Abstract
Complex traits such as susceptibility to diseases are determined in part by variants at multiple genetic loci. Genome-wide association studies can identify these loci, but most phenotype-associated variants lie distal to protein-coding regions and are likely involved in regulating gene expression. Understanding how these genetic variants affect complex traits depends on the ability to predict and test the function of the genomic elements harboring them. Community efforts such as the ENCODE Project provide a wealth of data about epigenetic features associated with gene regulation. These data enable the prediction of testable functions for many phenotype-associated variants.
Keywords: Epigenetics, Gene Regulation, Genetic Diseases, Human Genetics, Molecular Genetics, Complex Trait, Genome-wide Association Study, Genotype-Phenotype Association
Complex Genetics of Disease Susceptibility
Genetics informs us about human disease in at least two major ways, one through Mendelian diseases and the other through complex traits. Mutations that lead to Mendelian inheritance of disease usually alter the function of single genes (1), reducing or modifying the function of the protein product by changing the encoded amino acid sequence (2, 3). In addition, some Mendelian diseases are caused by debilitating mutations in promoters or enhancers of a gene, resulting in a deficiency of the protein product and the consequent pathological phenotype (4, 5). Genetic variants causing Mendelian disease are rare in the human population (6) because selective pressure against their deleterious effects keeps their allele frequency low. Because the genetic variants causing monogenic diseases are rare, mapping studies are confined to detailed analyses of affected families and kindreds (6). Such studies have mapped genetic variants at the heart of many monogenic disorders. The Online Mendelian Inheritance in Man® (OMIM®) database (1, 7) currently lists almost 3400 phenotypes for which the molecular basis is known.
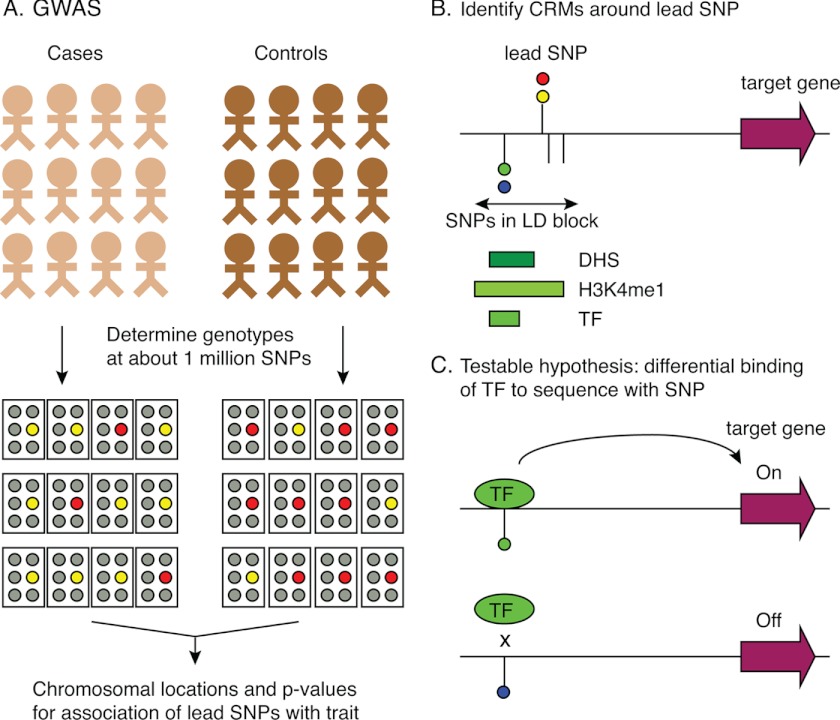
Susceptibilities to many common diseases such as coronary artery disease and many forms of cancer and type 2 diabetes have substantial genetic components, but in contrast to the Mendelian diseases, these phenotypes are affected by variants at multiple loci (6, 8, 9). Thus, susceptibility to a common disease is a complex trait. Mapping the multiple loci that contribute to these important traits usually follows a case-control design (Fig. 1A). The mapping experiments examine SNPs to ascertain the genotypes that are significantly more prevalent in the affected group than in the non-affected group; these genotypes are associated with the trait of interest. When genotypes are determined at SNPs throughout the genome of each individual, the study is called a genome-wide association study (GWAS).2
FIGURE 1.
Epigenetic data link GWAS results to hypotheses about how specific SNPs affect a phenotype. A, in a GWAS, individuals are grouped into cases or controls, denoted by different colors for stick figures. DNA samples from each person are genotyped at a large number of polymorphic sites, illustrated as panels of gray or colored circles. SNPs for which the frequency of one allele is significantly different between the groups (e.g. the yellow allele at one SNP is more frequent in cases, whereas red is more frequent in controls) are identified, and those passing stringent filters and replication are analyzed further as lead SNPs. B, epigenetic features in chromosomal regions containing the lead SNP and linked SNPs are examined for evidence of CRMs. In this illustration, a SNP in LD with the lead SNP is in chromatin that is hypersensitive to DNase, monomethylated at histone H3 lysine 4, and bound by a transcription factor (TF). C, combining the genetic and epigenetic information leads to testable hypotheses such as allele-specific binding by a transcription factor causes differential expression of a target gene.
GWASs
Conducting a GWAS for complex traits has been a formidable challenge because the contribution of any one locus to the phenotype is expected to be small compared with the sizable effects of variants causing monogenic disorders. Furthermore, the mapping experiments need to cover the entire human genome at a sufficiently high resolution for discovery. Of course, the fact that the diseases are common means that large cohorts of individuals can be recruited for case-control studies, with thousands of affected and non-affected persons enrolled in a study, thus providing substantial power.
Recent advances such as the HapMap Project (10) have enabled effective mapping of multiple loci for complex traits in humans. A driving assumption for GWASs is that common diseases are likely caused by common variants (9, 11). Because the phenotypic effect of any one variant is expected to be small, these alleles may reach sufficiently high frequencies to be considered common (at least 5%). The HapMap Project determined combinations of allelic configurations of loci (haplotypes) that are common in several human populations. This allowed the development of high-throughput approaches to ascertain the genotype for individuals at ∼1 million SNPs across the genome, giving a good resolution for GWASs (Fig. 1A). High-throughput high-resolution genotyping coupled with the large cohorts available for many complex traits enabled the completion of the first GWAS in 2005 (12), and the number of completed GWASs has increased each subsequent year. The National Human Genome Research Institute maintains a catalog of published GWAS results (13), and as of mid-2011, it contained the results of 1449 GWASs for 237 traits.
A critical step in interpreting the results of the GWAS is moving from maps of loci associated with a trait to identifying the genetic variants that actually contribute to the trait (14, 15). In the case of Mendelian disorders, once a locus was strongly implicated in the disease, attention was rightly focused on the protein-coding genes in the region because many of the causative variants impact the structure of the encoded protein. However, it is likely that a substantial fraction of genetic variants contributing to complex traits in humans are involved in gene regulation, just as has been observed in model organisms (8, 16). Most phenotype-associated variants discovered in GWASs are far from protein-coding regions, even appearing in gene deserts (13), which is similar to the genomic distribution of most cis-regulatory modules (CRMs) such as promoters and enhancers. Strikingly, trait-associated variants from GWASs are more likely to be associated with quantitative variation in gene expression than are other variants on the genotyping arrays (17, 18), further supporting the expectation that many variants associated with complex traits affect gene expression.
Thus, what is needed is to match the high-resolution maps of phenotype-associated variants to reliable information about the locations of CRMs (Fig. 1, B and C). Enhancers can be located very far from their target genes (19), and virtually any noncoding sequence in the human genome could potentially be a CRM. Although both interspecies conservation and direct assays for biochemical features of chromatin associated with CRMs can be used to predict the locations of regulatory regions (15, 20, 21), interpretation of the results from GWASs requires an extensive mapping of CRMs in multiple human tissues.
This minireview will cover recent advances in building a more comprehensive catalog of CRMs in humans and the use of that catalog to predict functions that are altered by phenotype-associated SNPs discovered through GWASs.
Epigenetic Features Associated with Expression and Regulation
The expression levels of genes in humans (and eukaryotes in general) are determined by both chromatin structure and the transcription factors that are bound to the CRMs (22–24). The molecular mechanisms regulating gene expression are a complex interplay among enzymes and factors that catalyze hundreds of reactions, including covalent modification of histones; alteration of nucleosomal structure and stability; binding of transcription factors to specific DNA sequences; recruitment of coactivators, repressors, and polymerases; and initiation, pausing, and elongation of transcription. The details of how these reactions lead to appropriate levels of expression at the right time and place are specific for each gene, and a full understanding of regulation requires intensive study of each locus.
However, some features of the biochemical machinery employed in regulated expression are common to most genes and their CRMs (25). The most obvious feature is the presence of transcripts in the steady-state RNA. Measurement of RNA levels using any of a variety of methods provides a good monitor of the expression of a gene (26–30).
CRMs have consistent features as well. Most are in regions of the chromatin that are accessible to macromolecules, reflecting the need for the CRM to interact with proteins such as transcription factors. These accessible regions can be mapped by treating nuclei with a DNase and determining the sites of cleavage. Such DNase-hypersensitive sites (DHSs) are a general feature of almost all active CRMs (31).
Particular histone modifications can distinguish categories of CRMs and expression states of genes (24, 32). Antibodies specific to individual histone modifications are used to immunoprecipitate chromatin (histones and DNA) bearing that modification. DNA isolated by ChIP is then assayed for the presence of segments of interest (33). Some features have substantial diagnostic importance. The chromatin around active promoters has high levels of trimethylation at lysine 4 of histone H3 (H3K4me3), whereas the chromatin around enhancers has high levels of monomethylation at the same position (H3K4me1) (25, 34, 35). Acetylation of histone H3 at lysine 27 is associated with active promoters and enhancers (32). The chromatin of transcribed regions is marked by di- and trimethylation at lysine 79 of H3 (H3K79me2/me3) in the initial portion of the transcription unit, followed by methylation at lysine 36 (H3K36me3) in the distal portion (36). Other histone H3 methylations mark distinct portions of the repressed chromatin, with trimethylation at lysine 27 (H3K27me3) or lysine 9 (H3K9me3) covering different sets of repressed genes (37, 38).
CRMs such as enhancers and promoters are clusters of binding sites for transcription factors (39), and thus, occupancy by transcription factors is a good indicator of potential regulatory regions. Using a ChIP approach but with antibodies against individual transcription factors, one can obtain reliable maps of transcription factor occupancy (40–42). However, because a distinct battery of factors is bound at each CRM and because many transcription factors are present in a limited number of cell types, maps of binding by many transcription factors are needed to find a broad range of CRMs. Conversely, once a region has been identified as a CRM, the set of bound proteins can be used to better understand regulation and the impact of genetic variation on that regulation (Fig. 1C).
Transcripts, DHSs, histone modifications, and transcription factor occupancy can all be considered epigenetic features (43). They are biochemical attributes that lie on top of (epi, “on” or “above”) the genetic material (DNA), and they reflect or influence the expression of genes. The epigenetic features are dynamic: RNA is made and degraded, histone modifications are added and removed, and transcription factors bind to and dissociate from DNA. However, the steady-state levels of these epigenetic features are characteristic of the chromatin containing a given segment of DNA in a given cell type, and that steady-state level can be inherited at least in somatic cells. This is thought to be a cellular memory for expression status (44), and the epigenetic features can be used as a monitor of gene activity and CRM location.
Genome-wide Determination of Informative Epigenetic Features
Detailed studies of the molecules and biochemical events that regulate expression of individual genes in chromatin led to the discovery of the connections between epigenetic features and regulation. Recent advances in genomic technology allow these features to be determined quantitatively throughout genomes. DNA that is highly enriched for the feature of interest can be mapped comprehensively, most commonly using second-generation sequencing methods (42, 45, 46). Transcriptomes can be determined by sequencing RNA after fragmentation and conversion to complementary DNA; this is called RNA-seq (30). DNA in chromatin with a certain modification or bound by a particular transcription factor can be determined by sequencing the DNA enriched by ChIP; this is called ChIP-seq (47, 48). DNA in exposed regions of chromatin, i.e. DHSs, can be identified by enriching for DNA cut by nucleases in chromatin and sequencing from the cleaved ends; this is called DNase-seq (49, 50).
A few community projects are assaying a broad collection of epigenetic features across a wide spectrum of cell types in humans and model organisms. In these consortia, complementary work in multiple laboratories is coordinated to cover a substantial portion of the matrix of features and cell types. Consistent data standards are established, and the data are released as soon as it is replicated. One of the major community projects is the ENCODE Project, which aims to establish an ENCyclopedia Of DNA Elements (51). The various branches of this consortium are determining transcriptomes, mapping histone modifications and transcription factor occupancy, and identifying accessible chromatin, in addition to manually curating the annotation of genes (42, 46). In the production phase culminating this year, all assays are being run on a set of human cell lines that represent some important human tissues, and some assays such as DNase-seq are being conducted on a wide range of cell types, including primary cells. Almost 3000 data sets have been released to date. Parallel work is being done in Caenorhabditis elegans (52) and Drosophila melanogaster (53) as the modENCODE Project.
Another community project is the NIH Roadmap Epigenomics Mapping Consortium (54), which is part of the International Human Epigenome Consortium. The NIH Roadmap Epigenomics Mapping Consortium is mapping histone modifications by ChIP-seq and accessible chromatin by DNase-seq in many human tissues and cell types, with an emphasis on primary cells from healthy individuals. Over 250 data sets have been released to date.
Several studies have shown that epigenetic data such as those being generated in these community projects can be highly effective at predicting CRMs. DNA segments in chromatin with the H3K4me1 modification are validated as enhancers in cell transfection assays at a high rate (55), and DNA segments bound by the coactivator p300 are frequently validated as enhancers in transgenic mice (56). Hence, it is reasonable to expect the epigenetic data from these consortia to be good predictors of gene regulatory function (46).
Connecting GWASs and Epigenetics
The extensive epigenetic data, although not comprehensive, are already proving to be useful for finding potential regulatory regions that could be affected by genetic variants (Fig. 1B). Several recent studies have shown that SNPs associated with complex traits are enriched in regions implicated in gene regulation based on epigenetic features. One study used statistical modeling to integrate information about several histone modifications in multiple cell lines, generating a segmentation, or partitioning, of the human genome into classes associated with different functions (37). The segmentation classes with properties of enhancers were significantly enriched for phenotype-associated SNPs. The integrative analysis of ENCODE data (46) showed that the phenotype-associated SNPs in the GWAS Catalog are enriched in DNase-sensitive regions and in DNA segments bound by transcription factors. These studies initially examined the lead SNPs from the GWAS, i.e. the SNPs on the genotyping arrays that are most highly associated with the trait of interest. Although these need not be the functional SNPs, a notable fraction of them (34%) are in DHSs (46). Of course, in many cases, the functional SNP is not the lead SNP, but rather another variant in linkage disequilibrium (LD) with the lead SNP is the functional one (Fig. 1B). When all SNPs in LD with the lead SNPs are included as phenotype-associated, then for a large fraction of the phenotype associations, at least one SNP is found in a DNA segment associated with regulatory function via the epigenetic data. For example, the lead or linked SNP is found in a DHS for 70–80% of the phenotype associations reported in the GWAS Catalog (46, 57). This strong correspondence between phenotype-associated variants and function-associated DNA indicates that current epigenetic data are already useful for interpretation of GWAS SNPs.
Examples of the Use of ENCODE Data to Interpret GWAS Results
Biochemical indicators of gene regulatory regions have long been used to interpret heritable phenotypes in humans, starting with Mendelian traits. Early examples are the use of DHSs to understand the impact of large deletions in the complex of genes encoding β-globins (5, 58, 59) and α-globins (60). DHSs distal to the genes were shown to be enhancers that when deleted led to thalassemias, which are inherited anemias resulting from inadequate production of one or more globin polypeptides.
Epigenetic features also provide insights into the interpretation of complex traits. A gene desert located upstream of the MYC gene contains genetic variants that are associated with breast and prostate cancers (61, 62). The MYC gene is an intriguing candidate for the target of the SNPs, given its role in cell cycle control, but all of the phenotype-associated variants are distal to the MYC gene, and from position alone, it is not clear how they may work. However, high-resolution mapping of several epigenetic features shows that some of the phenotype-associated variants are in transcription factor-binding sites in enhancers. The binding affinity is allele-specific, and the different alleles affect chromatin looping to the presumptive target MYC (63–65). Thus, the genetic variants do affect regulated expression of a target gene that could help explain cancer predisposition. Importantly, alignment of the ENCODE data in this region with the significant variants from the GWAS also reveals that key variants are found in the transcription factor-occupied DNA segments mapped by this consortium (42). This is true even though neither prostate nor breast tissue was used in the analysis at that time. Even without complete coverage of all tissues and factors, informative insights are gleaned from examining the GWAS results in the context of epigenetic features.
Recently, investigators employed ENCODE epigenetic data as an initial guide to discover regulatory regions in which genetic variation is affecting a complex trait. For example, Farrell et al. (66) used ENCODE data to help find likely causative variants in an enhancer in the HBS1L-MYB locus, one of three loci associated with quantitative levels of “fetal” hemoglobin in adult red blood cells. Their fine-mapping showed that the most strongly associated variants are clustered in the intergenic region (Fig. 2A), and a scan of ENCODE data showed that the variants are in DNA segments with epigenetic features expected for enhancers (Fig. 2, B and C). Guided by the initial ENCODE data, the authors focused further analysis in patients and controls and showed that the variants affect a transcriptional enhancer (66). Other recent examples of the use of ENCODE or other epigenetic data as guides for functional studies of trait-associated variants are studies of the TCF7L2 intronic enhancer strongly associated with type 2 diabetes (67), the gene desert at chromosome 9p21 associated with coronary artery disease (68), and a locus associated with susceptibility to colorectal cancer (69).
FIGURE 2.
GWAS variants associated with high levels of fetal hemoglobin in adults found in an enhancer marked by epigenetic features. A, fine mapping of genetic variants between the genes HBS1L and MYB on human chromosome 6 (chr6), with the position of SNPs along the x axis (assembly GRCh37/hg19) and the logarithm (base 10) of the 1/p value for the association with the trait on the y axis. This figure was adapted with permission from Fig. 4 in Ref. 66. B, current view of genomic data for the same 148-kb interval as in A, showing from top to bottom the position of the 3-bp deletion implicated in the trait (HMIPdel); genetic variants from the GWAS Catalog (vertical green lines); gene annotation; and signal tracks for DHSs in K562 cells, GATA1 occupancy in peripheral blood-derived erythroblasts, TAL1 occupancy in K562 cells, and GATA2 occupancy in K562 cells. The signal tracks are from the ENCODE Consortium (42, 46, 78). C, view focused on a 3-kb region containing the 3-bp deletion implicated in the trait and an enhancer bound by GATA factors and TAL1. PBDE, peripheral blood derived erythroblasts.
Some links between specific epigenetic features and trait-associated variants are found at multiple loci affecting a complex trait, suggesting that common regulatory mechanisms could be operating at multiple loci. Each phenotype in the GWAS Catalog can be associated with multiple loci, and in several cases, the loci affecting a given trait are associated more frequently than expected with a particular feature such as occupancy by a particular transcription factor or appearance of a DHS in a given cell line (46, 57). For example, variants associated with Crohn disease are over-represented in DNA segments bound by GATA2 (in human umbilical vein endothelial cells (HUVECs)) and are sensitive to DNase in T-helper cells. One example is a 1.25-Mb gene desert on chromosome 5 (Fig. 3). High-resolution mapping reveals a cluster of variants in LD that are strongly associated with Crohn disease (Fig. 3A) (70). Within this cluster are variants that affect the level of expression of PTGER4, a gene located ∼300 kb away that encodes the EP4 prostaglandin receptor (70). Examination of selected ENCODE tracks within this region shows that the trait-associated variants are in or close to DHSs that are binding sites for a GATA transcription factor (Fig. 3, B and C). The data from the T-helper cells are likely to be more relevant to autoimmunity than those from HUVECs, and one could hypothesize that the genetic variation could be affecting affinity for GATA3, a related protein that regulates gene expression in T-cells (71). This is an example of a readily testable hypothesis grounded in the examination of the GWAS and ENCODE data (Fig. 1C).
FIGURE 3.
GWAS variants associated with Crohn disease and other autoimmune diseases found in potential regulatory regions marked by epigenetic features. A, fine mapping of genetic variants in a 2-Mb interval on human chromosome 5 (chr5). The red vertical lines demarcate a LD block. This figure was reprinted with permission from the supplement to Ref. 70. B, current view of genomic data for the same interval as in A, showing from top to bottom genetic variants from the GWAS Catalog, followed by signal tracks for GATA2 occupancy in HUVECs and DHSs in HUVECs and T-helper 1 (Th1) and T-helper 2 (Ths) cells, with gene annotation at the bottom. The signal tracks are from the ENCODE Consortium (42, 46). C, view focused on a 50-kb region containing a cluster of variants associated with autoimmune diseases. The different disease associations are marked by the color of the circle at the top of the vertical green lines. This figure was reprinted with permission from Ref. 46.
These examples illustrate an important principle. The data from community projects such as ENCODE and NIH Roadmap Epigenomics Mapping Consortium may not cover the tissues, developmental stages, or transcription factors of greatest relevance to a particular phenotype. However, in many cases, they provide initial insights that help guide more definitive experiments. These may be cases in which a regulatory region is active in multiple cell types or is bound by several different transcription factors.
Prospects for the Future
Application of genomic technologies continues to stimulate discovery in biochemistry and molecular biology as the networks of regulatory interactions begin to be understood not only in model organisms (72) but also in humans. This information is helping to translate molecular insights into settings of clinical relevance. The results from GWASs reveal genomic locations in which genetic variation impacts susceptibility to the most common diseases of humans. It is now clear that many of these loci are involved in gene regulation, and the deep knowledge of the biochemistry of gene regulation can be coupled with high-throughput genomic assays to help identify candidate regulatory regions in the loci identified by GWASs. No longer will finding a key genetic variant in a gene desert mean the end of a search for a molecular connection between genotype and phenotype.
This minireview has emphasized the use of epigenetic features mapped by ENCODE and other projects for interpreting GWAS results. This approach has considerable power now, and plans are in place to increase substantially the coverage of cell types, transcription factors, and other epigenetic features (described at www.genome.gov/10005107). The large scale of these community projects and their commitment to rapid data release will ensure that data sets of closer relevance to a wider range of phenotypes will become available. Assays with higher resolution for mapping regulatory elements will also be used more widely (73–75).
Of course, the genome-wide mapping of epigenetic features originated in individual laboratories, and they will continue to provide important data sets and insights for regulation. Indeed, it is likely that individual or small groups of laboratories will explore epigenetic features in the cell types most closely related to the phenotypes of interest. The capacity of second-generation sequencing machines is increasing, which means that many more laboratories will be generating and analyzing genome-wide epigenetic data. This diversity of investigator-initiated projects should complement the community projects and likely fill gaps in coverage that are most relevant to the phenotypes of interest.
Several challenges must be met to effectively harvest the insights from the plethora of genome-scale genetic and epigenetic results. One of the most exciting challenges is integration of the data. Initial efforts using statistical modeling (76, 77) are being applied to some of the data from the community projects. Opportunities abound for novel approaches that have the capacity for even larger numbers of data sets and that provide more accurate predictions. These opportunities should engage not only biochemists and geneticists but also statisticians and bioinformaticians. Strong collaborations among teams of these investigators should lead to insights into connections between multiple genotypes and complex phenotypes of even greater importance to medicine.
Acknowledgments
I thank Aleksandar Milosavljevic for a unifying definition of epigenetics, J. Farrell and D. Chui for the primary data used in Fig. 2A, and M. Georges for permission to reproduce the data in Fig. 3A.
This work was supported, in whole or in part, by National Institutes of Health Grants R01 DK065806, RC HG005573, and U01 HG004695. This is the sixth article in the Thematic Minireview Series on Results from the ENCODE Project: Integrative Global Analyses of Regulatory Regions in the Human Genome.
- GWAS
- genome-wide association study
- CRM
- cis-regulatory module
- DHS
- DNase-hypersensitive site
- LD
- linkage disequilibrium
- HUVEC
- human umbilical vein endothelial cell.
REFERENCES
- 1. McKusick V. A. (2007) Mendelian Inheritance in Man and its online version, OMIM. Am. J. Hum. Genet. 80, 588–604 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Giardine B., Riemer C., Hefferon T., Thomas D., Hsu F., Zielenski J., Sang Y., Elnitski L., Cutting G., Trumbower H., Kern A., Kuhn R., Patrinos G. P., Hughes J., Higgs D., Chui D., Scriver C., Phommarinh M., Patnaik S. K., Blumenfeld O., Gottlieb B., Vihinen M., Väliaho J., Kent J., Miller W., Hardison R. C. (2007) PhenCode: connecting ENCODE data with mutations and phenotype. Hum. Mutat. 28, 554–562 [DOI] [PubMed] [Google Scholar]
- 3. Stenson P. D., Mort M., Ball E. V., Howells K., Phillips A. D., Thomas N. S., Cooper D. N. (2009) The Human Gene Mutation Database: 2008 update. Genome Med. 1, 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Orkin S. H., Sexton J. P., Cheng T. C., Goff S. C., Giardina P. J., Lee J. I., Kazazian H. H., Jr. (1983) ATA box transcription mutation in β-thalassemia. Nucleic Acids Res. 11, 4727–4734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Forrester W. C., Epner E., Driscoll M. C., Enver T., Brice M., Papayannopoulou T., Groudine M. (1990) A deletion of the human β-globin locus activation region causes a major alteration in chromatin structure and replication across the entire β-globin locus. Genes Dev. 4, 1637–1649 [DOI] [PubMed] [Google Scholar]
- 6. Botstein D., Risch N. (2003) Discovering genotypes underlying human phenotypes: past successes for Mendelian disease, future approaches for complex disease. Nat. Genet. 33, 228–237 [DOI] [PubMed] [Google Scholar]
- 7. Amberger J., Bocchini C. A., Scott A. F., Hamosh A. (2009) McKusick's Online Mendelian Inheritance in Man (OMIM®). Nucleic Acids Res. 37, D793–D796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Glazier A. M., Nadeau J. H., Aitman T. J. (2002) Finding genes that underlie complex traits. Science 298, 2345–2349 [DOI] [PubMed] [Google Scholar]
- 9. Hirschhorn J. N., Daly M. J. (2005) Genome-wide association studies for common diseases and complex traits. Nat. Rev. Genet. 6, 95–108 [DOI] [PubMed] [Google Scholar]
- 10. International HapMap Consortium (2005) A haplotype map of the human genome. Nature 437, 1299–1320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bodmer W., Bonilla C. (2008) Common and rare variants in multifactorial susceptibility to common diseases. Nat. Genet. 40, 695–701 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Klein R. J., Zeiss C., Chew E. Y., Tsai J. Y., Sackler R. S., Haynes C., Henning A. K., SanGiovanni J. P., Mane S. M., Mayne S. T., Bracken M. B., Ferris F. L., Ott J., Barnstable C., Hoh J. (2005) Complement factor H polymorphism in age-related macular degeneration. Science 308, 385–389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hindorff L. A., Sethupathy P., Junkins H. A., Ramos E. M., Mehta J. P., Collins F. S., Manolio T. A. (2009) Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl. Acad. Sci. U.S.A. 106, 9362–9367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Frazer K. A., Murray S. S., Schork N. J., Topol E. J. (2009) Human genetic variation and its contribution to complex traits. Nat. Rev. Genet. 10, 241–251 [DOI] [PubMed] [Google Scholar]
- 15. Cooper G. M., Shendure J. (2011) Needles in stacks of needles: finding disease-causal variants in a wealth of genomic data. Nat. Rev. Genet. 12, 628–640 [DOI] [PubMed] [Google Scholar]
- 16. Mackay T. F. (2001) Quantitative trait loci in Drosophila. Nat. Rev. Genet. 2, 11–20 [DOI] [PubMed] [Google Scholar]
- 17. Nicolae D. L., Gamazon E., Zhang W., Duan S., Dolan M. E., Cox N. J. (2010) Trait-associated SNPs are more likely to be eQTLs: annotation to enhance discovery from GWAS. PLoS Genet. 6, e1000888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Zhong H., Beaulaurier J., Lum P. Y., Molony C., Yang X., Macneil D. J., Weingarth D. T., Zhang B., Greenawalt D., Dobrin R., Hao K., Woo S., Fabre-Suver C., Qian S., Tota M. R., Keller M. P., Kendziorski C. M., Yandell B. S., Castro V., Attie A. D., Kaplan L. M., Schadt E. E. (2010) Liver and adipose expression-associated SNPs are enriched for association to type 2 diabetes. PLoS Genet. 6, e1000932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lettice L. A., Heaney S. J., Purdie L. A., Li L., de Beer P., Oostra B. A., Goode D., Elgar G., Hill R. E., de Graaff E. (2003) A long-range Shh enhancer regulates expression in the developing limb and fin and is associated with preaxial polydactyly. Hum. Mol. Genet. 12, 1725–1735 [DOI] [PubMed] [Google Scholar]
- 20. Wasserman W. W., Sandelin A. (2004) Applied bioinformatics for the identification of regulatory elements. Nat. Rev. Genet. 5, 276–287 [DOI] [PubMed] [Google Scholar]
- 21. Elnitski L., Jin V. X., Farnham P. J., Jones S. J. (2006) Locating mammalian transcription factor-binding sites: a survey of computational and experimental techniques. Genome Res. 16, 1455–1464 [DOI] [PubMed] [Google Scholar]
- 22. Felsenfeld G., Groudine M. (2003) Controlling the double helix. Nature 421, 448–453 [DOI] [PubMed] [Google Scholar]
- 23. Maston G. A., Evans S. K., Green M. R. (2006) Transcriptional regulatory elements in the human genome. Annu. Rev. Genomics Hum. Genet. 7, 29–59 [DOI] [PubMed] [Google Scholar]
- 24. Rando O. J., Chang H. Y. (2009) Genome-wide views of chromatin structure. Annu. Rev. Biochem. 78, 245–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. ENCODE Project Consortium (2007) Identification and analysis of functional elements in 1% of the human genome by the ENCODE Pilot Project. Nature 447, 799–816 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Galau G. A., Klein W. H., Davis M. M., Wold B. J., Britten R. J., Davidson E. H. (1976) Structural gene sets active in embryos and adult tissues of the sea urchin. Cell 7, 487–505 [DOI] [PubMed] [Google Scholar]
- 27. Thomas P. S. (1980) Hybridization of denatured RNA and small DNA fragments transferred to nitrocellulose. Proc. Natl. Acad. Sci. U.S.A. 77, 5201–5205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Eisen M. B., Spellman P. T., Brown P. O., Botstein D. (1998) Cluster analysis and display of genome-wide expression patterns. Proc. Natl. Acad. Sci. U.S.A. 95, 14863–14868 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Kapranov P., Cawley S. E., Drenkow J., Bekiranov S., Strausberg R. L., Fodor S. P., Gingeras T. R. (2002) Large-scale transcriptional activity in chromosomes 21 and 22. Science 296, 916–919 [DOI] [PubMed] [Google Scholar]
- 30. Mortazavi A., Williams B. A., McCue K., Schaeffer L., Wold B. (2008) Mapping and quantifying mammalian transcriptomes by RNA-seq. Nat. Methods 5, 621–628 [DOI] [PubMed] [Google Scholar]
- 31. Gross D. S., Garrard W. T. (1988) Nuclease hypersensitive sites in chromatin. Ann. Rev. Biochem. 57, 159–197 [DOI] [PubMed] [Google Scholar]
- 32. Kouzarides T. (2007) Chromatin modifications and their function. Cell 128, 693–705 [DOI] [PubMed] [Google Scholar]
- 33. Schones D. E., Zhao K. (2008) Genome-wide approaches to studying chromatin modifications. Nat. Rev. Genet. 9, 179–191 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Barski A., Cuddapah S., Cui K., Roh T. Y., Schones D. E., Wang Z., Wei G., Chepelev I., Zhao K. (2007) High-resolution profiling of histone methylations in the human genome. Cell 129, 823–837 [DOI] [PubMed] [Google Scholar]
- 35. Heintzman N. D., Stuart R. K., Hon G., Fu Y., Ching C. W., Hawkins R. D., Barrera L. O., Van Calcar S., Qu C., Ching K. A., Wang W., Weng Z., Green R. D., Crawford G. E., Ren B. (2007) Distinct and predictive chromatin signatures of transcriptional promoters and enhancers in the human genome. Nat. Genet. 39, 311–318 [DOI] [PubMed] [Google Scholar]
- 36. Steger D. J., Lefterova M. I., Ying L., Stonestrom A. J., Schupp M., Zhuo D., Vakoc A. L., Kim J. E., Chen J., Lazar M. A., Blobel G. A., Vakoc C. R. (2008) DOT1L/KMT4 recruitment and H3K79 methylation are ubiquitously coupled with gene transcription in mammalian cells. Mol. Cell. Biol. 28, 2825–2839 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ernst J., Kheradpour P., Mikkelsen T. S., Shoresh N., Ward L. D., Epstein C. B., Zhang X., Wang L., Issner R., Coyne M., Ku M., Durham T., Kellis M., Bernstein B. E. (2011) Mapping and analysis of chromatin state dynamics in nine human cell types. Nature 473, 43–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Wu W., Cheng Y., Keller C. A., Ernst J., Kumar S. A., Mishra T., Morrissey C., Dorman C. M., Chen K. B., Drautz D., Giardine B., Shibata Y., Song L., Pimkin M., Crawford G. E., Furey T. S., Kellis M., Miller W., Taylor J., Schuster S. C., Zhang Y., Chiaromonte F., Blobel G. A., Weiss M. J., Hardison R. C. (2011) Dynamics of the epigenetic landscape during erythroid differentiation after GATA1 restoration. Genome Res. 21, 1659–1671 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Maniatis T., Goodbourn S., Fischer J. A. (1987) Regulation of inducible and tissue-specific gene expression. Science 236, 1237–1245 [DOI] [PubMed] [Google Scholar]
- 40. Boyd K. E., Farnham P. J. (1997) Myc versus USF: discrimination at the cad gene is determined by core promoter elements. Mol. Cell. Biol. 17, 2529–2537 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Ren B., Robert F., Wyrick J. J., Aparicio O., Jennings E. G., Simon I., Zeitlinger J., Schreiber J., Hannett N., Kanin E., Volkert T. L., Wilson C. J., Bell S. P., Young R. A. (2000) Genome-wide location and function of DNA-binding proteins. Science 290, 2306–2309 [DOI] [PubMed] [Google Scholar]
- 42. ENCODE Project Consortium (2011) A user's guide to the Encyclopedia of DNA Elements (ENCODE). PLoS Biol. 9, e1001046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Goldberg A. D., Allis C. D., Bernstein E. (2007) Epigenetics: a landscape takes shape. Cell 128, 635–638 [DOI] [PubMed] [Google Scholar]
- 44. Ringrose L., Paro R. (2004) Epigenetic regulation of cellular memory by the Polycomb and Trithorax group proteins. Annu. Rev. Genet. 38, 413–443 [DOI] [PubMed] [Google Scholar]
- 45. Wold B., Myers R. M. (2008) Sequence census methods for functional genomics. Nat. Methods 5, 19–21 [DOI] [PubMed] [Google Scholar]
- 46. ENCODE Project Consortium (2012) An integrated Encyclopedia of DNA Elements in the Human Genome. Nature, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Robertson G., Hirst M., Bainbridge M., Bilenky M., Zhao Y., Zeng T., Euskirchen G., Bernier B., Varhol R., Delaney A., Thiessen N., Griffith O. L., He A., Marra M., Snyder M., Jones S. (2007) Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat. Methods 4, 651–657 [DOI] [PubMed] [Google Scholar]
- 48. Albert I., Mavrich T. N., Tomsho L. P., Qi J., Zanton S. J., Schuster S. C., Pugh B. F. (2007) Translational and rotational settings of H2A.Z nucleosomes across the Saccharomyces cerevisiae genome. Nature 446, 572–576 [DOI] [PubMed] [Google Scholar]
- 49. Boyle A. P., Davis S., Shulha H. P., Meltzer P., Margulies E. H., Weng Z., Furey T. S., Crawford G. E. (2008) High-resolution mapping and characterization of open chromatin across the genome. Cell 132, 311–322 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Hesselberth J. R., Chen X., Zhang Z., Sabo P. J., Sandstrom R., Reynolds A. P., Thurman R. E., Neph S., Kuehn M. S., Noble W. S., Fields S., Stamatoyannopoulos J. A. (2009) Global mapping of protein-DNA interactions in vivo by digital genomic footprinting. Nat. Methods 6, 283–289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. ENCODE Project Consortium (2004) The ENCODE (ENCyclopedia Of DNA Elements) Project. Science 306, 636–640 [DOI] [PubMed] [Google Scholar]
- 52. Gerstein M. B., Lu Z. J., Van Nostrand E. L., Cheng C., Arshinoff B. I., Liu T., Yip K. Y., Robilotto R., Rechtsteiner A., Ikegami K., Alves P., Chateigner A., Perry M., Morris M., Auerbach R. K., Feng X., Leng J., Vielle A., Niu W., Rhrissorrakrai K., Agarwal A., Alexander R. P., Barber G., Brdlik C. M., Brennan J., Brouillet J. J., Carr A., Cheung M. S., Clawson H., Contrino S., Dannenberg L. O., Dernburg A. F., Desai A., Dick L., Dosé A. C., Du J., Egelhofer T., Ercan S., Euskirchen G., Ewing B., Feingold E. A., Gassmann R., Good P. J., Green P., Gullier F., Gutwein M., Guyer M. S., Habegger L., Han T., Henikoff J. G., Henz S. R., Hinrichs A., Holster H., Hyman T., Iniguez A. L., Janette J., Jensen M., Kato M., Kent W. J., Kephart E., Khivansara V., Khurana E., Kim J. K., Kolasinska-Zwierz P., Lai E. C., Latorre I., Leahey A., Lewis S., Lloyd P., Lochovsky L., Lowdon R. F., Lubling Y., Lyne R., MacCoss M., Mackowiak S. D., Mangone M., McKay S., Mecenas D., Merrihew G., Miller D. M., Muroyama A., Murray J. I., Ooi S. L., Pham H., Phippen T., Preston E. A., Rajewsky N., Rätsch G., Rosenbaum H., Rozowsky J., Rutherford K., Ruzanov P., Sarov M., Sasidharan R., Sboner A., Scheid P., Segal E., Shin H., Shou C., Slack F. J., Slightam C., Smith R., Spencer W. C., Stinson E. O., Taing S., Takasaki T., Vafeados D., Voronina K., Wang G., Washington N. L., Whittle C. M., Wu B., Yan K. K., Zeller G., Zha Z., Zhong M., Zhou X., modENCODE Consortium, Ahringer J., Strome S., Gunsalus K. C., Micklem G., Liu X. S., Reinke V., Kim S. K., Hillier L. W., Henikoff S., Piano F., Snyder M., Stein L., Lieb J. D., Waterston R. H. (2010) Integrative analysis of the Caenorhabditis elegans genome by the modENCODE project. Science 330, 1775–1787 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. modENCODE Consortium, Roy S., Ernst J., Kharchenko P. V., Kheradpour P., Negre N., Eaton M. L., Landolin J. M., Bristow C. A., Ma L., Lin M. F., Washietl S., Arshinoff B. I., Ay F., Meyer P. E., Robine N., Washington N. L., Di Stefano L., Berezikov E., Brown C. D., Candeias R., Carlson J. W., Carr A., Jungreis I., Marbach D., Sealfon R., Tolstorukov M. Y., Will S., Alekseyenko A. A., Artieri C., Booth B. W., Brooks A. N., Dai Q., Davis C. A., Duff M. O., Feng X., Gorchakov A. A., Gu T., Henikoff J. G., Kapranov P., Li R., MacAlpine H. K., Malone J., Minoda A., Nordman J., Okamura K., Perry M., Powell S. K., Riddle N. C., Sakai A., Samsonova A., Sandler J. E., Schwartz Y. B., Sher N., Spokony R., Sturgill D., van Baren M., Wan K. H., Yang L., Yu C., Feingold E., Good P., Guyer M., Lowdon R., Ahmad K., Andrews J., Berger B., Brenner S. E., Brent M. R., Cherbas L., Elgin S. C., Gingeras T. R., Grossman R., Hoskins R. A., Kaufman T. C., Kent W., Kuroda M. I., Orr-Weaver T., Perrimon N., Pirrotta V., Posakony J. W., Ren B., Russell S., Cherbas P., Graveley B. R., Lewis S., Micklem G., Oliver B., Park P. J., Celniker S. E., Henikoff S., Karpen G. H., Lai E. C., MacAlpine D. M., Stein L. D., White K. P., Kellis M. (2010) Identification of functional elements and regulatory circuits by Drosophila modENCODE. Science 330, 1787–1797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Bernstein B. E., Stamatoyannopoulos J. A., Costello J. F., Ren B., Milosavljevic A., Meissner A., Kellis M., Marra M. A., Beaudet A. L., Ecker J. R., Farnham P. J., Hirst M., Lander E. S., Mikkelsen T. S., Thomson J. A. (2010) The NIH Roadmap Epigenomics Mapping Consortium. Nat. Biotechnol. 28, 1045–1048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Heintzman N. D., Hon G. C., Hawkins R. D., Kheradpour P., Stark A., Harp L. F., Ye Z., Lee L. K., Stuart R. K., Ching C. W., Ching K. A., Antosiewicz-Bourget J. E., Liu H., Zhang X., Green R. D., Lobanenkov V. V., Stewart R., Thomson J. A., Crawford G. E., Kellis M., Ren B. (2009) Histone modifications at human enhancers reflect global cell type-specific gene expression. Nature 459, 108–112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Visel A., Blow M. J., Li Z., Zhang T., Akiyama J. A., Holt A., Plajzer-Frick I., Shoukry M., Wright C., Chen F., Afzal V., Ren B., Rubin E. M., Pennacchio L. A. (2009) ChIP-seq accurately predicts tissue-specific activity of enhancers. Nature 457, 854–858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Schaub M. A., Boyle A. P., Kundaje A., Batzaglou S., Snyder M. (2012) Linking disease associations with regulatory information in the human genome. Genome Research, in press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Forrester W. C., Takegawa S., Papayannopoulou T., Stamatoyannopoulos G., Groudine M. (1987) Evidence for a locus-activating region: the formation of developmentally stable hypersensitive sites in globin-expressing hybrids. Nucleic Acids Res. 15, 10159–10177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Grosveld F., van Assendelft G. B., Greaves D. R., Kollias G. (1987) Position-independent, high-level expression of the human β-globin gene in transgenic mice. Cell 51, 975–985 [DOI] [PubMed] [Google Scholar]
- 60. Higgs D. R., Wood W. G., Jarman A. P., Sharpe J., Lida J., Pretorius I. M., Ayyub H. (1990) A major positive regulatory region located far upstream of the human α-globin gene locus. Genes Dev. 4, 1588–1601 [DOI] [PubMed] [Google Scholar]
- 61. Wokolorczyk D., Gliniewicz B., Sikorski A., Zlowocka E., Masojc B., Debniak T., Matyjasik J., Mierzejewski M., Medrek K., Oszutowska D., Suchy J., Gronwald J., Teodorczyk U., Huzarski T., Byrski T., Jakubowska A., Górski B., van de Wetering T., Walczak S., Narod S. A., Lubinski J., Cybulski C. (2008) A range of cancers is associated with the rs6983267 marker on chromosome 8. Cancer Res. 68, 9982–9986 [DOI] [PubMed] [Google Scholar]
- 62. Al Olama A. A., Kote-Jarai Z., Giles G. G., Guy M., Morrison J., Severi G., Leongamornlert D. A., Tymrakiewicz M., Jhavar S., Saunders E., Hopper J. L., Southey M. C., Muir K. R., English D. R., Dearnaley D. P., Ardern-Jones A. T., Hall A. L., O'Brien L. T., Wilkinson R. A., Sawyer E., Lophatananon A., The UK Genetic Prostate Cancer Study Collaborators/British Association of Urological Surgeons' Section of Oncology, The UK Prostate Testing for Cancer and Treatment Study (ProtecT Study) Collaborators, Horwich A., Huddart R. A., Khoo V. S., Parker C. C., Woodhouse C. J., Thompson A., Christmas T., Ogden C., Cooper C., Donovan J. L., Hamdy F. C., Neal D. E., Eeles R. A., Easton D. F. (2009) Multiple loci on 8q24 associated with prostate cancer susceptibility. Nat. Genet. 41, 1058–10560 [DOI] [PubMed] [Google Scholar]
- 63. Tuupanen S., Turunen M., Lehtonen R., Hallikas O., Vanharanta S., Kivioja T., Björklund M., Wei G., Yan J., Niittymäki I., Mecklin J. P., Järvinen H., Ristimäki A., Di-Bernardo M., East P., Carvajal-Carmona L., Houlston R. S., Tomlinson I., Palin K., Ukkonen E., Karhu A., Taipale J., Aaltonen L. A. (2009) The common colorectal cancer predisposition SNP rs6983267 at chromosome 8q24 confers potential to enhanced Wnt signaling. Nat. Genet. 41, 885–890 [DOI] [PubMed] [Google Scholar]
- 64. Pomerantz M. M., Ahmadiyeh N., Jia L., Herman P., Verzi M. P., Doddapaneni H., Beckwith C. A., Chan J. A., Hills A., Davis M., Yao K., Kehoe S. M., Lenz H. J., Haiman C. A., Yan C., Henderson B. E., Frenkel B., Barretina J., Bass A., Tabernero J., Baselga J., Regan M. M., Manak J. R., Shivdasani R., Coetzee G. A., Freedman M. L. (2009) The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat. Genet. 41, 882–884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Jia L., Landan G., Pomerantz M., Jaschek R., Herman P., Reich D., Yan C., Khalid O., Kantoff P., Oh W., Manak J. R., Berman B. P., Henderson B. E., Frenkel B., Haiman C. A., Freedman M., Tanay A., Coetzee G. A. (2009) Functional enhancers at the gene-poor 8q24 cancer-linked locus. PLoS Genet. 5, e1000597. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Farrell J. J., Sherva R. M., Chen Z. Y., Luo H. Y., Chu B. F., Ha S. Y., Li C. K., Lee A. C., Li R. C., Yuen H. L., So J. C., Ma E. S., Chan L. C., Chan V., Sebastiani P., Farrer L. A., Baldwin C. T., Steinberg M. H., Chui D. H. (2011) A 3-bp deletion in the HBS1L-MYB intergenic region on chromosome 6q23 is associated with HbF expression. Blood 117, 4935–4945 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Gaulton K. J., Nammo T., Pasquali L., Simon J. M., Giresi P. G., Fogarty M. P., Panhuis T. M., Mieczkowski P., Secchi A., Bosco D., Berney T., Montanya E., Mohlke K. L., Lieb J. D., Ferrer J. (2010) A map of open chromatin in human pancreatic islets. Nat. Genet. 42, 255–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Harismendy O., Notani D., Song X., Rahim N. G., Tanasa B., Heintzman N., Ren B., Fu X. D., Topol E. J., Rosenfeld M. G., Frazer K. A. (2011) 9p21 DNA variants associated with coronary artery disease impair interferon-γ signaling response. Nature 470, 264–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Carvajal-Carmona L. G., Cazier J. B., Jones A. M., Howarth K., Broderick P., Pittman A., Dobbins S., Tenesa A., Farrington S., Prendergast J., Theodoratou E., Barnetson R., Conti D., Newcomb P., Hopper J. L., Jenkins M. A., Gallinger S., Duggan D. J., Campbell H., Kerr D., Casey G., Houlston R., Dunlop M., Tomlinson I. (2011) Fine-mapping of colorectal cancer susceptibility loci at 8q23.3, 16q22.1, and 19q13.11: refinement of association signals and use of in silico analysis to suggest functional variation and unexpected candidate target genes. Hum. Mol. Genet. 20, 2879–2888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Libioulle C., Louis E., Hansoul S., Sandor C., Farnir F., Franchimont D., Vermeire S., Dewit O., de Vos M., Dixon A., Demarche B., Gut I., Heath S., Foglio M., Liang L., Laukens D., Mni M., Zelenika D., Van Gossum A., Rutgeerts P., Belaiche J., Lathrop M., Georges M. (2007) Novel Crohn disease locus identified by genome-wide association maps to a gene desert on 5p13.1 and modulates expression of PTGER4. PLoS Genet. 3, e58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Orkin S. H. (1992) GATA-binding transcription factors in hematopoietic cells. Blood 80, 575–581 [PubMed] [Google Scholar]
- 72. Davidson E. H., Erwin D. H. (2006) Gene regulatory networks and the evolution of animal body plans. Science 311, 796–800 [DOI] [PubMed] [Google Scholar]
- 73. Rhee H. S., Pugh B. F. (2011) Comprehensive genome-wide protein-DNA interactions detected at single-nucleotide resolution. Cell 147, 1408–1419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Boyle A. P., Song L., Lee B. K., London D., Keefe D., Birney E., Iyer V. R., Crawford G. E., Furey T. S. (2011) High-resolution genome-wide in vivo footprinting of diverse transcription factors in human cells. Genome Res. 21, 456–464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Degner J. F., Pai A. A., Pique-Regi R., Veyrieras J. B., Gaffney D. J., Pickrell J. K., De Leon S., Michelini K., Lewellen N., Crawford G. E., Stephens M., Gilad Y., Pritchard J. K. (2012) DNase I sensitivity QTLs are a major determinant of human expression variation. Nature 482, 390–394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Ernst J., Kellis M. (2010) Discovery and characterization of chromatin states for systematic annotation of the human genome. Nat. Biotechnol. 28, 817–825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Hoffman M. M., Buske O. J., Wang J., Weng Z., Bilmes J. A., Noble W. S. (2012) Unsupervised pattern discovery in human chromatin structure through genomic segmentation. Nat. Methods 9, 473–476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Fujiwara T., O'Geen H., Keles S., Blahnik K., Linnemann A. K., Kang Y. A., Choi K., Farnham P. J., Bresnick E. H. (2009) Discovering hematopoietic mechanisms through genome-wide analysis of GATA factor chromatin occupancy. Mol. Cell 36, 667–681 [DOI] [PMC free article] [PubMed] [Google Scholar]