Background: We previously showed that BRAG2, a known activator of Arf6, regulates cell surface levels of β1 integrin.
Results: BRAG2 can also activate Arf4 and Arf5, but it is Arf5 that regulates integrin endocytosis.
Conclusion: BRAG2 interacts with clathrin and activates Arf5 to enhance integrin internalization.
Significance: This is the first evidence of a role for Arf5 at the plasma membrane in the regulation of endocytosis.
Keywords: Adhesion, ARF, Endocytosis, Guanine Nucleotide Exchange Factor (GEF), Integrin, Clathrin
Abstract
ADP ribosylation factors (Arfs) are small GTP-binding proteins known for their role in vesicular transport, where they nucleate the assembly of coat protein complexes at sites of carrier vesicle formation. Similar to other GTPases, Arfs require guanine nucleotide exchange factors to catalyze GTP loading and activation. One subfamily of ArfGEFs, the BRAGs, has been shown to activate Arf6, which acts in the endocytic pathway to control the trafficking of a subset of cargo proteins including integrins. We have previously shown that BRAG2 modulates cell adhesion by regulating integrin surface expression. Here, we show that, in addition to Arf6, endogenous BRAG2 also activates the class II Arfs, Arf4 and Arf5, and that surprisingly, it is Arf5 that mediates integrin internalization. We observed that cell spreading on fibronectin is enhanced upon inhibition of BRAG2 or Arf5 but not Arf6. Similarly, spreading in BRAG2-depleted cells is reverted by expression of a rapid cycling Arf5 mutant (T161A) but not by a corresponding Arf6 construct (T157A). We also show that BRAG2 binds clathrin and the AP-2 adaptor complex and that both BRAG2 and Arf5 localize to clathrin-coated pits at the plasma membrane. Consistent with these observations, depletion of Arf5, but not Arf6 or Arf4, slows internalization of β1 integrins without affecting transferrin receptor uptake. Together, these findings indicate that BRAG2 acts at clathrin-coated pits to promote integrin internalization by activating Arf5 and suggest a previously unrecognized role for Arf5 in clathrin-mediated endocytosis of specific cargoes.
Introduction
Arfs2 are grouped into three classes based on sequence homology. Arf1, Arf2, and Arf3 (class I) function primarily in the Golgi and on endosomal membranes, where they promote the assembly of COPI, AP-1, AP-3, and AP-4 coat complexes (1). Arf4 and Arf5 (class II) localize primarily to the ER/Golgi intermediate compartment and the cis-Golgi, where they regulate ER to Golgi transport (2, 3). Arf6, the only class III member, is present throughout the endosomal system and regulates the transport of a subset of cargo through the endocytic pathway (4).
Similar to other GTPases, the Arfs require guanine nucleotide exchange factors (GEFs) to stimulate the exchange of GDP for GTP for activation. Mammalian genomes encode 15 GEFs with specificity for Arf family proteins (5). Of these, BRAG1, BRAG2, and BRAG3 (brefeldin-resistant Arf GEFs) comprise a subfamily that is characterized by the presence of an IQ motif N-terminal to the catalytic domain. Although BRAG1 (6) and BRAG3 (7) are predominantly expressed in the brain, BRAG2 appears to be ubiquitously expressed (8).
The biological functions of the BRAGs are diverse. In Drosophila, BRAG orthologs have been shown to mediate both myoblast fusion (9) and neuronal pathfinding (10). BRAG2 also regulates myoblast fusion in mammals (11). In addition, BRAG2 has been reported to mediate the internalization of cell adhesion molecules such as integrins (12) and E-cadherin (13), as well as AMPA receptors in excitatory synapses (14). Additionally, BRAG2 was found to be a major determinant involved in adenocarcinoma invasion, the exact mechanism of which remains elusive but involves a ligand-induced interaction with the EGF receptor and subsequent activation of Arf6 (26). All three BRAGs have been shown to activate Arf6 (7, 15, 16); however, whether they also act on other Arfs is unknown.
Here, we show that, in addition to Arf6, BRAG2 activates the class II Arfs, Arf4 and Arf5. Knockdown of endogenous BRAG2 in HeLa cells reduces activation of both Arf5 and Arf6 by ∼50% while having little effect on Arf1. We have previously shown that BRAG2 depletion enhances cell spreading on fibronectin-coated substrates, which is correlated with accumulation of β1 integrins at the cell surface (12). Surprisingly, we found that this spreading phenotype is mimicked by knockdown of Arf5, but not Arf6, and is reversed by expression of a constitutively active, rapid cycling mutant of Arf5 but not a corresponding mutant of Arf6. Moreover, knockdown of Arf5, but not Arf4 or Arf6, impairs β1 integrin internalization without affecting the kinetics of transferrin receptor endocytosis. Finally, we show that BRAG2 interacts with both clathrin and the AP-2 adaptor complex at the plasma membrane where a fraction of Arf5 is also localized. Together, these data suggest a model in which BRAG2 is recruited to endocytic sites during clathrin coat assembly, where it activates Arf5 to promote internalization of a subset of clathrin-dependent cargoes. This is the first demonstration of a role for Arf5 in the endocytic pathway.
EXPERIMENTAL PROCEDURES
Cells and Reagents
HeLa cells were cultured at 37 °C with 5% CO2 in high glucose DMEM supplemented with 10% FBS, antibiotics/antimycotics, l-glutamine, and non-essential amino acids. The following antibodies were used in this study: anti-β1 integrin monoclonal antibody (610467), anti-clathrin heavy chain (610499), anti-γ adaptin (610385), and anti-β adaptin (610381) antibodies were from BD Transduction Laboratories; anti-HA 16B12 monoclonal antibody was from Covance; anti-tubulin monoclonal antibody (T6199) was from Sigma; anti-actin monoclonal (AANO1) was from Cytoskeleton; anti-transferrin receptor (T56/14) and MEM-101A anti-β1 integrin phycoerythrin conjugates were from Invitrogen; anti-β1 integrin cytoplasmic domain antibody was a gift from Doug DeSimone (University of Virginia) and anti-transferrin receptor monoclonal antibody HB-84 was a gift from David Castle (University of Virginia). Anti-α5 integrin (CD49e) antibody was from BD Pharmigen (555651). X-22 anti-clathrin and AP-6 anti-AP-2 antibodies were gifts from Frances Brodsky (University of California, San Francisco); Alexa Fluor 647-conjugated anti-mouse antibody (A21235) was from Invitrogen Molecular Probes; anti-Arf6, anti-Arf1, and anti-BRAG2 polyclonal antibodies have been described previously (12). Anti-GFP (B2) antibody was from Santa Cruz Biotechnology (SC9996).
siRNA and Plasmids
siRNA oligonucleotide duplexes targeting Arf4, Arf5, Arf6, BRAG2, and clathrin heavy chain were from Invitrogen and transfected on consecutive days into cells using RNAi MAX reagent (Invitrogen protocol) and maintained in the media for the duration of the 4-day experiments. Sequences used in this study were as follows: sense siRNA sequence for Arf4, AGAUAGCAACGAUCGUGAA; sense siRNA for Arf5–1, UGAGCGAGCUGACUGACAA; siRNA for Arf5–2, UCUACAUUGAAGCCUAUGG; siRNA for Arf6–1, AACCCAUUCAUAGGAUUAU; siRNA for Arf6–2, GCACCGCAUUAUCAAUGACCGUU; siRNA for BRAG2–1, AGAACUCGGUGACGUACAGUU; siRNA for BRAG2–2, ACUCGGCCAGUGAUUUCACUU; siRNA for clathrin, AAGCUGGGAAAACUCUUCAGA; and siRNA for non-target luciferase gene, UAAGGCUAUGAAGAGAUAC. Plasmid encoding dsRed clathrin light chain was a kind gift from Tomas Kirchhausen (Harvard University). GST-fusion constructs containing BRAG2b clathrin boxes were generated by inserting cDNA corresponding to residues 1–50 (CB2) or 360–408 (CB1) into pGEX-5X-2 (GE Healthcare) using EcoRI and XhoI sites.
GGA and PBD Pulldown Experiments
Pulldowns to detect active Arf or active Rac1 were performed as described previously (18).
Spreading Assays
HeLa cells were suspended by incubation in phosphate buffered saline containing 5 mm EDTA/1 mm EGTA, washed, and replated for 1.5 h in serum-free DMEM on coverslips coated with 5 μg/ml fibronectin and blocked with 5 mg/ml heat-inactivated BSA. For rescue experiments, on the third day of siRNA treatment, cells were transfected with rescue plasmids for 24 h before the onset of the spreading assay. For imaging at the end of the 1.5-h spreading time course, cells were fixed and stained with fluorescent phalloidin and imaged on a Nikon E800 fluorescence microscope using a 40× 1.0 Plan Apo objective. Measurement of spread cell surface area was performed using Nikon Elements software. For Rac1 pulldown assays, cells were allowed to spread for 45 min prior to assay.
Flow Cytometry Assays for Surface Expression and Endocytosis
Cells were washed with phosphate buffered saline at 4 °C and then incubated at 4 °C with PE-conjugated anti-β1 integrin (MEM-101A) or anti-transferrin receptor (T56/14) antibodies diluted in Hank's buffered saline solution containing 0.2% BSA, 10 mm HEPES, pH 7.0, for 1 h. After washing to remove unbound antibody, internalization was initiated by adding pre-warmed Hank's buffered saline solution/BSA and incubation at 37 °C for the indicated times. At each time point, cells were returned to 4 °C and washed with cold PBS. After all time points were collected, cells were incubated for 30 min with Alexa Fluor 647 anti-mouse antibody in cold Hank's buffered saline solution to detect the remaining cell surface antibody. After washing, cells were detached non-enzymatically using Accutase (Innovative Cell Technologies) and kept chilled until analysis by flow cytometry (FACSCalibur, Becton Dickenson). Analysis was performed using FloJo software (Tree Star, Inc.). Internalization was calculated as the ratio of intracellular/cell surface antibody at each time point. Steady-state surface expression measurements were determined from the mean fluorescence intensity of the primary antibodies prior to internalization.
Clathrin Pulldown Experiments
Subconfluent HeLa cells (1 million cells) were lysed in 4 ml of clathrin extraction buffer (100 mm MES, pH 6.8, 0.1% Triton X-100, 1 mm EGTA, 0.5 mm MgCl2, protease inhibitors). Clarified lysates (1 ml) were incubated for 1.5 h at 4 °C with 25 μg of either GST alone or GST fused to BRAG2 clathrin box domain 1 or 2, prebound to glutathione-Sepharose beads. Beads were then pelleted at 2000 rpm, washed in clathrin extraction buffer, and resolved by SDS-PAGE before immunoblotting with anti-clathrin and anti-AP-2 antibodies.
Total Internal Reflection Microscopy
Cultured HeLa cells were plated on FN-coated glass-bottomed dishes and transfected overnight with plasmids encoding either GFP, BRAG2b-GFP, Arf5-GFP, or Arf6-GFP along with dsRed-clathrin LC. Cells were imaged at 1 frame/second on an Olympus IX70 TIRF microscope using a 60× objective.
RESULTS
BRAG2 Activates Both Arf5 and Arf6
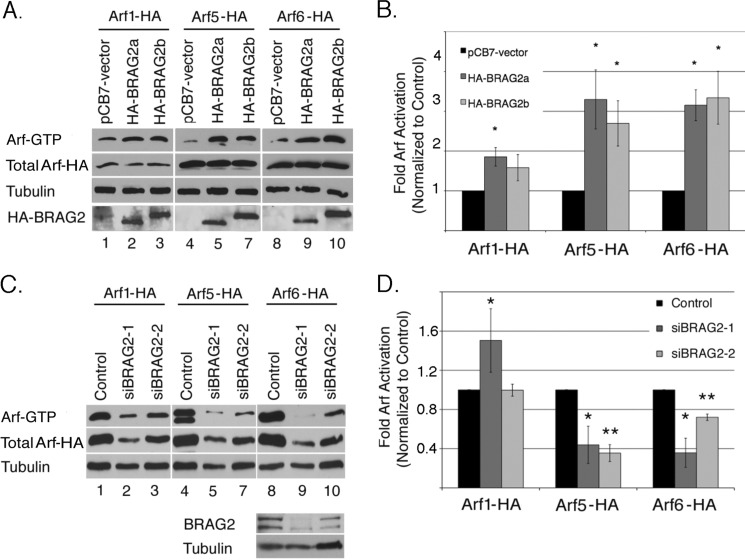
The original characterization of GEP100/BRAG2 using in vitro nucleotide exchange assays with purified recombinant proteins indicated strong activity toward Arf6, modest activity toward Arf5, and negligible activity on Arf1 (8). To examine the activity of BRAG2 in intact cells, we used a well characterized pulldown assay that takes advantage of the affinity of GTP-bound Arfs for the effector protein GGA3 (18). Initially, we coexpressed BRAG2 with C-terminally HA-tagged Arf1, Arf5, or Arf6 in HeLa cells, and GTP-bound Arf was precipitated from cell lysates by incubation with GST-GGA3 (Fig. 1A). Because all three Arf constructs contained the same epitope tag, it was possible to directly compare their relative activation by probing immunoblots with an anti-HA antibody. As shown in Fig. 1B, both BRAG2a and the longer isoform BRAG2b were capable of activating Arf1, Arf5, and Arf6, although Arf1 activation was less robust.
FIGURE 1.
BRAG2 activates Arf1, Arf5, and Arf6. A, HeLa cells were transfected overnight with plasmids encoding C-terminally tagged Arf1-HA, Arf5-HA, or Arf6-HA in the presence of either empty vector (pCB7-vector), HA-BRAG2a, or HA-BRAG2b to stimulate Arf activation. Cell lysates were incubated with GST-GGA3 to precipitate active GTP-bound Arf. The combined results of seven independent experiments are quantified in B. For each Arf, basal activity (in the absence of BRAG2) was normalized to a value of 1, and data are expressed as fold activation (±S.E.) relative to control. C, HeLa cells treated with non-target RNAi (control) or BRAG2 oligonucleotides (siBRAG2–1 and siBRAG2–2) were transfected as described above with either Arf1-HA, Arf5-HA, or Arf6-HA plasmids to determine changes in Arf activity after depletion of endogenous BRAG2. Results are quantified in D (n = 3). Using a t test, p < 0.05 is denoted by a single asterisk, and p < 0.005 is denoted by a double asterisk.
We next determined whether depletion of endogenous BRAG2 selectively affected activation of any individual Arf. Because HeLa cells express both BRAG2a and BRAG2b isoforms (see Fig. 6A) (12), we selected two separate oligonucleotide sequences each capable of depleting both isoforms (supplemental Fig. S1). For this assay, cells depleted of BRAG2 were transfected with C-terminally HA-tagged Arf1, Arf5, or Arf6, and activity was measured as described above. Representative immunoblots of each pulldown experiment are shown in Fig. 1C. As shown in Fig. 1D, BRAG2 depletion had little discernible effect on Arf1 activity. (One of the two oligonucleotides actually increased Arf1 activity.) In contrast, BRAG2 knockdown resulted in a ∼60% inhibition of Arf5 activity and a ∼30–60% reduction in Arf6 activity (Fig. 1D). Importantly, we observed the same trends when activation of endogenous Arf1 and Arf6 was analyzed (supplemental Fig. S2). The poor quality of Arf5 antibodies prevented us from examining activation of endogenous Arf5.
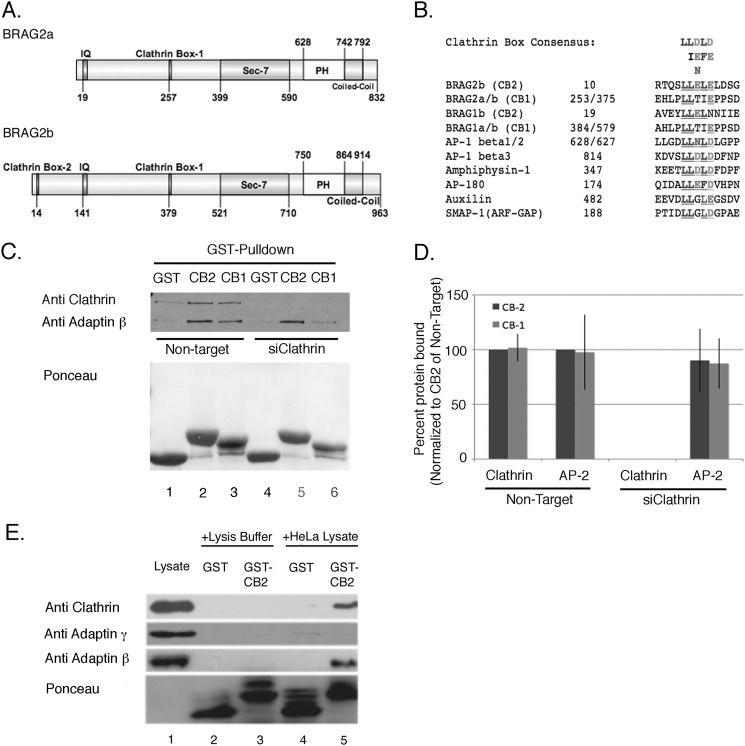
FIGURE 6.
BRAG2 interacts with clathrin in vitro. A, domain organization of BRAG2a and BRAG2b (GenBankTM accession numbers JF432314 and NM_014869.5). Positions of clathrin boxes are indicated. B, alignment of clathrin boxes from BRAG2 with related sequences from other clathrin-binding proteins. C, HeLa cell lysates were incubated with either GST alone (GST), GST-fusion proteins containing BRAG2 clathrin box 1 (CB1), or clathrin box 2 (CB2). The resulting precipitates were immunoblotted for clathrin heavy chain and β-adaptin. In lanes 4–6, cells were depleted of clathrin by siRNA to demonstrate clathrin-independent binding of β-adaptin to BRAG2. Ponceau staining was used to demonstrate equal levels of GST fusion proteins in each sample. D, quantitation of the results shown in C (n = 6). E, HeLa cell lysates or lysis buffer alone were incubated with GST or GST-CB2, and precipitates were immunoblotted for clathrin heavy chain, γ-adaptin (AP-1), or β-adaptin (AP-2). Note that only β-adaptin co-precipitates with GST-CB2.
To test the possibility that BRAG2 depletion may effect Arf protein expression, lysates from siRNA-treated cells were probed with antibodies for Arf1, Arf6, and a pan-antibody (1D9) that recognizes all Arf isoforms. As shown in supplemental Fig. S3, A and B, there was a trend toward higher expression of both Arf1 and Arf6 in BRAG2-depleted cells, but this did not reach statistical significance. Blotting with 1D9 did reveal a significant increase in total Arf expression, suggesting that compensatory mechanisms may exist to increase Arf expression when activation levels are low. Taken together, these data indicate that endogenous BRAG2 activates both Arf5 and Arf6 but not Arf1.
BRAG2 and Arf5 Regulate Cell Spreading
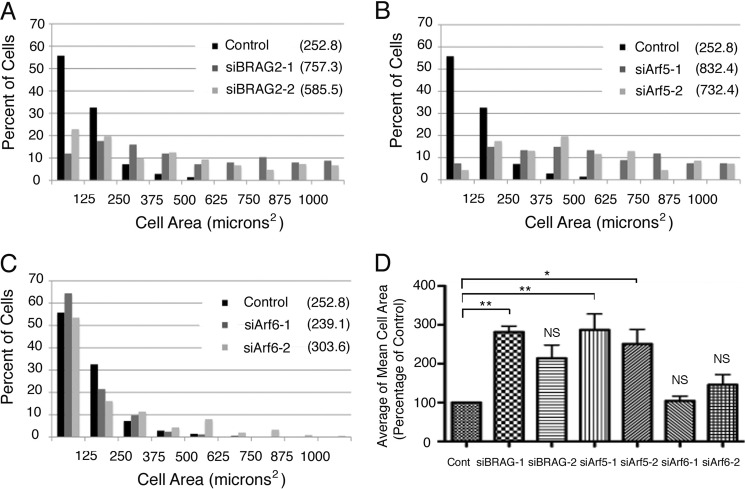
Our previous studies demonstrated that BRAG2 knockdown led to increased cell attachment and spreading on fibronectin coated-substrates, and this correlated with an increase in the level of cell surface β1 integrins (12). However, knockdown of Arf6 actually impaired spreading, suggesting that BRAG2 may be acting on another Arf in this context. Because we found that Arf5 is also a BRAG2 substrate, we depleted cells individually of BRAG2, Arf5, or Arf6 using siRNA and analyzed the phenotypic effects on cell spreading. Briefly, cells were detached using EDTA and replated onto fibronectin-coated coverslips for 1.5 h to permit spreading. As shown in Fig. 2A, BRAG2 knockdown resulted in a 2–3-fold increase in spread surface area (757.3 μm2 for oligonucleotide 1 and 585.5 μm2 for oligonucleotide 2) relative to controls (252.8 μm2). Importantly, a quantitatively similar effect was observed in cells depleted of Arf5 (832.4/732.4 μm2 for the two oligonucleotides) (Fig. 2B) but not Arf6 (239.1/303.6 μm2) (Fig. 2C). Histograms showing the relative distribution of cell areas for each condition are shown in Fig. 2, A–C. The mean cell areas of three experimental replicates were averaged to demonstrate statistical significance and are shown in Fig. 2D. Representative depletion efficiencies for Arf6 and BRAG2 are shown in supplemental Fig. S1. These results suggest that Arf5, and not Arf6, is the relevant BRAG2 target that is involved in the regulation of cell spreading.
FIGURE 2.
BRAG2 and Arf5 regulate cell spreading. Cells were depleted of endogenous BRAG2 (A), Arf5 (B), or Arf6 (C) using two individual siRNA oligonucleotides for each target. Cells were then suspended and replated on FN-coated coverslips (5 μg/ml) for 1.5 h to allow spreading. Cells were then fixed and stained with rhodamine phalloidin to visualize actin. Random fields were imaged, and the surface area of each cell was measured for 100 cells per condition. Because surface area varied considerably among cells in each population, measurements were binned to more accurately indicate variance. Mean surface area for each population is indicated in parentheses for each condition (A–C). Mean cell areas from three replicates (n = 3) of each experiment were averaged to generate D, and a one-way parametric analysis of variance (with Bonferroni's multiple comparison test) was performed to determine statistical significance between the indicated populations (*, p < 0.05; **, p < 0.005). Cont, control; NS, not significant.
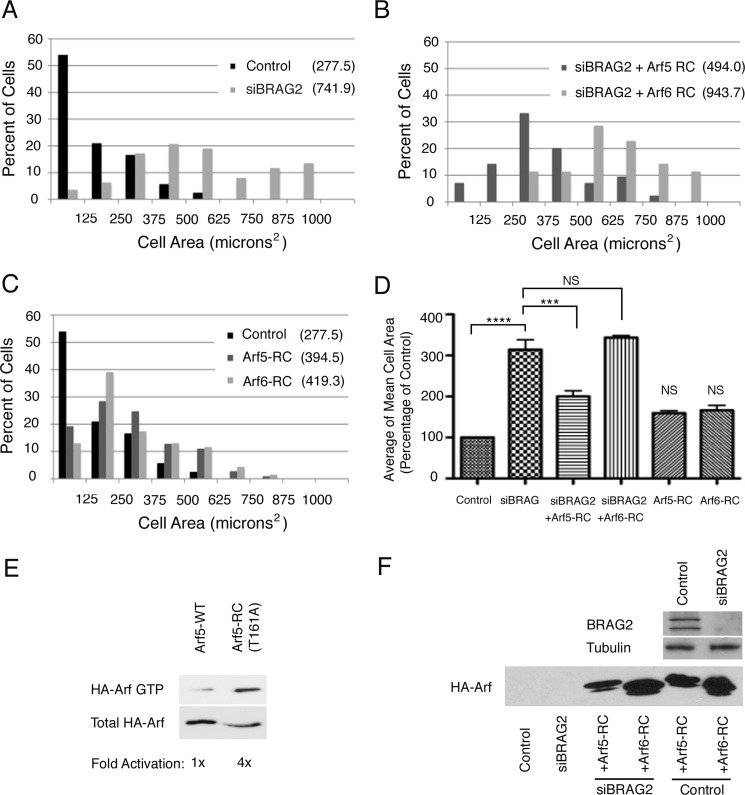
To confirm this is the case, we performed rescue experiments in which cells depleted of BRAG2 were transfected with constructs encoding rapid cycling mutants of Arf5 or Arf6. Such mutants exhibit a high rate of spontaneous (GEF-independent) nucleotide exchange and thus are primarily active. They behave like more traditional constitutively active mutants, with the exception that they can complete the GTPase cycle by hydrolyzing GTP. Thus, they do not sequester effectors and are less prone to artifacts that may affect the interpretation of results. A rapid cycling Arf6 mutant, T157A, has been previously characterized (19). For this study, we generated a corresponding Arf5 mutant, Arf5(T161A), which also exhibits enhanced binding to the effector protein GGA3, similar to its Arf6 counterpart (Fig. 3E) (19).
FIGURE 3.
Arf5 but not Arf6 restores normal spreading in BRAG2-depleted cells. A, surface area of spread cells was measured for control (mock-depleted) or BRAG2-depleted cells as in Fig. 2. B, BRAG2-depleted cells were transfected with plasmids encoding rapid-cycling mutants of either Arf5-HA (Arf5-RC) or Arf6-HA (Arf6-RC), and spreading was measured as described in A. C, neither rapid cycling Arf5 nor Arf6 affects spreading in control, non-BRAG2-depleted cells. Surface areas of spread cells were measured as indicated in the legend to Fig. 2. D, mean surface areas ± S.E. were calculated for each condition from three replicate (n = 3) experiments. A one-way parametric analysis of variance (with Bonferroni's multiple comparison test) was performed to demonstrate statistical significance, where p < 0.0005 are denoted by ***; and p < 0.00001 are denoted by ****. E, relative binding of wild-type Arf5-HA or Arf5-T161A-HA rapid cycling mutant to GST-GGA3 was determined in a pulldown assay. F, a representative immunoblot showing the extent of BRAG2 depletion and relative expression of rapid-cycling Arf5 and Arf6. NS, not significant.
As described in Fig. 2A, knockdown of BRAG2 increased cell spreading by nearly 3-fold (741.9 μm2 versus 277.5 for control) (Fig. 3, A and D). Interestingly, this effect was significantly reversed by expression of the rapid cycling Arf5 mutant Arf5(T161A) (494.0 μm2), whereas Arf5(T161A) expression (943.7 μm2) had no effect (Fig. 3, B and D). Neither mutant had a statistically significant effect on spreading when expressed on its own (Fig. 3, C and D). Representative immunoblots depicting depletion and overexpression efficiencies are shown in Fig. 3F. Taken together, these observations suggest that although BRAG2 can activate both Arf5 and Arf6, it is Arf5 that controls spreading.
Depletion of Either BRAG2 or Arf5 Slows Integrin Endocytosis
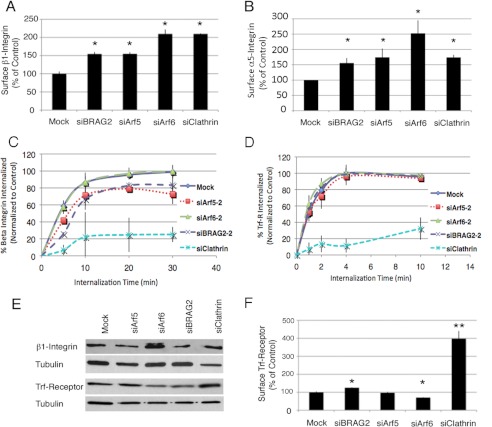
Our previous work indicated that the enhanced spreading observed in BRAG2-depleted cells correlated with increased surface expression of β1 integrins (12). To determine whether a similar up-regulation of surface integrin occurs in response to Arf5 depletion, cells were analyzed by flow cytometry. As shown in Fig. 4A, knockdown of Arf5 resulted in a 1.5-fold increase in surface β1 integrin, equivalent to the change in BRAG2-depleted cells. A similar increase was observed for α5 integrin (Fig. 4B), indicating that both subunits of the major fibronectin-binding integrin in HeLa cells accumulate on the surface of cells depleted of BRAG2 or Arf5.
FIGURE 4.
BRAG2 and Arf5 regulate β1 integrin endocytosis. Cells depleted of BRAG2, Arf5, Arf6, or clathrin were stained with antibody specific for β1 integrin (A), α5 integrin (B), or transferrin receptor (Trf-R; F), and the amount of surface-bound antibody in each sample was detected by flow cytometry. Values are normalized to control (mock-depleted) cells. C and D, internalization of β1 integrin (C) or transferrin receptor (D) antibody/receptor complexes was measured by flow cytometry (n = 3) as described under “Experimental Procedures.” E, total β1 integrin and transferrin receptor expression in clathrin-, Arf-, or BRAG2-depleted cells was measured by immunoblot. F, cell surface transferrin receptor was measured by flow cytometry (n = 3 for each condition). Using a t test, p < 0.05 is denoted by *; and p < 0.005 is denoted by **.
Knockdown of clathrin heavy chain had a similar effect to Arf5 depletion, suggesting that the accumulation of β1 integrin at the cell surface may be due to impaired clathrin-mediated endocytosis (Fig. 4, A and B). To test this possibility, we performed a flow cytometry-based internalization assay. Briefly, cells were labeled at 4 °C with PE-conjugated monoclonal antibody to either β1 integrin (MEM101A) or transferrin receptor (T56/14). Cells were then either kept at 4 °C (t = 0) or warmed to 37 °C for various times to allow receptor internalization. At each time point, cells were cooled again to 4 °C and then labeled with an Alexa Fluor 647-conjugated secondary antibody to detect primary antibody remaining on the cell surface. Cells were then washed, and the ratio of surface (Alexa Fluor 647) to total (PE) fluorescence was determined by flow cytometry. In agreement with recent observations that β1 integrin internalization is largely clathrin-dependent (20), clathrin depletion virtually abrogated integrin uptake (Fig. 4C). Depletion of Arf6, a known regulator of clathrin-independent cargo, had no effect on the rate of β1 integrin internalization. (Note that the lines depicting control and Arf6-depleted cells are superimposable (Fig. 4C).) In contrast, knockdown of either BRAG2 or Arf5 resulted in a quantitatively similar decrease in the rate of integrin endocytosis. Together, these observations support the hypothesis that BRAG2 acts on Arf5 to promote clathrin-dependent α5β1 integrin internalization. Importantly, none of the knockdowns (except for clathrin) significantly affected either internalization or steady-state surface levels of transferrin receptor (Fig. 4, D and F), demonstrating that the effects of BRAG2 and Arf5 depletion are cargo-specific.
Although knockdown of Arf6 did not impair integrin uptake, we did observe an increase in cell surface α5β1 integrin in Arf6-depleted cells (Fig. 4, A and B). However, this could be accounted for by a 2-fold increase in total protein expression (Fig. 4E) rather than redistribution to the cell surface. Despite an increase in surface integrin expression, no corresponding changes to cell spreading were observed. This suggests that in Arf6-depleted cells, adhesion-mediated signals may be uncoupled from the actin-dependent machinery that drives cell spreading.
Arf5 and Arf6 Differentially Affect Rac1 Activation
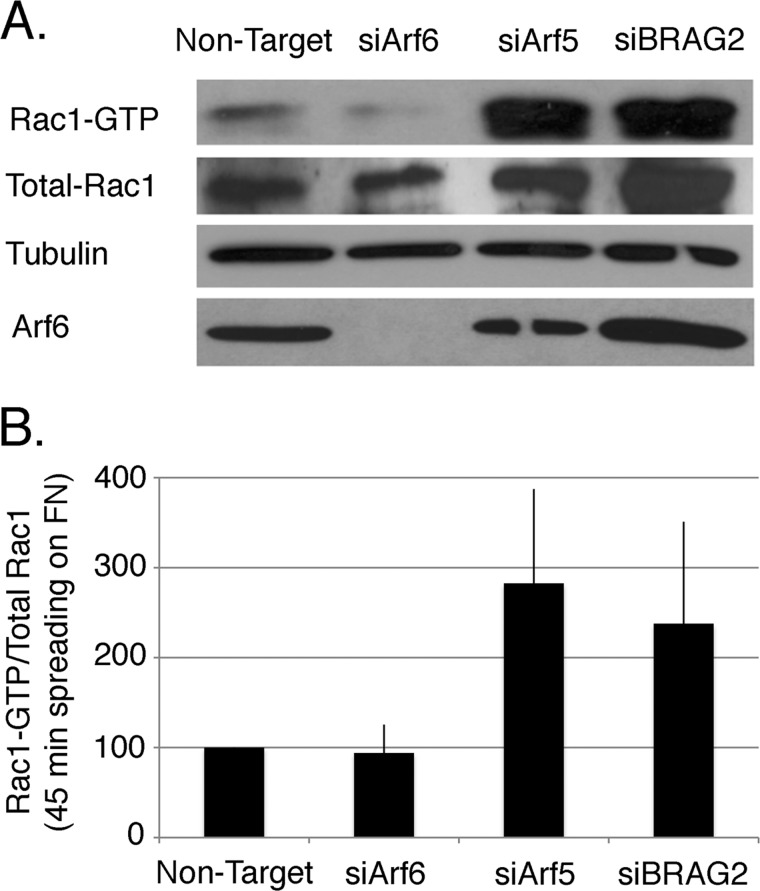
Integrin-mediated adhesion triggers the activation of Rac1, which drives the formation of lamellipodia that promote both cell spreading and migration. Arf6 has been shown to regulate delivery of Rac1 to the plasma membrane and its activation in response to integrin ligation (18, 21). We considered it likely that although Arf6-depleted cells express more β1 integrin on their surface, their spreading is attenuated because Arf6 depletion impairs the activation of Rac1. To determine whether this is the case, we measured Rac1 activation in spreading cells using a standard pulldown assay (18). As shown in Fig. 5, Rac1 activity remains low in Arf6-depleted cells relative to non-depleted controls, despite the more than 2-fold increase in surface integrin. In contrast, activation of Rac1 is enhanced 2–3-fold in cells depleted of either Arf5 or BRAG2, consistent with their more rapid spreading on fibronectin-coated substrates.
FIGURE 5.
Arf6 but not Arf5 regulates Rac1 activation during cell spreading. A, cells depleted of Arf6, Arf5, or BRAG2 were plated on FN-coated coverslips for 45 min in serum-free DMEM to allow spreading, and Rac1 activation was measured using a standard PBD pulldown assay as described under “Experimental Procedures.” B, quantification of the ratio of Rac1-GTP to total Rac1 (n = 3). Values are normalized to control cells treated with non-targeting siRNA.
Arf4 Is also Activated by BRAG2 but Does Not Control Integrin Trafficking
The class II Arfs, Arf4 and Arf5, are closely related (3), and we considered it possible that BRAG2 might activate Arf4 in addition to Arf5 and Arf6. Pulldown experiments using exogenously expressed Arf4 demonstrated that this is indeed the case (supplemental Fig. S4, A and B).
To determine whether Arf4 also modulates integrin trafficking, we compared the rates of β1 integrin internalization in cells depleted of either Arf4 or Arf5, using the flow cytometry assay. As described above, Arf5 knockdown slowed β1 integrin endocytosis (supplemental Fig. S4C) and increased the steady-state level of β1 integrin on the cell surface (supplemental Fig. S4D). In contrast, Arf4 knockdown neither inhibited internalization nor affected surface integrin levels. These observations demonstrate that BRAG2 can activate both class II Arfs, but that it is Arf5, not Arf4 or Arf6, which regulates the endocytic trafficking of β1 integrins.
BRAG2 Contains Multiple Clathrin Interaction Motifs
The data described above suggested that BRAG2 acts to regulate the clathrin-mediated endocytosis of integrins. Inspection of the BRAG2 sequence revealed the presence of two consensus motifs for clathrin binding (Fig. 6A). Clathrin boxes are pentapeptide, dileucine-containing motifs that mediate interaction with the β propeller groove of the clathrin-heavy chain (22). One clathrin box (CB1) is located in a central region of both BRAG2a and BRAG2b (BRAG2a residues 258–263, BRAG2b residues 379–384) (Fig. 6A). The more N-terminal clathrin box (CB2) is present only in the long variant BRAG2b, at residues 14–19. Both boxes share high sequence similarity to those contained in other clathrin-binding adaptor proteins (Fig. 6B), including SMAP1, an Arf-GAP that binds clathrin, and is reported to regulate transferrin receptor trafficking (23).
BRAG2 Interacts with Clathrin Heavy Chain and AP-2
To determine whether the putative clathrin boxes from BRAG2 do indeed bind clathrin, we performed pulldown experiments using GST fusions containing either the N-terminal (CB2) or central (CB1) clathrin boxes. Each construct contains ∼50 residues of BRAG2 sequence that flank a centrally located clathrin box. Detergent lysates of HeLa cells were incubated with either GST alone, GST-CB1, or GST-CB2, and associated proteins were probed for the presence of clathrin heavy chain by immunoblotting.
As shown in Fig. 6, C and D, both CB1 and CB2 bound clathrin in equivalent amounts, whereas no clathrin was precipitated by GST alone. Interestingly, GST-CB2 also coprecipitated β-adaptin, a subunit of the AP-2 adaptor complex that functions in clathrin-mediated endocytosis (Fig. 6E). This association was specific for AP-2, as γ-adaptin, a component of the AP-1 adaptor complex, did not coprecipitate with CB2 (Fig. 6E). This observation suggests that, although clathrin-coated vesicles form at multiple locations within the cell, BRAG2 associates selectively with AP-2-containing structures at the plasma membrane.
We could not identify any of the currently known AP-2 binding motifs in close proximity to the clathrin boxes, suggesting that this interaction may be indirect. To determine whether AP-2 binding to BRAG2 is a consequence of clathrin interaction, each GST fusion construct was incubated with control or clathrin-depleted HeLa cell lysates. As shown in Fig. 6, C and D, both constructs bind AP-2 in equivalent amounts, and this interaction persists in clathrin-depleted cell lysates, indicating that AP-2 binds BRAG2 independently of clathrin.
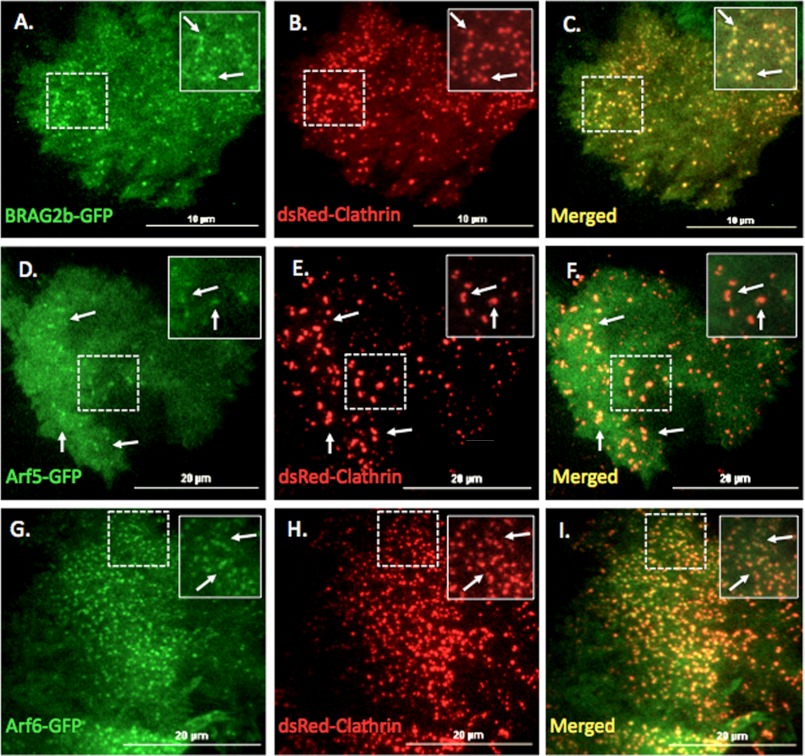
BRAG2 and Arf5 Localize to Clathrin-coated Pits
To confirm that BRAG2 and Arf5 localize to clathrin-coated structures, HeLa cells were co-transfected with either Arf5-GFP, Arf6-GFP, or BRAG2b-GFP and dsRed-clathrin light chain and then imaged live using TIRF microscopy. As shown in Fig. 7, A–C, BRAG2b-GFP was concentrated in puncta at the plasma membrane where it clearly colocalized with clathrin. Arf5-GFP was more diffusely distributed on the plasma membrane but could also be observed in puncta that colocalized with clathrin. In agreement with a recent report (24), Arf6-GFP also displayed colocalization with clathrin-positive structures (Fig. 6, G–I). BRAG2b-GFP was also found to colocalize with endogenous AP-2 in fixed cells (supplemental Fig. S5, A–C). Importantly, GFP alone did not associate with any clathrin-containing structures (supplemental Fig. S5, D–F). Together, these data support the hypothesis that BRAG2 interacts with clathrin in coated pits and activates Arf5 to mediate integrin internalization. This is the first evidence of an endocytic role for Arf5 at the plasma membrane.
FIGURE 7.
BRAG2, Arf5, and Arf6 localize to clathrin-coated pits at the plasma membrane. HeLa cells plated on FN (5 μg/ml) were transfected with plasmids encoding dsRed-clathrin and either BRAG2b-GFP (A–C), Arf5-GFP (D–F), or Arf6-GFP (G–I). Live cells were imaged using TIR-FM. Areas indicated with dashed boxes are enlarged and shown as insets. Arrows indicate colocalization with clathrin.
DISCUSSION
Early in vitro analysis of BRAG2 catalytic activity suggested a strong preference for Arf6 and only modest activity on Arf5 (8). Subsequent studies in which BRAG2 was overexpressed in cells have confirmed its ability to activate Arf6 (7, 14, 15, 26–28). In this study, we report that BRAG2 activates both class II Arfs, Arf4 and Arf5 and the single class III Arf, Arf6, in intact cells. In cells depleted of endogenous BRAG2 by siRNA, we found that the activity of Arf6 and, surprisingly, Arf5 was reduced by ∼50%. Although most Arfs can be activated by more than one GEF, this observation suggests that BRAG2 supplies roughly half of the total cellular pools of active Arf5 and Arf6. In contrast, Arf1 activation is not significantly affected by BRAG2 knockdown. Arf1 has been shown to be activated by at least three other GEFs, GBF1, BIG1, and BIG2 (5, 30), and it is therefore possible that the contribution of BRAG2 to the total cellular pool of active Arf1 is relatively minor.
Overall, very little is known about the function of class II Arfs. Both Arf4 and Arf5 have been localized to the cis-Golgi and ER-Golgi intermediate compartment (2, 3, 31). However, Kahn and colleagues (3) found that knockdown of either Arf4 or Arf5 alone had no effect on ER-Golgi transport. Instead, they found that combinatorial depletion of Arf4 and Arf1 caused fragmentation of Golgi stacks and the retention of secretory cargo (VSV-G) in the ER/Golgi intermediate compartment (3). In contrast, simultaneous knockdown of Arf5 with Arf1 had no effect on secretory cargo but did cause accumulation of KDEL receptors in structures resembling ER/Golgi intermediate compartment, suggesting a role for Arf5 in retrograde transport from the Golgi to the ER. Surprisingly, knockdown of both Arf4 and Arf5 had a distinct effect on the recycling of internalized transferrin, causing its accumulation in a perinuclear compartment resembling recycling endosomes. Although this suggests a role for class II Arfs in the endocytic pathway, neither Arf4 nor Arf5 have been localized to endosomal compartments or the plasma membrane, nor has their involvement in endocytosis been tested.
Here, we show that both Arf5 and Arf6 associate with clathrin-coated structures at the plasma membrane and that knockdown of endogenous Arf5, but not Arf6, slows internalization of β1 integrins. Importantly, this inhibition is selective, as internalization of transferrin receptors is completely unaffected. Although both cargoes require clathrin for efficient internalization, β1 integrins also require monomeric adaptors of the Dab2/ARH/Numb family, through which they are coupled to both clathrin and AP-2 (20, 25, 32). Another member of this adaptor family, GULP/Ced6, has been shown to bind Arf6 (33), and it is therefore possible that Arfs also interact with Dab2/ARH/Numb to modulate their activities.
A quantitatively similar inhibition of β1 integrin endocytosis was observed upon depletion of either Arf5 or BRAG2, resulting in an accumulation of β1 integrin on the cell surface and a corresponding enhancement of cell spreading on fibronectin-coated substrates. Importantly, spreading induced by BRAG2 depletion was reversed by expression of a rapid cycling Arf5 mutant but not a corresponding mutant of Arf6, further supporting a role for Arf5 in integrin endocytosis.
It is interesting to note that knockdown of Arf6 also resulted in increased surface expression of β1 integrin; however, we found that this correlates with increased integrin expression rather than a reduced rate of internalization. The reason for this increased expression remains unknown but may result from decreased integrin turnover in Arf6-deficient cells. Moreover, despite a 2-fold increase in surface β1 integrin levels (Fig. 4A), Arf6-depleted cells did not spread more rapidly than controls on fibronectin-coated substrates. One explanation for this is that Arf6 is involved in membrane targeting and activation of Rac1 that occurs upon integrin ligation (18, 21, 34) and that impaired Rac1 activation prevents the formation of lamellipodia that promote spreading. Here, we show that Rac1 activation is significantly enhanced in cells depleted of BRAG2 or Arf5 and that this correlates with more rapid spreading. In contrast, Rac activity is not detectably altered in Arf6-depleted cells, which correlates with their reduced rate of spreading. Together with the results described above, these findings indicate that Arf5 and Arf6 affect integrin function at different levels. Arf5 promotes integrin internalization and therefore regulates the amount of surface receptors capable of eliciting intracellular signals that reorganize the actin cytoskeleton. Conversely, Arf6 couples integrin ligation to downstream Rac1 activation but does not affect integrin internalization.
Sequence analysis revealed that all BRAG2 isoforms contain a canonical clathrin-binding motif (clathrin box) located between the IQ motif and the catalytic domain. BRAG2b contains an additional clathrin box near its N terminus. Here, we show that these sequences do indeed bind clathrin, and that BRAG2 localizes to clathrin-coated structures at the plasma membrane.
Additionally, we found that constructs containing each of the two clathrin boxes also bind AP-2 in a manner that does not require the presence of clathrin, suggesting the existence of independent AP-2 interaction sites. Previously identified AP-2 interaction motifs range from small tripeptide (DPF/W) sequences (17) to as many as 16 residues found in ARH or arrestins (29). None of the previously characterized motifs are present in the regions flanking the BRAG2 clathrin boxes, and it is therefore likely that other, unidentified motifs mediate interactions with AP-2.
Montagnac et al. (24) recently reported that Arf6 concentrates in clathrin-coated pits but that it is not necessary for endocytosis of transferrin receptors. Using a fluorescently tagged fragment of the Arf6 effector protein, JIP3, these authors demonstrated that Arf6 is active in clathrin-coated vesicles, but is not accessible to JIP3 until a later time when the vesicles uncoat (24). Additionally, the authors found that Arf6 regulates fast microtubule-dependent recycling of transferrin receptors. In the current study, we confirm the presence of Arf6 in clathrin-coated pits and extend this observation to include Arf5. Similar to Arf6, Arf5 does not appear to be necessary for transferrin receptor uptake; however, it does enhance the efficiency of β1 integrin internalization.
Interestingly, we found that knockdown of Arf5, BRAG2, or clathrin led to a quantitatively similar increase in cell surface β1 integrin, despite the fact that clathrin knockdown inhibited integrin internalization more potently. One possible explanation is that Arf5 and BRAG2 also control postendocytic trafficking and that β1integrin is recycled more efficiently in their absence. However, we have been unable to demonstrate any increase in integrin recycling in cells depleted of Arf5 or BRAG2 (data not shown), suggesting that the primary defect in these cells is in endocytosis.
Another recent study reported that BRAG2 catalytic activity is enhanced by interaction of its PH domain with phosphatidylinositol 4,5 bis-phosphate, which is abundant in coated pits (28). Consistent with this, BRAG2 has been shown to regulate the internalization of AMPA-type glutamate receptors in neurons through a direct interaction with GluA2, which stimulates Arf6 activation (14).
We hypothesize that BRAG2 is recruited to nascent coated pits coincident with clathrin/AP-2 assembly, where it is activated by PIP2, and subsequently mediates the activation of both Arf5 and Arf6 at these sites. This is the first evidence of a role for a class II Arf in endocytosis and provides a mechanism for Arf activation by BRAG2 during the uptake of specific clathrin-dependent cargo.
Supplementary Material
Acknowledgments
We thank Kenneth Myers, Emily Billings, Katherine Owen, and Corey Meyer for careful reading of the manuscript. We also acknowledge Karen Litwa, Jessica Zareno, and Rick Horwitz for invaluable help with TIRF microscopy and the University of Virginia Flow Cytometry Facility for outstanding service.
This work was supported, in whole or in part, by National Institutes of Health Grant GM078585 (to J. E. C.). This work was also supported by the Commonwealth Health Research Board of Virginia (42-10 to A. B. A.).

This article contains supplemental Figs. S1–S5.
- Arf
- ADP ribosylation factor
- GEF
- guanine-nucleotide exchange factor
- BRAG2
- brefeldin-resistant Arf-GEF
- ER
- endoplasmic reticulum
- FN
- fibronectin.
REFERENCES
- 1. Robinson M. S. (2004) Adaptable adaptors for coated vesicles. Trends Cell Biol. 14, 167–174 [DOI] [PubMed] [Google Scholar]
- 2. Chun J., Shapovalova Z., Dejgaard S. Y., Presley J. F., Melançon P. (2008) Characterization of class I and II ADP-ribosylation factors (Arfs) in live cells: GDP-bound class II Arfs associate with the ER-Golgi intermediate compartment independently of GBF1. Mol. Biol. Cell 19, 3488–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Volpicelli-Daley L. A., Li Y., Zhang C. J., Kahn R. A. (2005) Isoform-selective effects of the depletion of ADP-ribosylation factors 1–5 on membrane traffic. Mol. Biol. Cell 16, 4495–4508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. D'Souza-Schorey C., Chavrier P. (2006) ARF proteins: Roles in membrane traffic and beyond. Nat. Rev. Mol. Cell Biol. 7, 347–358 [DOI] [PubMed] [Google Scholar]
- 5. Casanova J. E. (2007) Regulation of Arf activation: The Sec7 family of guanine nucleotide exchange factors. Traffic 8, 1476–1485 [DOI] [PubMed] [Google Scholar]
- 6. Sakagami H., Sanda M., Fukaya M., Miyazaki T., Sukegawa J., Yanagisawa T., Suzuki T., Fukunaga K., Watanabe M., Kondo H. (2008) IQ-ArfGEF/BRAG1 is a guanine nucleotide exchange factor for Arf6 that interacts with PSD-95 at postsynaptic density of excitatory synapses. Neurosci. Res. 60, 199–212 [DOI] [PubMed] [Google Scholar]
- 7. Fukaya M., Kamata A., Hara Y., Tamaki H., Katsumata O., Ito N., Takeda S., Hata Y., Suzuki T., Watanabe M., Harvey R. J., Sakagami H. J. (2011) Neurochem. 116, 1122–1137 [DOI] [PubMed] [Google Scholar]
- 8. Someya A., Sata M., Takeda K., Pacheco-Rodriguez G., Ferrans V. J., Moss J., Vaughan M. (2001) ARF-GEP(100), a guanine nucleotide-exchange protein for ADP-ribosylation factor 6. Proc. Natl. Acad. Sci. U.S.A. 98, 2413–2418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Chen E. H., Pryce B. A., Tzeng J. A., Gonzalez G. A., Olson E. N. (2003) Control of myoblast fusion by a guanine nucleotide exchange factor, loner, and its effector ARF6. Cell 114, 751–762 [DOI] [PubMed] [Google Scholar]
- 10. Johnson R. I., Sedgwick A., D'Souza-Schorey C., Cagan R. L. (2011) Role for a Cindr-Arf6 axis in patterning emerging epithelia. Mol. Biol. Cell 22, 4513–4526 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Pajcini K. V., Pomerantz J. H., Alkan O., Doyonnas R., Blau H. M. (2008) Myoblasts and macrophages share molecular components that contribute to cell-cell fusion. J. Cell Biol. 180, 1005–1019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Dunphy J. L., Moravec R., Ly K., Lasell T. K., Melancon P., Casanova J. E. (2006) The Arf6 GEF GEP100/BRAG2 regulates cell adhesion by controlling endocytosis of beta1 integrins. Curr. Biol. 16, 315–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Hiroi T., Someya A., Thompson W., Moss J., Vaughan M. (2006) GEP100/BRAG2: Activator of ADP-ribosylation factor 6 for regulation of cell adhesion and actin cytoskeleton via E-cadherin and α-catenin. Proc. Natl. Acad. Sci. 103, 10672–10677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Scholz R., Berberich S., Rathgeber L., Kolleker A., Köhr G., Kornau H. C. (2010) AMPA receptor signaling through BRAG2 and Arf6 critical for long term synaptic depression. Neuron. 66, 768–780 [DOI] [PubMed] [Google Scholar]
- 15. Someya A., Moss J., Nagaoka I. (2010) The guanine nucleotide exchange protein for ADP-ribosylation factor 6, ARF-GEP100/BRAG2, regulates phagocytosis of monocytic phagocytes in an ARF6-dependent process. J. Biol. Chem. 285, 30698–30707 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Shoubridge C., Walikonis R. S., Gécz J., Harvey R. J. (2010) Subtle functional defects in the Arf-specific guanine nucleotide exchange factor IQSEC2 cause non-syndromic X-linked intellectual disability. Small GTPases 1, 98–103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Bonifacino J. S., Traub L. M. (2003) Signals for sorting of transmembrane proteins to endosomes and lysosomes. Annu. Rev. Biochem. 72, 395–447 [DOI] [PubMed] [Google Scholar]
- 18. Santy L. C., Casanova J. E. (2001) Activation of ARF6 by ARNO stimulates epithelial cell migration through downstream activation of both Rac1 and phospholipase D. J. Cell Biol. 154, 599–610 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Santy L. C. (2002) Characterization of a fast cycling ADP-ribosylation factor 6 mutant. J. Biol. Chem. 277, 40185–40188 [DOI] [PubMed] [Google Scholar]
- 20. Nishimura T., Kaibuchi K. (2007) Numb controls integrin endocytosis for directional cell migration with aPKC and PAR-3. Developmental Cell 13, 15–28 [DOI] [PubMed] [Google Scholar]
- 21. Goldfi L. E., Ptak C., Jeffrey E. D., Shabanowitz J., Hunt D. F., Ginsberg M. H. (2006) Cell 174, 877–888 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Mousavi S. A., Malerød L., Berg T., Kjeken R. (2004) Clathrin-dependent endocytosis. Biochem. J. 377, 1–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tanabe K., Torii T., Natsume W., Braesch-Andersen S., Watanabe T., Satake M. (2005) A novel GTPase-activating protein for ARF6 directly interacts with clathrin and regulates clathrin-dependent endocytosis. Mol. Biol. Cell 16, 1617–1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Montagnac G., de Forges H., Smythe E., Gueudry C., Romao M., Salamero J., Chavrier P. (2011) Decoupling of activation and effector binding underlies ARF6 priming of fast endocytic recycling. Curr. Biol. 21, 574–579 [DOI] [PubMed] [Google Scholar]
- 25. He G., Gupta S., Yi M., Michaely P., Hobbs H. H., Cohen J. C. (2002) ARH is a modular adaptor protein that interacts with the LDL receptor, clathrin, and AP-2. J. Biol. Chem. 277, 44044–44049 [DOI] [PubMed] [Google Scholar]
- 26. Morishige M., Hashimoto S., Ogawa E., Toda Y., Kotani H., Hirose M., Wei S., Hashimoto A., Yamada A., Yano H., Mazaki Y., Kodama H., Nio Y., Manabe T., Wada H., Kobayashi H., Sabe H. (2008) GEP100 links epidermal growth factor receptor signaling to Arf6 activation to induce breast cancer invasion. Nat. Cell Biol. 10, 85–92 [DOI] [PubMed] [Google Scholar]
- 27. Menju T., Hashimoto S., Hashimoto A., Otsuka Y., Handa H., Ogawa E., Toda Y., Wada H., Date H., Sabe H. (2011) Engagement of overexpressed Her2 with GEP100 induces autonomous invasive activities and provides a biomarker for metastases of lung adenocarcinoma. PLoS One 6, e25301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Sakurai A., Jian X., Lee C. J., Manavski Y., Chavakis E., Donaldson J., Randazzo P. A., Gutkind J. S. (2011) Phosphatidylinositol-4-phosphate 5-kinase and GEP100/Brag2 protein mediate antiangiogenic signaling by semaphorin 3E-plexin-D1 through Arf6 protein. J. Biol. Chem. 286, 34335–34345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Mishra S. K., Keyel P. A., Edeling M. A., Dupin A. L., Owen D. J., Traub L. M. (2005) Functional dissection of an AP-2 β2 appendage-binding sequence within the autosomal recessive hypercholesterolemia protein. J. Biol. Chem. 280, 19270–19280 [DOI] [PubMed] [Google Scholar]
- 30. Bui Q. T., Golinelli-Cohen M. P., Jackson C. L. (2009) Large Arf1 guanine nucleotide exchange factors: Evolution, domain structure and roles in membrane trafficking and human disease. Mol. Genet. Genomics 282, 329–350 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Duijsings D., Lanke K. H., van Dooren S. H., van Dommelen M. M., Wetzels R., de Mattia F., Wessels E., van Kuppeveld F. J. (2009) Differential membrane association properties and regulation of class I and class II Arfs. Traffic 10, 316–323 [DOI] [PubMed] [Google Scholar]
- 32. Teckchandani A., Toida N., Goodchild J., Henderson C., Watts J., Wollscheid B., Cooper J. A. (2009) Quantitative proteomics identifies a Dab2/integrin module regulating cell migration. J. Cell Biol. 186, 99–111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Ma Z., Nie Z., Luo R., Casanova J. E., Ravichandran K. S. (2007) Regulation of Arf6 and ACAP1 signaling by the PTB-domain-containing adaptor protein GULP. Curr. Biol. 17, 722–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Balasubramanian N., Scott D. W., Castle J. D., Casanova J. E., Schwartz M. A. (2007) Arf6 and microtubules in adhesion-dependent trafficking of lipid rafts. Nat. Cell Biol. 9, 1381–1391 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.