Background: Nuclear translocation and activity of EGFR are correlated with radioresistance of cancer.
Results: PNPase, identified as a novel EGFR interacting partner, is inactivated by DNAPK-mediated Ser-776 phosphorylation and contributes to radiosensitivity of cancer.
Conclusion: An entirely new mechanism of nuclear EGFR in radioresistance is discovered.
Significance: This study provides new insight into resistance of breast cancer to radiotherapy.
Keywords: Epidermal Growth Factor Receptor (egfr), Nuclear Translocation, Phosphorylase, Phosphorylation, Protein-Protein Interactions, Breast Cancer, c-MYC, Nuclear EGFR, p-PNPase, Radioresistance
Abstract
Nuclear existence of epidermal growth factor receptor (EGFR) has been documented for more than two decades. Resistance of cancer to radiotherapy is frequently correlated with elevated EGFR expression, activity, and nuclear translocation. However, the role of nuclear EGFR (nEGFR) in radioresistance of cancers remains elusive. In the current study, we identified a novel nEGFR-associated protein, polynucleotide phosphorylase (PNPase), which possesses 3′ to 5′ exoribonuclease activity toward c-MYC mRNA. Knockdown of PNPase increased radioresistance. Inactivation or knock-down of EGFR enhanced PNPase-mediated c-MYC mRNA degradation in breast cancer cells, and also increased its radiosensitivity. Interestingly, the association of nEGFR with PNPase and DNA-dependent protein kinase (DNAPK) increased significantly in breast cancer cells after exposure to ionizing radiation (IR). We also demonstrated that DNAPK phosphorylates PNPase at Ser-776, which is critical for its ribonuclease activity. The phospho-mimetic S776D mutant of PNPase impaired its ribonuclease activity whereas the nonphosphorylatable S776A mutant effectively degraded c-MYC mRNA. Here, we uncovered a novel role of nEGFR in radioresistance, and that is, upon ionizing radiation, nEGFR inactivates the ribonuclease activity of PNPase toward c-MYC mRNA through DNAPK-mediated Ser-776 phosphorylation, leading to increase of c-MYC mRNA, which contributes to radioresistance of cancer cells.
Introduction
Epidermal growth factor receptor (EGFR)4 is a member of the ErbB family of receptor-tyrosine kinases (RTKs) comprised of four structurally related receptors, namely EGFR (ErbB1/HER1), ErbB2 (HER2/Neu), ErbB3/HER3, and ErbB4/HER4 (1). Overexpression of EGFR is a key frequent event of human tumors that acquire aggressive tumor behaviors, such as increased proliferation, metastasis, and therapeutic resistance (2, 3). Hence, EGFR is considered as an important therapeutic target for human cancers, and many anti-EGFR therapies have been developed with promising pre-clinical and clinical outcomes (4–6).
Emerging evidence demonstrates the existence of a new model of nuclear EGFR (nEGFR) signaling pathway, which subsequently controls gene expression and regulates different cellular functions (7–9). Various laboratories have shown that nEGFR correlates with patient survival in multiple cancer types, such as oropharyngeal cancer (10, 11), esophageal carcinoma (12), and breast cancer (9, 13). Moreover, resistance of cancer to radiotherapy is frequently correlated with elevated EGFR expression, activity, and nuclear translocation (14). While studies have shown that ionizing radiation (IR) triggers caveolin-1-driven internalization and nuclear transport of EGFR to activate DNA-dependent protein kinase (DNAPK) in response to DNA double-strand break (DSB) repair (15–17) and that blocking IR-induced nuclear transport of EGFR by a cyclooxygenase-2 (COX-2) inhibitor, celecoxib, induces tumor cell radiosensitization (18), the mechanism by which nEGFR drives radioresistance in cancers remains elusive.
Polynucleotide phosphorylase (PNPase) is an evolutionary conserved 3′-5′ exoribonuclease that uses the phosphorolytic mechanism to degrade RNA (19–22). It maps to 2p15–2p16.1 in which genomic alterations like deletion and amplification have been implicated in human cancers, such as diffuse large B-cell lymphoma (23) and various genetic disorders (24). PNPase consists of five motifs, including two catalytic RNase PH domains separated by an α-helical domain and two RNA-binding domains, KH and S1, at the C terminus (25, 26). A homotrimeric complex of PNPase forms a ring-shaped enzyme with the hexameric RNase PH domains that surround a central channel to accommodate a single-stranded RNA molecule (27). In the cytoplasm, PNPase has been shown to directly and specifically degrade c-MYC mRNA by its exoribonuclease property, resulting in cell growth arrest (28).
Here, we identify PNPase as a novel nEGFR-associated protein upon IR. Inactivation of EGFR attenuates the interaction of EGFR/PNPase/DNAPK and results in enhanced degradation of c-MYC mRNA by PNPase in breast cancer cells. nEGFR-mediated phosphorylation of PNPase at Ser-776 by DNAPK abolishes the ribonuclease activity of PNPase toward c-MYC mRNA and increases its mRNA level. The elevated level of c-MYC mRNA caused by inactivation of PNPase contributes to radioresistance of cancer cells.
EXPERIMENTAL PROCEDURES
Materials
The following antibodies and chemicals were purchased from commercial companies: anti-EGFR (Lab Vision); anti-PNPase (Santa Cruz Biotechnology); anti-DNAPK (Thermo); anti-α-tubulin (Sigma); anti-lamin B1 (Calbiochem); anti-phosphoserine (Upstate); anti-V5 (GeneTex); recombinant human EGF, DAPI (Sigma); 4-(3-chloroanilino)-6,7-dimethoxyquinazoline (AG1478) and 4,5-dimethoxy-2-nitrobenzaldehyde (DMNB) (Calbiochem). Iressa was a gift from Dr. Chia-Jui Yen (National Cheng Kung University Hospital, Tainan, Taiwan).
Cell Culture
Human cell lines were grown in DME/F12 media with 10% fetal bovine serum, 100 units/ml penicillin, and 100 μg/ml streptomycin in a humidified incubator with 5% CO2 at 37 °C.
Cell Extraction and Cell Lines
For total cell lysate extraction, cells were washed twice with phosphate buffer saline (PBS, containing 137 mm NaCl, 2.7 mm KCl, 10 mm Na2HPO4, 2 mm KH2PO4), and then lysed in RIPA Buffer (50 mm Tris at pH 7.5, 150 mm NaCl, 1 mm EDTA, 0.25% Na-deoxycholate, 1% Nonidet P-40, 1 mm NaF, 1 mm Na3VO4, 1 mm PMSF, 1 μg/ml aprotinin) by sonication. The soluble extraction was collected from the supernatant after centrifugation at 15,000 × g for 10 min. The cytosolic and nuclear fractions were extracted as follows: cells were lysed in buffer A (20 mm HEPES at pH 7, 10 mm KCl, 2 mm MgCl2, 0.5% Nonidet P-40, 1 mm NaF, 1 mm Na3VO4, 1 mm PMSF, 1 μg/ml aprotinin) on ice, ground in a glass dounce homogenizer, and centrifuged at 1500 × g for 10 min. The supernatant is the cytosolic fraction. The nuclear pellet was isolated and washed. The nuclei were lysed in NETN buffer (20 mm Tris at pH 8.0, 150 mm NaCl, 1 mm EDTA, 0.5% Nonidet P-40, 1 mm NaF, 1 mm Na3VO4, 1 mm PMSF, 1 μg/ml aprotinin) by sonication. Both clones of radio-sensitive (231S) and -resistant (231) MDA-MB 231 cell lines were obtained from The Center for Molecular Medicine, China Medical University Hospital, Taichung, Taiwan.
INM and NP Purification
INM and NP purification were performed as described previously with a slight modification (29). The isolated nuclei in the nuclear fractions extracted using cellular fractionation were resuspended in buffer A (0.25 m sucrose, 50 mm Tris-HCl, pH 7.4, 10 mm MgCl2, 1 mm dithiothreitol, and protease inhibitor mixture), incubated with 1% (w/v) sodium citrate at 4 °C with gentle rotation for 30 min, and centrifuged at 500 × g for 15 min. The pellet was resuspended in buffer A and digested with DNase I (250 mg/ml; Sigma) at 4 °C for 14 h. After centrifugation at 10,000 × g for 2 h, the supernatant was collected as an NP fraction, and the digested pellet was then recentrifuged at 100,000 × g for 20 min in a sucrose gradient to obtain purified INM fractions (30, 31). The membrane fraction collected at the 0.25–1.60 m sucrose interface contained the purified INM.
Immunoprecipitation and Western Blotting
Cell lysates were pre-cleared with protein A/G-agarose (Pierce) in NETN buffer with gentle mixing for 1 h at 4 °C. The pre-cleared lysate were added with appropriate antibody and incubated overnight at 4 °C. Afterward, protein A/G-agarose was added into the mixture and incubated with gentle agitation for another 2 h at 4 °C to capture antibody and its associated proteins. For immunoblotting (IB), cell lysates were boiled at 100 °C for 5 min, separated by SDS-PAGE, and transferred to a PVDF membrane. The membrane was blocked with 5% skim milk in PBST buffer (PBS containing 0.1% Tween-20) for 1 h at room temperature and then hybridized with primary antibody with gentle agitation overnight at 4 °C. After washing with PBST, the membrane was incubated with HRP-conjugated secondary antibody (Chemicon) for 1 h at room temperature. The immunoreactive band was visualized by the enhanced chemiluminescence (ECL) detection reagent (GE Healthcare). The immunoprecipitated nEGFR was confirmed by sequencing (supplemental Fig. S1).
Confocal Microscopy
Cultured cells were washed three times with PBS, fixed in 4% paraformaldehyde for 15 min, permeabilized with 0.5% Triton X-100 for 15 min, and incubated with 5% bovine serum albumin (BSA) for 1 h. Cells were then incubated with the primary antibodies overnight at 4 °C. Cells were washed with PBS and then further incubated with the appropriate secondary fluorescein isothiocyanate- or Texas red-conjugated antibody (Molecular Probes) (1:500 dilution) for 45 min at room temperature. Nuclei were stained with DAPI before mounting. Confocal fluorescence images were captured using a LSM 710 laser microscope (Zeiss).
RNA Extraction and Reverse Transcriptase-Polymerase Chain Reaction (RT-PCR)
Total RNA was extracted with TRIZOL reagent (Invitrogen) according to the manufacturer's instruction. The complementary DNA (cDNA) was synthesized from 5 μg of total RNA in a reaction mixture containing 2.5 μm oligo (dT) primer, 0.5 mm dNTP mixture, 200 units of SuperScript III reverse transcriptase, 40 units of RNaseOUT, an RNase inhibitor (all from Invitrogen). After incubation at 50 °C for 50 min, the reaction mixture was heat inactivated at 85 °C for 5 min and then treated with 2 units of RNase H at 37 °C for 20 min. The quantitative PCR (q-PCR) was performed by detection of the hydrolyzed fluorescent probes from Universal ProbeLibrary (UPL; Roche) using the LightCycler 480 equipment (Roche). Primers and UPL probes were designed using the ProbeFinder software. The primer sets and the matched UPL probe numbers are as following: c-MYC: 5′-CACCAGCAGCGACTCTGA-3′ (forward), 5′-GATCCAGACTCTGACCTTTTGC-3′ (reverse), and UPL probe 34; β-actin (ACTB): 5′-ATTGGCAATGAGCGGTTC-3′ (forward), 5′-GGATGCCACAGGACTCCAT-3′ (reverse), and UPL probe 11. The q-PCR was examined by incubating the cDNA in a reaction mixture containing, 0.5 μm of each primer, 0.1 μm UPL probe, and 1-fold concentration of Probes Master reagent (Roche). The amplification conditions were initial denaturation at 95 °C for 10 min, followed by 45 cycles of 95 °C for 10 s, 55 °C for 30 s, and 72 °C for 1 s. The fluorescent signal was detected at 72 °C step of each cycle. The relative quantification of interested gene was normalized by β-actin, and calculated by the value of cross-point (CP) in each fluorescence curve of each gene. For traditional PCR, the reaction mixture containing 2 μl of cDNA, 0.2 mm dNTP mixture, 2 μm of each primers, 1 unit of TaqDNA polymerase, and 1-fold concentration of ThermalPol Buffer (New England BioLabs) was started by denatured 95 °C for 5 min, followed by amplification of indicated cycles of 95 °C for 30 s, 55 °C for 30 s, and 72 °C for 30 s. The numbers of cycles for amplification of c-MYC and β-actin were 35 and 25, respectively. The primer sequences for these genes are as follows: c-MYC: 5′-GGTCCTCAAGAGGTGCCACG-3′ (forward), 5′-TTACGCACAAGAGTTCCGTAGCTG-3′ (reverse), and β-actin: 5′-GCACTCTTCCAGCCTTCCTTCC-3′ (forward), 5′-TCACCTTCACCGTTCCAGTTTTT-3′ (reverse).
Gene Knock-down by Short-hairpin RNA (shRNA)
Knock-down of genes were performed with the specific shRNAs delivered by the lentivirus system from National RNAi Core Facility (Academia Sinica) according to the protocol. Briefly, to generate the lentivirus containing specific shRNA, 293T cells were co-transfected with 2.25 μg of pCMV-ΔR8.91 plasmid harboring Gag and Pol genes, 0.25 μg of pMD.G plasmid containing VSV-G gene for expression of envelope glycoprotein, and 2.5 μg of pLKO.1 plasmid bearing the specific shRNA for 16 h, and then cultured in growth medium containing 1% BSA for another 24 h. The cultured medium containing lentivirus was collected and stored at −80 °C as aliquots for further use. To deliver the specific shRNA construct, approximate 80% confluent cells were infected with the lentivirus bearing specific shRNA in growth medium containing 8 μg/ml polybrene and incubated at 37 °C for 24 h. Afterward, cells were sub-cultured and selected with 4 μg/ml puromycin. The specific shRNAs used are as below, EGFR: TRCN0000039633 (shEGFR-1) and TRCN0000039634 (shEGFR-2) corresponding to the sequences, 5′-GCTGAGAATGTGGAATACCTA-3′ and 5′-GCTGGATGATAGACGCAGATA-3′, respectively; PNPase: TRCN0000035908 (shPNPase-2) and TRCN0000312599 (shPNPase-4) corresponding to the sequences, 5′-GCAAGAGACTTCATTACTGAA-3′ and 5′-GCAAGAGACTTCATTACTGAA-3′, respectively; DNAPK: TRCN0000006257 (shDNAPK-1) and TRCN0000006259 (shDNAPK-2) corresponding to the sequences, 5′-CCAGTGAAAGTCTGAATCATT-3′ and 5′-GCAGCCTTATTACAAAGACAT-3′, respectively; c-MYC: TRCN0000039642 (shMYC-2) and TRCN0000174055 (shMYC-5) corresponding to the sequences, 5′-CCTGAGACAGATCAGCAACAA-3′ and 5′-GAATGTCAAGAGGCGAACACA-3′, respectively. The shRNA construct against luciferase (shLuc), TRCN0000072244 referring to the sequence, 5′-ATCACAGAATCGTCGTATGCA-3′ was used as negative control.
Mass Spectrometry
Tandem mass (MS/MS) experiments were performed with an LTQ-FT ICR MS (Thermo Electron) equipped with a nanoelectrospray ion source (New Objective), an Agilent 1100 Series binary HPLC pump (Agilent Technologies) and a Famos autosampler (LC Packings). Tryptic peptide mixtures were injected at a 10 μl/min flow rate into a self packed pre-column with reverse phase C18 nano-column (75 μm I.D. × 200 mm) that used Magic C18AQ resin (particle size, 5 μm; pore size, 200 Å; Michrom Bioresources). The analytic program was set at a linear gradient from 10 to 50% buffer B with a 60 min running cycle and a split flow rate 300 nl/min. The full-scan survey MS experiment (m/z 320–2000) was executed in FT ICR MS with mass resolution of 100,000 at m/z 400. The top ten most abundant multiply charged ions detected in the scan, which is above a minimum threshold of 1,000 counts, were sequentially isolated for MS/MS by LTQ. Singly charged ions were rejected for MS/MS sequencing.
Plasmids Construction, Protein Expression, and Protein Purification
Wild-type PNPase was cloned into the pcDNA3.1D/V5-His-TOPO vector (Invitrogen). PNPaseS776A or PNPaseS776D mutant was generated by QuikChange Site-directed Mutagenesis kit (Stratagene) according to the instruction manual. To purify recombinant proteins by the bacteria expression system, each construct was subcloned into pET29a vector containing His-tag (Novagen). The constructs were transformed into BL21 (DE3) competent cells. The recombinant proteins were induced by 0.2 mm IPTG in bacteria culture. After lysing the bacteria, the recombinant proteins were purified with Ni-NTA resin (Invitrogen).
In Vitro Kinase Assay
DNAPK was isolated by immunoprecipitation with specific antibody from MDA-MB 468 cells. Each recombinant PNPase protein was incubated with immunoprecipitated DNAPK in the kinase buffer (60 mm HEPES at pH 7.5, 5 mm MgCl2, 5 mm MnCl2, 3 μm Na3VO4, 1.25 mm DTT, 200 μm ATP) at 30 °C for 30 min. The mixture was then separated on SDS-PAGE. The serine phosphorylation was detected by Western blotting with anti-phosphoserine antibody.
In Vitro Translation and in Vitro mRNA Degradation Assay (PNPase Activity Assay)
The in vitro translation was performed with the TNT Quick-coupled Transcription/Translation system (Promega) according to the manufacturer's instruction. In brief, V5-tagged pPNPase, pPNPaseS776A, or pPNPaseS776D plasmid was individually incubated with the reaction mixture containing 20 μm methionine and 1× TNT Quick Master Mix at 30 °C for 90 min. The in vitro mRNA degradation assay was carried out as previously described (32) with a slight modification. Five micrograms of total RNA from MDA-MB 468 cells were incubated with 5 μl of each in vitro-translated or immunoprecipitated PNPase protein in the buffer containing 50 mm Tris-HCl (pH 7.5), 5 mm potassium phosphate (pH 7.5), 1 mm ATP, 5 mm DTT, 5 mm MgCl2, 80 mm KCl, and 100 ng/μl bovine serum albumin at 37 °C for 0 to 2 h. After incubation, the RNA was purified and the amount of c-MYC mRNA was determined by RT-PCR and q-PCR as aforementioned. The PNPase activity was measured by the relative mRNA intensity of c-MYC/β-actin. The activity of PNPasae in normal cell culture condition was defined as 100%.
Cell Viability Assay
Cell viability was determined by WST-1 (4-[3-(4-Iodophenyl)-2-(4-nitrophenyl)-2H-5-tetrazolio]-1,3-benzene disulfonate) assay (Roche). Cells were irradiated with or without indicated dosage of IR. After culturing for another 2 days, one-tenth volume of WST-1 was added at 4 h before harvest, and the absorbance was detected at 450 nm. Cell viability was normalized by the absorbance from the cells without IR exposure.
RESULTS
Nuclear EGFR Associates with PNPase
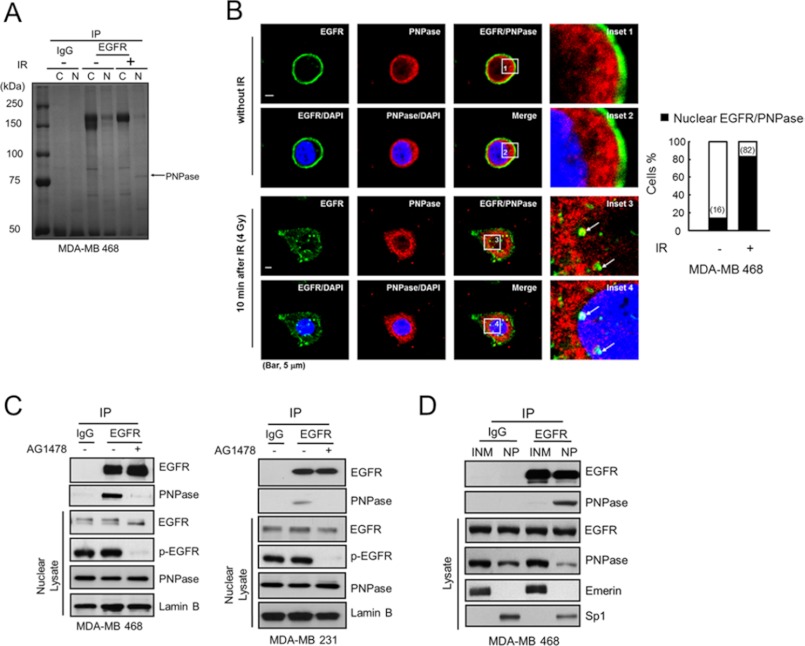
To address the roles of nuclear EGFR (nEGFR) in response to radioresistance in breast cancer, we compared the interactome of EGFR upon ionizing radiation (IR) from cytosolic and nuclear lysates of MDA-MB 468 cells by immunoprecipitation with an anti-EGFR monoclonal antibody. A nuclear EGFR-associated protein around 75 kDa was increased after IR exposure and identified as PNPase by electrospray ionization tandem mass (MS/MS) spectrometry analysis (Fig. 1A). Moreover, the co-localization of EGFR and PNPase in the nucleus was also increased after IR exposure (Fig. 1B; white spots in inset 4). To further confirm the nuclear location of EGFR-PNPase complexes, we examined the sequential photosections of a nucleus and showed that the EGFR-PNPase complexes were clearly detected in middle sections in MDA-MB 468 cells (planes 10–14, supplemental Fig. S2). We further validated their interaction by immunoblotting against PNPase in the EGFR-immunoprecipitated complexes. As shown in Fig. 1C, the interaction between EGFR and PNPase in the nucleus of MDA-MB 468, MDA-MB 231, and A431 cells (Fig. 1C and supplemental Fig. S3) was reduced by the addition of AG1478, an EGFR tyrosine kinase inhibitor (TKI). In addition, we further demonstrated that nEGFR associated with PNPase in the non-membrane-embedded form as their interaction occurred mainly in the nucleoplasm (NP) but not the inner nuclear membrane (INM) fraction (Fig. 1D). Taken together, these results suggest that EGFR and PNPase interact mainly in the NP, and their interaction requires the tyrosine kinase activity of EGFR and is enhanced upon IR exposure.
FIGURE 1.
Nuclear EGFR interacts with PNPase. A, MDA-MB 468 cells were treated with or without 4 Gy ionizing radiation (IR). After 10 min, the cytosolic or nuclear lysate was extracted and immunoprecipitated with IgG or anti-EGFR antibody and separated by SDS-PAGE. The major band near 75k Da as indicated was identified as PNPase by tandem mass (MS/MS) spectrometry. B, left, co-localization of EGFR (green) and PNPase (red) in the nucleus (blue) at 10 min post-IR was examined by immunofluorescence staining and observed under a confocal microscopy. The arrows in insets 3 and 4 indicate the colocalized EGFR and PNPase in the nucleus, which is shown in yellow in the merged image. Scale bar, 5 μm. Right, bar graph shows the percentage of the 50 counted cells with colocalized EGFR and PNPase. C, interaction between EGFR and PNPase in nuclear lysate from MDA-MB 468 and MDA-MB 231 cells were verified by IP (immunoprecipitation)/IB (immunoblotting) with anti-EGFR/anti-PNPase antibody, respectively. Cells were treated with or without 10 μm AG1478 for 12 h, and the interaction of EGFR and PNPase in the nucleus was examined. D, purified inner nuclear membrane (INM) and nucleoplasm (NP) portions were immunoprecipitated using the indicated antibodies followed by IB in MDA-MB 468 cells. Eemerin, and Sp1 were used as markers for the INM, and NP, respectively. IP performed with IgG was used as a negative control.
DNAPK Phosphorylates PNPase at Ser-776
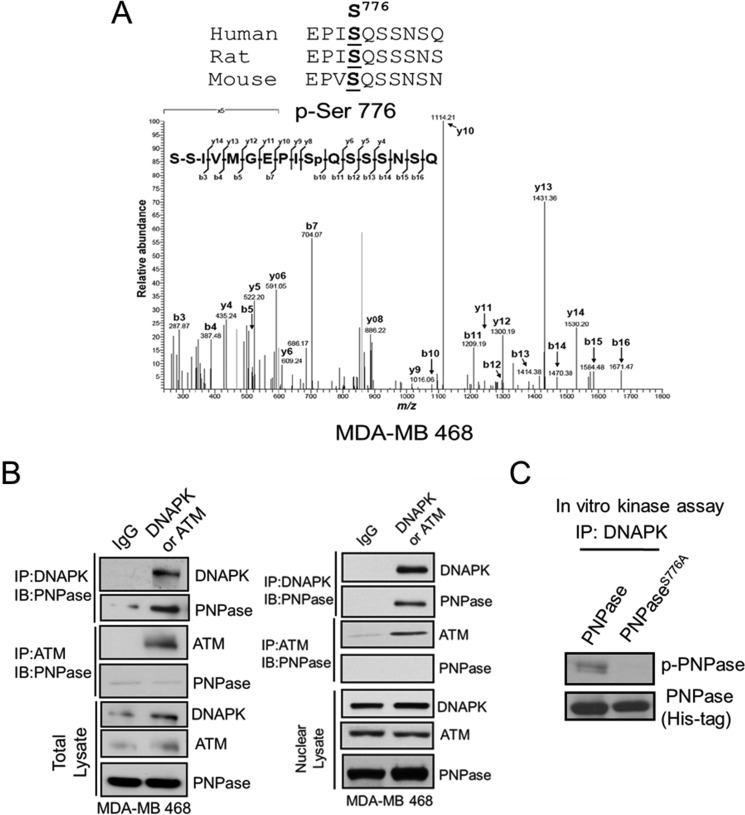
From above, we showed that nEGFR interacts with PNPase. Next, to investigate if and how EGFR regulates PNPase activity, we first determined whether EGFR phosphorylates PNPase. The endogenous PNPase protein was isolated by immunoprecipitation with a monoclonal anti-PNPase antibody from the nucleus of MDA-MB 468 cells, and its phosphorylation sites in vivo was identified by tandem mass (MS-MS) spectrometric analysis. Although no tyrosine phosphorylation was identified in vivo, we reproducibly identified specific peptides containing phosphorylated serine residue at Ser-776 of PNPase, and this serine residue was conserved among mouse, rat, and human (Fig. 2A), suggesting that Ser-776 phosphorylation of PNPase is an evolutionarily conserved event and associated with important functions. We then used the NetworKIN software (33) to predict the potential kinase toward Ser-776 of PNPase and found that both ATM and DNAPK are possible candidates. To determine whether ATM or DNAPK phosphorylates Ser-776 of PNPase, we carried out immunoprecipitation and showed that DNAPK but not ATM associated with PNPase in the nucleus (Fig. 2B). Moreover, to further validate that Ser-776 is the substrate for DNAPK phosphorylation, we created a S776A mutant of PNPase. We found that only the wild-type but not the S776A mutant PNPase can be phosphorylated by DNAPK (Fig. 2C), indicating that DNAPK has specific serine kinase activity toward Ser-776 of PNPase.
FIGURE 2.
Phosphorylation of PNPase at Ser-776 is mediated by DNAPK. A, PNPase in the nuclear lysate was isolated by IP from MDA-MB 468 cells. The phosphorylated residue of PNPase was identified at Ser-776 by tandem mass (MS/MS) spectrometric analysis. The top panel shows the alignment of the flanking regions of Ser-776 of PNPase among different species. B, total and nuclear lysates were immunoprecipitated with IgG, anti-DNAPK (top panel) or anti-ATM (middle panel) antibody, and PNPase in the IP complex was determined by IB. The expression of indicated protein in lysate was also examined by IB (bottom panel). C, purified recombinant His-tagged PNPase or PNPaseS776A proteins were incubated with immunoprecipitated DNAPK and applied to in vitro kinase assay. The reaction mixture was separated by SDS-PAGE, followed by IB with anti-p-Ser antibody (top). The Coomassie Blue-stained gel (bottom) served as loading control.
EGFR Regulates DNAPK-mediated Serine Phosphorylation of PNPase upon IR but Not Nuclear Translocation of PNPase
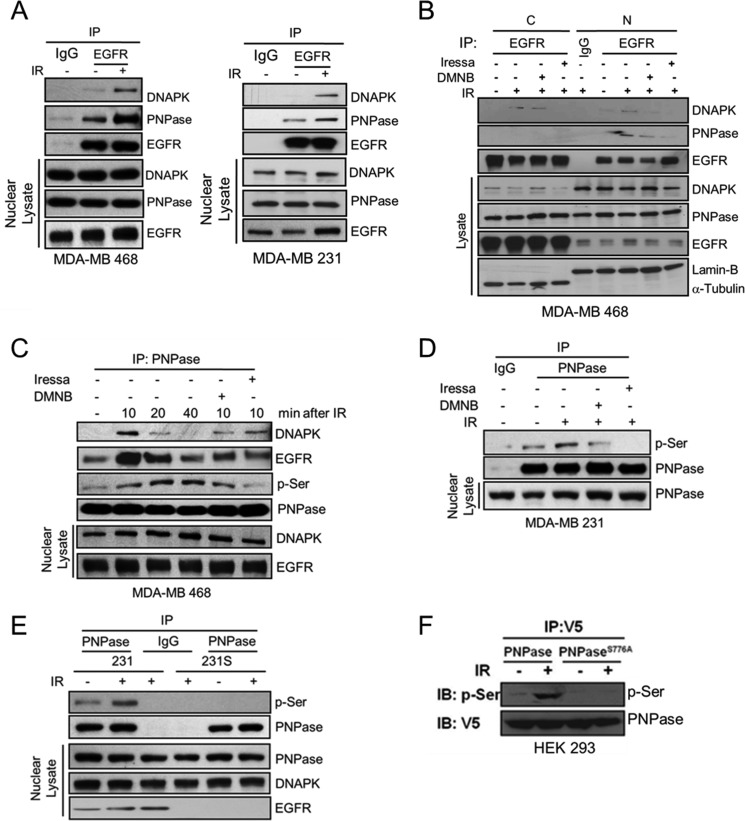
It has been reported that IR triggers caveolin-1-driven internalization and nuclear transport of EGFR to activate DNAPK (15, 16). Thus, we asked whether EGFR affects DNAPK-mediated serine phosphorylation of PNPase after IR. We exposed cells to 4 Gy IR for 10 min and then collected cell lysate for immunoprecipitation followed by immunoblotting (Fig. 3A and supplemental Fig. S4). As shown in Fig. 3, A and B, IR significantly enhanced the association of PNPase with both EGFR and DNAPK in the nucleus. However, when we added the inhibitor of DNAPK (DNMB) or EGFR (Iressa), the IR-enhanced nuclear interaction was reduced but not the nuclear translocation of PNPase (Fig. 3B), indicating that the association of the PNPase/EGFR or PNPase/DNAPK complex requires the kinase activity of EGFR or DNAPK and is mainly in the nucleus. Moreover, serine phosphorylation of PNPase was significantly increased upon IR exposure but attenuated by Iressa or DNMB (Fig. 3, C and D). We used a pair of radio-sensitive (231S) and -resistant (231) MDA-MB 231 cell lines to further show that serine phosphorylation of PNPase was induced by IR in radio-resistant MDA-MB 231 (231; Fig. 3E). Moreover, IR-induced serine phosphorylation was also detected in the ectopically expressed V5-tagged wild-type PNPase but not in the S776A nonphosphorylatable mutant (Fig. 3F). Altogether, these results indicated that IR triggers the association of EGFR, DNAPK, and PNPase, leading to phosphorylation of PNPase at Ser-776 by EGFR-mediated activation of DNAPK.
FIGURE 3.
EGFR regulates DNAPK-mediated serine phosphorylation of PNPase at Ser-776 upon ionizing radiation. A, MDA-MB 468 and MDA-MB 231 cells were irradiated with or without 4 Gy IR. After 10 min, nuclear lysates were harvested and individually immunoprecipitated with antibodies against IgG or PNPase, followed by SDS-PAGE separation and IB (upper panel). The endogenous expression levels of the indicated protein were examined by IB (lower panel). B, MDA-MB 468 cells were irradiated with or without 4 Gy IR in the presence or absence 10 μm DMNB or 10 μm Iressa. After indicated treatment, cytosolic (C) and nuclear (N) lysates were harvested and individually immunoprecipitated with antibody against IgG or EGFR, followed by SDS-PAGE separation and IB (upper panel). The endogenous levels of DNAPK and PNPase are shown in the lower panel. C, same as B, at the indicated time interval, The nuclear lysates of MDA-MB 468 cells were harvested and individually IP with antibody against IgG or PNPase, followed by SDS-PAGE separation and IB (upper panel). The endogenous levels of DNAPK and EGFR were shown in the lower panel. D, MDA-MB 231 cells were irradiated with or without 4 Gy IR in the presence or absence 10 μm DMNB or 10 μm Iressa. After 10 min, nuclear lysates were harvested and individually immunoprecipitated with antibody against IgG or PNPase, followed by IB for phospho-serine (p-Ser) (upper panel). The endogenous expression level of PNPase was shown in the lower panel. E, MDA-MB 231 radio-resistant (231) and radio-sensitive (231S) cells were irradiated with or without 4 Gy IR. After 10 min, nuclear lysates were harvested and individually immunoprecipitated with antibody against IgG or PNPase, followed by IB for phospho-serine (p-Ser) (upper panel). The endogenous expression level of PNPase was shown in the lower panel. F, HEK-293 cells were transfected with plasmid harboring V5-tagged PNPase or PNPaseS776A. These transfectants were exposed to 0 or 4 Gy IR. After 10 min, cell lysate was extracted and immunoprecipitated with anti-V5 antibody, followed by IB for p-Ser (upper panel) or V5 (lower panel).
Phosphorylation of PNPase at Ser-776 Abolishes Its Ribonuclease Activity
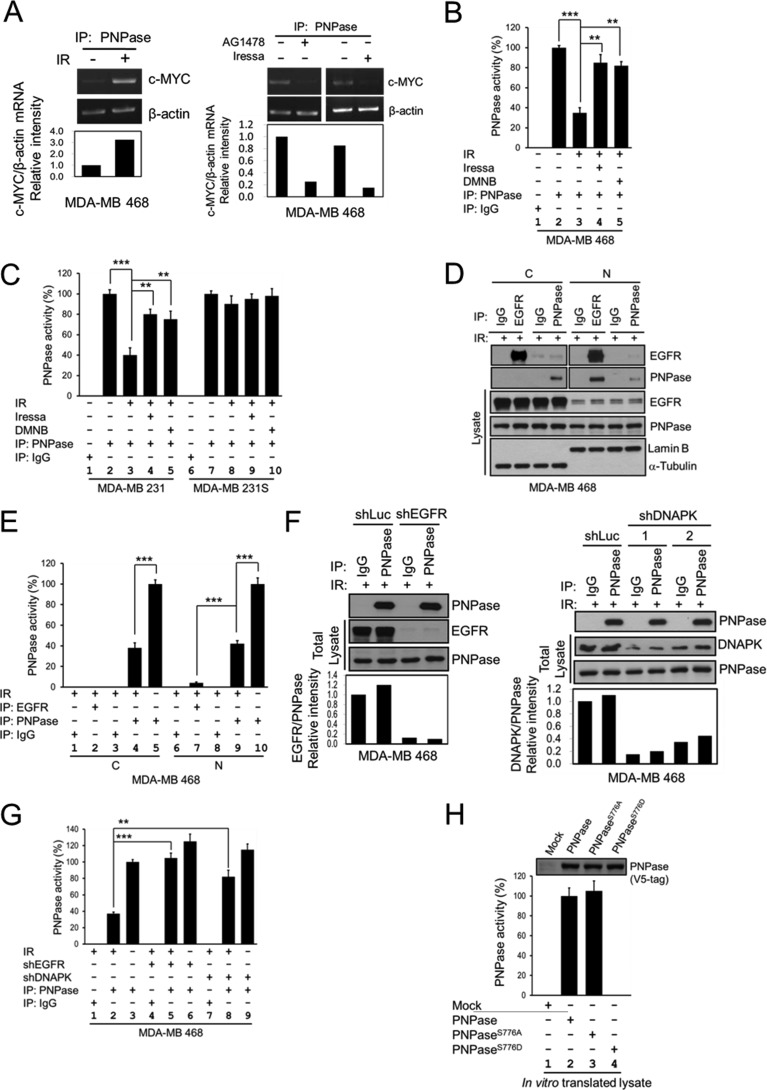
The 3′-5′ exoribonuclease activity of PNPase is known to degrade c-MYC mRNA (28). Therefore, we investigated whether phosphorylation at Ser-776 of PNPase affects its ribonuclease activity toward c-MYC mRNA. To this end, we first established an in vitro c-MYC mRNA degradation assay of PNPase to measure the PNPase activity. The PNPase activity was significantly decreased in IR-treated cells (Fig. 4A, left panel; Fig. 4B, lanes 2 and 3) but was restored by the addition of EGFR (Iressa) or DNAPK (DMNB) inhibitor (Fig. 4A, right panel; Fig. 4B, lanes 4 and 5). We further compared the PNPase activity in the radio-sensitive and -resistant MDA-MB 231 cell lines (Fig. 4C). Similarly, inhibition of PNPase activity by IR was restored by the addition of Iressa or DMNB in radio-resistant MDA-MB 231 cells (Fig. 4C, lanes 4 and 5). However, in MDA-MB 231 radio-sensitive cell line, the PNPase activity was not affected by IR, Iressa, or DMNB (Fig. 4C, lanes 8–10).
FIGURE 4.
Phosphorylation of PNPase at Ser-776 abolishes its ribonuclease activity. MDA-MB 468 cells were exposed with or without 4 Gy IR (A, left panel), or treated with or without 10 μm AG1478 or 10 μm Iressa for 12 h (A, right panel). PNPase was isolated by IP with anti-PNPase antibody from total lysates. The degradation of c-MYC mRNA after each treatment was determined by the in vitro c-MYC mRNA degradation assay with each immunoprecipitated PNPase. B, MDA-MB 468 cells were exposed with or without 4 Gy IR, or treated with or without 10 μm DMNB or 10 μm Iressa for 12 h. PNPase was isolated by individual IP with antibody against IgG or PNPase from total lysates. The ribonuclease activity of PNPase toward c-MYC mRNA after each treatment was determined by the in vitro mRNA degradation assay with each immunoprecipitated IgG or PNPase. C, MDA-MB 231 radio-resistant (231) and radio-sensitive (231S) cells were exposed with or without 4 Gy IR, or treated with or without 10 μm DMNB or 10 μm Iressa for 12 h. PNPase was isolated by individually IP with antibody against IgG or PNPase from total lysates. The ribonuclease activity of PNPase toward c-MYC mRNA after each treatment was determined by the in vitro mRNA degradation assay. D, MDA-MB 468 cells were exposed with 4 Gy IR. PNPase was isolated by individual IP with antibody against IgG or PNPase or EGFR from cytosolic (C) and nuclear (N) lysates. E, ribonuclease activity of PNPase or bound to EGFR toward c-MYC mRNA after each treatment was determined by the in vitro mRNA degradation assay by using same amount of immunoprecipitated PNPase. F, MDA-MB 468 cells were infected with lentivirus containing shRNAs against luciferase (Luc, as negative control), EGFR (shEGFR-1), or DNAPK (shDNAPK-1 and shDNAPK-2) to knock down specific gene expression. PNPase was isolated by individually IP with antibody against IgG or PNPase from total lysates. G, ribonuclease activity of PNPase toward c-MYC mRNA after each treatment was determined by the in vitro mRNA degradation assay by using same amount of immunoprecipitated PNPase. H, each PNPase variant was in vitro translated by TNT-coupled reticulocyte lysate system, and the expression of each recombinant PNPase protein was determined by IB (top). The ribonuclease activity of each in vitro translated PNPase was examined as aforementioned (lower panel). The PNPase activity was measured by the relative mRNA intensity of c-MYC/β-actin through qPCR. The activity of PNPase in normal cell culture condition or wild type from in vitro translated will be defined as 100%. ***, p < 0.005; **, p < 0.01 by t test.
As shown in Fig. 4D, a majority of nuclear PNPase can be pulled down by EGFR after IR exposure, which rendered the protein less active as indicated by the loss of PNPase activity (Fig. 4E). These results suggest that PNPase activity is negatively regulated by both EGFR and DNAPK. We also validated the effect of EGFR and DNAPK on the PNPase activity by using shEGFR and shDNAPK (Fig. 4F). We found that knocking down EGFR and DNAPK restored the activity of PNPase after IR (Fig. 4G, lanes 5 and 8 versus 2). We further constructed and expressed V5-tagged wild-type, S776A (nonphosphorylatable mutant), and S776D (phospho-mimetic mutant) of PNPase. We first validated the localization of these mutants and showed that each of the PNPase variant can be detected in both cytosolic and nuclear fractions. The distribution among all PNPase variants was similar, suggesting that phosphorylation at Ser-776 of PNPase does not alter its localization within the cells. To determine whether Ser-776 phosphorylation affects the ribonuclease activity of PNPase, we expressed the recombinant protein of each PNPase variant by in vitro translation and then applied it to an in vitro RNA degradation assay. We showed that all these constructs gave rise to proteins of the expected molecular weight with similar expression levels Fig. 4H, top; supplemental Fig. S5B, top). As expected, the nonphosphorylatable S776A mutant exhibited similar activity compared with wild-type PNPase whereas the phospho-mimetic S776D mutant inactivated the ribonuclease activity of PNPase (Fig. 4H). Incubation of RNA with the in vitro-expressed wild-type PNPase or PNPaseS776A protein resulted in significant degradation of c-MYC mRNA but not with control (mock) or PNPaseS776D (supplemental Fig. S5B, bottom). These results indicated that phosphorylation at Ser-776 of PNPase is important for its ribonuclease activity.
EGFR Negatively Regulates the Function of PNPase on c-MYC mRNA Degradation
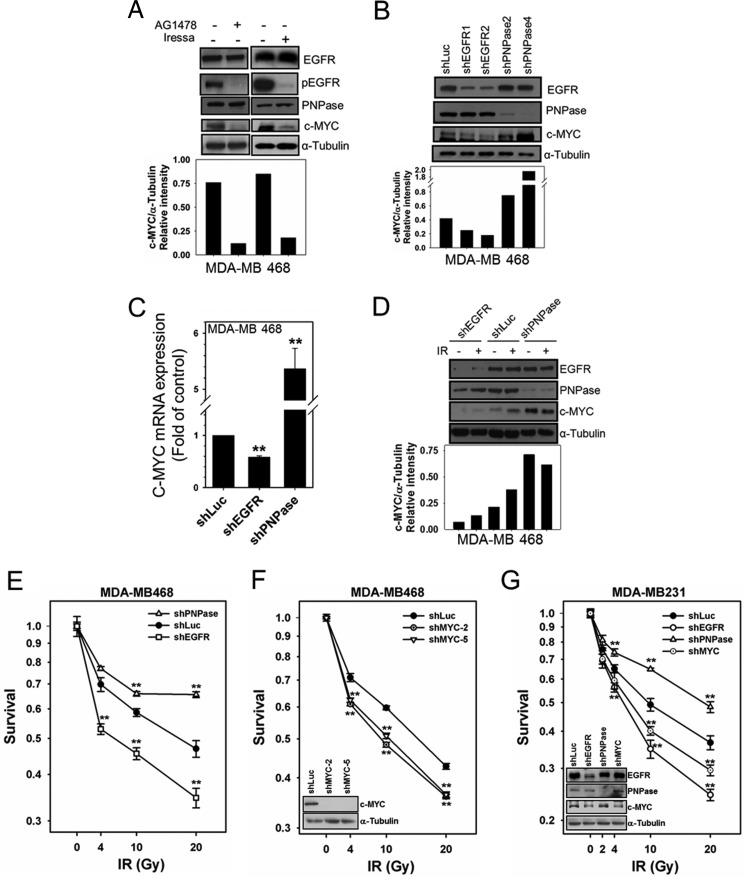
To determine whether EGFR regulates the function of PNPase, we suppressed tyrosine kinase activity of EGFR by TKIs or knocked down its expression by shRNA and determined the expression of PNPase and its downstream target, c-MYC. Inactivation of EGFR by its TKIs, AG1478, or Iressa, did not affect the expression of PNPase but reduced the level of its downstream target, c-MYC (Fig. 5A). Moreover, knocking down EGFR decreased c-MYC protein (Fig. 5B) and mRNA (Fig. 5C) levels without altering the level of PNPase protein. On the other hand, knocking down PNPase significantly increased both protein (Fig. 5B) and mRNA (Fig. 5C) levels of c-MYC. These results suggest that EGFR negatively regulates PNPase activity toward c-MYC mRNA without affecting the protein expression of PNPase.
FIGURE 5.
PNPase function is negatively regulated by EGFR. A, MDA-MB 468 cells were treated with or without 10 μm AG1478 or 10 μm Iressa for 12 h. The expressions of c-MYC and indicated proteins were examined. MDA-MB 468 cells were infected with lentivirus containing shRNAs against luciferase (Luc, as negative control), EGFR (shEGFR-1 and shEGFR-2), PNPase (shPNPase-2 and shPNPase-4) or c-MYC (shMYC-2 and shMYC-5) to knock down specific gene expression. The protein (B) and mRNA (C; shEGFR-1 and shPNPase-4) expression of the PNPase target, c-MYC, were determined by IB and qRTPCR, respectively. D, each shRNA-infected cell line (shEGFR-1 and shPNPase-4) was irradiated with or without 4 Gy IR. The expression of c-MYC and indicated proteins were examined by IB. E to G, survival of indicated shRNA-infected cells was determined after 2 days post-irradiation in MDA-MB 468 cells (E; shEGFR-1 and shPNPase-4 and F; shMYC-2 and shMYC-5) and in MDA-MB 231 cells (G; shEGFR-1, shPNPase-4 and shMYC-5). The blots in F and G represent the indicated protein expression in each shRNA-infected cells. **, p < 0.01 by t test.
Knock-down of PNPase Enhances c-MYC Level and Enhances Radioresistance in Breast Cancer Cells
We next determined whether PNPase is involved in modulation of radiosensitivity. In MDA-MB 468 breast cancer cells, we found that IR enhanced the protein level of c-MYC without altering the level of PNPase (middle two lanes, Fig. 5D), suggesting that IR repressed PNPase activity toward c-MYC mRNA. Knocking down PNPase enhanced radioresistance of both MDA-MB 468 and MDA-MB 231 breast cancer cells (Fig. 5, E and G). In contrast, knocking down EGFR decreased the levels of c-MYC protein (Fig. 5D) and increased radiosensitivity of these cells (Fig. 5, E, G, and supplemental Fig. S6). Based on our findings and previous studies (34, 35), radioresistance via silencing of PNPase is likely due to elevated levels c-MYC. Indeed, we further showed that knocking down c-MYC increased the radiosensitivity of these cells after IR (Fig. 5, F and G).
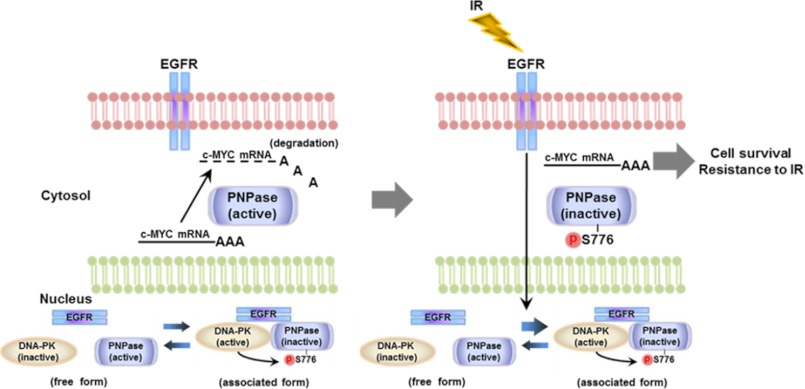
In summary, we provided evidence in this study to demonstrate a mechanism of EGFR-mediated radioresistance through inactivation of PNPase. IR triggers EGFR translocation into the nucleus. Then, nEGFR associates with DNAPK and activates its serine kinase activity toward Ser-776 of PNPase. This in turn represses the ribonuclease activity of PNPase toward c-MYC mRNA, resulting in cell survival and resistance to IR (Fig. 6).
FIGURE 6.
The proposed model of EGFR in the regulation of PNPase. A schematic illustrating the regulation of PNPase activity toward c-MYC mRNA by EGFR and DNAPK. Left: in normal condition, PNPase actively regulates the homeostasis of c-MYC mRNA. Right: Exposure to IR increases translocation of EGFR into the nucleus and increases its association with DNAPK and PNPase. EGFR then activates the serine kinase activity of DNAPK toward Ser-776 of PNPase to suppress its ribonuclease activity, leading to increase of c-MYC mRNA. The elevated c-MYC contributes to cell survival and radioresistance.
DISCUSSION
It has been showed that IR induces the kinase activity of EGFR, which contributes to protection of cancer cells from killing by radiotherapy (36, 37). Somatic mutations in the tyrosine kinase domain of EGFR, which is defective in nuclear translocation, abrogate EGFR-mediated radioresistance in non-small cell lung carcinoma (38). IR-induced nuclear translocation of EGFR through protein kinase C- (39) and caveolin-1-(16) dependent mechanisms activates DNAPK (15) to launch DNA DSB repair, leading to radioprotection of cancer cells. Treatment of the anti-EGFR antibody, C225 (cetuximab), blocks nuclear translocation of EGFR and consequently inhibits DNAPK activity, resulting in increased radiosensitivity of cancer (40). Deletion of nuclear localization signal sequence of EGFR impairs its nuclear localization and association with DNAPK, leading to reduced DNAPK kinase activity and deficient DNA repair (17). Altogether, IR-induced nuclear transport of EGFR is essential for activation of DNAPK activity and DNA DSB repair. In this study, we uncover a novel role of IR-induced nEGFR/DNAPK complex in radioresistance through association with and inactivation of PNPase exoribonuclease activity (Fig. 4).
Although PNPase has been known to have a 3′-5′ exoribonuclease activity (19–22), its regulation upon IR is still unclear. The study of gene structure of PNPase revealed that its promoter contains an interferon stimulated regulatory element (ISRE) and one SP1 element, which are crucial for the expression of PNPase in human melanoma HO-1 cells. Treating cells with interferon results in transcriptional up-regulation of PNPase through JAK/STAT signaling pathway (41). Here, we identified DNAPK, which is activated by IR-induced nEGFR, as a serine kinase toward Ser-776 of PNPase (Fig. 2). IR induced Ser-776 phosphorylation of PNPase without altering its protein level (Fig. 3, C–F), and IR-induced phosphorylation could be abolished by addition of a clinical used EGFR TKI, Iressa (Fig. 3, C and D). Phosphorylation at Ser-776 of PNPase disrupts its ribonuclease activity on c-MYC mRNA degradation without alteration of its cellular localization (supplemental Fig. S5A). These results provide new regulatory mechanism of PNPase through post-translational suppression via Ser-776 phosphorylation by nEGFR activated DNAPK upon IR.
Human PNPase was originally cloned from a subtracted cDNA library prepared from cells with terminal differentiation and senescence, suggesting that PNPase may play important roles in these physiological functions (21). Overexpression of PNPase results in suppression of cell growth by degradation of c-MYC mRNA (28) through the RNase PH domain of PNPase (32). c-MYC has been characterized as a proto-oncogene, of which expression is crucial in cell growth and malignancy of cancers including breast cancer (42, 43), and up-regulated expression or gene amplification of c-MYC upon IR is observed in different types of cancers, such as breast cancer (44–46) and acute myeloid leukemia (47). Moreover, elevated protein levels of c-MYC protects cancer cells from radiotherapy (34, 35), which could be a result of direct activation of transcription of a DNA double-stranded break repair gene, NBS1, by c-MYC protein (48).
Here, we identified a novel nEGFR-associated protein, PNPase, which is inactivated via Ser-776 phosphorylation by DNAPK. Upon IR, nEGFR-activated DNAPK suppresses PNPase ribonuclease activity toward c-MYC mRNA. The up-regulated c-MYC mRNA resulted from inactivation of PNPase may contribute to nEGFR-mediated radioresistance (Fig. 6). Our studies provide valuable insights on how nEGFR mediates cell survival in response to radiation in breast cancer through PNPase.
Supplementary Material
Acknowledgments
We thank the National RNAi Core Facility (Academia Sinica, Taipei, Taiwan) for providing the shRNAs and Dr. Jennifer L. Hsu for editorial assistance.
This study was supported, in whole or in part, by the following grants: the National Institutes of Health (RO1 CA109311, PO1 CA099031), The University of Texas MD Anderson-China Medical University and Hospital Sister Institution Fund (DMR-101-115, to M.-C. H. and Y.-L. Y.), Cancer Research Center of Excellence (DOH101-TD-C-111-005, Taiwan), Private University grant (NSC99-2632-B-039-001-MY3, Taiwan), Program for Stem Cell and Regenerative Medicine Frontier Research (NSC100-2321-B-039-002, Taiwan), International Research-Intensive Centers of Excellence in Taiwan (NSC101-2911-I-002-303, Taiwan), Personal Research Grant for New Faculty (to R.-H. C., CMU100-N2-09; Taiwan), and Project for Junior Researcher (to R.-H. C., NSC 101-2311-B-039-001, Taiwan).

This article contains supplemental Figs. S1–S6.
- EGFR
- epidermal growth factor receptor
- nEGFR
- nuclear EGFR
- RTK
- receptor-tyrosine kinase
- PNPase
- polynucleotide phosphorylase
- IR
- ionizing radiation
- DNAPK
- DNA-dependent protein kinase
- DSB
- double-strand break
- COX-2
- cyclooxygenase-2
- AG1478
- 4-(3-chloroanilino)-6,7-dimethoxyquinazoline
- DMNB
- 4,5-dimethoxy-2-nitrobenzaldehyde
- WST-1
- (4-[3-(4-Iodophenyl)-2-(4-nitrophenyl)-2H-5-tetrazolio]-1,3-benzene disulfonate).
REFERENCES
- 1. Citri A., Yarden Y. (2006) EGF-ERBB signaling: towards the systems level. Nat. Rev. Mol. Cell Biol. 7, 505–516 [DOI] [PubMed] [Google Scholar]
- 2. Navolanic P. M., Steelman L. S., McCubrey J. A. (2003) EGFR family signaling and its association with breast cancer development and resistance to chemotherapy (Review). Int. J. Oncol. 22, 237–252 [PubMed] [Google Scholar]
- 3. Dutta P. R., Maity A. (2007) Cellular responses to EGFR inhibitors and their relevance to cancer therapy. Cancer Lett. 254, 165–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Huang S. M., Harari P. M. (2000) Modulation of radiation response after epidermal growth factor receptor blockade in squamous cell carcinomas: inhibition of damage repair, cell cycle kinetics, and tumor angiogenesis. Clin. Cancer Res. 6, 2166–2174 [PubMed] [Google Scholar]
- 5. Mendelsohn J., Baselga J. (2003) Status of epidermal growth factor receptor antagonists in the biology and treatment of cancer. J. Clin. Oncol. 21, 2787–2799 [DOI] [PubMed] [Google Scholar]
- 6. Arteaga C. (2003) Targeting HER1/EGFR: a molecular approach to cancer therapy. Semin Oncol. 30, 3–14 [PubMed] [Google Scholar]
- 7. Wang S. C., Hung M. C. (2009) Nuclear translocation of the epidermal growth factor receptor family membrane tyrosine kinase receptors. Clin. Cancer Res. 15, 6484–6489 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Huo L., Wang Y. N., Xia W., Hsu S. C., Lai C. C., Li L. Y., Chang W. C., Wang Y., Hsu M. C., Yu Y. L., Huang T. H., Ding Q., Chen C. H., Tsai C. H., Hung M. C. (2010) RNA helicase A is a DNA-binding partner for EGFR-mediated transcriptional activation in the nucleus. Proc. Natl. Acad. Sci. U.S.A. 107, 16125–16130 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Hadzisejdic I., Mustac E., Jonjic N., Petkovic M., Grahovac B. (2010) Nuclear EGFR in ductal invasive breast cancer: correlation with cyclin-D1 and prognosis. Mod. Pathol. 23, 392–403 [DOI] [PubMed] [Google Scholar]
- 10. Psyrri A., Yu Z., Weinberger P. M., Sasaki C., Haffty B., Camp R., Rimm D., Burtness B. A. (2005) Quantitative determination of nuclear and cytoplasmic epidermal growth factor receptor expression in oropharyngeal squamous cell cancer by using automated quantitative analysis. Clin. Cancer Res. 11, 5856–5862 [DOI] [PubMed] [Google Scholar]
- 11. Psyrri A., Egleston B., Weinberger P., Yu Z., Kowalski D., Sasaki C., Haffty B., Rimm D., Burtness B. (2008) Correlates and determinants of nuclear epidermal growth factor receptor content in an oropharyngeal cancer tissue microarray. Cancer Epidemiol Biomarkers Prev. 17, 1486–1492 [DOI] [PubMed] [Google Scholar]
- 12. Hoshino M., Fukui H., Ono Y., Sekikawa A., Ichikawa K., Tomita S., Imai Y., Imura J., Hiraishi H., Fujimori T. (2007) Nuclear expression of phosphorylated EGFR is associated with poor prognosis of patients with esophageal squamous cell carcinoma. Pathobiology 74, 15–21 [DOI] [PubMed] [Google Scholar]
- 13. Lo H. W., Xia W., Wei Y., Ali-Seyed M., Huang S. F., Hung M. C. (2005) Novel prognostic value of nuclear epidermal growth factor receptor in breast cancer. Cancer Res. 65, 338–348 [PubMed] [Google Scholar]
- 14. Chen D. J., Nirodi C. S. (2007) The epidermal growth factor receptor: a role in repair of radiation-induced DNA damage. Clin. Cancer Res. 13, 6555–6560 [DOI] [PubMed] [Google Scholar]
- 15. Dittmann K., Mayer C., Fehrenbacher B., Schaller M., Raju U., Milas L., Chen D. J., Kehlbach R., Rodemann H. P. (2005) Radiation-induced epidermal growth factor receptor nuclear import is linked to activation of DNA-dependent protein kinase. J. Biol. Chem. 280, 31182–31189 [DOI] [PubMed] [Google Scholar]
- 16. Dittmann K., Mayer C., Kehlbach R., Rodemann H. P. (2008) Radiation-induced caveolin-1 associated EGFR internalization is linked with nuclear EGFR transport and activation of DNA-PK. Mol. Cancer 7, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Liccardi G., Hartley J. A., Hochhauser D. (2011) EGFR nuclear translocation modulates DNA repair following cisplatin and ionizing radiation treatment. Cancer Res. 71, 1103–1114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Dittmann K. H., Mayer C., Ohneseit P. A., Raju U., Andratschke N. H., Milas L., Rodemann H. P. (2008) Celecoxib induced tumor cell radiosensitization by inhibiting radiation induced nuclear EGFR transport and DNA-repair: a COX-2 independent mechanism. Int. J. Radiat Oncol. Biol. Phys. 70, 203–212 [DOI] [PubMed] [Google Scholar]
- 19. Mohanty B. K., Kushner S. R. (2000) Polynucleotide phosphorylase functions both as a 3′ → 5′ exonuclease and a poly(A) polymerase in Escherichia coli. Proc. Natl. Acad. Sci. U.S.A. 97, 11966–11971 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yehudai-Resheff S., Hirsh M., Schuster G. (2001) Polynucleotide phosphorylase functions as both an exonuclease and a poly(A) polymerase in spinach chloroplasts. Mol. Cell Biol. 21, 5408–5416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Leszczyniecka M., Kang D. C., Sarkar D., Su Z. Z., Holmes M., Valerie K., Fisher P. B. (2002) Identification and cloning of human polynucleotide phosphorylase, hPNPase old-35, in the context of terminal differentiation and cellular senescence. Proc. Natl. Acad. Sci. U.S.A. 99, 16636–16641 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Sarkar D., Park E. S., Fisher P. B. (2006) Defining the mechanism by which IFN-beta dowregulates c-Myc expression in human melanoma cells: pivotal role for human polynucleotide phosphorylase (hPNPaseold-35). Cell Death Differ. 13, 1541–1553 [DOI] [PubMed] [Google Scholar]
- 23. Fukuhara N., Tagawa H., Kameoka Y., Kasugai Y., Karnan S., Kameoka J., Sasaki T., Morishima Y., Nakamura S., Seto M. (2006) Characterization of target genes at the 2p15–16 amplicon in diffuse large B-cell lymphoma. Cancer Sci. 97, 499–504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Kirschner L. S., Taymans S. E., Pack S., Pak E., Pike B. L., Chandrasekharappa S. C., Zhuang Z., Stratakis C. A. (1999) Genomic mapping of chromosomal region 2p15-p21 (D2S378-D2S391): integration of Genemap'98 within a framework of yeast and bacterial artificial chromosomes. Genomics 62, 21–33 [DOI] [PubMed] [Google Scholar]
- 25. Leszczyniecka M., DeSalle R., Kang D. C., Fisher P. B. (2004) The origin of polynucleotide phosphorylase domains. Mol. Phylogenet Evol. 31, 123–130 [DOI] [PubMed] [Google Scholar]
- 26. Chen H. W., Koehler C. M., Teitell M. A. (2007) Human polynucleotide phosphorylase: location matters. Trends Cell Biol. 17, 600–608 [DOI] [PubMed] [Google Scholar]
- 27. Symmons M. F., Williams M. G., Luisi B. F., Jones G. H., Carpousis A. J. (2002) Running rings around RNA: a superfamily of phosphate-dependent RNases. Trends Biochem. Sci. 27, 11–18 [DOI] [PubMed] [Google Scholar]
- 28. Sarkar D., Leszczyniecka M., Kang D. C., Lebedeva I. V., Valerie K., Dhar S., Pandita T. K., Fisher P. B. (2003) Down-regulation of Myc as a potential target for growth arrest induced by human polynucleotide phosphorylase (hPNPaseold-35) in human melanoma cells. J. Biol. Chem. 278, 24542–24551 [DOI] [PubMed] [Google Scholar]
- 29. Klein C., Gensburger C., Freyermuth S., Nair B. C., Labourdette G., Malviya A. N. (2004) A 120 kDa nuclear phospholipase Cγ1 protein fragment is stimulated in vivo by EGF signal phosphorylating nuclear membrane EGFR. Biochemistry 43, 15873–15883 [DOI] [PubMed] [Google Scholar]
- 30. Wang Y. N., Yamaguchi H., Huo L., Du Y., Lee H. J., Lee H. H., Wang H., Hsu J. M., Hung M. C. (2010) The translocon Sec61β localized in the inner nuclear membrane transports membrane-embedded EGF receptor to the nucleus. J. Biol. Chem. 285, 38720–38729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Wang Y. N., Lee H. H., Lee H. J., Du Y., Yamaguchi H., Hung M. C. (2012) Membrane-bound trafficking regulates nuclear transport of integral epidermal growth factor receptor (EGFR) and ErbB-2. J. Biol. Chem. 287, 16869–16879 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Sarkar D., Park E. S., Emdad L., Randolph A., Valerie K., Fisher P. B. (2005) Defining the domains of human polynucleotide phosphorylase (hPNPaseOLD-35) mediating cellular senescence. Mol. Cell Biol. 25, 7333–7343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Linding R., Jensen L. J., Ostheimer G. J., van Vugt M. A., Jørgensen C., Miron I. M., Diella F., Colwill K., Taylor L., Elder K., Metalnikov P., Nguyen V., Pasculescu A., Jin J., Park J. G., Samson L. D., Woodgett J. R., Russell R. B., Bork P., Yaffe M. B., Pawson T. (2007) Systematic discovery of in vivo phosphorylation networks. Cell 129, 1415–1426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pfeifer A., Mark G., Leung S., Dougherty M., Spillare E., Kasid U. (1998) Effects of c-raf-1 and c-myc expression on radiation response in an in vitro model of human small-cell-lung carcinoma. Biochem. Biophys. Res. Commun. 252, 481–486 [DOI] [PubMed] [Google Scholar]
- 35. Chiang C. S., Sawyers C. L., Mcbride W. H. (1998) Oncogene Expression and Cellular Radiation Resistance: A Modulatory Role for c-Myc. Mol. Diagn. 3, 21–27 [DOI] [PubMed] [Google Scholar]
- 36. Schmidt-Ullrich R. K., Mikkelsen R. B., Dent P., Todd D. G., Valerie K., Kavanagh B. D., Contessa J. N., Rorrer W. K., Chen P. B. (1997) Radiation-induced proliferation of the human A431 squamous carcinoma cells is dependent on EGFR tyrosine phosphorylation. Oncogene 15, 1191–1197 [DOI] [PubMed] [Google Scholar]
- 37. Rodemann H. P., Dittmann K., Toulany M. (2007) Radiation-induced EGFR-signaling and control of DNA-damage repair. Int. J. Radiat. Biol. 83, 781–791 [DOI] [PubMed] [Google Scholar]
- 38. Das A. K., Chen B. P., Story M. D., Sato M., Minna J. D., Chen D. J., Nirodi C. S. (2007) Somatic mutations in the tyrosine kinase domain of epidermal growth factor receptor (EGFR) abrogate EGFR-mediated radioprotection in non-small cell lung carcinoma. Cancer Res. 67, 5267–5274 [DOI] [PubMed] [Google Scholar]
- 39. Wanner G., Mayer C., Kehlbach R., Rodemann H. P., Dittmann K. (2008) Activation of protein kinase Cϵ stimulates DNA-repair via epidermal growth factor receptor nuclear accumulation. Radiother Oncol. 86, 383–390 [DOI] [PubMed] [Google Scholar]
- 40. Dittmann K., Mayer C., Rodemann H. P. (2005) Inhibition of radiation-induced EGFR nuclear import by C225 (Cetuximab) suppresses DNA-PK activity. Radiother Oncol. 76, 157–161 [DOI] [PubMed] [Google Scholar]
- 41. Leszczyniecka M., Su Z. Z., Kang D. C., Sarkar D., Fisher P. B. (2003) Expression regulation and genomic organization of human polynucleotide phosphorylase, hPNPase(old-35), a Type I interferon inducible early response gene. Gene. 316, 143–156 [DOI] [PubMed] [Google Scholar]
- 42. Shiu R. P., Watson P. H., Dubik D. (1993) c-Myc oncogene expression in estrogen-dependent and -independent breast cancer. Clin. Chem. 39, 353–355 [PubMed] [Google Scholar]
- 43. Liao D. J., Dickson R. B. (2000) c-Myc in breast cancer. Endocr. Relat Cancer 7, 143–164 [DOI] [PubMed] [Google Scholar]
- 44. Calaf G. M., Hei T. K. (2004) Ionizing radiation induces alterations in cellular proliferation and c-Myc, c-Jun, and c-Fos protein expression in breast epithelial cells. Int. J. Oncol. 25, 1859–1866 [DOI] [PubMed] [Google Scholar]
- 45. Imaoka T., Nishimura M., Teramoto A., Nishimura Y., Ootawara M., Osada H., Kakinuma S., Maekawa A., Shimada Y. (2005) Cooperative induction of rat mammary cancer by radiation and 1-methyl-1-nitrosourea via the oncogenic pathways involving c-Myc activation and H-ras mutation. Int. J. Cancer 115, 187–193 [DOI] [PubMed] [Google Scholar]
- 46. Miura S., Nakashima M., Ito M., Kondo H., Meirmanov S., Hayashi T., Soda M., Matsuo T., Sekine I. (2008) Significance of HER2 and C-MYC oncogene amplifications in breast cancer in atomic bomb survivors: associations with radiation exposure and histologic grade. Cancer 112, 2143–2151 [DOI] [PubMed] [Google Scholar]
- 47. Hirouchi T., Takabatake T., Yoshida K., Nitta Y., Nakamura M., Tanaka S., Ichinohe K., Oghiso Y., Tanaka K. (2008) Upregulation of c-Myc gene accompanied by PU.1 deficiency in radiation-induced acute myeloid leukemia in mice. Exp. Hematol. 36, 871–885 [DOI] [PubMed] [Google Scholar]
- 48. Chiang Y. C., Teng S. C., Su Y. N., Hsieh F. J., Wu K. J. (2003) c-Myc directly regulates the transcription of the NBS1 gene involved in DNA double-strand break repair. J. Biol. Chem. 278, 19286–19291 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.