Background: TIR domains mediate TLR signaling and are hijacked for immunosuppression by bacteria.
Results: Tethering of TIR domains or their dimerization by fusion with coiled-coil segment strongly improved inhibition of TLR signaling.
Conclusion: The presence of a coiled-coil segment in bacterial TCPs potentiates inhibition while preventing the constitutive activity of TIR domain dimer.
Significance: Dimeric TIR domain represents the platform for the formation of Myddosome.
Keywords: Immunosuppression, Innate Immunity, MyD88, Toll IL-1 Receptor (TIR) Domain, Toll-like Receptors (TLR), Bacterial TCPs, TcpB
Abstract
TIR (Toll/IL-1 receptor) domains mediate interactions between TLR (Toll-like) or IL-1 family receptors and signaling adapters. While homotypic TIR domain interactions mediate receptor activation they are also usurped by microbial TIR domain containing proteins for immunosuppression. Here we show the role of a dimerized TIR domain platform for the suppression as well as for the activation of MyD88 signaling pathway. Coiled-coil dimerization domain, present in many bacterial TCPs, potently augments suppression of TLR/IL-1R signaling. The addition of a strong coiled-coil dimerization domain conferred the superior inhibition against the wide spectrum of TLRs and prevented the constitutive activation by a dimeric TIR platform. We propose a molecular model of MyD88-mediated signaling based on the dimerization of TIR domains as the limiting step.
Introduction
Toll-like receptors (TLRs)2 are a family of pattern recognition receptors (PRRs) whose stimulation results in the activation of innate immunity. Downstream signaling of TLRs is mediated through homotypic interactions between Toll-interleukin-1 (TIR) domains of TLR/IL-1R receptors and adapter proteins such as MyD88 and TRIF, which mediate transduction of the signaling pathway cascade (1, 2). TLRs are activated by ligand-induced dimerization of their ectodomain (3) and the TLR cytoplasmic TIR domain signaling dimer interface was proposed as the scaffold for the recruitment of signaling co adapters (4, 5). Signaling adapter MyD88, which mediates activation of all TLRs except TLR3 is composed of an N-terminal death domain (DD), intermediate domain (INT) and a C-terminal TIR domain (6, 7) where the TIR domain is required for coupling with the receptor TIR domains (8) and DD for the recruitment of IRAK kinases (9). In the crystal structure of DD Myddosome, death domains of MyD88, IRAK4 and IRAK2 assemble into a tower in a 6:4:4 stoichiometry based on the multiple DD:DD interactions (10). The molecular mechanisms of MyD88 recruitment to the activated receptor thus remain an open question. MyD88 is monomeric in nonstimulated cells (11) while at higher level of expression it can self-associate either through its DD or TIR domain (12, 13). The isolated TIR domain of MyD88 is on the other hand monomeric even at high concentration (14) and its expression in the cytosol inhibits TLR activation (8). This inhibitory property of TIR domain has been hijacked by several pathogenic bacteria where the inhibitory TIR domain containing proteins (TCPs) are important virulence factors as they enable pathogens to suppress the TLR-mediated host response. Bacterial TCPs share a common structural organization comprising TIR domain and typically an extended N-terminal domain (15–18). TcpB of Brucella is an important virulence factor (15) which suppresses TLR4 and TLR2 signaling (16) and interferes with dendritic cell maturation (17).
In this study, we investigated the effect of a dimerization state of TIR domains on TLR signaling. Tethered TIR dimers exhibited augmented inhibition of TLR signaling but on the other hand demonstrated constitutive activity at high expression level. We demonstrate that TIR domain dimerization strategy is employed by bacterial immunosuppressive virulence factors TCPs, where the N-terminal coiled-coil segment of TcpB from B. melitensis strongly improved inhibition of TLR signaling with a simultaneous decrease of the constitutive activity. The functional role of the N-terminal coiled-coil was additionally corroborated by the addition of an artificial strong coiled-coil dimerization domain to a MyD88 TIR domain which conferred potent inhibition over the broad range of TLRs and IL-1R, which could be potentially used for the therapeutic suppression of TLR activation.
EXPERIMENTAL PROCEDURES
Cell Culture and Reagents
The human embryonic kidney HEK293 cell lines were gifts from J. Weiss (University of Iowa). The MyD88 deficient HEK293 I3A was a gift from G. Stark (Department of Molecular Genetics, Cleveland Clinic) and A. Weber (German Cancer Research Centre) and HEK293 stably expressing TLR4-CFP/MD-2 was from T. Espevik (NTNU, Norway). Plasmids expressing TLR4, MD-2, CD-14, AU1-MyD88, and pELAM-1 firefly luciferase plasmid were a gift from Dr. C. Kirschning (Institute of Medical Microbiology, University Duisburg-Essen). pUNO-hTLR3, pUNO-hTLR9-HA, pUNO1-hTLR05-HA3x, pUNO2-hTRIF, and pUNO1-hUNC93B1 were from Invivogen. The codon optimized nucleotide sequences for 25 amino acids long peptide linker (amino acid sequence GSEGKSSGSGSESKVTDSGSETGSS) and GCN4-p1 peptide (18) were from GeneArt (Regensburg, Deutschland). The Renilla luciferase phRL-TK plasmid was from Promega. The IFN-β luciferase reporter plasmid was from J. Hiscott (McGill University). S-LPS (from Salmonella abortus equi) was gift from K. Brandenburg (Research Center Borstel, Germany), poly (I:C) and recombinant flagellin from S. typhimurium (Rec FLA) were from Invivogen and ODN 10104 from Coley Pharmaceutical Group.
Molecular Modeling
Molecular model of MyD88 dimer was prepared by the superposition of MyD88 TIR NMR structure (PDB code: 2Z5V) (14) to the TIR dimer of TLR10 (4) (PDB code: 2J67). Docking of MyD88 TIR dimers to TLR10 TIR dimer was performed using Gramm docking (19). Coiled-coil prediction was performed by program COILS (20).
DNA Constructs Preparation
Fusion DNA products were created with PCR overlap extension technique. TIR domain of MyD88 (mTIR) was PCR amplified from plasmid pDeNy-hMyD88 (InvivoGen), transmembrane segment (TM) and TIR domain of TLR4 (mTIR TLR4) were from plasmid pUNO-hTLR4 (InvivoGen). DNA coding for TcpB was amplified from genomic DNA of Brucella abortus (gift from I. Moriyon, University of Navarra, Pamplona), FLAG tag nucleotide sequence at the N terminus of TcpB was introduced with PCR. mCitrine, a gift from O. Griesbeck (LMU München), was linked to mTIR, dTIR, or GCN-mTIR by a linker peptide GGSGGGSGGSGG. Prepared DNA fusions were ligated into pcDNA3 vector (Invitrogen) or pFLAG-CMV-3 expression vector (Sigma). All chimeric DNA constructs were sequenced.
Luciferase Reporter Assay
NF-κB or IFN-β-dependent firefly luciferase and constitutive Renilla luciferase reporter were used to analyze the cell activation using a dual luciferase assay as described before (21). In the experiments with ligand stimulation, the RLU = RLU (stimulated cells)- RLU (unstimulated cells), unless stated otherwise.
Immunoblotting
HEK293T cells transfected with DNA constructs were lysed in the buffer (50 mm HEPES, pH 7.6, 0.5% Triton X-100, 150 mm NaCl, 20 mm β-glycerophosphate, 2 mm EDTA, 50 mm NaF, 1 mm Na3VO4, 1 mm DTT, 1 mm PMSF) with Protease Inhibitor Mixture (Sigma). Immunoblotting was performed as described (21). The antibodies used were polyclonal MyD88 Ab 1:500 dilution (PRS2127, Sigma), GFP antibody 1:1000 dilution (A11122, Invitrogen), αβ-tubulin rabbit polyclonal Ab 1:1000 dilution (2148, NEB), polyclonal anti-Flag antibodies 1:1000 dilution (F7425, Sigma), anti-HA antibody 1:1000 dilution (H6908, Sigma). Detection was performed with secondary goat anti-rabbit horseradish peroxidase-labeled antibody 1:5000 dilution (ab6721, Abcam) and blots were developed by ECL Western blotting detection reagent (Amersham Biosciences). For quantification analysis of Western blot bands the ImageJ program was used.
Cross-linking
Cross linking was performed on HEK293T cells transfected with DNA plasmids for 48 h by the addition of DSS in 1,5 mm final concentration to cells resuspended at ∼15 × 106 cells/ml in PBS. The reaction mixture was incubated for 30 min and quenched with 20 mm Tris, pH 7.5.
Immunoprecipitation
HEK293T cells were 48 h after transfection washed and lysed. Immunoprecipitation was performed using anti-FLAG antibody 1:500 dilution (F7425, Sigma), anti-HA antibody 1:200 dilution (H6908, Sigma), and protein G-Sepharose beads (GE Healthcare) as described before (22).
Confocal Microscopy
A Leica TCS SP5 laser scanning microscope mounted on a Leica DMI 6000 CS inverted microscope (Leica Microsystems, Germany) with an HCX plan apo 63× (NA 1.4) oil immersion objective was used for imaging. ECFP was excited with 405 nm diode laser and detected in the range of 470 nm to 500 nm, mCitrine was excited with 514 nm laser line and detected in the range of 520 nm to 580 nm.
LPS Labeling
LPS (0111:B4) from Escherichia coli was purchased from Invivogen. LPS was labeled using Cy5 labeling kit (GE Healthcare).
Fluorescence Resonance Energy Transfer (FRET)
HEK293 cells stably expressing TLR4-ECFP/MD-2 were seeded at a density of 4.5 × 104 cells per well in an 8-well microscope chamber slides, the next day transfected with plasmids for 48 h and stimulated with LPS-Cy5 for 3 h. Cells were fixed with 4% paraformaldehyde, washed with PBS and visualized. FRET was calculated by measuring donor de-quenching in the presence of an acceptor after acceptor bleaching using the FRET AB wizard. The FRET efficiency was quantified as: FRET eff = (Dpost-Dpre)/Dpost where Dpost is the fluorescence intensity of the Donor after acceptor photo bleaching, and Dpre the fluorescence intensity of the Donor before acceptor photo bleaching. The FRET efficiencies were calculated using ImageJ software (23).
RESULTS
Covalently Linked TIR Dimers Augment Inhibition of TLR Signaling
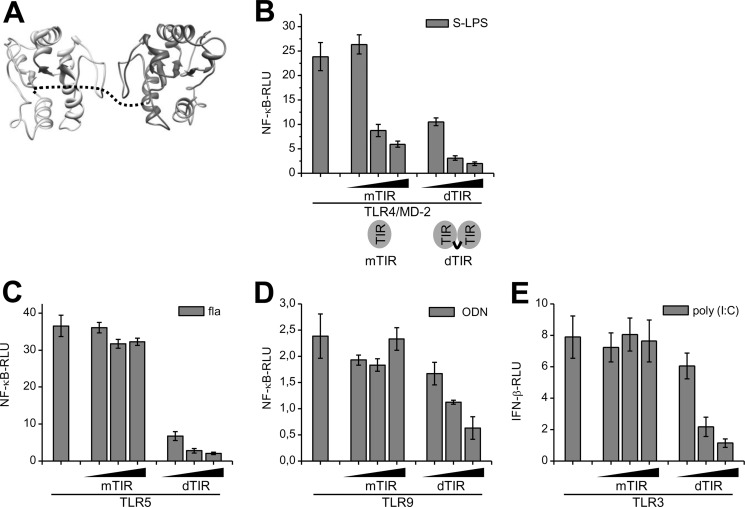
TIR domain of MyD88 is responsible for mediating signal transduction through TLRs but the isolated TIR domain acts as a TLR signaling inhibitor (8). We reasoned that the affinity of a dimerized TIR domain of the MyD88 for an activated receptor must be higher than the monomeric TIR domain due to the increased interaction interface of dimeric TIR domain. We designed a tethered TIR domain dimer (dTIR) based on a molecular model of a MyD88 TIR dimer (Fig. 1A) with a flexible peptide linker connecting the two TIR domains. The inhibition of a signaling response induced by stimulation of the receptor complex TLR4/MD-2 by LPS was markedly improved with dTIR in comparison to the monomeric TIR (mTIR) at the comparable molar protein expression level (Fig. 1B; supplemental Figs. S1 and S2). TLR5 signaling was suppressed by dTIR already at concentrations where the mTIR did not show any effect (Fig. 1C; supplemental Fig. S2B). Inhibition was also observed for the ODN triggered TLR9 signaling (Fig. 1D; supplemental Fig. S2C) and surprisingly even for the poly (I:C)-induced TRIF-dependent TLR3 activation (Fig. 1E; supplemental Fig. S2D), which were not inhibited by mTIR. Activation of TLR9 in absolute terms is lower than activation of other receptors so the relative effect of the constitutive activation by dTIR is more pronounced, while the difference between the TLR9 stimulated and nonstimulated experiment becomes readily distinguishable. Inhibition with dTIR is specific for the TIR domain-mediated signaling as the TNFα-R signaling was not inhibited with dTIR (supplemental Fig. S3).
FIGURE 1.
Covalent dimerization of MyD88 TIR domains improves TLR inhibition. A, Structural model of MyD88 TIR dimer based on the crystal structure of dimeric TLR10 TIR domains including the 25 amino acid linker (dashed line). B–E, improved inhibition of TLRs with dTIR in comparison to mTIR on HEK293 cells. B, cells were transfected with TLR4 and MD-2 plasmids (1 ng) and mTIR or dTIR plasmids (1, 5, 10 ng) and stimulated with S-LPS (10 ng/ml). C, cells were transfected with TLR5 (20 ng) and mTIR or dTIR plasmids (1, 5, 10 ng) and stimulated with flagellin (10 ng/ml). D, cells were transfected with TLR9 (10 ng) and Unc93B1 plasmids (1 ng) and mTIR or dTIR plasmids (1, 10, 20 ng) and stimulated with ODN (3 μm). E, cells were transfected with TLR3 (20 ng) and mTIR or dTIR plasmids (1, 5, 10 ng) and stimulated with poly(I:C) (10 μg/ml). Activation of TLR signaling pathway was determined by the dual luciferase assay. Representative graphs are shown from three separate experiments. Data are represented as mean ± S.D.
Constitutive Activity and Myddosome Induction of dTIR
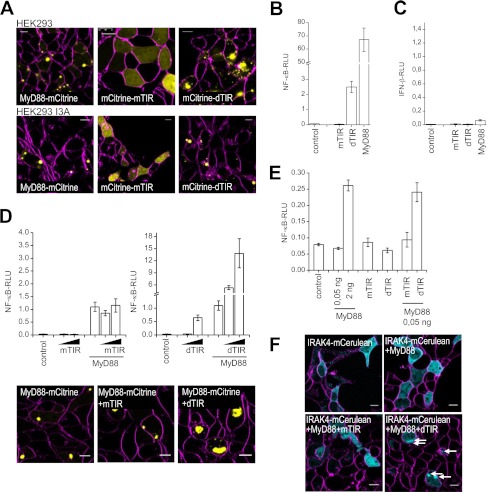
Overexpressed MyD88 is localized in discrete foci throughout the cytosol (11). We examined cellular localization of MyD88-YFP, YFP-mTIR and YFP-dTIR, transiently transfected in HEK293T or HEK293 I3A MyD88-deficient cell line (Fig. 2A). While wt MyD88 forms aggregates in the cytoplasm, mTIR was localized diffusely throughout the cytosol, as reported previously (11). dTIR, in contrast to mTIR, formed large punctated aggregates similar to the wild type MyD88, however its localization did not depend on the presence of MyD88 (Fig. 2A, lower panel), demonstrating molecular clustering triggered by TIR domain dimerization. While TLR activation was efficiently inhibited by dTIR at lower expression level (Fig. 1), higher expression level of dTIR exhibited constitutive activation of NF-κB (Fig. 2B). The activation of TRIF-dependent IFN-β promoter was on the other hand not affected by dTIR (Fig. 2C) demonstrating the selective activation of MyD88 signaling pathway. The constitutive activity of dTIR was unexpected since MyD88 requires DD and INT domain for the signaling function (9, 12). Coexpression of dTIR with MyD88 further potentiated activation of the wild type MyD88 (Fig. 2D, upper right) while mTIR had no effect (Fig. 2D, upper left). Expression of dTIR with MyD88-YFP resulted in the increased size of MyD88-YFP aggregates suggesting that dTIR recruits MyD88 into large complex through multivalent interactions (Fig. 2D lower panel), which was further confirmed by the co-localization of YFP-dTIR with MyD88-CFP (supplemental Fig. S4, right). On the other hand, mTIR did not affect activation by MyD88 and formation of MyD88 aggregates (Fig. 2D lower panel), although YFP-mTIR colocalized with MyD88-CFP (supplemental Fig. S4, left). No NF-κB activation was observed in HEK293 I3A transfected with dTIR while cotransfection of dTIR (but not mTIR) with wt MyD88 reconstituted signaling (Fig. 2E) thus confirming our hypothesis that dTIR-induced activation depends on MyD88.
FIGURE 2.
Covalently linked TIR MyD88 dimers exhibit constitutive MyD88-dependent activity. A, dTIR forms similar clusters within cells as MyD88 but independent of it. HEK293T or HEKI3A cells were transfected with MyD88-YFP, YFP-mTIR or YFP-dTIR (10 ng) and stained with SynaptoRed. Fusion constructs (yellow); SynaptoRed (magenta). B, dTIR exhibits constitutive activity. HEK293 cells were transfected with indicated plasmids (20 ng). C, dTIR does not activate IFN-β promoter. HEK293 cells were transfected with indicated plasmids (100 ng). D, dTIR potentiates the activity triggered by MyD88 and increases the size of MyD88 aggregates. Upper panel, cells were transfected with empty vector, mTIR (20, 50 ng) (left), dTIR (20, 50 ng) (right), and/or MyD88 plasmid (0.25 ng). Lower panel, HEK293T cells were transfected with indicated plasmids (MyD88-YFP 2.5 ng; mTIR 25 ng; dTIR 25 ng). MyD88-YFP (yellow); SynaptoRed (magenta). E, MyD88 is indispensable for constitutive activity of dTIR. HEK293 I3A cells were transfected with MyD88 (0.05, 2 ng), mTIR or dTIR plasmids (20 ng each) alone or MyD88 plasmid (0.05 ng) was co-transfected with mTIR or dTIR plasmid (20 ng each). F, dTIR triggers IRAK-4 assembly in the presence of MyD88. HEK293T cells were transfected with IRAK4-CFP (70 ng), MyD88 plasmids (1 ng) and mTIR or dTIR plasmids (150 ng each). IRAK4-CFP (cyan); Synaptored (magenta). White arrows indicate IRAK4-CFP aggregation. Activation of TLR signaling pathway was determined by the dual luciferase assay. Representative graphs and micrographs are shown from three separate experiments. Data on graphs are represented as mean ± S.D. Scale bars on micrographs represent 10 μm.
Further, we demonstrate that dTIR but not mTIR scaffold enables IRAK4 aggregation in the presence of MyD88, required for the Myddosome formation. IRAK4 is diffusely distributed in the cytoplasm but its coexpression with MyD88 recruits it into distinct foci (24). In our study, we cotransfected fluorescently tagged IRAK4 with an amount of MyD88 that is insufficient to trigger IRAK4 aggregation. Upon addition of the dTIR platform IRAK4 was found in aggregates and clustered into distinct foci (Fig. 2F) which were not observed in the absence of MyD88. Our results thus show that dimeric TIR of MyD88 forms a platform for the recruitment of additional MyD88 molecules through oligomerization of TIR domains that lead to the assembly of the signaling Myddosome.
Dimerization of TLR4 TIR Domain Improves the Inhibition of TLR Signaling
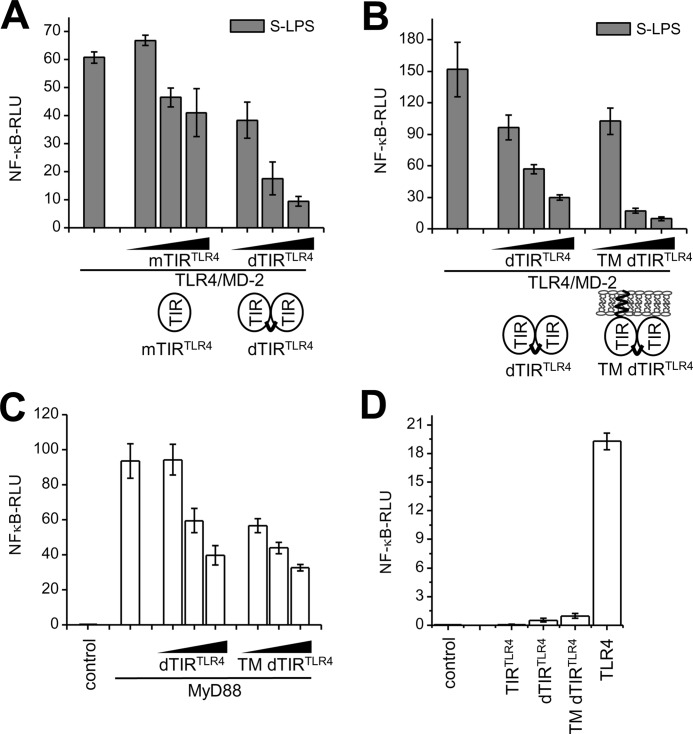
In analogy to covalently linked MyD88 TIR dimers, we prepared covalently linked TIR domains of TLR4 (dTIR TLR4) tethered by the 25 amino acid linker. Similar to dTIR, the dTIR TLR4 exhibited enhanced inhibition of TLR4/MD-2 signaling in comparison to mTIR TLR4 (Fig. 3A; supplemental Fig. S5). We reasoned that association of the TIR domain of TLR4 to the membrane could increase its interaction with the target. The addition of a transmembrane segment (TM) of TLR4 to dTIR TLR4 further improved the inhibition (Fig. 3B; supplemental Fig. S5). dTIR TLR4 and TM dTIR TLR4 also inhibited cell signaling triggered by overexpression of MyD88 (Fig. 3C) suggesting the sequestration of MyD88 as the mechanism of inhibition. On the other hand, membrane anchored cytoplasmic portion of TLR4 forms active homodimers (25, 26) and the dimeric TLR4 TIR domain was proposed as a scaffold for the recruitment of adapters (5). Therefore it was not surprising that in addition to inhibition at lower concentrations, dTIR TLR4 and TM dTIR TLR4 also exhibited low constitutive activity at very high concentrations (Fig. 3D).
FIGURE 3.
Covalent dimerization of TLR4 TIR domains improves TLR inhibition and leads to low constitutive activity. A and B, dimeric TIR TLR4 domain inhibits TLR4/MD-2 signaling better than monomeric TIR TLR4. HEK293 cells were transfected with TLR4 and MD-2 plasmids (1 ng) and the indicated plasmids (1, 60, 140 ng) and stimulated with S-LPS (10 ng/ml). C, dTIR TLR4 and TM dTIR TLR4 inhibit NF-κB activation of MyD88. HEK293 cells were transfected with control or MyD88 (5 ng) and dTIR TLR4 or TM dTIR TLR4 plasmids (10, 50, 100 ng). D, dTIR TLR4 and TM dTIR TLR4 exhibit low constitutive activity. HEK293 cells were transfected with indicated plasmids (140 ng). Activation of TLR signaling pathway was determined by the dual luciferase assay. Representative graphs are shown from three separate experiments. Data on graphs are represented as mean ± S.D.
Enhancement of TLR Inhibition by Coiled-coil Dimerizing Segments of Bacterial TIR Domain Virulence Factors
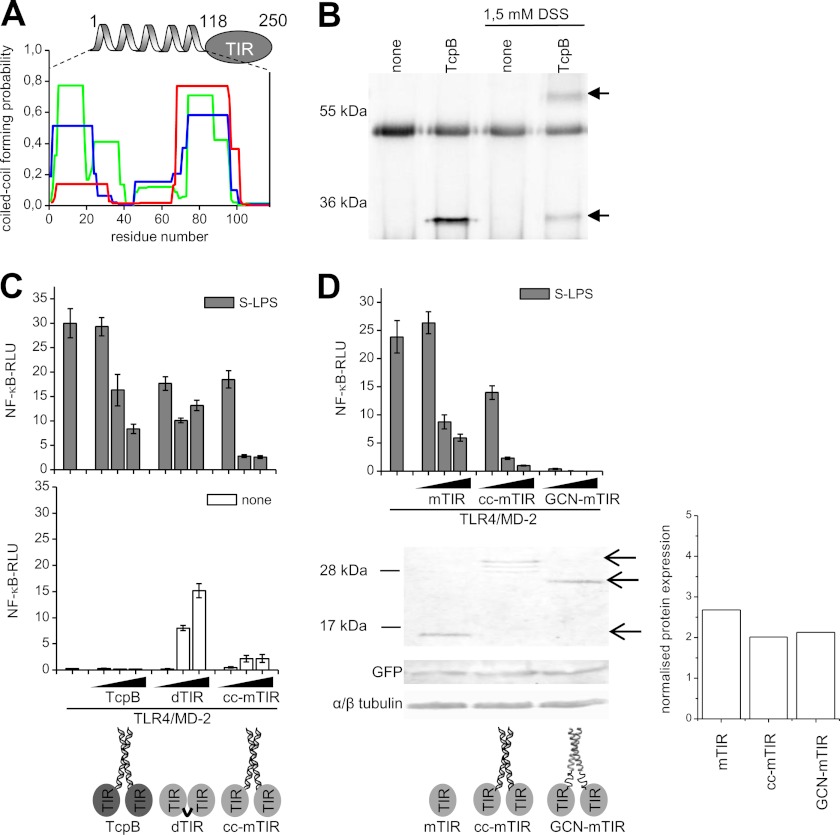
Inhibitory TIR domain-containing proteins are used by several bacteria to subvert host TLR signaling (15–17, 27). Bioinformatic analysis reveals that TcpB from Brucella contains a strong coiled-coil motif in the N-terminal segment (Fig. 4A). Based on the potentiation of TLR inhibition by dimeric TIR domain, we considered whether Brucella uses a coiled-coil mediated TIR domain dimerization to create a more efficient TLR immunosuppressor. We confirmed the existence of TcpB dimers in the cytosol of cells expressing TcpB (Fig. 4B) using cross-linking. PdTLP from P. denitrificans was also reported to form a dimer, while its isolated TIR domain is monomeric (28). Comparison of the inhibition (Fig. 4C, upper) and activation (Fig. 4C, middle) of TLR4/MD-2 signaling caused by TcpB versus dTIR revealed a remarkable difference. While the TcpB is less potent inhibitor than dTIR it shows no constitutive activity at higher expression level in contrast to dTIR. By grafting the coiled-coil segment of the TcpB (cc) to the TIR domain of MyD88 we gained a protein (cc-mTIR) with improved inhibitory effect even in comparison to dTIR (Fig. 4C, upper) while it almost completely lacked the constitutive activity (supplemental Fig. S6).
FIGURE 4.
Fusion of coiled-coil dimerizing segment with TIR domain potentiates the suppression of TLR activation. A, prediction of a coiled-coil formation within an N-terminal segment of TcpB. The coiled-coil-forming propensity is presented for frames of 14 (green), 21 (blue), and 28 (red) residues. B, TcpB dimerizes in cells. HEK293T cells transfected with control or TcpB plasmid were treated with PBS or 1.5 mm cross-linker DSS. Proteins were immunoprecipitated from cell lysates, separated on SDS-PAGE and immunoblotted using anti FLAG antibodies. Monomeric (29 kDa) and dimeric (58 kDa) TcpB are indicated with arrows. C, grafting of TcpB coiled-coil segment to TIR MyD88 enhances inhibition of TLR signaling. HEK293 cells were transfected with TLR4 and MD-2 plasmids (1 ng) and TcpB, dTIR or cc-mTIR (1, 30, 60 ng). Cells were stimulated with S-LPS (10 ng/ml). Data represented are RLUs of unstimulated and stimulated cells. D, dimerization of MyD88 TIR by a strong coiled-coil maximizes inhibitory potential. Upper, HEK293 cells were transfected with TLR4 and MD-2 plasmids (1 ng) and mTIR, cc-mTIR or GCN-mTIR plasmids (1, 5, 10 ng). Cells were stimulated with S-LPS (10 ng/ml). Middle, control of protein expression levels. HEK293T cells were transfected with mTIR, cc-mTIR or GCN-mTIR (3 μg) plasmids together with GFP (5 ng) plasmid. Proteins from cell lysates were separated on SDS-PAGE and immunoblotted using antibodies against MyD88, GFP, and tubulin. Arrows indicate the positions of cc-mTIR (30 kDa), GCN-mTIR (27 kDa), and mTIR (17 kDa) from top to bottom. Right, quantification of Western blot bands of mTIR, cc-mTIR, and GCN-mTIR. The normalization was done against the level GFP. Activation of TLR signaling pathway was determined by the dual luciferase assay. Representative graphs and micrographs are shown from three separate experiments. Data on graphs are represented as mean ± S.D. Scale bars on micrographs represent 10 μm.
Mal/TIRAP-like membrane association of the N-terminal segment of TcpB has been proposed as the underlying cause of its TLR inhibition based on the decreased inhibition of a TcpB variant with five mutated basic residues (15). To establish the functional role of the coiled-coil mediated dimerization of bacterial TCPs we prepared an artificial TCP variant based on the coiled-coil segment (GCN4-p1) from the yeast transcriptional activator GCN4. GCN4-p1 forms a highly stable parallel dimeric coiled-coil leucine zipper (29). GCN4-p1 segment was coupled by a flexible peptide linker to the mTIR, resulting in the fusion construct GCN-mTIR. Inhibition of TLR signaling was exceptionally improved by this modification (Fig. 4D, upper) at comparable protein expression level (Fig. 4D, middle). Additionally, the constitutive activation observed in other types of dimeric TIR was completely abolished (supplemental Fig. S6). The cellular localization of GCN-mTIR fused to YFP was MyD88-independent and very alike to YFP-dTIR and MyD88-YFP (supplemental Fig. S7) indicating that coiled-coil induced dimeric TIR domains aggregate and localize similar to wild type MyD88 and that formation of punctated aggregates therefore does not necessary coincide with cell activation. Fusion of a coiled-coil dimerization segment and TIR domain is thus an efficient microbial strategy adopted to improve suppression of TIR domain-mediated innate immune response, combining strong inhibition while preventing the constitutive activity.
Combination of a Designed Coiled-coil Forming Segment with TIR Domain Creates a Broad Spectrum Inhibitor Specific for TLR/IL-1R Signaling
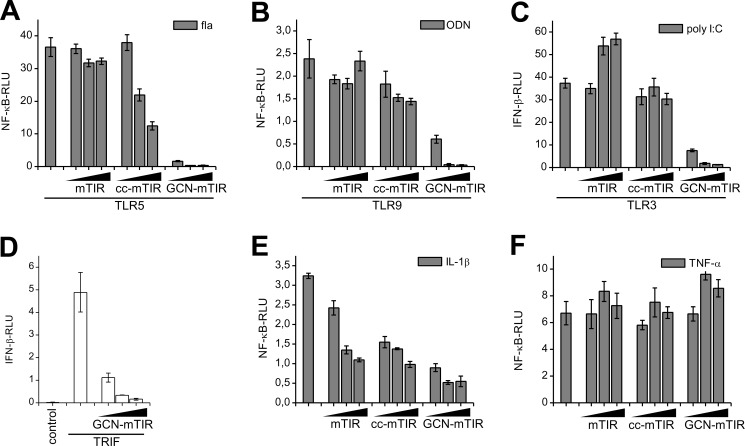
Pathogens often engage several TLRs and inhibitors with an expanded spectrum for TLR targets are of potential therapeutic interest for different inflammatory and autoimmune diseases which involve activation of TLRs/IL-1R, e.g. SLE (30), rheumatoid arthritis (31, 32) or gout (33). GCN-mTIR based on the designed coiled-coil exhibited the most efficient inhibition of all the tested TLRs (Fig. 5, A–C; supplemental Fig. S8, A–C), inhibiting all tested TLRs including TLR3. Inhibition of TRIF-dependent TLR3 signaling by GCN-mTIR probably occurs due to the sequestration of TRIF as this designed TCP also suppressed activation triggered by overexpression of TRIF (Fig. 5D). In addition to TLRs GCN-mTIR also inhibited IL1-R signaling (Fig. 5E; supplemental Fig. S8D) while it had no effect on the TNFα-R signaling pathway (Fig. 5F; supplemental Fig. S8E) confirming the specificity for the TIR domain mediated TLR/IL-1R signaling pathways.
FIGURE 5.
Fusion of a designed coiled-coil with mTIR creates inhibitor with broad TLR and IL-1R inhibitory spectrum. A–C, GCN-mTIR is a strong inhibitor of TLR5, TLR9 and TLR3 signaling. HEK293 cells were transfected with mTIR, cc-mTIR or GCN-mTIR plasmids (1, 5, 10 ng) and either TLR5 plasmid (20 ng) (A), TLR9 plasmid (10 ng) and Unc93B1 plasmid (1 ng) (B) or TLR3 plasmid (20 ng) (C). Cells were stimulated with flagellin (10 ng/ml), ODN (3 μm) or poly(I:C) (10 μg/ml). D, GCN-mTIR inhibits signaling triggered by overexpression of TRIF. Cells were transfected with control or TRIF plasmid (2 ng) or cotransfected with TRIF (2 ng) and GCN-mTIR plasmid (10, 30, 50 ng). E, coiled-coils linked TIR proteins improve inhibition of IL-1R signaling. HEK293 cells were transfected with mTIR, cc-mTIR or GCN-mTIR plasmids (1, 5, 10 ng) and stimulated with IL-1β (50 ng/ml). F, TNFα-R signaling is unaffected by TIR fusion proteins. HEK293 cells were transfected with mTIR, cc-mTIR or GCN-mTIR plasmids (5, 35, 75 ng) and stimulated with TNF-α (100 ng/ml). Activation of TLR signaling pathway was determined by the dual luciferase assay. Representative graphs are shown from three separate experiments. Data on graphs are represented as mean ± S.D.
TLR4 as the Target for Inhibition by Dimeric TIR Inhibition
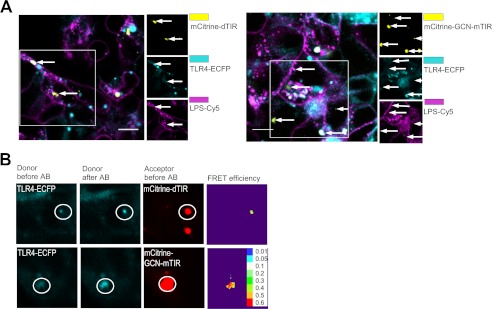
Our results show that neither TIR dimers nor monomeric TIR inhibited the constitutive activity of MyD88 which acts downstream of TLRs. We therefore surmised that dimerized TIR domains of the activated TLRs represent the target for inhibition. We observed colocalization of YFP-dTIR or YFP-GCN-mTIR with TLR4-CFP (Fig. 6A). YFP-mTIR was diffusely localized as in unstimulated cells which therefore prevented the unambiguous determination of its colocalization with TLR4-CFP.3 To confirm the close physical association of TIR dimers with TLR4 we used the FRET experiment. A strong FRET effect determined by the donor dequenching upon the acceptor bleaching was observed in the spots where TLR4-CFP colocalized with YFP-dTIR or YFP-GCN-mTIR (Fig. 6B). Results thus demonstrate that activated TLR4 forms a complex with TIR dimers and represents the target for binding and inhibition.
FIGURE 6.
TLR4 binds inhibitory MyD88 TIR dimers. A and B, TLR4-CFP/MD-2 cells were transfected with CD-14 (40 ng) and the indicated YFP-tagged constructs (5 ng). Cells were stimulated with LPS-Cy5 (5 μg/ml) for 3 h and fixed. A, co-localization of dimeric TIR of MyD88 with TLR4 is indicated on images with white arrows. B, MyD88 TIR dimers bind to TLR4. FRET between YFP-tagged constructs and TLR4-CFP was calculated from donor de-quenching in the presence of an acceptor after acceptor bleaching and the FRET efficiency is shown as a color-coded scale. Representative micrographs are shown from three separate experiments. Scale bars on micrographs represent 10 μm.
DISCUSSION
Signaling pathway activation of TLRs is triggered by the ligand-induced receptor dimerization bringing the cytosolic TIR domains of the receptor into close proximity, which is a prerequisite for the subsequent binding of the adapters mediated by TIR domains. MyD88 oligomerization is required as the first step for the binding of IRAK4 which is recruited to the composite binding site composed of the associated MyD88 DD via three different types of DD interactions. We demonstrated for the first time the important consequences of TIR domain dimerization on the inhibition and activation of the TLR signaling pathway.
Enhanced Inhibition of TLR Signaling by a Dimeric TIR Domains
We showed that a MyD88 TIR dimer platform of covalently linked TIR domains (dTIR) improved inhibition of TLR signaling compared with TIR monomers against all TLRs tested. Improved inhibition probably results from the strong interaction between dimeric TIR domains of MyD88 that interact with a dimer of TIR domains of TLRs. The dTIR TLR4 exhibited increased inhibition of TLR4/MD-2 signaling similar to dimeric TIR of MyD88 but in addition also inhibited the activation triggered by MyD88 overexpression by sequestration of MyD88. Suppression of TLR activation through TIR domain containing proteins has been adopted by bacteria as well as by viruses to diminish the immune response. Our results demonstrate that bacteria take advantage of the coiled-coil domains for creating dimeric TIR inhibitors with enhanced potency and devoid of the constitutive activity. Coiled-coil fused to the TIR domain probably interferes with the assembly of IRAK4 into the complex, which is required for the TLR signaling, thereby preventing constitutive activation. All of the bacterial TIR domains that have been so far demonstrated to suppress host immune response including TcpC of E. coli, TcpB of B. melitensis (16), and TlpA of S. enterica serovar Enteriditis (27) are composed of the C-terminal TIR domain and an N-terminal segment that includes a coiled-coil motif. While it has been proposed before that the N-terminal segment of TCPs may also contain other functional motifs, such as the ability to associate with the membrane (15), we demonstrated that the addition of an artificial coiled-coil domain is sufficient to strongly augment the inhibition of TLR signaling. Our results demonstrate an enhanced TLR inhibition potency in proteins with a strong coiled-coil dimerization propensity. TCPs are widely distributed in bacteria (34). While it may be premature to generalize but it is interesting to note that TCPs of some pathogenic bacteria where the role of TCPs for the suppression of the innate immune response has not yet been established contain a strong coiled-coil signal (Fig. S9), e.g. TCPs of Staphylococcus aureus, Helicobacter pylori, Porphyromonas gingivalis etc. Interestingly also bacteria invading plants contain TCPs with TIR and coiled-coil domains such as in Agrobacterium tumefaciens, which is an important plant pathogen and an efficient biotechnological vector for the introduction of DNA into plants. Plant homologues of TLRs (resistance proteins) contain either TIR or coiled-coil domain to mediate downstream signal (35), which may have facilitated the creation of the coiled-coil-TIR domain fusions through the horizontal gene transfer.
The combination of a strong coiled-coil and TIR domain exhibited superior inhibition of all TLRs tested as well as IL1-R. Dominant negative form of MyD88 (mTIR) has already been tested for the therapy of inflammation (36, 37). Inhibition of TLR3 pathway by dimeric TIR MyD88 domains indicates that dimeric TIR domains are probably more promiscuous and that there is a cross-talk between MyD88 and TRIF-dependent pathways, which has also been demonstrated with inhibition of TLR3 signalization by Mal (38).
Activation of Signaling by a Dimeric TIR Platform
Besides the enhanced inhibition dTIR also exhibited the constitutive activation of MyD88 signaling, leading to a biphasic concentration-dependent response. The constitutive activity of a dimeric TIR domains depended on MyD88 as the dTIR cannot directly recruit IRAK4 due to the absence of DD but can serve as the scaffold for the assembly of functional MyD88 molecules. TIR domains dimerized either by a covalent tether or through a fusion with a coiled-coil dimerization domain formed aggregates in the cytosol, which demonstrates that TIR domains can form oligomers beyond dimers. This is in agreement with the proposition that TIR domain comprises several homotypic interacting surfaces (14, 39), similar to death domain (10). Although the mTIR co localized with MyD88, it had no effect on the formation of MyD88 aggregates in contrast to dTIR. Additionally, only dimeric TIR was able to trigger the recruitment of IRAK4 into the assembly depending on MyD88.
Molecular Mechanism of the TLR Activation by the Dimeric TIR Platform
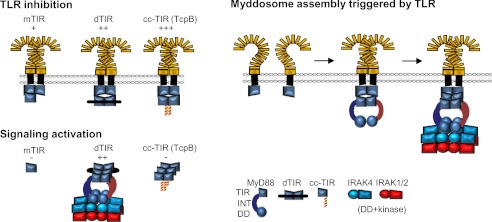
Dimeric TIR domain of MyD88 creates the large interaction surface which has higher affinity for TIR dimers of activated TLRs than monomeric TIR domain, which augments the inhibition (Fig. 7). The ability of MyD88 TIR dimer to activate signaling and form clusters demonstrates that TIR domains of the adapters can form multivalent aggregates through multiple interaction surfaces that support formation of the active Myddosome complex, which requires at least four and probably six MyD88 molecules. TLRs cannot tetramerize at the membrane via their TIR domains due to the steric hindrance of their large TLR ectodomains and topology within the membrane. The crystal structure of the DD Myddosome may differ in some details from the structure of the integral MyD88/IRAK complex. We have already demonstrated in our paper on the role of the INT domain of MyD88 (21) that this segment, although missing from the crystallized domain plays a role in interaction of MyD88 with IRAK4. However, this structure identified the large interaction surfaces between DD, which are most likely relevant and are supported by the mutagenesis data We propose that the dimerization of MyD88 via TIR domains is the trigger that initiates the assembly of the signaling complex downstream of TLR while the exact stoichiometry of MyD88 binding to the activated TLRs may not be that important. The addition of a coiled-coil dimerizing segment to the TIR domain created a TIR dimer with the interaction surface that can still bind to TIR dimers of the activated TLRs, while at least one of the other interaction surfaces is occluded, probably by steric hindrance of the coiled-coil segment. This prevents the recruitment of additional MyD88 molecules to the dimeric TIR complex in an arrangement that is compatible with formation of the Myddosomal complex.
FIGURE 7.
Model of the role of dimeric TIR platform for the inhibition and activation of MyD88 signaling pathway. Left, monomeric and dimeric MyD88 TIR domains inhibit TLR signaling by binding to cytoplasmic TIR dimer of activated TLRs, preventing binding of MyD88. Tethered TIR dimer induce MyD88 oligomerization, which triggers assembly of the Myddosome complex. Right, model of the assembly of Myddosome induced by TLR dimerization: sequential addition of MyD88 through interactions with TIR of TLRs and TIRs of MyD88 assembles the DD complex and triggers phosphorylation of IRAK kinases.
The delicate balance between the silent monomeric MyD88 and its dimeric form is responsible for the high sensitivity of sensing pathogens though TLR dimerization while preventing the constitutive activation of MyD88 that may lead to the persistent inflammation and cancer (40). At the physiological level of MyD88 expression the equilibrium is shifted toward the monomer, while the presence of activated TLRs shifts it toward MyD88 dimers which initiate the assembly of a Myddosome (supplemental Fig. S10). Molecular docking supports the proposal that the dimeric TIR domain interaction surface induces dimerization of MyD88, which further recruits additional MyD88 molecules via the secondary and tertiary TIR:TIR interaction sites (supplemental Fig. S11). Mal adapter, required for the strong responsiveness through TLR2 and TLR4 probably acts primarily as a chaperone that facilitates the transport of MyD88 to the cell membrane (41) and may additionally stabilize its interaction with a dimerized TIR domain of TLRs. In the docked arrangement (supplemental Fig. S11) all N termini of TIR domains of MyD88 which are connected to intermediary linker segments and DD, point away from the membrane, allowing association of DD of those MyD88 molecules, assembly of the Myddosome and activation of downstream IRAK kinases.
Supplementary Material
Acknowledgments
We thank colleagues who generously shared reagents (described under “Experimental Procedures”).
This work was supported by the Slovenian Research Agency (to R. J. and M. B.) and Centre of Excellence EN-FIST, Ljubljana, Slovenia supported in part by the EU structural funds.

This article contains supplemental Figs. S1–S11.
R. Jerala, unpublished observations.
- TLR
- Toll-like receptor
- TIR
- Toll/IL-1 receptor
- IL-1
- interleukin-1
- MyD88
- myeloid differentiation primary response gene (88)
- TCP
- TIR domain-containing protein
- TLP
- TIR-like protein
- TRIF
- TIR domain-containing adapter-inducing interferon β
- DD
- death domain; INT-intermediary domain of MyD88
- IRAK
- IL-1 receptor-associated kinase
- MD-2
- myeloid differentiation protein-2
- Mal/TIRAP
- MyD88-adapter-like/TIR domain-containing adapter protein
- mTIR
- monomeric TIR domain of MyD88
- dTIR
- covalently linked dimeric TIR domain of MyD88; mTIR TLR4 monomeric TIR domain of TLR4
- dTIR TLR4
- covalently linked dimeric TIR domain of TLR4
- TM
- transmembrane segment of TLR4
- cc
- coiled-coil segment of TcpB
- GCN
- GCN4-p1 peptide
- FRET
- fluorescence resonance energy transfer.
REFERENCES
- 1. Akira S., Takeda K. (2004) Toll-like receptor signaling. Nat. Rev. Immunol. 4, 499–511 [DOI] [PubMed] [Google Scholar]
- 2. O'Neill L. A., Bowie A. G. (2007) The family of five: TIR domain-containing adaptors in Toll-like receptor signaling. Nat. Reviews Immunol. 7, 353–364 [DOI] [PubMed] [Google Scholar]
- 3. Gay N. J., Gangloff M. (2007) Structure and function of Toll receptors and their ligands. Annu. Rev. Biochem. 76, 141–165 [DOI] [PubMed] [Google Scholar]
- 4. Nyman T., Stenmark P., Flodin S., Johansson I., Hammarström M., Nordlund P. (2008) The crystal structure of the human toll-like receptor 10 cytoplasmic domain reveals a putative signaling dimer. J. Biol. Chem. 283, 11861–11865 [DOI] [PubMed] [Google Scholar]
- 5. Nuñez Miguel R., Wong J., Westoll J. F., Brooks H. J., O'Neill L. A., Gay N. J., Bryant C. E., Monie T. P. (2007) A dimer of the Toll-like receptor 4 cytoplasmic domain provides a specific scaffold for the recruitment of signaling adaptor proteins. PLoS One 2, e788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Bonnert T. P., Garka K. E., Parnet P., Sonoda G., Testa J. R., Sims J. E. (1997) The cloning and characterization of human MyD88: a member of an IL-1 receptor related family. FEBS Lett. 402, 81–84 [DOI] [PubMed] [Google Scholar]
- 7. Hardiman G., Rock F. L., Balasubramanian S., Kastelein R. A., Bazan J. F. (1996) Molecular characterization and modular analysis of human MyD88. Oncogene 13, 2467–2475 [PubMed] [Google Scholar]
- 8. Medzhitov R., Preston-Hurlburt P., Kopp E., Stadlen A., Chen C., Ghosh S., Janeway C. A., Jr. (1998) MyD88 is an adaptor protein in the hToll/IL-1 receptor family signaling pathways. Mol. Cell 2, 253–258 [DOI] [PubMed] [Google Scholar]
- 9. Burns K., Janssens S., Brissoni B., Olivos N., Beyaert R., Tschopp J. (2003) Inhibition of interleukin 1 receptor/Toll-like receptor signaling through the alternatively spliced, short form of MyD88 is due to its failure to recruit IRAK-4. J. Exp. Med. 197, 263–268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Lin S. C., Lo Y. C., Wu H. (2010) Helical assembly in the MyD88-IRAK4-IRAK2 complex in TLR/IL-1R signaling. Nature 465, 885–890 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Nishiya T., Kajita E., Horinouchi T., Nishimoto A., Miwa S. (2007) Distinct roles of TIR and non-TIR regions in the subcellular localization and signaling properties of MyD88. FEBS Lett. 581, 3223–3229 [DOI] [PubMed] [Google Scholar]
- 12. Burns K., Martinon F., Esslinger C., Pahl H., Schneider P., Bodmer J. L., Di Marco F., French L., Tschopp J. (1998) MyD88, an adapter protein involved in interleukin-1 signaling. J. Biol. Chem. 273, 12203–12209 [DOI] [PubMed] [Google Scholar]
- 13. Ulrichts P., Peelman F., Beyaert R., Tavernier J. (2007) MAPPIT analysis of TLR adaptor complexes. FEBS Lett. 581, 629–636 [DOI] [PubMed] [Google Scholar]
- 14. Ohnishi H., Tochio H., Kato Z., Orii K. E., Li A., Kimura T., Hiroaki H., Kondo N., Shirakawa M. (2009) Structural basis for the multiple interactions of the MyD88 TIR domain in TLR4 signaling. Proc. Natl. Acad. Sci. U.S.A. 106, 10260–10265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Radhakrishnan G. K., Yu Q., Harms J. S., Splitter G. A. (2009) Brucella TIR domain-containing protein mimics properties of the Toll-like receptor adaptor protein TIRAP. J. Biol. Chem. 284, 9892–9898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cirl C., Wieser A., Yadav M., Duerr S., Schubert S., Fischer H., Stappert D., Wantia N., Rodriguez N., Wagner H., Svanborg C., Miethke T. (2008) Subversion of Toll-like receptor signaling by a unique family of bacterial Toll/interleukin-1 receptor domain-containing proteins. Nat. Med. 14, 399–406 [DOI] [PubMed] [Google Scholar]
- 17. Salcedo S. P., Marchesini M. I., Lelouard H., Fugier E., Jolly G., Balor S., Muller A., Lapaque N., Demaria O., Alexopoulou L., Comerci D. J., Ugalde R. A., Pierre P., Gorvel J. P. (2008) Brucella control of dendritic cell maturation is dependent on the TIR-containing protein Btp1. PLoS Pathog 4, e21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lumb K. J., Carr C. M., Kim P. S. (1994) Subdomain folding of the coiled coil leucine zipper from the bZIP transcriptional activator GCN4. Biochemistry 33, 7361–7367 [DOI] [PubMed] [Google Scholar]
- 19. Tovchigrechko A., Vakser I. A. (2006) GRAMM-X public web server for protein-protein docking. Nucleic Acids Res. 34, W310–314 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Lupas A., Van Dyke M., Stock J. (1991) Predicting coiled coils from protein sequences. Science 252, 1162–1164 [DOI] [PubMed] [Google Scholar]
- 21. Avbelj M., Horvat S., Jerala R. (2011) The role of intermediary domain of MyD88 in cell activation and therapeutic inhibition of TLRs. J. Immunol. 187, 2394–2404 [DOI] [PubMed] [Google Scholar]
- 22. Divanovic S., Trompette A., Atabani S. F., Madan R., Golenbock D. T., Visintin A., Finberg R. W., Tarakhovsky A., Vogel S. N., Belkaid Y., Kurt-Jones E. A., Karp C. L. (2005) Negative regulation of Toll-like receptor 4 signaling by the Toll-like receptor homolog RP105. Nat. Immunol. 6, 571–578 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Roszik J., Szöllosi J., Vereb G. (2008) AccPbFRET: an ImageJ plugin for semi-automatic, fully corrected analysis of acceptor photobleaching FRET images. BMC Bioinformatics 9, 346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Nagpal K., Plantinga T. S., Sirois C. M., Monks B. G., Latz E., Netea M. G., Golenbock D. T. (2011) Natural loss-of-function mutation of myeloid differentiation protein 88 disrupts its ability to form Myddosomes. J. Biol. Chem. 286, 11875–11882 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Zhang H., Tay P. N., Cao W., Li W., Lu J. (2002) Integrin-nucleated Toll-like receptor (TLR) dimerization reveals subcellular targeting of TLRs and distinct mechanisms of TLR4 activation and signaling. FEBS Lett. 532, 171–176 [DOI] [PubMed] [Google Scholar]
- 26. Panter G., Jerala R. (2011) The ectodomain of the Toll-like receptor 4 prevents constitutive receptor activation. J. Biol. Chem. 286, 23334–23344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Newman R. M., Salunkhe P., Godzik A., Reed J. C. (2006) Identification and characterization of a novel bacterial virulence factor that shares homology with mammalian Toll/interleukin-1 receptor family proteins. Infect. Immun. 74, 594–601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Pascual J., Low L. Y., Mukasa T., Reed J. C. (2007) Characterization of a TIR-like protein from Paracoccus denitrificans. Biochem. Biophys. Res. Commun. 356, 481–486 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. O'Shea E. K., Klemm J. D., Kim P. S., Alber T. (1991) X-ray structure of the GCN4 leucine zipper, a two-stranded, parallel coiled coil. Science 254, 539–544 [DOI] [PubMed] [Google Scholar]
- 30. Boulé M. W., Broughton C., Mackay F., Akira S., Marshak-Rothstein A., Rifkin I. R. (2004) Toll-like receptor 9-dependent and -independent dendritic cell activation by chromatin-immunoglobulin G complexes. J. Exp. Med. 199, 1631–1640 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Brentano F., Schorr O., Gay R. E., Gay S., Kyburz D. (2005) RNA released from necrotic synovial fluid cells activates rheumatoid arthritis synovial fibroblasts via Toll-like receptor 3. Arthritis Rheum. 52, 2656–2665 [DOI] [PubMed] [Google Scholar]
- 32. Sacre S. M., Andreakos E., Kiriakidis S., Amjadi P., Lundberg A., Giddins G., Feldmann M., Brennan F., Foxwell B. M. (2007) The Toll-like receptor adaptor proteins MyD88 and Mal/TIRAP contribute to the inflammatory and destructive processes in a human model of rheumatoid arthritis. Am. J. Pathol. 170, 518–525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Chen C. J., Shi Y., Hearn A., Fitzgerald K., Golenbock D., Reed G., Akira S., Rock K. L. (2006) MyD88-dependent IL-1 receptor signaling is essential for gouty inflammation stimulated by monosodium urate crystals. J. Clin. Invest. 116, 2262–2271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Spear A. M., Loman N. J., Atkins H. S., Pallen M. J. (2009) Microbial TIR domains: not necessarily agents of subversion? Trends Microbiol. 17, 393–398 [DOI] [PubMed] [Google Scholar]
- 35. Burch-Smith T. M., Dinesh-Kumar S. P. (2007) The functions of plant TIR domains. Sci. STKE 2007, pe46. [DOI] [PubMed] [Google Scholar]
- 36. Dupraz P., Cottet S., Hamburger F., Dolci W., Felley-Bosco E., Thorens B. (2000) Dominant negative MyD88 proteins inhibit interleukin-1β /interferon-γ-mediated induction of nuclear factor κB-dependent nitrite production and apoptosis in beta cells. J. Biol. Chem. 275, 37672–37678 [DOI] [PubMed] [Google Scholar]
- 37. Hua F., Ha T., Ma J., Gao X., Kelley J., Williams D. L., Browder I. W., Kao R. L., Li C. (2005) Blocking the MyD88-dependent pathway protects the myocardium from ischemia/reperfusion injury in rat hearts. Biochem. Biophys. Res. Commun. 338, 1118–1125 [DOI] [PubMed] [Google Scholar]
- 38. Kenny E. F., Talbot S., Gong M., Golenbock D. T., Bryant C. E., O'Neill L. A. (2009) MyD88 adaptor-like is not essential for TLR2 signaling and inhibits signaling by TLR3. J. Immunol. 183, 3642–3651 [DOI] [PubMed] [Google Scholar]
- 39. Jiang Z., Georgel P., Li C., Choe J., Crozat K., Rutschmann S., Du X., Bigby T., Mudd S., Sovath S., Wilson I. A., Olson A., Beutler B. (2006) Details of Toll-like receptor:adapter interaction revealed by germ-line mutagenesis. Proc. Natl. Acad. Sci. U.S.A. 103, 10961–10966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Ngo V. N., Young R. M., Schmitz R., Jhavar S., Xiao W., Lim K. H., Kohlhammer H., Xu W., Yang Y., Zhao H., Shaffer A. L., Romesser P., Wright G., Powell J., Rosenwald A., Muller-Hermelink H. K., Ott G., Gascoyne R. D., Connors J. M., Rimsza L. M., Campo E., Jaffe E. S., Delabie J., Smeland E. B., Fisher R. I., Braziel R. M., Tubbs R. R., Cook J. R., Weisenburger D. D., Chan W. C., Staudt L. M. (2011) Oncogenically active MYD88 mutations in human lymphoma. Nature 470, 115–119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Kagan J. C., Medzhitov R. (2006) Phosphoinositide-mediated adaptor recruitment controls Toll-like receptor signaling. Cell 125, 943–955 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.