Background: The mouse mutant spastic carries a retrotransposon insertion in the Glrb gene leading to missplicing.
Results: Glrb missplicing in the spastic allele results from an exonic SNP amplified by retrotransposon insertion.
Conclusion: The consequences of retrotransposon insertions depend on the properties of the element and on its genomic environment.
Significance: SNPs without transcriptional relevance might contribute to disease phenotypes after additional gene alteration.
Keywords: Glycine Receptors, Mouse Genetics, mRNA, RNA Splicing, Spliceosome
Abstract
The glycine receptor-deficient mutant mouse spastic carries a full-length long interspersed nuclear element (LINE1) retrotransposon in intron 6 of the glycine receptor β subunit gene, Glrbspa. The mutation arose in the C57BL/6J strain and is associated with skipping of exon 6 or a combination of the exons 5 and 6, thus resulting in a translational frameshift within the coding regions of the GlyR β subunit. The effect of the Glrbspa LINE1 insertion on pre-mRNA splicing was studied using a minigene approach. Sequence comparison as well as motif prediction and mutational analysis revealed that in addition to the LINE1 insertion the inactivation of an exonic splicing enhancer (ESE) within exon 6 is required for skipping of exon 6. Reconstitution of the ESE by substitution of a single residue was sufficient to prevent exon skipping. In addition to the ESE, two regions within the 5′ and 3′ UTR of the LINE1 were shown to be critical determinants for exon skipping, indicating that LINE1 acts as efficient modifier of subtle endogenous splicing phenotypes. Thus, the spastic allele of the murine glycine receptor β subunit gene is a two-hit mutation, where the hypomorphic alteration in an ESE is amplified by the insertion of a LINE1 element in the adjacent intron. Conversely, the LINE1 effect on splicing may be modulated by individual polymorphisms, depending on the insertional environment within the host genome.
Introduction
Glycine receptors (GlyRs)3 belong to the superfamily of Cys-loop containing ligand-gated ion channels and mediate fast inhibitory neurotransmission preferentially in spinal cord and brainstem (1). GlyRs are a pentameric assembly of developmentally regulated proteins composed of two α1 and three β subunits (2, 3). Mutations in GlyR genes are one of the major causes of the hereditary neuromotor disorder hyperekplexia (STHE, OMIM accession no. 149000) in humans and mice (4). In the GlyR mutant mouse spastic (Glrbspa mice), a full-length long interspersed nuclear element (LINE1) has been inserted in antisense orientation in intron 6 of the GlyR β subunit gene, Glrb (5, 6). Although exons 5 and 6 of the Glrb transcripts are constitutively spliced in wild-type mice, exon 6 or both exons 5 and 6, are skipped in homozygous Glrbspa/spa mice (5, 6). This exon skipping results in a translational frameshift and, as a consequence, leads to a profound numerical reduction of functional GlyRs (6, 7).
LINE1 elements are the most abundant autonomous retrotransposons in mammalian genomes (8, 9). All full-length LINE1 elements share an identical structural organization, comprised of two ORF, ORF1, and ORF2 which are flanked by 5′ and 3′ UTRs, respectively. ORF1 encodes a RNA binding protein (pORF1, 40 kDa), and ORF2 encodes a protein exerting endonuclease as well as reverse transcriptase activities (pORF2, 150 kDa). Most of the full-length LINE1 elements present in mammalian genomes are rendered retrotransposition-incompetent through nonsense or frameshift mutations, 5′ truncations, or internal rearrangements (10, 11). In humans and mice, LINE1-associated diseases are most frequently caused by insertions into either exons or gene regulatory sequences, resulting in gene dysfunction (9, 12). Moreover, homologous recombination of LINE1 at non-allelic chromosomal sites are thought to underlie genomic rearrangements reflected by deletions or insertions (13, 14). The high frequency of LINE1 elements within intronic sequences is contrasted by a relatively low number of known pathological phenotypes. In some human genetic disorders (15, 16) and in the mutant mouse spastic (5, 6), however, intronic insertions of LINE1 elements are associated with aberrant splicing. The mechanism resulting in missplicing is not yet fully understood. LINE1-dependent interference has been attributed to a variety of mechanisms, including disruption of consensus splice sites or RNA regulatory motifs such as intronic splicing enhancers or silencers (8). Moreover, bioinformatic analysis indicated that intronic LINE1 insertion in sense orientation are underrepresented compared with antisense insertions, suggesting an underlying negative selection (12, 17). In this study, we used the Glrbspa gene, and recombinant variations thereof, as a model system for studying the molecular mechanism by which a full-length intronic LINE1 insertion affects pre-mRNA splicing.
To elucidate the pathomechanism by which the LINE1 affects splicing in Glrbspa mice, in vivo splicing assays were conducted in human embryonic kidney (HEK293) cells using minigenes encompassing Glrb exons 4–7 with intervening partial or full-length intronic sequences. Because the Glrbspa allele was first discovered in a B6C3Fe hybrid background, we used DNA from the parental inbred lines C57BL/6J Glrb+/+ and C3H/HeJ Glrb+/+ and from the inbred spastic line C57BL/6J Glrbspa/spa. Although minigenes constructed on a C57BL/6J genetic background exhibited robust exon skipping, either in the presence of LINE1 sequences or specific splicing regulatory proteins, this missplicing was not observed in minigenes prepared from genomic DNA of C3H/HeJ mice. A polymorphic short nucleotide polymorphism (SNP) localized in Glrb exon 6 was found to function as an exonic splicing enhancer and thus regulating exon skipping by influencing binding of the essential splicing regulatory protein SRSF1 (formerly also known as ASF/SF2). These observations suggest that the missplicing observed in Glrbspa mice results from a splicing signal mutation amplified by insertion of a LINE1 retrotransposon.
EXPERIMENTAL PROCEDURES
Generation of Minigene Constructs
Exon nomenclature was based on ensembl release 55 (18). Mouse Glrb (NM_010298) exon 4, 5′ intron 4 (up to IVS4272), 3′ intron 4 (from IVS4 + 15720), exon 5, intron 5, exon 6, intron 6 and exon 7 were amplified from C57BL/6J Glrb+/+, C3H/HeJ Glrb+/+, or C57BL/6J Glrbspa/spa genomic DNA using long range PCR (Triple Master, Eppendorf, Germany). For in vivo splicing assays, inserts were cloned into the eukaryotic expression vector pRK7, which contains a CMV promoter, creating the minigenes B-WT (from C57BL/6J Glrb+/+ genomic DNA), C-WT (from C3H/HeJ Glrb+/+ genomic DNA), or Spa (from C57BL/6J Glrbspa/spa genomic DNA). For analyzing protein expression, ORFs were created by adding to the respective Spa and B-WT minigenes cDNA sequences derived from the exons 1–3, including a sequence encoding for an N-terminal Myc tag as well as cDNA sequences encoding the sequences from exons 8 and 9, yielding a Spa plasmid of 15.84 kb. For comparable transfection conditions, the B-WT (7.02 kb) minigene was extended to a similar size (15.38 kb) by adding 6.8 kb of IVS4. The inclusion of this additional sequence did not affect splicing efficiencies (data not shown).
In Vivo Splicing Assays
HEK293 cells were grown in MEM supplemented with 10% (v/v) fetal calf serum and penicillin/streptomycin. Cells were plated at 2 × 105 cells/6 well and grown until 60% confluency. Transient transfection was carried out using calcium phosphate precipitation employing a total of 6 μg of plasmid DNA/well. Minigenes were transfected at 2 μg/well and splicing factors at increasing concentrations (up to 4 μg). With total plasmid DNA <6 μg/well, pEGFP-N1 (Clontech, Mountain View, CA) was co-transfected for filling in. In vivo splicing assays were performed as described (19). For protein expression analysis, HEK293 cells were transfected with the indicated constructs, and membrane preparations were performed as described previously (20). For RT-PCR analysis, RNA was isolated from transfected cells using standard GTC/phenol extraction method (PEQGold-RNA Pure, PEQLab, Erlangen, Germany). Total RNA (1–2 μg) was reverse transcribed using a vector specific primer (pRK-Cis: 5′-AACCATTATAAGCTGCAATAAAC-3′) and M-MuLV reverse transcriptase (New England Biolabs). 1–3 μl of cDNA were used for PCR (GoTaq, Promega, Madison, WI; primers: E4 + 246-F, 5′-GTAGTCAACATTTTTATTAATAG-3′; E7 + 656-R, 5′-AGGATCTCCTGACTGCCAGATGAA-3′).
Western Blot Analysis
Western blot analysis was performed as described (20). The following antibodies were used: monoclonal anti-SRSF1 (32-4500, Invitrogen), monoclonal anti-GFP (Roche Applied Science); polyclonal anti-ATPA1 (sc-28801, Santa Cruz Biotechnology, Santa Cruz, CA), monoclonal anti-c-Myc (9E10; Santa Cruz Biotechnology); HRP conjugated goat anti-mouse F(ab)2 fragments (Dianova, Hamburg, Germany).
In Vitro Transcription and Affinity Purification of Glrb Exon 6 RNA Binding Proteins
Procedures were performed as described previously (21). In brief, to generate an RNA probe of a Glrb exon 6 fragment (E6.13–E6.61), the corresponding linearized pBluescript II KS plasmid was transcribed in vitro. One nanomole (∼7.9 μg) of RNA was placed in a reaction mixture containing fresh 0.1 m NaOAc, pH 5.0, and 5 mm sodium m-periodate (Sigma). The reaction mixture was incubated for 1 h in the dark at room temperature. The RNA was ethanol-precipitated and resuspended. Then, prewashed adipic acid dehydrazide-agarose bead 50% slurry (Sigma) was mixed with the periodate-treated RNA sample and incubated for 12 h at 4 °C on a rotator. The RNA-bound beads were washed with RNA washing buffer. They were incubated in 1× RNA binding buffer with 0.3 mg of HeLa cell nuclear extract (CilBiotech) for 20 min at 30 °C, pelleted by centrifugation, and washed five times in RNA washing buffer. After the final centrifugation, 60 μl of SDS-PAGE sample buffer were added to the beads and heated to 90 °C before loading onto a 10% SDS-PAGE gel, transferred to PVDF membranes, and probed with a monoclonal antibody directed against an N-terminal epitope of SRSF1 (Invitrogen).
siRNA Knockdown
The siRNA knockdown of SRSF1 in HEK293 cells was performed using the reverse transfection procedure according to the manufacturer's instructions (Qiagen, Hilden, Germany) on six-well plates. A sequence within the human SRSF1 coding region was selected for designing a siRNA (Dharmacon, Chicago, IL). Transfections with scrambled siRNAs were used as a control (sc-37007; Santa Cruz Biotechnology). After 48 h, 1000 ng/well of the Spa minigene was transfected as described earlier. RNA was isolated after 72 h of siRNA treatment (16–18 h after minigene transfection).
RESULTS
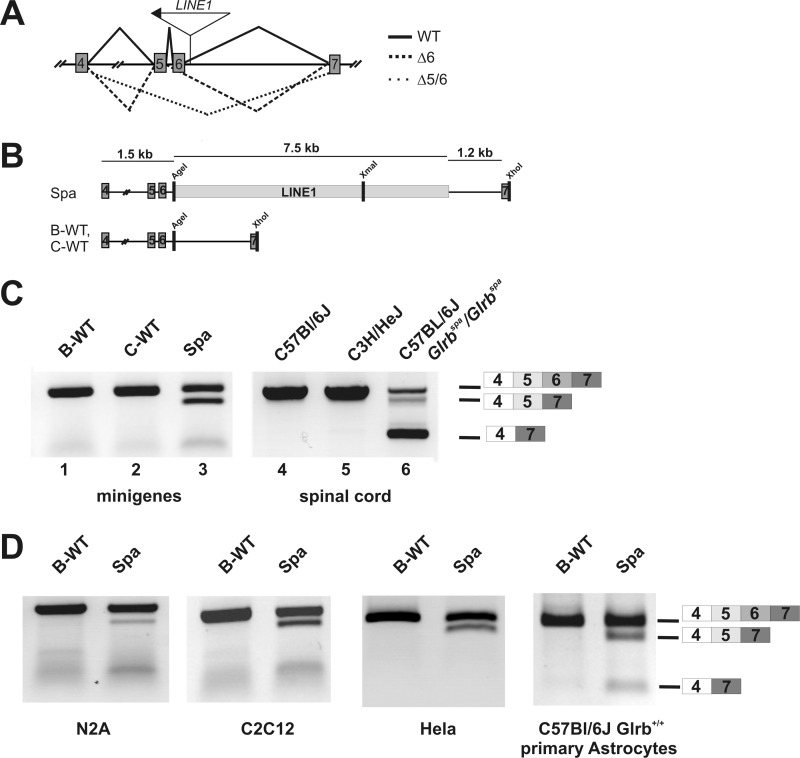
In the mutant mouse spastic, exon skipping from Glrb transcripts has been associated with an intronic insertion of a full-length LINE1 element into intron 6 of the Glrb gene (Fig. 1A, see also Refs. 5 and 6). To study Glrbspa pre-mRNA missplicing, in vivo splicing assays were performed (19), using minigene constructs derived from genomic DNA from C57BL/6J Glrbspa/spa (Spa), C57BL/6J Glrb+/+ (B-WT), and C3H/HeJ Glrb+/+ (C-WT) mice. Plasmids encompassing Glrb exons 4 to 7 (Fig. 1B) were transiently transfected into HEK293 cells, and RNA transcripts were analyzed by semiquantitative RT-PCR using primers binding in the exons 4 and 7, respectively. For both wild-type minigene constructs, i.e. B-WT and C-WT, only one amplimer was obtained which corresponded to the full-length transcript composed of exons 4–7 (Fig. 1C, lanes 1 and 2), consistent with the Glrb transcripts observed in cDNA preparations from spinal cord tissue of wild-type mice (Fig. 1C, lanes 4 and 5). In samples derived from cells transfected with the Spa minigene, additional shorter amplimers, corresponding to RNAs lacking either exon 6 (Δ6) or both, exons 5 and 6 (Δ5/6), (Fig. 1C, lane 3) were detected. Amplimers of identical size were observed in samples derived from spinal cord tissue of mutant mice carrying the Glrbspa allele (Fig. 1C, lane 6, see also Ref. 6). Interestingly, although skipping of exon 6 was reliably detected in samples from transfected HEK293 cells only a faint band representing the amplimer from a Δ5/6 mRNA was visible. Testing of other cell lines including neuroblastoma cells as well as primary mouse astrocytes revealed that the combined skipping of exons 5/6 was highly variable between experiments and in addition depended on the cell type used for analysis (Fig. 1D). Therefore, we focused on missplicing of exon 6, which was reliably detected in all cell types tested.
FIGURE 1.
A, structure of the Glrb gene surrounding the LINE1 insertion site. Exons are indicated as gray boxes, position as well as orientation of the LINE1 insertion is indicated by the arrow. Splicing of the pre mRNA derived from the wild-type Glrb allel is indicated as solid line, splicing events observed in Glrbspa mice resulting in skipping of exon 6 (Δ6) as a dashed line or exons 5 and 6 (Δ5/6) as a dotted line, respectively. B, structure of the Glrb minigenes. Exons are depicted as gray boxes, and LINE1 is represented as an open box. The Spa minigene was generated from genomic DNA of a C57BL/6J spastic mouse (Glrbspa/spa). The wild-type minigenes B-WT and C-WT were generated from genomic DNA of C57BL/6J and C3H/HeJ mice, respectively. C, RT-PCR analysis of HEK293 cells transfected with the minigenes indicated in B or spinal cord mRNA preparations from mice with the indicated genomic background and genotype. For amplification, primers specific for a Glrb amplimer containing the exons 4–7 were used. Expected sizes for the full-length amplimer, the Δ6 amplimer and the Δ5–6 amplimer are indicated. Note that skipping of exon 6 or the exons 5 and 6 was only observed in samples from Spa minigene expressing cells or Glrbspa/spa mice. D, RT-PCR analysis from RNA preparations of N2A (mouse neuroblastoma cells, differentiated after 12 h of serum withdrawal), C2C12 (a mouse myoblast cell line, undifferentiated), HeLa cells and primary astrocytes derived from P0 C57BL/6J Glrb+/+ animals after transfection with the indicated minigenes. In all cell lines investigated, skipping of exon 6 could be observed reliably after transfection of the Spa minigene, whereas the combined skipping of exons 5 and 6 was highly variable.
LINE1-associated Glrb Missplicing Depends on Genetic Context
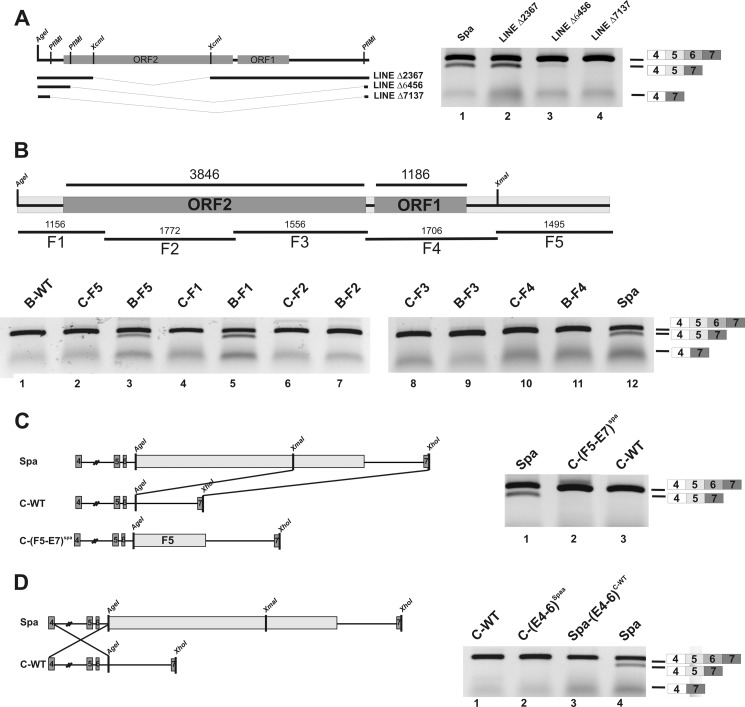
To determine whether a full-length LINE1 insertion is necessary to induce missplicing, we performed deletion analysis of the LINE1 sequence. When most of the LINE1 sequence was missing, exon skipping was nearly absent (Fig. 2A, lanes 3 and 4), whereas deletion of a fragment from ORF2 alone did not prevent missplicing (Fig. 2A, lane 2). To test which LINE1 segments were necessary for missplicing, five overlapping fragments (Fig. 2B, F1–F5) were cloned individually into both wild-type minigenes (B-WT and C-WT) at position IVS6 +193, matching the site of LINE1 integration into intron 6 of the Glrbspa allele. Exon skipping similar to the Spa minigene was only observed for constructs B-F1 and B-F5, encompassing the 3′ UTR and adjacent 370 bp of ORF2, or the 5′ UTR of the LINE1, respectively (Fig. 2B, lanes 3 and 5). Surprisingly, aberrant splicing was completely absent in similar minigenes generated from genomic DNA of C3H/HeJ mice (Fig. 2B, lanes 2 and 4). Apparently, Glrb missplicing was suppressed in the C3H/HeJ genomic context. To evaluate which parts of the C-WT sequence were necessary for suppression of exon 6 skipping, the F5 fragment of the LINE1 sequence and adjoining Glrb sequences, including exon 7, were excised from the Spa minigene (Fig. 2C) and cloned into C-WT downstream of the AgeI site, thereby generating the hybrid C-(F5-E7)Spa minigene (Fig. 2C). Exon skipping was also absent in cells transfected with this minigene, indicating that the upstream exons 4–6 from C-WT were sufficient for suppression of missplicing. Moreover, swapping of exons 4–6 from the C-WT minigene with Spa, yielding the construct Spa-(E4–6)C-WT, prevented aberrant splicing (Fig. 2D, lane 3), although this minigene contained the full-length LINE1. Similarly, no missplicing was observed with the C-(E4–6)Spa minigene (Fig. 2D, lane 2). Taken together, these results suggest that in addition to the insertion of the LINE1 element, a second sequence element present in the fragment containing the exons 4–6 is required for the missplicing observed in Glrbspa mice.
FIGURE 2.
LINE1 associated exon skipping depended on the genetic context. A, deletion constructs containing only fragments of the LINE1 element were generated from the Spa minigene by internal restriction digest. ORFs within the LINE1 are indicated as gray boxes. After transfection in HEK293 cells and RNA extraction from the transfected cells, RT-PCR analysis was performed. Deletion of 2.3 kb from ORF2 did not change splicing, whereas exon skipping was diminished with larger deletions (Δ6456 bp, Δ7137 bp; lanes 3 and 4). B, minigenes containing the indicated LINE1 fragments F1 to F5 at the position of the original LINE1 insertion were generated on the basis of genomic DNA from C57BL/6J and C3H/HeJ mice. The respective minigenes were transfected in HEK293 cells and mRNA extracts analyzed by RT-PCR analysis. Of note, only the 3′ UTR plus 370 bps of ORF2 and 5′ UTR, respectively, promoted exon skipping when inserted into Glrb minigenes derived from C57BL/6J. C, schematic drawing of the construction of a chimeric C3H/HeJ Spa minigene containing exons 4–6 based on the C3HJ/HeJ genomic DNA and the fragment F5–E7 sequence containing the region IVS6 + 194-exon 7 from the Spa construct. After transfection in HEK293 cells and RNA extraction from the transfected cells, RT-PCR analysis was performed. D, schematic drawing of the construction of a Spa C3H/HeJ minigene. A fragment containing E4-IVS5 + 193 from C3H/HeJ genetic background was introduced in the Spa minigene, replacing the homologous region within the Spa minigene suppresses exon skipping despite the presence of a full-length LINE1. For analysis, HEK293 cells were transfected with the indicated minigenes, and exon skipping was determined by RT-PCR.
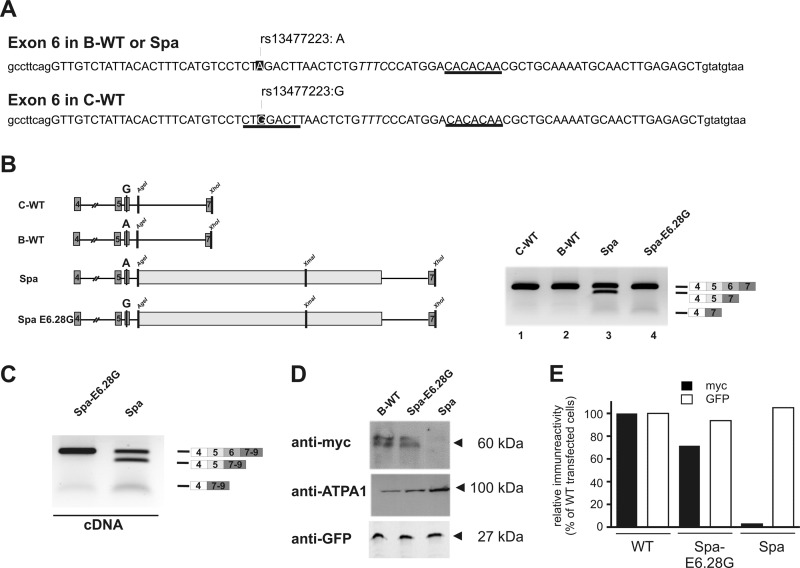
Using RNA regulatory motif prediction (22–24), we searched for sites within Glrb exons 4–6 able to modulate exon inclusion, e.g. by providing binding sites for splicing regulatory factors such as serine/arginine-rich (SR) or heterogeneous ribonucleoproteins. In particular, we focused on sequence elements that were polymorphic between C57BL/6J and C3H/HeJ. A single nucleotide polymorphism (Glrbrs13477223) was found to coincide with an exonic splicing enhancer (ESE) motif in exon 6, predicted to bind the SR protein SRSF1 (Fig. 3A, boxed; Table 1). Here, an A allele is present at position E6.28 in C57BL/6J (E6.28A, B-type), whereas a G allele is found in C3H/HeJ (E6.28G, C-type). Based on our motif prediction, the G-allele present in the C-WT minigene contributes to an SRSF1 binding site that was absent in Spa (B-type). A second SRSF1 binding motif was predicted 23 bp downstream of this site, which was identical in both C57BL/6J and C3H/HeJ (Fig. 3A, underlined). To test whether differences in splicing could be attributed to the polymorphism rs13477223, residues at position E6.28 were swapped, thereby generating the minigenes C-E6.28A and Spa-E6.28G. Upon expression, skipping of exon 6 could be observed only with the original Spa construct, containing the B-type nucleotide E6.28A (Fig. 3B, lane 3). In contrast, a G-residue at this position (C-type) was sufficient to prevent skipping of exon 6 despite the presence of a full-length LINE1 (Fig. 3B, lane 4). Similarly, no missplicing was observed in cells transfected with the C-E6.28A minigene (data not shown). These findings suggest that the polymorphism at position E6.28 significantly contributes to the missplicing observed in Spa mice. To test whether this modulation of exon skipping by nucleotide E6.28 was also evident at the level of the full-length GlyR β subunit protein, we created translatable ORFs from both Spa and Spa-E6.28G minigenes by adding cDNA sequences containing the sequences encoded by the exons 1–3 at the 5′ end, as well as exons 8–9 at the 3′ end of both Spa constructs. For detection, a sequence encoding for an N-terminal Myc tag was incorporated. Whereas the B-WT and Spa E6.28G minigenes produced only full-length mRNA (data not shown and Fig. 3C), robust skipping of exon 6 was observed in minigenes derived from the Spa sequence (Fig. 3C, left panel). Western blot analysis of detergent extracts from B-WT transfected cells revealed a doublet of Myc immunoreactive bands of ∼60 kDa that might result from different glycosylation forms of the full-length GlyR β protein in this cell system. Consistent with our cDNA data similar immunoreactive signals were observed in samples from cells transfected with the Spa E6.28G minigenes (Fig. 3D) but not in samples from Spa minigene expressing cells.
FIGURE 3.
LINE1-associated skipping of exon 6 depended on a polymorphic residue at position E6.28. A, sequence of the Glrb exon 6 from C57BL/6J and C3H/HeJ mice, including the surrounding intronic regions. Exonic sequence is displayed in uppercase letters. Sequence motifs predicted to bind to SRSF1 are indicated. Note that one SRSF1 site predicted E6.23 in the C3H/HeJ exon 6 was not detected in the C57BL/6J due to a SNP at position E6.28 (A, strain C57BL/6J; G, strain C3H/HeJ; dbSNP, rs13477223). B, the effect of the SNP at position E6.28 was analyzed by introducing a G at E6.28 in the Spa minigene. The indicated minigenes were transfected into HEK293 cells and exon skipping was analyzed by RT-PCR. Note that mutation of E6.28 in the Spa minigene to a G (Spa-E6.28G) was sufficient to prevent skipping of exon 6. C, B-WT, Spa, and Spa E6.28G minigenes were complemented to full-length ORFs by adding coding sequences of the Glrb exons 1–3, including an N-terminal Myc tag and at the 5′ end and coding sequences of the exons 8–9 at the 3′ end of the minigene. The constructs were transfected into HEK293 cells and exon skipping was analyzed by RT-PCR using primers positioned in exons 4 and 9. D, membrane preparations form HEK293 cells transfected with the minigenes containing the full-length GlyR β ORF as indicated and for testing transfection efficiencies a plasmid encoding for GFP, were subjected to SDS-PAGE and Western blot analysis. The blots were probed with antibodies against Myc, ATPA1, and GFP. E, expression levels from the experiment shown in D as quantified by scanning of the blots and densitometric analysis using NIH ImageJ software. Note that in contrast to samples from WT and Spa-E6.28G transfected cells, samples from Spa transfected cells, showed almost no Myc immunoreactivity although the cells were transfected efficiently as indicated by GFP immunoreactivity.
TABLE 1.
Prediction of splicing factor binding at rs13477223
Default thresholds were used for all web-based prediction tools. For ESE finder and Altsplice, only factors predicted to bind at E6.28 are listed with scores obtained for E6.28 A versus E628.G.
| Modification | ESE finder | Altsplice | Rescue ESE | Observation |
|---|---|---|---|---|
| SRSF1 (10-mer), 5.13/5.93 | ||||
| E6.28 A>G | SRSF1, n.d./2.41 | SRSF1 (7-mer), n.d./3.05 | None found at position | A, exon 6 skipped |
| (c.555A>G; rs13477223) | SRSF2, 4.59/5.04 | SRSF5 (5-mer), n.d./3.24 | None found at position | G, no skipping |
| SRSF5, 4.10/4.67 | SRSF2 (8-mer), 4.70/5.13 |
Modulation of Exon Skipping by the Splicing Factor SRSF1
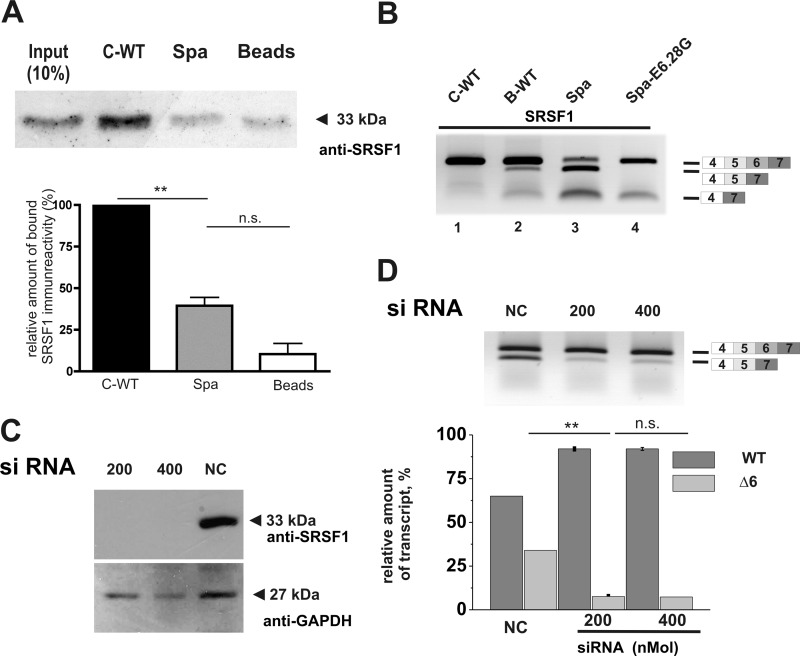
The ESE polymorphism between B-WT and C-WT resides within a putative binding site for splicing factor SRSF1. We assayed SRSF1 binding to Glrb exon 6 RNA using fragments from Spa and C-WT encompassing the polymorphic E6.28 and using them for pulldowns from HeLa nuclear extracts to biochemically test for a possible reduction of binding to E6.28A containing sequences. In pulldowns using C-WT RNA fragments as bait, strong SRSF1 binding was observed. On the other hand, in samples where a Spa RNA fragment was used as a bait, binding of SRSF1 was significantly reduced (Fig. 4A), consistent with the predicted loss of SRSF1 binding motif in Spa RNA. Upon overexpression of SRSF1, enhanced skipping of exon 6 was observed in RNA preparations from cells transfected with minigenes lacking the second SRSF1 binding site (E6.28A, constructs B-WT and Spa, Fig. 4B, lanes 2 and 3; compare with Fig. 3). Conversely, no skipping of exon 6 was detected in C-WT and Spa E6.28G (Fig. 4B, lanes 1 and 4). The effect of reduced SRSF1 levels on exon skipping was then analyzed by a siRNA-based approach. Here, transfection of HEK293 cells with a SRSF1-specific siRNA resulted in an efficient reduction of SRSF1 expression, whereas SRSF1 levels in cells transfected with control siRNA was comparable with untransfected cells (Fig. 4C and data not shown). Upon co-transfection of siRNAs and the Spa minigene, robust skipping of exon 6 was observed in cells transfected with control siRNA, whereas co-transfection of SRSF1 specific siRNA resulted in enhanced inclusion of exon 6 in a dose-dependent manner (Fig. 4D), suggesting that SRSF1 is an important splicing regulator for this exon. Taken together, these data suggest that the LINE1 sequences modifies splicing by binding and/or sequestering SR proteins, in particular SRSF1, in a sequence and/or position dependent manner. The possibility that transcription from an internal LINE1 promoter interfered with Glrb splicing was excluded, since no LINE1 specific amplicons were detected in samples from cells transfected with the Spa minigenes (data not shown).
FIGURE 4.
Modulation of exon skipping by splicing factor SRSF1. A, pulldown assays of HeLa nuclear extracts were conducted using in vitro-transcribed Glrb E6.13–E6.61 RNA containing either E6.28G (C-WT) or E6.28A (Spa). After pulldown, proteins were analyzed by Western blotting using a monoclonal antibody against SRSF1. Using a C-WT derived sequence, a strong signal for SRSF1 was detected at 33 kDa, which was diminished when a E6.28G RNA fragment was used as a bait. Lower panel, for quantification band band intensities were analyzed using ImageJ software. All values represent means ± S.E. (n = 3). **, p < 0.01 (one-way ANOVA followed by Bonferroni's multiple comparison test). B, HEK cells were cotransfected with an expression construct for SRSF1 and the minigenes as indicated. In RNA preparations from these cells, exon skipping was analyzed by RT-PCR using primers positioned in exons 4 and 7. C, HEK293 cells were transfected with 200 or 400 ng of an SRSF1 specific siRNA or 400 ng of scrambled siRNA. Efficiency of SRSF1 knockdown was determined in Western blot from protein extracts of transfected cells using an SRSF1-specific antibody. Comparable loading of the gel was assessed by probing the Western blot with antibodies against GAPDH (D); HEK293 cells were transfected with siRNA as described in C. After 24 h, cells were transfected additionally with the Spa minigene. Exon skipping was analyzed in RNA preparations from these cells using primers positioned in exons 4 and 7. For quantification, band intensities were determined on digital images of the gel using ImageJ software. All values represent means ± S.E. (n = 3). **, p < 0.01 (one-way ANOVA followed by Bonferroni's multiple comparison test).
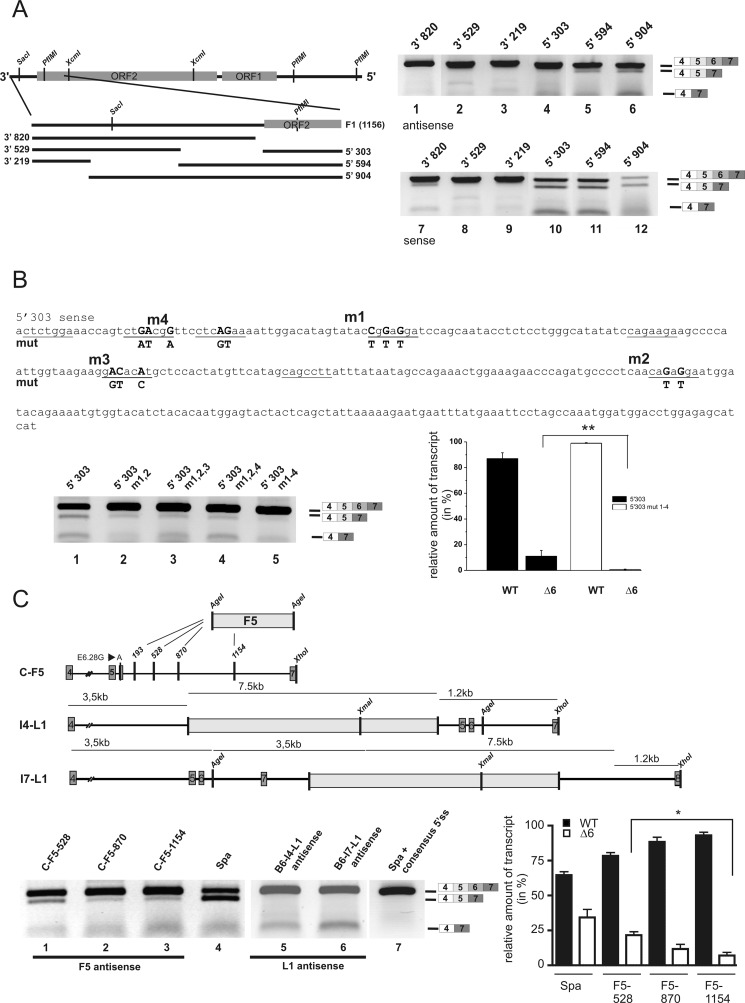
To determine a minimal sequence of the LINE1 sufficient to induce exon skipping in an E6.28A environment, we performed deletion analysis of constructs B-F1 (3′ UTR and adjacent 370 bp of ORF2) and B-F5 (5′ UTR) to obtain fragments amenable to mutational studies (Fig. 5, data not shown). As F1 is located closest to exon 6 in Spa, we conducted a detailed analysis of this fragment. Truncations from its 5′ and 3′ ends were obtained by PCR (Fig. 5A). Skipping of exon 6 and/or exons 5/6 was still observed, albeit weakly, in minigenes lacking the utmost 3′ sequences of the LINE1, indicating that the 303 bp at the 5′ end of F1 were the smallest LINE1-derived sequence sufficient to induce exon skipping (Fig. 5A, lanes 4–6). Smaller deletions of the minigene construct 5′-303 did not induce overt exon skipping (data not shown). As it is known that the direction of LINE1 insertion contributes to the severity of the LINE1-induced effects on mRNA levels (11, 20), we tested corresponding minigenes with sense inserts (with respect to Glrb), three of which exhibited pronounced skipping (Fig. 5A, lanes 10–12).
FIGURE 5.
Delineating the LINE1 minimal sequence required to induce exon skipping. A, truncations of F1 (corresponding to the 3′ UTR and adjacent 370 bp of ORF2) from either its 5′ end (constructs F1 3′-820, 3′-529, 3′-219) or its 3′ end (F1 5′-303, 5′-594, 5′-904) were generated by PCR and inserted into C-6.28A via AgeI (schematic in left panel). The respective constructs were transfected in HEK293 cells. RNA extracts from these cells were analyzed by RT-PCR using primers binding in exons 4 and 7, respectively. Skipping was more apparent in minigenes containing LINE1 3′ deletions (right panel, lanes 4–6) and when inserts were oriented in sense with respect to Glrb sequences (right panel, lanes 10–12). B, mutations (m1–m4) in fragment F1–5′-303 of predicted SRSF1 binding sites were introduced to reduce SRSF1 binding to the fragment. Putative SRSF1 binding sites are underscored, mutated residues in boldface type. Left panel, statistical analysis comparing 5′-303 and 5′-303m1–4. All values represent means ± S.E. (n = 3). Note that the combined mutations m1–4 resulted in a significant reduction of exon skipping (lanes 1–5). **, p > 0.001, one-way ANOVA followed by Bonferroni's multiple comparison test. C, skipping of exon 6 depends on the distance of the LINE1 insertion to the skipped exon. Upper panel, plasmid constructs were derived from the C-E6.28A minigene. A fragment of the LINE1 corresponding to its 5′ UTR (F5) was inserted at intronic positions IVS6 + 528, +870, +1154 into the C-E6.28A minigene via an AgeI site. The full-length LINE1 sequence was inserted into “skipping-permissive” B-WT introns 4 (IVS4.15513) and 7 (IVS7.873; this minigene also contains an exon 8 and adjacent intronic sequence IVS7–3720-4223) via a PmlI and SalI site, respectively. Lower panel, exon 6 skipping was significantly reduced with increasing distance from the exon (lanes 1–3, graph); for comparison, see Spa (lane 4). No skipping of exon 6 could be observed with insertions in introns 4 and 7 (lanes 5 and 6). Restoring the weak 5′ splice donor site (5′ ss) of intron 6 to consensus (replacing ..GCTgtatgt.. with ..CAGgtaagt … ) in the Spa construct prevented skipping despite the presence of the full-length LINE1 insertion (lane 7). All values represent means ± S.E. (n = 3). *, p < 0.05 (one-way ANOVA followed by Bonferroni's multiple comparison test).
To evaluate the hypothesis that sequestration of SRSF1 by the short LINE1 fragment would enhance skipping, SRSF1 binding motif prediction was performed using the 5′-303-bp fragment oriented in antisense (Spa) or sense with respect to Glrb, and sequences were mutated to abolish predicted binding sites (Table 2). Although neither single nor combined mutations of the fragment oriented in Spa direction diminished skipping further (data not shown), combined mutations in sense orientation lead to a significant increase in exon 6 inclusion, suggesting that interaction of SRSF1 with the LINE1-derived sequence is important for splicing modulation (Fig. 5B, lanes 2–5). In known examples of SR protein-mediated inhibition of exon inclusion, an interaction of the respective factors bound to intronic regulatory elements and the essential snRNPs attached at the 5′ and 3′ splice sites has been postulated (21, 25). To disrupt these putative short range interaction, we moved the F5 sequence of the LINE1, which has been shown to be sufficient for the induction of exon skipping downstream of its insertion site at IVS6.193 (F5 in Fig. 5C). This resulted in a diminished skipping of exon 6 with increasing distance from the exon (for quantification, see lower right panel of Fig. 5C). Similarly, the moving of the entire LINE1 into introns 4 or 7, respectively in a B-WT background, did not result in missplicing, suggesting that the proximity of the LINE1 insertion to the skipped exon is important. Furthermore, computational prediction revealed a weak 5′ splice donor site (5′ ss) of Glrb intron 6 (score < 0.15; NNSPLICE (26)), enabling SR proteins bound to a downstream intronic site to compete for binding of the essential small nuclear RNA protein U1. Accordingly, mutating the Glrb donor site to a perfect splicing consensus sequence abolished E6 skipping in the presence of a full-length LINE1 (Fig. 5C, lane 7). Thus, the Glrbspa allele is characterized by an intronic antisense LINE1 insertion, which amplifies the impairment of pre-mRNA splicing in a weak neighboring exon.
TABLE 2.
Prediction of SRSF1 binding in LINE1 fragment 303
Default thresholds were used for all web-based prediction tools. Binding sites mutated are indicated in boldface type. Predictions derived for the mutated sites are indicated after the slash. Mut, mutation.
| Position (start) sense | ESE finder (7-mer) 5′-303/5′-303 Mut | Altsplice (7-mer) 5′-303/5′-303 Mut | Rescue ESE (6-mer) 5′-303/5′-303 Mut |
|---|---|---|---|
| 2 | 2.411/2.411 | 3.057/3.057 | 4/4 |
| 16 | 4.493/n.d. | 4.664/n.d. (12:2.717) | |
| 26 | 2.284/n.d. | 2.745/n.d. | 27, 6/34, 2 |
| 40 | 2.240/2.240 | ||
| 48 | 2.040/n.d. | ||
| 51 | 5.291/n.d. | 5.390/n.d. | 3/56, 1 |
| 87 | 3.819/3.819 | 4.152/4.152 | 8/8 |
| 113 | 2.080/n.d. | 107, 3/107, 2 | |
| 138 | 2.876/2.876 | 3.083/3.083 | 1/1 |
| 156 | 12/8 | ||
| 175 | 2/3 | ||
| 189 | 5.738/n.d. | 6.058/n.d. | 187, 12/194, 8 |
| 215 | 3/3 | ||
| 248 | 5/5 | ||
| 265 | 1/1 | ||
| 279 | 3/3 | ||
| 291 | 1/1 |
DISCUSSION
In this study, we analyzed the molecular details resulting in LINE1-induced missplicing in the Glrb mutant mouse spastic, using a minigene approach. Comparison of splicing products obtained from the respective WT minigenes and with minigenes engineered from the Spa gene revealed differences in splicing similar to those found in vivo. We could show that skipping of exon 6 as seen in RNA preparations from spinal cords of Spa mice was reliably detectable in cells transfected with the Spa minigene, thus allowing a detailed analysis of the mechanisms leading to this missplicing phenotype. Interestingly, the combined skipping of exon 5 and 6 as seen in samples from GlrbSpa mice was highly variable between cell types or preparations and thus precluded further analysis. Using truncations of the inserted LINE1 as well as reconstruction of a Spa minigene on a C3H/HeJ background, we demonstrated that the splicing defect of Glrb pre-mRNA in Spa mice results from the interaction of a SNP affecting an ESE site with the adjacent intronic LINE1. In our assay system, the substitution of a single nucleotide restored normal wild-type splicing at the level of mRNA and allowed for the transcription of full-length protein, despite the presence of the full-length LINE1 insertion. The fact that the substitution of a single nucleotide was sufficient to significantly alter the LINE1 associated missplicing points to the importance of SNPs in the context of gene regulation. Glrb SNP rs13477223 belongs to the class of coding polymorphisms that do not alter protein sequence or splicing of the wild-type Glrb pre-mRNA. Our findings indicate, however, that the SNP can modulate pre-mRNA splicing and thereby alters the physiological function of the encoded protein when placed in a different genetic environment. As retrotransposition events are thought to be rare (one LINE1 insertion per 212 births (11, 27)) and point mutations occur at a much higher frequency (28), tissue-specific genetic variation might result from the interaction of intronic DNA repetitive elements or fragments thereof and an individual set of exonic SNPs within defined regulatory sequences. The strength of the splicing regulatory sequences, in our case an ESE localized within exon 6, then becomes critical to determine whether the adjacent retroelement becomes apparent phenotypically.
We have previously shown that the intronic insertion of an antisense LINE1 into Glrb intron 6 is associated with exon skipping (6). The exact mechanism, however, by which LINE1 insertions induce exon skipping had not yet been fully characterized. An overall decrease of mRNA levels due to LINE sequences has been observed for exonic and splice site insertions (12, 29). Although the disruption of consensus splice sites at the exon-intron border and of exonic splicing regulatory sites can account for missplicing events and the consecutive decrease in full-length transcripts, intronic insertions also result in a reduction of mature mRNA. The occurrence of length-dependent elongation defects (17) and premature polyadenylation (30) were found causative in experimental minigene systems and in vivo. The Glrbspa allele, however, is associated with exon skipping, but no LINE1-Glrb chimeric transcripts indicative of LINE1 cryptic splice site usage were detected in affected animals by Northern blot analysis (6).4 Moreover, by using in vivo splicing assays and analysis of full-length protein, we found comparable levels of correctly spliced cDNA and of full-length protein for both, Spa E6.28G with a LINE1 antisense insertion and WT minigenes. These findings suggest that LINE1 DNA is transcribed completely and subsequently spliced out. In the Glrbspa allele, distinct sequence fragments of the LINE1 appear to act analogous to intronic splicing silencers and a number of intronic silencer elements are known to promote exon skipping in the presence of SR proteins (21). In the Glrbspa allele, inclusion of exon 6 is hampered by a weak splice donor site and, in the C57BL/6J genomic background, a missing ESE. When the respective sites were improved by either substituting a residue at position 5 of the heptameric ESE predicted to bind SRSF1 (22, 31) or by restoring the 5′ splice donor site of Glrb intron 6 to consensus renders the adjacent LINE1 ineffective.
Our findings suggest that in Glrbspa, the LINE1 is interfering with the splicing machinery. We excluded the possibility that the LINE1 is transcribed from an internal antisense promotor. Furthermore, only defined segments of the LINE1 were associated with exon skipping: its 5′ and 3′ UTR and smaller fragments thereof, which were unlikely to function as promotor elements in this system. One possibility, by which an LINE1 element could interfere with pre-mRNA splicing, is the binding and sequestration of SR molecules and the interaction with defined regulatory proteins bound to adjacent exons, as in our study SRSF1. Because such interactions are limited with respect to distance, the impact of intronic LINE1 insertions most likely depends on the proximity to exons. Consistently, increasing the distance of the LINE1 to exon 6 ameliorated the skipping of the exon in our minigene assays. Moreover, not all exons in the vicinity appear to be affected similarly, as we have demonstrated by moving the full-length LINE1 to neighboring introns. The observation that the degree of exon skipping seen in in vivo minigene assays also depended on the cell type and diverged partially from the splicing seen in spinal cord tissue suggest that the effect of the LINE1 insertion is a combination of cis- and trans-acting factors that can thus be attenuated by cellular factors such as splicing regulatory proteins, even when sequence determinants are unfavorable, as in C57BL/6J Glrbspa.
In summary, we have shown that missplicing of the murine glycine receptor β subunit observed in the mutant mouse spastic is the result of a two-hit mutation consisting of a hypomorphic SNP that leads to the destruction of an ESE within exon 6 becoming functionally amplified by the insertion of a LINE1 in the adjacent intron of the Spa allele. Further aggravation comes from a weak splice donor site delineating the exon upstream of the LINE1 insertion site. Taken together, our findings provide insights in the molecular mechanisms of the LINE-induced changes in splicing observed in the mutant mouse spastic.
Supplementary Material
Acknowledgments
We thank Marina Wenzel, Annette Serwotka, and Rosa Weber for expert technical assistance and Drs. Katrin Schiebel and Pamela Strissel for a critical reading of the manuscript.
This work was supported by Deutsche Forschungsgemeinschaft (BE 1138/5-3), the Bundesministerium für Forschung und Bildung/Deutsch Israelische Projektkooperation (BMBF/DIP G3.2), the NIH (GM083187), and Fonds der Chemischen Industrie (160-43).
K. Becker, unpublished observations.
- GlyR
- glycine receptor
- LINE1
- long interspersed nuclear element 1
- ESE
- exonic splicing enhancer
- SRSF1
- SR protein splicing factor 1.
REFERENCES
- 1. Betz H., Laube B. (2006) Glycine receptors: Recent insights into their structural organization and functional diversity. J. Neurochem. 97, 1600–1610 [DOI] [PubMed] [Google Scholar]
- 2. Grudzinska J., Schemm R., Haeger S., Nicke A., Schmalzing G., Betz H., Laube B. (2005) The β subunit determines the ligand binding properties of synaptic glycine receptors. Neuron 45, 727–739 [DOI] [PubMed] [Google Scholar]
- 3. Oertel J., Villmann C., Kettenmann H., Kirchhoff F., Becker C. M. (2007) A novel glycine receptor β subunit splice variant predicts an unorthodox transmembrane topology. Assembly into heteromeric receptor complexes. J. Biol. Chem. 282, 2798–2807 [DOI] [PubMed] [Google Scholar]
- 4. Harvey R. J., Topf M., Harvey K., Rees M. I. (2008) The genetics of hyperekplexia: More than startle! Trends Genet. 24, 439–447 [DOI] [PubMed] [Google Scholar]
- 5. Kingsmore S. F., Giros B., Suh D., Bieniarz M., Caron M. G., Seldin M. F. (1994) Glycine receptor β-subunit gene mutation in spastic mouse associated with LINE-1 element insertion. Nat. Genet. 7, 136–141 [DOI] [PubMed] [Google Scholar]
- 6. Mülhardt C., Fischer M., Gass P., Simon-Chazottes D., Guénet J. L., Kuhse J., Betz H., Becker C. M. (1994) The spastic mouse: Aberrant splicing of glycine receptor β subunit mRNA caused by intronic insertion of L1 element. Neuron 13, 1003–1015 [DOI] [PubMed] [Google Scholar]
- 7. Becker C. M., Hermans-Borgmeyer I., Schmitt B., Betz H. (1986) The glycine receptor deficiency of the mutant mouse spastic: Evidence for normal glycine receptor structure and localization. J. Neurosci. 6, 1358–1364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Babushok D. V., Ostertag E. M., Kazazian H. H., Jr. (2007) Current topics in genome evolution: Molecular mechanisms of new gene formation. Cell Mol. Life Sci. 64, 542–554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Han J. S., Boeke J. D. (2005) LINE-1 retrotransposons: Modulators of quantity and quality of mammalian gene expression? Bioessays 27, 775–784 [DOI] [PubMed] [Google Scholar]
- 10. Brouha B., Schustak J., Badge R. M., Lutz-Prigge S., Farley A. H., Moran J. V., Kazazian H. H., Jr. (2003) Hot L1s account for the bulk of retrotransposition in the human population. Proc. Natl. Acad. Sci. U.S.A. 100, 5280–5285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kazazian H. H., Jr., Moran J. V. (1998) The impact of L1 retrotransposons on the human genome. Nat. Genet. 19, 19–24 [DOI] [PubMed] [Google Scholar]
- 12. Chen J., Rattner A., Nathans J. (2006) Effects of L1 retrotransposon insertion on transcript processing, localization, and accumulation: Lessons from the retinal degeneration 7 mouse and implications for the genomic ecology of L1 elements. Hum. Mol. Genet. 15, 2146–2156 [DOI] [PubMed] [Google Scholar]
- 13. Becker K., Hohoff C., Schmitt B., Christen H. J., Neubauer B. A., Sandrieser T., Becker C. M. (2006) Identification of the microdeletion breakpoint in a GLRA1null allele of Turkish hyperekplexia patients. Hum. Mutat. 27, 1061–1062 [DOI] [PubMed] [Google Scholar]
- 14. Burwinkel B., Kilimann M. W. (1998) Unequal homologous recombination between LINE-1 elements as a mutational mechanism in human genetic disease. J. Mol. Biol. 277, 513–517 [DOI] [PubMed] [Google Scholar]
- 15. Meischl C., Boer M., Ahlin A., Roos D. (2000) A new exon created by intronic insertion of a rearranged LINE-1 element as the cause of chronic granulomatous disease. Eur. J. Hum. Genet. 8, 697–703 [DOI] [PubMed] [Google Scholar]
- 16. Narita N., Nishio H., Kitoh Y., Ishikawa Y., Ishikawa Y., Minami R., Nakamura H., Matsuo M. (1993) Insertion of a 5′-truncated L1 element into the 3′ end of exon 44 of the dystrophin gene resulted in skipping of the exon during splicing in a case of Duchenne muscular dystrophy. J. Clin. Invest. 91, 1862–1867 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Han J. S., Szak S. T., Boeke J. D. (2004) Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature 429, 268–274 [DOI] [PubMed] [Google Scholar]
- 18. Hubbard T. J., Aken B. L., Ayling S., Ballester B., Beal K., Bragin E., Brent S., Chen Y., Clapham P., Clarke L., Coates G., Fairley S., Fitzgerald S., Fernandez-Banet J., Gordon L., Graf S., Haider S., Hammond M., Holland R., Howe K., Jenkinson A., Johnson N., Kahari A., Keefe D., Keenan S., Kinsella R., Kokocinski F., Kulesha E., Lawson D., Longden I., Megy K., Meidl P., Overduin B., Parker A., Pritchard B., Rios D., Schuster M., Slater G., Smedley D., Spooner W., Spudich G., Trevanion S., Vilella A., Vogel J., White S., Wilder S., Zadissa A., Birney E., Cunningham F., Curwen V., Durbin R., Fernandez-Suarez X. M., Herrero J., Kasprzyk A., Proctor G., Smith J., Searle S., Flicek P. (2009) Ensembl 2009. Nucleic Acids Res. 37, D690–697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Stoss O., Stoilov P., Hartmann A. M., Nayler O., Stamm S. (1999) The in vivo minigene approach to analyze tissue-specific splicing. Brain Res. Brain Res. Protoc. 4, 383–394 [DOI] [PubMed] [Google Scholar]
- 20. Humeny A., Bonk T., Becker K., Jafari-Boroujerdi M., Stephani U., Reuter K., Becker C. M. (2002) A novel recessive hyperekplexia allele GLRA1 (S231R): Genotyping by MALDI-TOF mass spectrometry and functional characterization as a determinant of cellular glycine receptor trafficking. Eur. J. Hum. Genet. 10, 188–196 [DOI] [PubMed] [Google Scholar]
- 21. Buratti E., Stuani C., De Prato G., Baralle F. E. (2007) SR protein-mediated inhibition of CFTR exon 9 inclusion: Molecular characterization of the intronic splicing silencer. Nucleic Acids Res. 35, 4359–4368 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cartegni L., Wang J., Zhu Z., Zhang M. Q., Krainer A. R. (2003) ESEfinder: A web resource to identify exonic splicing enhancers. Nucleic Acids Res. 31, 3568–3571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Houdayer C., Dehainault C., Mattler C., Michaux D., Caux-Moncoutier V., Pagès-Berhouet S., d'Enghien C. D., Laugé A., Castera L., Gauthier-Villars M., Stoppa-Lyonnet D. (2008) Evaluation of in silico splice tools for decision making in molecular diagnosis. Hum. Mutat 29, 975–982 [DOI] [PubMed] [Google Scholar]
- 24. Thanaraj T. A., Stamm S., Clark F., Riethoven J. J., Le Texier V., Muilu J. (2004) ASD: The Alternative Splicing Database. Nucleic Acids Res. 32, D64–69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Cartegni L., Chew S. L., Krainer A. R. (2002) Listening to silence and understanding nonsense: Exonic mutations that affect splicing. Nat. Rev. Genet. 3, 285–298 [DOI] [PubMed] [Google Scholar]
- 26. Reese M. G., Eeckman F. H., Kulp D., Haussler D. (1997) Improved splice site detection in Genie. J. Comput. Biol. 4, 311–323 [DOI] [PubMed] [Google Scholar]
- 27. Graham T., Boissinot S. (2006) The genomic distribution of L1 elements: The role of insertion bias and natural selection. J. Biomed. Biotechnol. 2006, 75327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. López-Bigas N., Audit B., Ouzounis C., Parra G., Guigó R. (2005) Are splicing mutations the most frequent cause of hereditary disease? FEBS Lett. 579, 1900–1903 [DOI] [PubMed] [Google Scholar]
- 29. Takahara T., Ohsumi T., Kuromitsu J., Shibata K., Sasaki N., Okazaki Y., Shibata H., Sato S., Yoshiki A., Kusakabe M., Muramatsu M., Ueki M., Okuda K., Hayashizaki Y. (1996) Dysfunction of the Orleans reeler gene arising from exon skipping due to transposition of a full-length copy of an active L1 sequence into the skipped exon. Hum. Mol. Genet. 5, 989–993 [DOI] [PubMed] [Google Scholar]
- 30. Perepelitsa-Belancio V., Deininger P. (2003) RNA truncation by premature polyadenylation attenuates human mobile element activity. Nat. Genet. 35, 363–366 [DOI] [PubMed] [Google Scholar]
- 31. Liu H. X., Zhang M., Krainer A. R. (1998) Identification of functional exonic splicing enhancer motifs recognized by individual SR proteins. Genes Dev. 12, 1998–2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.