Acad. Sci. U.S.A. 105, 19756–19761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106. Hu G., Kim J., Xu Q., Leng Y., Orkin S. H., Elledge S. J. (2009) A genome-wide RNAi screen identifies a new transcriptional module required for self-renewal. Genes Dev. 23, 837–848 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Dejosez M., Levine S. S., Frampton G. M., Whyte W. A., Stratton S. A., Barton M. C., Gunaratne P. H., Young R. A., Zwaka T. P. (2010) Ronin/Hcf-1 binds to a hyperconserved enhancer element and regulates genes involved in the growth of embryonic stem cells. Genes Dev. 24, 1479–1484 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Cole M. F., Johnstone S. E., Newman J. J., Kagey M. H., Young R. A. (2008) Tcf3 is an integral component of the core regulatory circuitry of embryonic stem cells. Genes Dev. 22, 746–755 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Chia N. Y., Chan Y. S., Feng B., Lu X., Orlov Y. L., Moreau D., Kumar P., Yang L., Jiang J., Lau M. S., Huss M., Soh B. S., Kraus P., Li P., Lufkin T., Lim B., Clarke N. D., Bard F., Ng H. H. (2010) A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. Nature 468, 316–320 [DOI] [PubMed] [Google Scholar]
- 110. Kim S. W., Yoon S. J., Chuong E., Oyolu C., Wills A. E., Gupta R., Baker J. (2011) Chromatin and transcriptional signatures for Nodal signaling during endoderm formation in hESCs. Dev. Biol. 357, 492–504 [DOI] [PubMed] [Google Scholar]
- 111. Wu H., D'Alessio A. C., Ito S., Xia K., Wang Z., Cui K., Zhao K., Sun Y. E., Zhang Y. (2011) Dual functions of Tet1 in transcriptional regulation in mouse embryonic stem cells. Nature 473, 389–393 [DOI] [PMC free article] [PubMed] [Google Scholar]