Background: Fbxl10 is a member of the JHDM family.
Results: We show that Fbxl10 functions as an H3K4me3 demethylase. The PHD domain recognizes H3K4me3 and H3K36me2 and shows E3 ligase activity. Using a microarray approach we identified target genes for Fbxl10.
Conclusion: Our data reveal a regulatory role of Fbxl10 in cell morphology, chemokine expression, and the metabolic control.
Significance: Fbxl10 plays a novel role of in the regulation of target genes.
Keywords: Chromatin Immunoprecipitation (ChiP), Chromatin Modification, Gene Expression, Histone Methylation, Transcription, Fbxl10, PHD Domain, Histone Demethylases
Abstract
Fbxl10 (Jhdm1b/Kdm2b) is a conserved and ubiquitously expressed member of the JHDM (JmjC domain-containing histone demethylase) family. Fbxl10 was implicated in the demethylation of H3K4me3 or H3K36me2 thereby removing active chromatin marks and inhibiting gene transcription. Apart from the JmjC domain, Fbxl10 consists of a CxxC domain, a PHD domain, and an Fbox domain. By purifying the JmjC and the PHD domain of Fbxl10 and using different approaches we were able to characterize the properties of these domains in vitro. Our results suggest that Fbxl10 is rather a H3K4me3 than a H3K36me2 histone demethylase. The PHD domain exerts a dual function in binding H3K4me3 and H3K36me2 and exhibiting E3 ubiquitin ligase activity. We generated mouse embryonic fibroblasts stably overexpressing Fbxl10. These cells reveal an increase in cell size but no changes in proliferation, mitosis, or apoptosis. Using a microarray approach we were able to identify potentially new target genes for Fbxl10 including chemokines, the noncoding RNA Xist, and proteins involved in metabolic processes. Additionally, we found that Fbxl10 is recruited to the promoters of Ccl7, Xist, Crabp2, and RipK3. Promoter occupancy by Fbxl10 was accompanied by reduced levels of H3K4me3 but unchanged levels of H3K36me2. Furthermore, knockdown of Fbxl10 using small interfering RNA approaches showed inverse regulation of Fbxl10 target genes. In summary, our data reveal a regulatory role of Fbxl10 in cell morphology, chemokine expression, and the metabolic control of fibroblasts.
Introduction
Histone modifications including acetylation, phosphorylation, and methylation are tightly regulated by a diverse set of site-specific enzymes (1). Chromatin-modifying enzymes conferring alterations to the chromatin by placing, removing, or interpreting modifications to establish active or repressive chromatin marks have been recently referred to as writers, erasers, and readers of the histone code, respectively (2–4).
Lysine methylation has important functions in many biological processes including heterochromatin formation, X-chromosome inactivation, and transcriptional regulation. Histone demethylases confer a hitherto unrecognized plasticity of epigenetic regulation to gene expression programs and cellular adaption in response to environmental signals. Whereas the class of FAD-dependent histone demethylases contains only two members, LSD1 and LSD2, the JmjC family of histone demethylases comprises >30 potential members in mammals (5). JmjC family members are able to specifically demethylate mono-, di-, and trimethylated lysines on the histone tails in a Fe(II) and α-ketoglutarate-dependent manner. Specifically, these enzymes catalyze the hydroxylation of mono-, di-, or trimethylated lysine on histone tails, giving rise to an unstable hydroxylmethyl group, which is released spontaneously as formaldehyde. JmjC proteins are described to be involved in multiple physiological and pathophysiological processes but due to the recent discovery of these enzymes, only little information is available for most of the family members. One of these members, Fbxl10, also known as Kdm2b or Jhmd1b, consists of a JmjC, a CxxC, a PHD, and a Fbox domain.
Whereas the functional characterization of the demethylation domain JmjC and the DNA binding domain CxxC are well described for at least two demethylases (5, 6), PHD domains are discussed as either histone modification reader domains or as E3 ligases. PHD domains were shown to bind different methylated lysines on the histone tail of H3 functioning as reader domains (7). Due to the high similarity to RING domains which are well known E3 ligase domains, the PHD domains were recently also analyzed for E3 ligase function (7, 8). Fbox domains function most likely as an anchor between a target protein and an E3 ubiquitin ligase (9).
The biological role of Fbxl10 is controversially discussed in recent reports focusing mainly on its role as a histone demethylase. Whereas He et al. demonstrate it to be an H3K36me1/2 demethylase like its ortholog Fbxl11 (10, 11), others characterized it as an H3K4me3 demethylase (12). In a recent work by Tzatsos and co-workers the authors postulate the option of Fbxl10 being a demethylase for both modifications (13). Whereas slight experimental differences in Fbxl10 expression and demethylase assay condition might explain different experimental results, it is still striking that similar approaches by overexpression of Fbxl10 in infected HeLa cells revealed in one study H3K36me2 and in the other study H3K4me3 as the Fbxl10 target (10, 12).
In addition, Fbxl10 is described as a nucleolar protein to repress rRNA transcription (12), to be involved in apoptosis (14) and inhibition of cellular senescence (10). Furthermore, Fbxl10 plays a role in hematopoietic stem cell self- renewal and proper generation of the neural tube in vivo (15, 16).
Therefore, we investigated in further detail the role of the JmjC domain and the yet uncharacterized PHD domain of Fbxl10. Our data clearly indicate that Fbxl10 functions as a H3K4me3 rather than a H3K36me1/2 demethylase. However, the PHD domain recognizes both H3K4me3 and H3K36me2. Notably, the PHD domain of Fbxl10 also shows E3 ligase activity in vitro, indicating a role for Fbxl10 in addition to histone demethylation. Overexpression of Fbxl10 in mouse embryonic fibroblasts (MEFs)3 resulted in an increase in both total cellular and nuclear size, but did not affect proliferation, mitosis, or apoptosis in these cells. Using a microarray approach we were able to identify 131 new putative target genes for Fbxl10 including a set of chemokines, noncoding RNA Xist and metabolic regulators. To verify selected target genes we showed direct binding of Fbxl10 to the promoters of Ccl7, Xist, Crabp2, and RipK3 accompanied by a significant decrease in H3K4me3 whereas H3K36me2 remained unchanged. Knockdown of Fbxl10 inversed the regulation of Ccl2, Ccl7, Cxcl10, and Xist. Our data indicate a novel function of Fbxl10 in regulating metabolic processes and cell shape.
EXPERIMENTAL PROCEDURES
Protein Purification
Domains of Fbxl10 were cloned into the pGEX-4T1 expression vector (GE Healthcare) using the following primers: JmjC, 5′-tccggaattcgaggcagagaaagactctgga and 5′-cgatgcggccgctcactgaacccgggtcctgtcctcgat; PHD, 5′-tccggaattccacaccgccgtgtgccttgtgtgt and 5′-cgatgcggccgctcacttgccggcatggttacacttcgg; RING, 5′-tccggaattcgagaagctttgcaaaatctgtatg and 5′-cgatgcggccgctcaagacataaaaattttttgcttgaa.
Plasmids were transformed in One Shot BL21 STAR (DE3)pLysS Escherichia coli (Invitrogen). Protein expression and purification were done using glutathione-Sepharose bead columns (GE Healthcare) according the manufacturer's instructions.
Western Blotting
Cells were harvested with radioimmuneprecipitation assay buffer, and samples were heated with Laemmli buffer at 95 °C for 5 min. Membranes were blocked in 5% milk or BSA and later washed with PBS-T or TBST-T. The following antibodies were used: β-actin (1:5000, A5441; Sigma-Aldrich), HA antibody (1:2500, 16B12; Hiss Diagnostics, Freiburg, Germany), PCNA (1:1000, ab29; Abcam, Cambridge, UK), histone H3 (1:5000, ab1791; Abcam), H3K4me2 (1:1000, pAb-035050; Diagenode, Liege, Belgium), H3K4me3 (1:1000, ab1012; Abcam), H3K9me2 (1:1000, 07-441; Upstate, Billerica), H3K27me3 (1:1000, pAb-069-050; Diagenode), H3K36me1 (1:1000, ab9048; Abcam), H3K36me2 (1:1000, 07-274; Upstate), H2Aub (1:500, 05-678; Upstate), GST (1:5000, 27-4577-01; Amersham Biosciences), ubiquitin antibody (1:2000, Z0458; Dako) and anti-mouse or anti-rabbit HRP antibodies (1:5000; Sigma-Aldrich).
PHD-Histone Binding Assay
Binding assays of 1 μg of GST-PHD and 10 μg of bulk histones (1:5000; Sigma-Aldrich) were performed as described by Taverna et al. (17). Samples were then analyzed by Western blotting with the indicated antibodies.
In Vitro Ubiquitination Assay
E3 ligase assay was performed in ubiquitination buffer (50 mm HEPES, pH 8.0, 4 mm ATP, 10 mm MgCl2, 0.2 mm ZnCl2, 50 mm NaCl, 11.4 μg of ubiquitin (Sigma-Aldrich), 200 nm (His)6UBE1 human recombinant protein (Boston Biochem, Boston, MA), 1 μm GST-UBC5a human recombinant protein (Boston Biochem) by adding purified PHD domain, equal amounts of GST or RING domain for 2 h at 30 °C. Samples were then analyzed by Western blotting.
Histone Demethylation Assay
Histone demethylation assay was performed in demethylation buffer (50 mm Tris-HCl, pH 7.9, 50 mm KCl, 10 mm MgCl2, 2 mm ascorbate, 1 mm α-ketoglutarate, 100 μm Fe2+). As substrate, 1 μg of bulk histone (1:5000; Sigma-Aldrich) or recombinant H3K4me3 or H3K36me2 (Active Motif, Rixensart, Belgium) was mixed with purified JmjC domain of Fbxl10. Samples were then incubated for 30 min at 37 °C and analyzed by Western blotting.
Generation of MEFs Stably Overexpressing Fbxl10
We used the Tet-Off system (Clontech) for inducible gene expression. The murine Fbxl10 cDNA was cloned into pUHD10-3 with an N-terminal HA tag and a Kozak sequence. Primers used for cloning pUHD10-3-HA-Fbxl10 were 5′-gctagatatcccaccatgtatccttatgatgttcctgattatgctgaggcagagaaagactctggaagaagattg and 5′-tgcggctagcctaacttagtttttgcaggagtttctcttccac. MEF cells (Clontech) were co-transfected with the pUHD10-3 plasmid and the hygromycin resistance plasmid pTKHyg 343 (Clontech) using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol. Cells were selected by hygromycin treatment (100 μg/ml), and cell clones were analyzed for HA-Fbxl10 expression.
Immunofluorescence
For immunofluorescence cells were fixed with 4% paraformaldehyde and stained with HA antibody (1:400, 16B12; Hiss Diagnostics, Freiburg, Germany), Alexa Fluor 594 phalloidin (1:2000; Invitrogen) and Hoechst. Alexa Fluor 488 rabbit anti-mouse (1:400; Invitrogen) was used as secondary antibody.
Apoptosis Induction and Cell Viability
Cells were treated with increasing concentrations of staurosporine, menadione, or thapsigargin (Sigma-Aldrich) for 6 h. Cell viability was measured 6 h after treatment by MTT assay (Roche Applied Science), as described previously (18).
Quantitative Real-time PCR (qRT-PCR)
RNA was extracted with PureLink RNA Micro scale kit (Invitrogen) according to the manufacturer's protocol. cDNA syntheses were performed with 0.5 μg of RNA employing SuperScriptIII (Invitrogen), and qRT-PCRs were done as described by Lim et al. (18). Control values were set to 1, and hypoxanthine-guanine-phosphoribosyltransferase (Hprt) was chosen as a housekeeping gene. The following primers were used for qRT-PCR: Hprt, 5′-tcctcctcagaccgctttt and 5′-cctggttcatcatcgctaatc; Col2A1, 5′-cggtcctacggtgtcagg and 5′-ttatacctctgcccattctgc; Lama5, 5′-ggccaggaagaaccagcta and 5′-gcaatcttctcactggtctcg; Xist, 5′-ctactgctcctccgttacatca and 5′-aggagcacaaaacagactcca; Sfrp2, 5′-gacaacgacctctgcatcc and 5′-tcacacaccttgggagctt; Ccl2, 5′-catccacgtgttggctca and 5′-gatcatcttgctggtgaatgagt; Ccl5, 5′-tgcagaggactctgagacagc and 5′-gagtggtgtccgagccata; Ccl7, 5′-ttctgtgcctgctgctcata and 5′-ttgacatagcagcatgtggat; Cxcl10, 5′-gctgccgtcattttctgc and 5′-tctcactggcccgtcatc; Cx3cl1, 5′-cgcgttcttccatttgtgta and 5′-catgatttcgcatttcgtca; Fbxl10, 5′-taccagggggacttcgtg and 5′-gaccctgagagcttctctctgta; Crabp2, 5′-ttgaggaaatgctaaaagctctg and 5′-tcctgtttgatctcgactgct; Mme3, 5′-gggaggctttatgtggaagc and 5′-ccggatttgtgcaatcaagt; RipK3, 5′-ggcttctaaagcgagtgatgtc and 5′-tgaagtcttgtctaccaactcagc; Xlr, 5′-tcttgaggaagccgagaagt and 5′-tcccatcttgcatttcatca; E2f5, 5′-gcgtcctggatctcaaagc and5′-gatatcataaattcttcgcttttgc; Tll1, 5′-aaagagtgcacgtgggtgat and 5′-aacctcaaactcattgaaggcta.
PCR
Genomic DNA was extracted from a 12-well plate by incubating cells in cell lysis buffer (10 mm NaCl, 10 mm Tris, pH 7.5, 10 mm EDTA, 0.5% sarkosyl, 1 mg/ml proteinase K) at 56 °C overnight. Pellets were washed with ethanol and resuspended in water. PCR was performed with 100 ng of DNA and recombinant Taq (Invitrogen) according to the manufacturer's protocol. Primers were 5′-atgtatcctttatgatgttcctga and 5′-catgaccatccaccaaacggcg.
Proliferation Assay
Proliferation was measured with the Click-iT EdU Alexa Fluor 488 Imaging Kit (Invitrogen) as recommended by the manufacturer. Percentage of proliferating cells was assessed by counting the number of positive cells compared with the total number of cells on 8 visual fields 3 and 24 h after EdU treatment.
Mitosis Assay
Cells were prepared for immunofluorescence as described above. Staining was done with the mitosis marker H3S10P antibody (1:400, ab5176; Abcam) and Alexa Fluor 594 goat anti-rabbit (1:400; Invitrogen). Ratio of mitotic to total cell number was measured by counting positive cells on 8 visual fields 24 and 48 h after plating.
Histone Preparation
Histone preparation was done using acidic extraction. Briefly, cells were incubated in Triton extraction buffer (PBS with 0.5% Triton X-200, 2 mm PMSF, and 0.02% NaN3), washed, and incubated overnight in 0.2 n HCl. Purified histones were analyzed by Western blotting as described above.
Microarray Analysis
Microarray experiments were performed with Affymetrix Mouse Gene St 1.0 arrays using 300 ng of total RNA. Analysis was performed according to Lim et al. (18).
Chromatin Immunprecipitation (ChIP)
ChIP was performed with SimpleChIP Enzymatic Chromatin IP Kit (Cell Signaling) according to the manufacturer's protocol. 10–15 μg of chromatin was immunoprecipitated with 3 μg of HA (ab9110; Abcam), H3K4me3 (ab1012; Abcam), or H3K36me2 (07-274, Upstate;) antibody. Equal amounts of IgG antibodies (kch-819-015; Diagenode) were used as control antibody. ChIP after knockdown of Fbxl10 was performed as described previously (19). Recovered DNA was analyzed by TaqMan qRT-PCR using the following loci-specific primers: Xist, 5′-tctggtcctcatttggagtacata and 5′-ggtttgacggatagtgagtgtctc; Ccl7, 5′-gattccaagatctgaatccttctg and 5′-acatgatgccttaaagtgtccag; Ccl2, 5′-gacttaaccaaaaggcatgtttct and 5′-agtagatgctgttactccccattc; Crabp2, 5′-ctccctgtcacccaaaagaa and 5′-aggtcccgtttcaccagttc; RipK3, 5′-tgccgcttagtaggcaaaat and 5′-gggtcagtttgttctctgctg; Cxcl10, 5′-tgtcacctctatgcgagatctatg and 5′-aagaggaattgcaaagctaattgt.
Ccl2 ELISA
D5 and control cells were seeded in 12-well plates in normal growth medium. After 24 h cells were washed twice with PBS, and medium was replaced by serum-reduced medium for another 24 h. Supernatant was collected and cleared from cellular debris by rotation. Cleared supernatant was then used in mouse Ccl2 ELISA Ready-SET-Go! (eBioscience, San Diego, CA) following the manufacturer's instructions.
siRNA
Untransfected MEF cells (controls) were transfected with Fbxl10-specific siRNA (EMU092751; Sigma-Aldrich) using HiPerfect (Qiagen) according to the manufacturer's protocol. All star negative siRNA (Qiagen) was used as a control.
PathScan RTK Signaling
PathScan RTK Signaling Antibody Array Kit (Cell Signaling) was used according to the manufacturer's instructions.
Statistical Analysis
For statistical analysis one-sampled t tests and Student's t tests were performed. Results were considered significant with a p value < 0.05 and marked with an asterisk.
RESULTS
Fbxl10 Is a H3K4me3 Rather Than a H3K36me2 Histone Demethylase
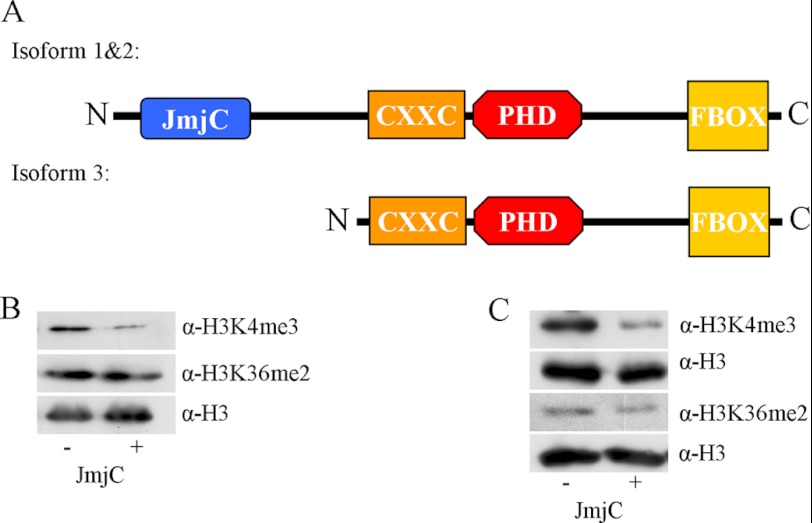
Fbxl10 is a protein of the JmjC domain family which is expressed in three different isoforms (Fig. 1A). Isoform 1 represents the full-length Fbxl10 protein containing the four main domains of the potential histone demethylase. Isoform 2 differs only slightly from isoform 1 in the 5′-UTR and coding region of the gene, still harboring all four main domains, whereas isoform 3 lacks the JmjC domain. The isoforms are known to be expressed in vivo as shown by Fukuda et al. (16). This is of particular interest because Fbxl10 research has focused mainly on its role as a histone demethylase requiring the presence of the JmjC domain. However, expression of isoform 3 suggests a potential function of the short isoform other than histone demethylation. With respect to the existing controversial data on the specificity of histone demethylation by Fbxl10, we first tested the capacity of a recombinant GST-tagged JmjC domain of Fbxl10 to demethylate H3K4me3 or H3K36me2 in vitro. Therefore, we purified the GST-tagged JmjC domain and incubated the recombinant protein and bulk histone as substrate. Changes in H3K4me3 and H3K36me2 were detected via Western blotting and compared with an input control. Incubation of the purified JmjC domain of Fbxl10 with bulk histones led to a strong reduction of the H3K4me3 signal compared with the input control. However, H3K36me2 methylation levels were not altered (Fig. 1B). To further validate the substrate specificity, we also tested a commercially available recombinant H3K4me3 or H3K36me2 as substrate. The strong decrease in H3K4me3 and the unchanged signals in H3K36me2 confirm that the JmjC domain of Fbxl10 functions as H3K4me3-specific demethylase (Fig. 1C).
FIGURE 1.
Fbxl10 is a H3K4me3 demethylase consisting of four distinct domains. Fbxl10 exists in three isoforms. Two isoforms contain the four major domains: JmjC, CxxC, PHD, and Fbox. A and B, the third isoform lacks the JmjC domain (A). Incubation of JmjC domain of Fbxl10 together with bulk histone (B) resulted in demethylation of H3K4me3 but not in demethylation of H3K36me2 in vitro. C, specificity of H3K4me3 demethylation was verified by incubation with recombinant H3K4me3 and H3K36me2.
The PHD Domain of Fbxl10 Binds to H3K4me3 and H3K36me2 and Shows E3 Ligase Activity
Recently, several reports described PHD domains beside bromo- and chromodomains as the main reader domains of histone modifications (7). To investigate the role and function of the PHD domain of Fbxl10, we purified the GST-tagged PHD domain of Fbxl10 and performed GST pulldown assays with bulk histones. We analyzed the binding of the PHD domain to histones by Western blotting using methylation state-specific antibodies.
Our results shown in Fig. 2A demonstrate binding of the Fbxl10 PHD domain to H3, H3K4me3, and H3K36me2. Other modifications like H3K4me2, H3K9me2, or H3K36me1 were not enriched by the pulldown experiments, indicating specificity of binding (lane 2). To exclude binding of the histone to the GST tag, GST-only protein was used as a control (lane 3). The input control in lane 1 showed comparable amounts of the different proteins.
FIGURE 2.
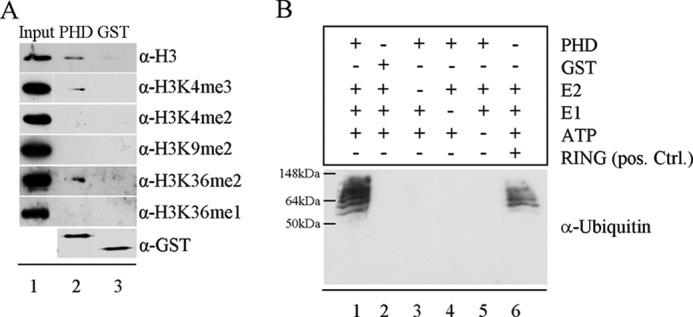
PHD domain of Fbxl10 shows binding affinity for H3K4me3 and H3K36me2 and exhibits E3 ligase activity. A, GST pulldown assays were performed with purified GST-tagged PHD domain of Fbxl10 and bulk histones. Lane 1 shows signals of input control for all histone marks. Lane 2 shows binding of PHD domain to H3, H3K4me3, and H3K36me2. Lane 3 indicates that GST does not bind to H3K4me3 and H3K36me2. B, incubation of PHD domain of Fbxl10 with E1, E2, ATP, and ubiquitin led to an E3 ligase typical pattern (lane 1) similar to that of RING domain from human XIAP used as positive control (lane 6). The exchange of PHD through GST (lane 2) or the removal of either E2, E1, or ATP (lanes 3–5) abolished ubiquitination of target proteins (lanes 2–5).
PHD domains are closely related to RING domains (20), which are known E3 ligases (21). Interestingly, recent publications also propose E3 ligase activity for some PHD domains (8, 22). Furthermore, Fbox domain proteins are also considered to be part of E3 ligase complexes (9). Therefore, we subjected the PHD domain of Fbxl10 to an E3 ligase activity assay. Incubation of the purified PHD domain of Fbxl10 together with ATP, recombinant E1, recombinant E2, and ubiquitin resulted in multiple bands reflecting ubiquitination of various proteins in the preparation by the PHD domain. This expected pattern clearly indicates E3 ligase activity similar to the GST-tagged RING domain of human XIAP that we used as a positive control in the same assay (Fig. 2B, lanes 1 and 6). Replacement of the PHD domain by GST (lane 2) or absence of either E2 (lane 3), E1 (lane 4), or ATP (lane 5) abolished the presence of ubiquitinated proteins (Fig. 2B). These results indicate a dual function for the PHD domain of Fbxl10, linking Fbxl10 not only to chromatin remodeling but also to ubiquitin-mediated protein degradation or signaling.
Stable Overexpression of Fbxl10 Leads to an Increase in Cell Size and Changes the Transcriptome
To investigate the role of Fbxl10 in cell physiology we generated a stable Fbxl10-overexpressing MEF cell line. This cell line was generated by co-transfection of a pUHD10–3 plasmid containing HA-tagged Fbxl10 cDNA and a pTKHyg 343 plasmid containing a hygromycin resistance gene. Screening for HA tag expression revealed several cell clones strongly overexpressing HA-tagged Fbxl10 (data not shown). One of these clones, D5, was used for further studies. Overexpression of Fbxl10 was verified by qRT-PCR. Western blotting indeed visualized the Fbxl10 protein of the expected size (144 kDa) which was not detected in the untransfected control cells (Fig. 3, A and B). In addition, stable integration of the HA-Fbxl10-cDNA was confirmed by PCR analysis on genomic DNA using the HA tag sequence as forward primer and the Fbxl10 sequence as reverse primer (Fig. 3C).
FIGURE 3.
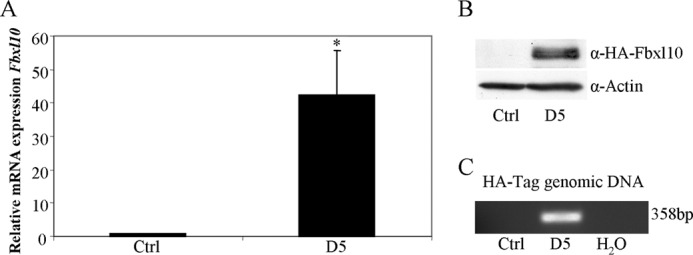
MEFs stably overexpressing Fbxl10. MEFs were transfected with pUHD10–3 containing HA-tagged Fbxl10 cDNA and a pTKHyg 343 selection plasmid. Cell clones were screened for HA-Fbxl10 expression by qRT-PCR (A) and Western blotting (B). In addition, stable integration of the HA-Fbxl10 cDNA was tested with PCR on genomic DNA and resulted in the amplification of a 358-bp band using primers derived from the HA tag and the Fbxl10 cDNA (C).
Interestingly, the Fbxl10-overexpressing cell line showed a significant increase in total cellular and nuclear size compared with control cells (Fig. 4A). We next asked whether the overexpression of Fbxl10 also affects other important cellular features, including proliferation, mitotic count, and apoptosis. Contradictory reports on Fbxl10 promoting or blocking proliferation (10, 12) prompted us to investigate proliferation of Fbxl10-overexpressing cells with Click-iT EdU Alexa Fluor 488 Imaging Kit which relies on the incorporation of EdU in the DNA of proliferating cells. The assay clearly demonstrated an equal amount of proliferating cells per time unit in D5 and control cells 3 and 24 h after treatment with EdU (Fig. 4B). These results were further validated by Western blotting of the proliferation marker PCNA (Fig. 4C) and identical mitotic rate of clone D5 and control cells determined by immunofluorescence against phosphorylated histone H3 24 and 48 h after plating (H3S10P; Fig. 4D).
FIGURE 4.
Stable overexpression of Fbxl10 leads to an increase in cellular and nuclear size but has no effect on proliferation, mitosis, and apoptosis or global histone marks. A, stable Fbxl10-overexpressing D5 cell clone shows an increase in nuclear and total cellular size. B and C, number of proliferative cells per time unit did not change by permanent overexpression, demonstrated by EdU incorporation assay 3 and 24 h after EdU addition to the media (B) and PCNA protein levels by Western blotting (C). D, mitotic events 24 and 48 h after plating were not altered between control cells and D5 cells measured by H3S10P-positive cells per visual field in immunofluorescence analysis. E, inducing apoptosis by staurosporine and measuring cell viability by MTT assays led to equal levels of cell viability indicating similar apoptosis responses. F, global histone marks were analyzed between control cells and D5 cells by Western blotting. No changes were observed.
To examine whether the stable overexpression of Fbxl10 has an effect on apoptosis we treated our cell lines with increasing concentrations of staurosporine, menadione, or thapsigargin known to induce apoptosis by different mechanisms. Cell viability was measured after 6 h of treatment using an MTT assay. At this time point the cell viability for both cell lines was decreased compared with untreated cell lines in a very similar manner. Results shown in Fig. 4E and supplemental Fig. 1 revealed an identical degree of apoptosis in both cell lines.
Fbxl10 was previously reported to confer specific changes in histone modification either directly via its JmjC domain or indirectly by its binding partners (10, 12, 23, 24). Whereas H3K4me3 and H3K36me2 were discussed as direct targets, H3K27me3 and H2Aub were reported as indirect targets because Fbxl10 was found in Polycomb complexes with H3K27 methylation activity and H2A ubiquitination capability (23, 24). H3K4me2 and H3K36me1 have not been described as Fbxl10 targets and were therefore used as negative controls. To investigate potential changes in global histone modification after Fbxl10 overexpression, we purified histones by acidic extraction and analyzed the histone modifications H3K4me2/3, H3K36me1/2, H3K27me3, and H2Aub (Fig. 4F). Indeed, global levels of histone modifications were not affected by overexpression of Fbxl10, indicating its function through gene locus-specific modifications.
Fbxl10 Regulates Genes Involved in the Cellular Metabolome and Anatomical Structures
To investigate gene expression patterns regulated by Fbxl10 we next analyzed transcriptome changes resulting from Fbxl10 overexpression using a microarray approach. Comparing three independently reproduced transcriptomes of untransfected control cells and the D5 cell clone, respectively, we identified 131 genes with a -fold change >2 (supplemental Table 1) and an additional 133 genes with a -fold change >1.72 (supplemental Table 2 and Supplemental Fig. 2). The microarray data were released into the GEO database for public access (accession number GSE34691).
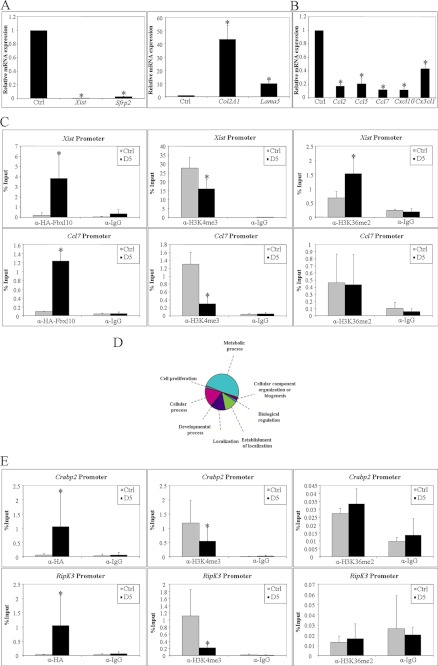
To confirm these data, we validated selected sets of both up- and down-regulated genes by qRT-PCR. Steady-state transcript levels of Col2A1 and Lama5 were increased and expression of the noncoding RNA Xist and Sfrp2 were decreased confirming the microarray analysis (Fig. 5A). Interestingly, among the 131 regulated genes we identified a group of chemokines (Ccl2, Ccl5, Ccl7, and Cxcl10), strongly down-regulated in the D5 cell clone. Even a fifth down-regulated chemokine, Cx3cl1, was identified among the 133 genes with the slightly reduced threshold of >1.72-fold change. Reduced expression levels of these chemokines were verified by qRT-PCR (Fig. 5B). We then further verified the microarray data on protein level using a Ccl2 ELISA, showing a significantly decrease of Ccl2 protein in supernatant of D5 cells compared with control cells (supplemental Fig. 3). As fibroblasts are well known to produce chemokines (25, 26) our observation suggests an important role for Fbxl10 in chemokine repression.
FIGURE 5.
Fbxl10 regulates chemokines and metabolic genes. A and B, microarray was validated by qRT-PCR for four strongly regulated genes (A) and a group of regulated chemokines (B). C, ChIP analysis at the Xist and Ccl7 promoters showed the direct binding of Fbxl10 to these promoters and changes in H3K4me3 levels, whereas H3K36me2 levels remains nearly unchanged. IgG was used as a control antibody. D, functional classification of differentially expressed genes upon Fbxl10 overexpression revealed strong involvement of Fbxl10 in metabolic processes. C and E, ChIP analysis at the Crabp2 and RipK3 promoters identified that direct binding of Fbxl10 to these promoters results in changes in H3K4me3 but not H3K36me2 (E). IgG antibody served as a control (C and E).
To assess whether the promoters of Xist and the chemokine Ccl7 are direct or rather indirect targets of Fbxl10, we performed ChIP assays using α-HA, α-H3K4me3, α-H3K36me2, and IgG control antibodies followed by qRT-PCR. Results shown in Fig. 5C clearly reveal direct binding of Fbxl10 to both of these promoters. In addition, we observed significant reduction of H3K4me3 levels, emphasizing the function of Fbxl10 as a gene locus-specific H3K4me3 demethylase at the promoters of these target genes. H3K36me2 levels remained unchanged at the Ccl7 and increased at the Xist promoter. IgG antibody was used as a control and confirmed specificity of the HA, H3K4me3, and H3K36me2 antibodies (Fig. 5C).
Functional classification of all differentially expressed genes revealed that ∼49% of all Fbxl10-regulated genes in fibroblasts are involved in metabolic processes indicating a strong involvement of Fbxl10 in metabolic regulation (Fig. 5D). From the 264 genes significantly changed by a threshold of -fold change >1.72, 101 genes are involved in metabolism (supplemental Table 3 and supplemental Fig. 4A). Considering that the entire gene list for metabolic processes contains only 1376 genes, we calculate that overexpression of Fbxl10 in fibroblasts alters expression of almost 8% of all genes involved in cell metabolism (supplemental Fig. 4A).
As illustrated in supplemental Fig. 2 the metabolic regulators Crabp2 and RipK3 were regulated by Fbxl10. We assessed whether the promoters of Crabp2 and RipK3 are targets of histone modification by Fbxl10. Indeed, we found that Fbxl10 binds to the promoter of these genes. We found significantly reduced levels of H3K4me3 and a nearly unchanged H3K36me2 level when Fbxl10 is overexpressed (Fig. 5E).
Biological pathway analysis of these metabolism genes indicated that they are strongly involved in phosphor metabolism and oxidation and reduction reactions (supplemental Fig. 4B). Due to the enrichment of phosphor processes in this analysis we asked whether main receptor tyrosine kinase pathways are altered in D5 cells. However, using the PathScan RTK Signaling Kit we could not detect different activation of these pathways (supplemental Fig. 4C).
Gene ontology analysis revealed a further 26 genes belonging to the cluster “anatomical structure morphogenesis.” In accordance with Fig. 5A we could show here that Fbxl10 regulates the expression of genes encoding cell adhesion molecules and extracellular matrix proteins including Col2A1 and Lama5. Further genes of this cluster like Efnb2 and Efna5 were previously described to alter cell morphology whereas Edn1 is known to influence cell size and is involved in cellular hypertrophy (27–29). These genes are of special interest regarding the altered morphology of D5 compared with the control cell line (supplemental Fig. 5).
To assess whether the knockdown of Fbxl10 leads to an inverse regulation of previously identified target genes, we used siRNA approaches against the demethylase in the control cell line. Interestingly, reduction of Fbxl10 levels resulted in increased Xist, Ccl2, Ccl5, Ccl7, and Cxcl10 RNA expression. Other genes differentially expressed in D5 cells were not affected by the knockdown of Fbxl10 (supplemental Fig. 6A). ChIP analysis at the promoters of Xist, Ccl2, Ccl7, and Cxcl10 confirmed the qRT-PCR data and showed significantly increased H3K4me3 levels and unchanged H3K36me2 levels at both promoters (supplemental Fig. 6B).
DISCUSSION
Fbxl10 Is a H3K4me3 Demethylase
In this report, we show that Fbxl10 functions as a H3K4me3 demethylase. Our data obtained from purified JmjC domain in vitro are confirmed by decreased gene locus-specific H3K4me3 levels upon binding of Fbxl10 to its direct targets Xist, Ccl7, RipK3, and Crabp2. In agreement with our data, Frescas et al. also observed H3K4me3 demethylase activity for Fbxl10 (12). This is further supported by a very recent publication analyzing the demethylase function of the Drosophila homolog dKdm2 which was described as a H3K4me3 demethylase in vivo (30). Nevertheless, it has to be considered that histone demethylases act in multienzyme complexes, and the specificity of histone modifiers may be dependent on interaction with other proteins or modifications on histone residues nearby (1, 5). Thus, we cannot completely exclude a potential H3K36me2 demethylase function of Fbxl10 in vivo. Distinct substrate specificity depending on the complex involved has already been described for the histone demethylase LSD1 (31, 32). We also do not know whether the demethylation reaction of Fbxl10 is dependent on other modifications on the histone tail. This has been described for H3K36me which depends on H3P38 isomerization (33) or for the influence of H3T6, which in consequence prevents removal of active methyl marks from H3K4 during AR/LSD1-stimulated gene expression (34).
The PHD Domain of Fbxl10 Binds to H3K4me3 and H3K36me2
In addition to bromo- and chromodomains, PHD domains are well known for their function as histone readers, recognizing and binding to methylated lysines on histones (7). Interestingly, we identified H3K4me3 as a binding partner for this PHD domain. Regarding the ability of Fbxl10 to demethylate the same mark, this finding clearly offers a trans mode action for Fbxl10. Although binding to H3K4me3 on one nucleosome, Fbxl10 might demethylate the same mark on a neighboring histone or nucleosome. In addition to binding H3K4me3, we also showed for the first time the binding of a PHD domain to H3K36me2. Binding to this mark would offer a cis mode for Fbxl10 in that it binds to H3K36me2 and demethylates H3K4me3 on the same histone tail. As mentioned above we cannot completely exclude the previously proposed role of Fbxl10 as a H3K36me2 demethylase. Under these circumstances binding to H3K36me2 would also offer a trans mode of action for Fbxl10 (35).
Fbxl10 Exhibits E3 Ubiquitin Ligase Activity
Besides binding to histones some PHD domains were also reported recently to function as E3 ligase in ubiquitination (8) as well as sumoylation reactions (22). PHD domains are closely related to RING domains (20) which have already been identified as E3 ubiquitin ligases (21). Furthermore, the Fbox domain of Fbxl10 has been regarded as an anchor protein in E3 ubiquitin ligase complexes (9). These data prompted us to analyze whether the PHD domain of Fbxl10 also possesses E3 ubiquitin ligase potential. The PHD domain, however, is present in the short isoform 3 of Fbxl10 and might mediate a demethylase-independent protein function. As the ubiquitin pattern mimicked that of the RING domain of human XIAP and was abolished after the replacement of PHD, E1, E2, or ATP, these data unambiguously reveal a functional E3 ubiquitin cascade in which the PHD domain serves as an E3 ligase.
Previous reports postulated that Fbxl10 contributes to the BCOR complex and may be indirectly involved in H2A ubiquitination (23, 24). Offering bulk histones as substrate in the in vitro ubiquitination assay, however, did not result in H2A ubiquitination, indicating that Fbxl10 cannot ligate H2A with ubiquitin or might require an essential co-factor (data not shown).
Fbxl10 Regulates Chemokines, the Metabolome, and the Cell Anatomy in Fibroblasts
To get a deeper insight into Fbxl10 function, we generated a cell line stably overexpressing Fbxl10 and investigated Fbxl10-regulated target genes. Notably, we did not detect any changes in proliferation or response to different proapoptotic stimuli. Comparing our results with published data (10, 12) regarding effects of Fbxl10 on proliferation, it is important to point out that results from transient transfection of Fbxl10 are of limited value and may explain the different data sets. Moreover, we cannot exclude that distinct multienzyme complexes in different cell lines are favored by Fbxl10 leading to a complete different outcome. However, our stably transfected cell may better reflect the in vivo situation than the relative short time of transient overexpression or siRNA approaches (12).
Here, we provide the first insights into Fbxl10 transcriptome regulation by identifying a set of 264 genes differentially regulated by permanent Fbxl10 overexpression. Consistently, Cdkn2b, the most frequently described Fbxl10 target gene, was among our gene set regulated in fibroblasts. Fbxl10 was recently linked to senescence by regulating Cdkn2b expression (10, 36). In addition, we identified an entire set of chemokines and verified that Fbxl10 indeed binds directly to and demethylates histones of the Ccl7 promoter in a gene locus-specific manner. Remarkably, a knockdown of Fbxl10 led to inverse regulation of these chemokines.
Most interestingly, pathway analysis of all significantly regulated transcripts revealed that nearly 50% of all putative Fbxl10 target genes are linked to metabolic processes including the direct targets RipK3 and Crabp2 (37, 38). Further analysis showed significant increase of phosphor metabolism genes among the putative Fbxl10 targets. This finding strongly suggests an important role of Fbxl10 in cellular metabolism. Moreover, the analysis also emphasized our observation that Fbxl10 overexpression does not alter cell proliferation and mitotic index because genes association were not enriched in the microarray. Strikingly, the permanent overexpression of Fbxl10 resulted in an increase in total cellular and nuclear size. Consistently, regulation through Fbxl10 may provide a pathway in response to external signals by changes in cell morphology and metabolome.
Supplementary Material
Acknowledgment
We thank Dr. Soyoung Lim for comments on the manuscript.

This article contains supplemental Figs. 1–6 and Tables 1–3.
- MEF
- murine embryonic fibroblast
- Hprt
- hypoxanthine-guanine-phosphoribosyltransferase
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- PCNA
- proliferating cell nuclear antigen
- qRT-PCR
- quantitative real-time PCR.
REFERENCES
- 1. Kouzarides T. (2007) Chromatin modifications and their function. Cell 128, 693–705 [DOI] [PubMed] [Google Scholar]
- 2. Upadhyay A. K., Cheng X. (2011) Dynamics of histone lysine methylation: structures of methyl writers and erasers. Prog. Drug Res. 67, 107–124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Yun M., Wu J., Workman J. L., Li B. (2011) Readers of histone modifications. Cell Res. 21, 564–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Gardner K. E., Allis C. D., Strahl B. D. (2011) Operating on chromatin, a colorful language where context matters. J. Mol. Biol. 409, 36–46 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Mosammaparast N., Shi Y. (2010) Reversal of histone methylation: biochemical and molecular mechanisms of histone demethylases. Annu. Rev. Biochem. 79, 155–179 [DOI] [PubMed] [Google Scholar]
- 6. Lee J. H., Voo K. S., Skalnik D. G. (2001) Identification and characterization of the DNA binding domain of CpG-binding protein. J. Biol. Chem. 276, 44669–44676 [DOI] [PubMed] [Google Scholar]
- 7. Musselman C. A., Kutateladze T. G. (2009) PHD fingers: epigenetic effectors and potential drug targets. Mol. Interv. 9, 314–323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Dul B. E., Walworth N. C. (2007) The plant homeodomain fingers of fission yeast Msc1 exhibit E3 ubiquitin ligase activity. J. Biol. Chem. 282, 18397–18406 [DOI] [PubMed] [Google Scholar]
- 9. Frescas D., Pagano M. (2008) Deregulated proteolysis by the F-box proteins SKP2 and β-TrCP: tipping the scales of cancer. Nat. Rev. Cancer 8, 438–449 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. He J., Kallin E. M., Tsukada Y., Zhang Y. (2008) The H3K36 demethylase Jhdm1b/Kdm2b regulates cell proliferation and senescence through p15(Ink4b). Nat. Struct. Mol. Biol. 15, 1169–1175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Tsukada Y., Fang J., Erdjument-Bromage H., Warren M. E., Borchers C. H., Tempst P., Zhang Y. (2006) Histone demethylation by a family of JmjC domain-containing proteins. Nature 439, 811–816 [DOI] [PubMed] [Google Scholar]
- 12. Frescas D., Guardavaccaro D., Bassermann F., Koyama-Nasu R., Pagano M. (2007) JHDM1B/FBXL10 is a nucleolar protein that represses transcription of ribosomal RNA genes. Nature 450, 309–313 [DOI] [PubMed] [Google Scholar]
- 13. Tzatsos A., Pfau R., Kampranis S. C., Tsichlis P. N. (2009) Ndy1/KDM2B immortalizes mouse embryonic fibroblasts by repressing the Ink4a/Arf locus. Proc. Natl. Acad. Sci. U.S.A. 106, 2641–2646 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Ge R., Wang Z., Zeng Q., Xu X., Olumi A. F. (2011) F-box protein 10, an NF-κB-dependent anti-apoptotic protein, regulates TRAIL-induced apoptosis through modulating c-Fos/c-FLIP pathway. Cell Death Differ. 18, 1184–1195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Konuma T., Nakamura S., Miyagi S., Negishi M., Chiba T., Oguro H., Yuan J., Mochizuki-Kashio M., Ichikawa H., Miyoshi H., Vidal M., Iwama A. (2011) Forced expression of the histone demethylase Fbxl10 maintains self-renewing hematopoietic stem cells. Exp. Hematol. 39, 697–709, e5 [DOI] [PubMed] [Google Scholar]
- 16. Fukuda T., Tokunaga A., Sakamoto R., Yoshida N. (2011) Fbxl10/Kdm2b deficiency accelerates neural progenitor cell death and leads to exencephaly. Mol. Cell. Neurosci. 46, 614–624 [DOI] [PubMed] [Google Scholar]
- 17. Taverna S. D., Ilin S., Rogers R. S., Tanny J. C., Lavender H., Li H., Baker L., Boyle J., Blair L. P., Chait B. T., Patel D. J., Aitchison J. D., Tackett A. J., Allis C. D. (2006) Yng1 PHD finger binding to H3 trimethylated at K4 promotes NuA3 HAT activity at K14 of H3 and transcription at a subset of targeted ORFs. Mol. Cell 24, 785–796 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lim S., Janzer A., Becker A., Zimmer A., Schüle R., Buettner R., Kirfel J. (2010) Lysine-specific demethylase 1 (LSD1) is highly expressed in ER-negative breast cancers and a biomarker predicting aggressive biology. Carcinogenesis 31, 512–520 [DOI] [PubMed] [Google Scholar]
- 19. Janzer A., Lim S., Fronhoffs F., Niazy N., Buettner R., Kirfel J. (2012) Lysine-specific demethylase 1 (LSD1) and histone deacetylase 1 (HDAC1) synergistically repress proinflammatory cytokines and classical complement pathway components. Biochem. Biophys. Res. Commun. 421, 665–670 [DOI] [PubMed] [Google Scholar]
- 20. Matthews J. M., Bhati M., Lehtomaki E., Mansfield R. E., Cubeddu L., Mackay J. P. (2009) It takes two to tango: the structure and function of LIM, RING, PHD and MYND domains. Curr. Pharm. Des. 15, 3681–3696 [DOI] [PubMed] [Google Scholar]
- 21. Chasapis C. T., Spyroulias G. A. (2009) RING finger E(3) ubiquitin ligases: structure and drug discovery. Curr. Pharm. Des. 15, 3716–3731 [DOI] [PubMed] [Google Scholar]
- 22. Ivanov A. V., Peng H., Yurchenko V., Yap K. L., Negorev D. G., Schultz D. C., Psulkowski E., Fredericks W. J., White D. E., Maul G. G., Sadofsky M. J., Zhou M. M., Rauscher F. J., 3rd (2007) PHD domain-mediated E3 ligase activity directs intramolecular sumoylation of an adjacent bromodomain required for gene silencing. Mol. Cell 28, 823–837 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Gearhart M. D., Corcoran C. M., Wamstad J. A., Bardwell V. J. (2006) Polycomb group and SCF ubiquitin ligases are found in a novel BCOR complex that is recruited to BCL6 targets. Mol. Cell. Biol. 26, 6880–6889 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Sánchez C., Sánchez I., Demmers J. A., Rodriguez P., Strouboulis J., Vidal M. (2007) Proteomics analysis of Ring1B/Rnf2 interactors identifies a novel complex with the Fbxl10/Jhdm1B histone demethylase and the Bcl6 interacting corepressor. Mol. Cell Proteomics 6, 820–834 [DOI] [PubMed] [Google Scholar]
- 25. Hembruff S. L., Jokar I., Yang L., Cheng N. (2010) Loss of transforming growth factor-β signaling in mammary fibroblasts enhances CCL2 secretion to promote mammary tumor progression through macrophage-dependent and -independent mechanisms. Neoplasia 12, 425–433 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Eyman D., Damodarasamy M., Plymate S. R., Reed M. J. (2009) CCL5 secreted by senescent aged fibroblasts induces proliferation of prostate epithelial cells and expression of genes that modulate angiogenesis. J. Cell Physiol. 220, 376–381 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bochenek M. L., Dickinson S., Astin J. W., Adams R. H., Nobes C. D. (2010) Ephrin-B2 regulates endothelial cell morphology and motility independently of Eph-receptor binding. J. Cell Sci. 123, 1235–1246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Davy A., Robbins S. M. (2000) Ephrin-A5 modulates cell adhesion and morphology in an integrin-dependent manner. EMBO J. 19, 5396–5405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. McWhinnie R., Pechkovsky D. V., Zhou D., Lane D., Halayko A. J., Knight D. A., Bai T. R. (2007) Endothelin-1 induces hypertrophy and inhibits apoptosis in human airway smooth muscle cells. Am. J. Physiol. Lung Cell Mol. Physiol. 292, L278–286 [DOI] [PubMed] [Google Scholar]
- 30. Kavi H. H., Birchler J. A. (2009) Drosophila KDM2 is a H3K4me3 demethylase regulating nucleolar organization. BMC Res. Notes 2, 217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Shi Y., Lan F., Matson C., Mulligan P., Whetstine J. R., Cole P. A., Casero R. A. (2004) Histone demethylation mediated by the nuclear amine oxidase homolog LSD1. Cell 119, 941–953 [DOI] [PubMed] [Google Scholar]
- 32. Metzger E., Wissmann M., Yin N., Müller J. M., Schneider R., Peters A. H., Günther T., Buettner R., Schüle R. (2005) LSD1 demethylates repressive histone marks to promote androgen-receptor-dependent transcription. Nature 437, 436–439 [DOI] [PubMed] [Google Scholar]
- 33. Nelson C. J., Santos-Rosa H., Kouzarides T. (2006) Proline isomerization of histone H3 regulates lysine methylation and gene expression. Cell 126, 905–916 [DOI] [PubMed] [Google Scholar]
- 34. Metzger E., Imhof A., Patel D., Kahl P., Hoffmeyer K., Friedrichs N., Müller J. M., Greschik H., Kirfel J., Ji S., Kunowska N., Beisenherz-Huss C., Günther T., Buettner R., Schüle R. (2010) Phosphorylation of histone H3T6 by PKCβ(I) controls demethylation at histone H3K4. Nature 464, 792–796 [DOI] [PubMed] [Google Scholar]
- 35. Hou H., Yu H. (2010) Structural insights into histone lysine demethylation. Curr. Opin. Struct. Biol. 20, 739–748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Wang T., Chen K., Zeng X., Yang J., Wu Y., Shi X., Qin B., Zeng L., Esteban M. A., Pan G., Pei D. (2011) The histone demethylases Jhdm1a/1b enhance somatic cell reprogramming in a vitamin-C-dependent manner. Cell Stem. Cell 9, 575–587 [DOI] [PubMed] [Google Scholar]
- 37. Zhang D. W., Shao J., Lin J., Zhang N., Lu B. J., Lin S. C., Dong M. Q., Han J. (2009) RIP3, an energy metabolism regulator that switches TNF-induced cell death from apoptosis to necrosis. Science 325, 332–336 [DOI] [PubMed] [Google Scholar]
- 38. Budhu A. S., Noy N. (2002) Direct channeling of retinoic acid between cellular retinoic acid-binding protein II and retinoic acid receptor sensitizes mammary carcinoma cells to retinoic acid-induced growth arrest. Mol. Cell. Biol. 22, 2632–2641 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.