Background: Expression of amyloid-β (Aβ) precursor protein (APP), implicated in Alzheimer disease (AD), is regulated by complex mechanisms involving microRNAs.
Results: miR-153 reduces APP and Aβ in human brain cultures and is dysregulated in AD.
Conclusion: miR-153 physiologically regulates human APP expression and Aβ and may contribute to AD pathoetiology.
Significance: miR-153 is a potential novel drug target in AD.
Keywords: Aging, Amyloid Precursor Protein, Cell Culture, Primary Neuronal Culture, Gene Regulation, Neurons, beta-Peptide, Dementia, Human Brain Tissue, Post-transcriptional, Small RNA
Abstract
Regulation of amyloid-β (Aβ) precursor protein (APP) expression is complex. MicroRNAs (miRNAs) are expected to participate in the molecular network that controls this process. The composition of this network is, however, still undefined. Elucidating the complement of miRNAs that regulate APP expression should reveal novel drug targets capable of modulating Aβ production in AD. Here, we investigated the contribution of miR-153 to this regulatory network. A miR-153 target site within the APP 3′-untranslated region (3′-UTR) was predicted by several bioinformatic algorithms. We found that miR-153 significantly reduced reporter expression when co-transfected with an APP 3′-UTR reporter construct. Mutation of the predicted miR-153 target site eliminated this reporter response. miR-153 delivery in both HeLa cells and primary human fetal brain cultures significantly reduced APP expression. Delivery of a miR-153 antisense inhibitor to human fetal brain cultures significantly elevated APP expression. miR-153 delivery also reduced expression of the APP paralog APLP2. High functional redundancy between APP and APLP2 suggests that miR-153 may target biological pathways in which they both function. Interestingly, in a subset of human AD brain specimens with moderate AD pathology, miR-153 levels were reduced. This same subset also exhibited elevated APP levels relative to control specimens. Therefore, endogenous miR-153 inhibits expression of APP in human neurons by specifically interacting with the APP 3′-UTR. This regulatory interaction may have relevance to AD etiology, where low miR-153 levels may drive increased APP expression in a subset of AD patients.
Introduction
Alzheimer disease (AD),2 the most common form of dementia (1), is thought to arise in part from excess accumulation of the amyloid-β peptide (Aβ) (2, 3). Aβ is derived from its parental molecule, Aβ precursor protein (APP), during a series of processing steps that result in the liberation of various soluble fragments (4). Internal cleavage by the β-secretase enzyme (β site APP-cleaving enzyme; BACE1) initiates Aβ production by cleaving APP at the Aβ N terminus, releasing a truncated secreted β form of APP. Cleavage by the γ-secretase complex at the Aβ C terminus results in the release of the soluble Aβ peptide and APP C-terminal fragments. Promiscuous cleavage by the γ-secretase complex results in the release of two C-terminal length variants of the Aβ peptide (Aβ(1–40) and Aβ(1–42)) that are the main constituents of one of the hallmark pathologies of the disease, extracellular neuritic plaques (5).
Overexpression of APP alone leads to AD in rare forms of the disease linked to specific genetic aberrations. These include Down syndrome (6), APP gene locus duplication events (7), and APP promoter polymorphisms (8, 9) that promote elevated expression. Therefore, therapeutic strategies that aim to reduce APP expression may be useful as a means to reduce Aβ production in sporadic AD and normalize APP expression in these more specific forms of the disease.
Given the centrality of APP and Aβ to AD pathology, it is imperative to elucidate the various mechanisms that regulate physiological expression of APP as a means to identify novel drug targets for modulating Aβ levels. The regulation of APP expression has been extensively studied, with controls being mediated at both the transcriptional and post-transcriptional level. The promoter structure is complex (10), containing various proximal (11–14) and distal promoter elements that mediate both constitutive and induced transcriptional regulation, including via elements located in the genomic 5′-untranslated region (5′-UTR) (15–18). Elements in the 5′-UTR and 3′-UTR also regulate transcript stability at the post-transcriptional level. Elements in the 5′-UTR include an IL-1-responsive element (19), an iron-responsive element (20), and an internal ribosomal entry site (21). In the 3′-UTR, several stability control elements bind various cytosolic proteins to stabilize the APP transcript (22–25). Alternative polyadenylation also regulates APP transcript stability through the inclusion or exclusion of a GG dinucleotide motif (26, 27).
MicroRNAs (miRNAs) are small (18–24 nucleotides) non-coding RNAs that interact with target mRNAs and mediate inhibitory controls on protein production (28). They generally base-pair to sites in the 3′-UTR of target mRNAs with imperfect complementarity, with the exception of a region at the 5′ end of an miRNA termed the seed sequence. Studies have shown that nearly perfect complementarity between the seed sequence and target mRNA is required for a functional interaction (29, 30). miRNAs exist in complex with protein mediators as part of the RNA-induced silencing complex (31), with AGO proteins serving as primary core proteins. Interactions between miRNAs and their target mRNAs bring the mRNA in close association with effector proteins that generally inhibit protein production either by transcript destabilization or translational inhibition (32), although recent studies suggest that transcript destabilization is the primary mechanism (33).
We and others have begun to describe the contributions that miRNAs bring to the post-transcriptional control of APP expression (34, 35). Specifically, we have recently described the negative regulatory control exerted by miR-101 on APP expression (34, 36). This finding has also been replicated in an independent laboratory (37). Several other miRNAs that modulate APP production have also been described (38–43). However, many additional miRNA target sites are predicted in the APP 3′-UTR. These miRNAs may mediate potent inhibitory effects and participate in the network of molecular regulators that control APP expression.
Here, we demonstrate that miR-153 inhibits expression of APP in human primary brain cultures via a specific target site in the APP 3′-UTR and is a participant in the endogenous molecular network that controls physiological APP expression. We further show that miR-153 is dysregulated in a subset of AD patients and that APP levels are inversely dysregulated, suggesting that the regulatory relationship may be relevant to AD pathology.
EXPERIMENTAL PROCEDURES
HeLa Cell Cultures and Transfection
HeLa cells were cultured in minimum essential medium (Mediatech) supplemented with 10% FBS (Atlanta Biologicals) and penicillin/streptomycin/amphotericin solution (Mediatech) at 37 °C in a 5% CO2 humidified incubator. Antibiotics and antimycotics were omitted from the media during all transfections. For co-transfections of DNA constructs and miRNA mimics (Dharmacon), HeLa cells were cultured in 96-well plates (5 × 104 cells/well) and transfected with 150 ng of DNA and 40 nm miRNA using 0.2 μl of Transfectin (Bio-Rad). For single transfections of siRNA (Applied Biosystems) or miRNA mimics, HeLa cells were cultured in 24-well plates (1.35 × 105 cells/well) and reverse transfected with 20 nm siRNA or 50 nm miRNA using 0.5 μl of Lipofectamine RNAiMAX (Invitrogen).
Generation of APP 3′-UTR Reporter Construct
The luciferase reporter construct was prepared as previously described (36). Briefly, the full-length APP 3′-UTR (1.2 kb) was PCR-amplified from the pGALA construct (20). The amplicon was then inserted into the XhoI site of psiCHECK-2 (Promega), immediately downstream of a Renilla luciferase coding sequence, using the InFusion cloning system (Clontech).
Site-directed Mutagenesis of Predicted miR-153 Target Site
The predicted target site in the APP 3′-UTR reporter construct was mutated using mutagenic primer-directed replication as implemented in the QuikChange Lightning site-directed mutagenesis kit (Agilent). The following primers were utilized to introduce seed sequence mutations in the APP 3′-UTR reporter: sense, 5′-cagctgcttctcttgcctaagtattcctttcctgatcaccgcatgttttaaagttaaacatttttaagtatttcagatgctttag-3′; antisense, 5′-ctaaagcatctgaaatacttaaaaatgtttaactttaaaacatgcggtgatcaggaaaggaatacttaggcaagagaagcagctg-3′.
Luciferase Reporter Assays
HeLa cells were transfected with the WT and mutant APP 3′-UTR reporter construct either alone or in combination with miRNA mimics, as described above. Forty-eight hours post-transfection, the Renilla and firefly luciferase activity was assayed using the Dual-Luciferase reporter system (Promega) on a Turner Biosystems Veritas luminomenter. Ratios of Renilla/firefly luminescence values were calculated and scaled relative to the value for the APP 3′-UTR reporter alone transfection.
Primary Human Fetal Brain Culture and Transfection
Primary cultures of mixed human fetal brain cells (HFB) were prepared from the brain parenchyma of aborted fetuses (80–100 days gestational age). The tissues were obtained from the Birth Defects Research Laboratory at the University of Washington with approval from the Indiana University Institutional Review Board. Fetal brain materials (10–20 g) were shipped overnight in chilled Hibernate-E medium (Invitrogen) supplemented with B27 (Invitrogen), GlutaMAX (Invitrogen), and antibiotic-antimycotic solution (Cellgro).
The culture procedures closely followed our previously described protocol (44) with some modifications. Briefly, the tissues were digested in 0.05% trypsin, 0.53 mm EDTA solution and incubated in a shaking water bath (150 rpm) at 37 °C for 15 min. The trypsin-digested tissues were transferred to Hibernate-E medium and triturated several times using a siliconized, fire-polished pipette followed by centrifugation at 400 × g for 15 min. The cell pellet was resuspended in Hibernate-E medium and triturated once more followed by centrifugation. The pellet was resuspended in culture medium (see below), and cells were counted by the trypan blue exclusion method.
The cells were plated on poly-d-lysine-coated 24-well plates in Neurobasal medium (Invitrogen), supplemented with 1× B27, 0.5 mm GlutaMAX, 5 ng/ml bFGF (Invitrogen), and antibiotic/antimycotic mixture. Half-medium changes were performed every fourth day of culture.
HFB cultures were transfected at day in vitro (DIV) 17 in 24-well plates. One culture batch was transfected with 20 nm siRNA and 150 nm miRNA mimics using 1.25 μl of RNAiMAX. A second culture batch was transfected with 1 μm LNA miRNA inhibitors (Exiqon) using 1.25 μl of RNAiMAX. Antibiotics and bFGF were omitted from media during transfections.
Immunohistochemical Analysis
Human fetal brain cultures were fixed in 4% paraformaldehyde for 15 min, washed three times with chilled PBS and then permeabilized with 0.12% Triton X-100 (Sigma) for 10 min. Permeabilized cells were blocked with 10% horse serum (Atlanta Biologicals) for 15 min followed by overnight incubation with primary antibodies.
Primary antibodies used in this study include a mouse pan-neuronal antibody mixture (Millipore) active against neurites, neuronal nuclei, and neuronal cell bodies; rabbit anti-GFAP (Sigma) for astrocyte labeling; and rabbit anti-nestin (Sigma) for neural stem cell labeling. Biotin-conjugated donkey anti-mouse (Jackson Immunoresearch) and Cy3-conjugated donkey anti-rabbit (Jackson Immunoresearch) secondary antibodies were used as described previously (44). Fluorescein-conjugated streptavidin (Jackson Immunoresearch) was employed to label biotin-conjugated secondary antibody. Nuclei were visualized using Hoechst stain (Sigma), and labeled cells were examined under a Leica DMIL HC inverted fluorescence microscope (Leica Microsystems). Images were captured using a SPOT RT-SE digital camera (Diagnostic Instruments).
Western Blotting Analysis
For protein analysis by Western blotting, cell lysates were prepared at various time points in culture as indicated in the figure legends. Briefly, cells were first washed with PBS and then lysed on plate with vigorous shaking using mammalian protein extraction reagent (Pierce) supplemented with 0.1% SDS and protease inhibitor mixture set III (Calbiochem). Lysates were centrifuged at 30,000 × g for 10 min at 4 °C, and cleared lysates were collected. Lysate protein concentrations were then assayed by BCA (Pierce) per the manufacturer's instructions. An equal amount of lysate protein (1–5 μg) was loaded onto BisTris XT denaturing 10% polyacrylamide gels containing SDS (Bio-Rad). Proteins were resolved by SDS-PAGE and transferred onto PVDF membranes. Membranes were blocked for 1 h in 5% nonfat milk and then incubated overnight with primary antibodies against APP (22C11, Chemicon), polyclonal C-terminal anti-APLP2 (Calbiochem), α-tubulin (B-5-1-2, Sigma), and β-actin (AC15, Sigma). Membranes were then incubated with HRP-conjugated goat anti-mouse secondary antibody (Rockland Immunochemicals) for 1 h. Bands were visualized using ECL reagent (Pierce), detected on film, and scanned.
ELISA of Aβ(1–40)
Levels of Aβ(1–40) were measured in the conditioned media (CM) of HFB cultures using a sensitive and specific commercially available ELISA (IBL America). Briefly, an equal volume of CM (25 μl) was loaded in a plate precoated with anti-human Aβ (35–40) antibody (clone 1A10) and incubated overnight. This kit uses anti-human Aβ (11–28) as a detection antibody. The overall assay was performed according to the manufacturer's instructions.
Absolute Aβ(1–40) values (in pg/ml CM) were measured. This value was normalized to the total lysate protein yield from each well to control for variability attributable to differences in cell number and scaled relative to mock transfection values.
Quantification of APP mRNA and Small RNA Expression Levels in Cell Culture
Total RNA was extracted from HeLa and HFB cultures using the miRVana miRNA Isolation kit (Ambion). RNA quantity and purity were assessed using a Nanodrop instrument (Thermo Scientific). RNA integrity was assessed on a Bioanalyzer (Agilent). All samples had acceptable A260/A280 ratios and RIN values greater than 8.5. Both mRNA and miRNA levels were quantified by reverse transcription quantitative PCR (RT-qPCR). Briefly, RNA was first converted to cDNA using the TaqMan microRNA Reverse Transcription kit (Applied Biosystems) for miRNA assays or High Capacity RNA-to-cDNA kit (Applied Biosystems) for mRNA assays. cDNA was then subjected to qPCR analysis using TaqMan hydrolysis probe assays (Applied Biosystems) for miRNA or mRNA on a 7300 real-time PCR instrument (Applied Biosystems). Relative quantification was performed using the ΔCq method and normalized to the geometric mean of at least three reference genes (45). For miRNA studies, RNU48, RNU6B, and hsa-miR-16 were used for normalization. For mRNA studies, GAPDH, B2M, β-actin, and TBP were used. Assay names and IDs are as follows: hsa-miR-153 (001191), RNU48 (001006), RNU6B (001093), RNU49 (001005), hsa-miR-16 (000391), human APP (all splice variants) (Hs01552283_m1), human GAPDH (4333764T), human B2M (4333766T), human ACTB (4333762T), and human TBP (4333769T).
Human Brain Specimens and Processing
Frozen brain specimens from the Harvard Brain Tissue Resource Center isolated from BA9 (Brodmann area 9) of the frontal cortex in age-matched control and AD patients were provided by Dr. P. H. Reddy. Non-control specimens were neuropathologically characterized as early (Braak stage I/II), definite (Braak stage III/IV), and severe (Braak stage V/VI) AD via Braak staging criteria (46). Sample demographics and post-mortem interval were the same as described previously (47). There were five specimens in each category. Specimens were initially pulverized using a stainless steel pulverizing chamber prechilled with liquid nitrogen and were quickly aliquoted, avoiding sample thawing.
One aliquot of each sample was processed for protein analysis. This frozen aliquot was immersed in mammalian protein extraction reagent (M-PER; Thermo) supplemented with 0.1% SDS and protease inhibitor mixture and immediately sonicated using a Sonifier Cell Disruptor 350 (Branson) until visible clumps disappeared. Lysates were then incubated with 50 units/ml Benzonase enzyme (Calbiochem) for 10 min at 37 °C to reduce nucleic acid content and associated viscosity. Lysates were then centrifuged down at 30,000 × g for 2 h to clear debris. Protein concentrations of the cleared lysates were determined by BCA. Western blot analyses were performed as described above.
A second aliquot was processed for RNA analysis. The frozen aliquot was immersed in cell disruption buffer from the miRVana miRNA isolation kit and immediately homogenized using a Polytron homogenizer (Kinematica). These homogenates were then processed per the manufacturer's instructions for tissue samples. RNA quality control was performed as described above. Several samples had low RIN values (<6), probably attributable to prior sample processing.
Quantification of Small RNA Expression Levels in Human Brain Specimens
RT-qPCR was performed on these RNA extracts as described above but with additional modifications. There was no detectable association between low RIN value and higher Ct values in this collection. Therefore, all samples were included in analyses.
Both relative and absolute quantification methods were employed for miRNA analyses in brain specimens. Relative quantification was performed using a modified ΔCq method as implemented in qBASE software (48). This modified ΔCq method determines relative amplicon levels by taking into account experimentally determined PCR amplification efficiencies for both the gene of interest and reference genes and by normalizing expression of the gene of interest to the geometric mean of multiple reference gene expression levels. In order to determine amplification efficiencies for each small RNA target, aliquots of each RNA sample in a given analysis were pooled and used to create a relative standard curve by serial dilution. These serial dilutions were then converted to cDNA and analyzed by qPCR in parallel with unknown samples. Amplification efficiencies were then determined by examining the slope of the standard curve for each small RNA target. For relative quantification studies, RNU48, RNU49, RNU6B, and hsa-miR-16 were used for normalization (assay IDs listed above).
To perform absolute quantification in miRNA analyses, an HPLC-purified synthetic oligoribonucleotide standard identical in sequence to hsa-miR-153 was obtained commercially (Sigma-Aldrich). The oligoribonucleotide was resuspended, and exact concentrations were measured by A260 measurements. Based on measured concentrations, standard curves with absolute copy counts were prepared by serial dilution. These serially diluted standards were converted to cDNA and analyzed by qPCR in parallel with unknown samples. Copy counts per reaction were then determined based upon standard curve analysis. Given that each unknown reaction was loaded with a known amount of total RNA (generally 10 ng), copy counts were then presented as copy counts/15 pg of total RNA. This serves as a rough estimate of copy counts per average human cell.
Data and Statistical Analysis
Densitometric analysis of Western blots was performed using ImageJ software. qPCR data analysis and normalization were performed using qbasePLUS software (48). Fluorescence micrograph and Western blot image processing was performed using Adobe Photoshop. Western blot images were adjusted for contrast and brightness, and some extraneous sections of blots between boxed regions were removed for the sake of clarity. No manipulations have been made to images that alter data quantification or interpretation. Statistical analyses were performed using Prism GraphPad and SPSS. For comparison between two categories, two-tailed Student's t test was performed. For comparison across multiple categories, ANOVA was performed followed by post hoc Dunnett's t test, Tukey's honest significant difference (HSD) test, or Student-Neuman-Keuls test for multiple comparison corrections. The α threshold for statistical significance was set at 0.05.
RESULTS
Bioinformatic Prediction of miR-153 Target Site in APP 3′-UTR
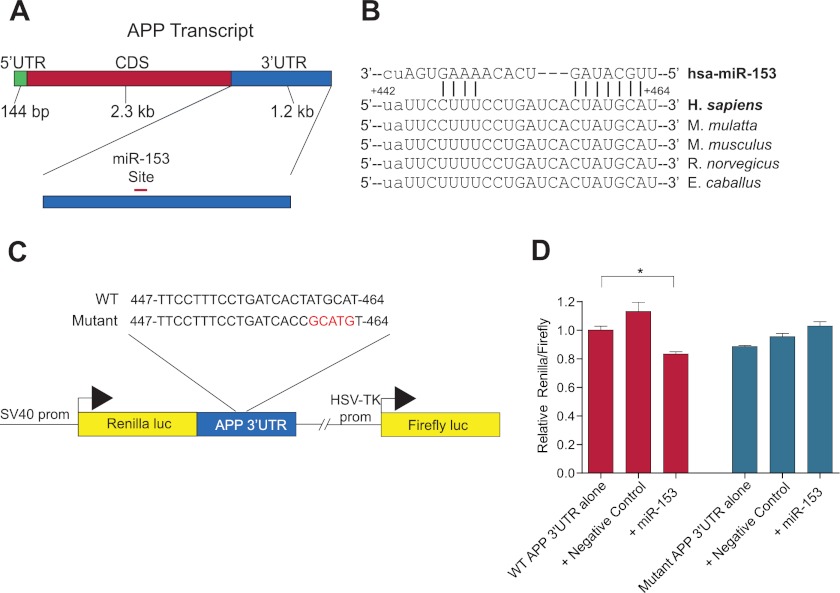
To identify novel miRNA target sites in the human APP 3′-UTR, we used multiple Web-based bioinformatics algorithms to predict favorable miRNA interactions. A target site for miR-153 was predicted by TargetScan version 6.0 (30, 49–51), PicTar (52), DIANA-MicroT version 4.0 (53, 54), miRanda-mirSVR (55–57), and rna22 (58) in the APP 3′-UTR at positions +442 to +464 relative to the start of the 3′-UTR (Fig. 1A). A previously validated miR-101 target site in the APP 3′-UTR (36, 37) was also predicted. Scores generated by the algorithms are compared between the validated miR-101 target site and predicted miR-153 target site in Table 1. The significant level of identity between the sequence of the predicted miR-153 target site in the human APP 3′-UTR and orthologous sequences from multiple mammalian species suggests that this specific site has been under evolutionary pressure to retain sequence conservation during mammalian speciation (Fig. 1B).
FIGURE 1.
Identification and reporter validation of the putative miR-153 target site in the APP 3′-UTR. A, schematic of the 3.6-kb APP mRNA transcript demonstrating the approximate location of the predicted miR-153 target site in the 3′-UTR. B, sequence and predicted base pairing of human miR-153 with its predicted target site in the human APP 3′-UTR, including the seed sequence interaction (red box). Sequences from rhesus macaque, mouse, rat, and horse from positions orthologous to the predicted miR-153 target site in the human APP 3′-UTR demonstrate strong sequence conservation at this site. C, schematic of the APP 3′-UTR reporter construct containing the APP 3′-UTR located downstream of the Renilla luciferase coding sequence. Firefly luciferase is independently transcribed and serves as an internal control. Both a WT and mutant construct containing a mutated seed sequence in the predicted miR-153 target site (in red) were prepared. D, reporter assay demonstrating functional activity of miR-153 against the APP 3′-UTR and specificity of predicted target site. The WT and mutant APP 3′-UTR reporter constructs were transfected into HeLa cells either alone or in combination with a negative control or miR-153 mimic (40 nm). Renilla and firefly luciferase assays were performed 48 h post-transfection and analyzed as relative ratios of Renilla to firefly luciferase activity. Co-transfection of miR-153 with WT reporter resulted in reduced Renilla luciferase expression relative to reporter alone or negative control (*, p = 0.015 by Tukey's HSD test; n = 6). No inhibitory effect of miR-153 on reporter expression was observed in co-transfections with the mutant reporter construct. Red bars, transfections with the WT reporter construct. Blue bars, transfections with the mutant reporter construct. CDS, coding sequence; luc, luciferase; prom, promoter. Error bars, S.E.
TABLE 1.
APP 3′-UTR miR-153 target site prediction and score summary
| Target site predictor | miR-153 sitea predicted? | miR-153 score | miR-101b score |
|---|---|---|---|
| TargetScanHuman 6.0c | Yes | −0.22 | −0.35 |
| PicTard | Yes | 2.50 | 5.21 |
| DIANA-microT version 4.0e | Yes | 0.414 | 0.307 |
| miRanda-mirSVRf | Yes | −1.259 | −1.78 |
| PITAg | Yes | −4.44 | −1.61 |
| rna22h | Yes | −21.3 | Not predicted |
a Site prediction based on seed sequence location as indicated in Fig. 1; duplex pairing varies among predictors.
b Scores for conserved miR-101 site with seed sequence at nucleotides 242–248.
c Context plus score.
d PicTar score.
e Site score.
f mirSVR score.
g ddG score.
h Folding energy (kcal/mol), M = 14, G = 0, E = −20 kcal/mol.
Reporter Validation of Predicted miR-153 Target Site
To confirm functionality and test efficacy of this putative target site, we constructed an APP 3′-UTR reporter construct containing the full-length APP 3′-UTR (1.2 kb) inserted downstream of a Renilla luciferase coding sequence. A separate firefly luciferase coding sequence under independent transcriptional control was also present in this construct to serve as an internal control (Fig. 1C).
Co-transfection of the reporter construct along with miR-153 mimic in HeLa cells resulted in significantly reduced expression of reporter relative to co-transfection with negative control mimic or transfection of reporter construct alone (83% of reporter alone), suggesting an inhibitory regulatory interaction between miR-153 and the APP 3′-UTR (Fig. 1D). To probe for the site of interaction between miR-153 and the APP 3′-UTR, we mutated the seed sequence of the putative miR-153 target site in the reporter construct (Fig. 1C). The criticality of seed sequence interactions for most functional miRNA-based regulation has been clearly demonstrated (28), and mutation at this position should eliminate effective interaction between miRNA and the target site. miR-153 mimic had no inhibitory effect on reporter expression when co-transfected with this mutant reporter construct in HeLa cells (Fig. 1D), thereby confirming that the predicted miR-153 target site is indeed responsible for mediating the inhibitory interaction between miR-153 and the WT reporter mRNA.
miR-153 Down-regulates Expression of Endogenous APP
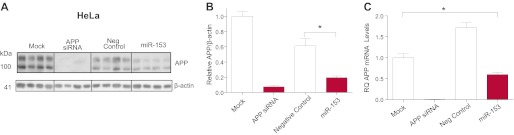
In order to confirm the reporter analysis data and check that miR-153 is capable of inhibiting the expression of endogenous APP, we transfected HeLa cells with miR-153 mimic and assayed APP levels directly by Western blot. Delivery of exogenous miR-153 was very effective in HeLa cells because miR-153 levels measured post-transfection were found to be dramatically increased as compared with mock-transfected cells (nearly 200,000-fold increase). Transfection of miR-153 mimic significantly reduced APP levels by nearly 70% compared with transfection with negative control mimic (Fig. 2, A and B). miRNAs may mediate their effects on protein output by inhibiting translation or promoting mRNA degradation. Transfection of miR-153 in HeLa significantly reduced APP mRNA levels by nearly 40% (Fig. 2D), suggesting that both mechanisms may be in play.
FIGURE 2.
miR-153 inhibits expression of endogenous APP mRNA and protein. HeLa cells were either mock-transfected or transfected with 20 nm APP siRNA, 50 nm negative control, or 50 nm miR-153 mimic. RNA was extracted, and protein cell lysates were prepared 48 and 72 h post-transfection, respectively, as described under “Experimental Procedures.” A, APP and β-actin protein levels were measured by Western blot analysis. B, densitometric analysis of APP normalized to β-actin revealed that miR-153 significantly reduced APP protein levels relative to mock or negative control mimic transfections (*, p = 0.002 by Tukey's HSD test; n = 4). C, APP mRNA levels were significantly decreased following miR-153 transfection as measured by RT-qPCR (*, p = 0.01 by Tukey's HSD test; n = 3). RT-qPCR expression levels were normalized to the geometric mean of β-actin, B2M, GAPDH and TBP expression levels and further scaled relative to mock-transfected levels. RQ, relative quantification. Error bars, S.E.
Characterization of APP and miR-153 Levels in Human Brain Cultures
HeLa and many other cancer cell lines express endogenous miR-153 levels at inherently low levels, making them poorly suited for studying the role of endogenous miR-153 in regulating putative mRNA targets, such as APP. As a more suitable model, we have recently been successful in preparing primary HFB cultures derived from aborted fetal brain parenchyma.
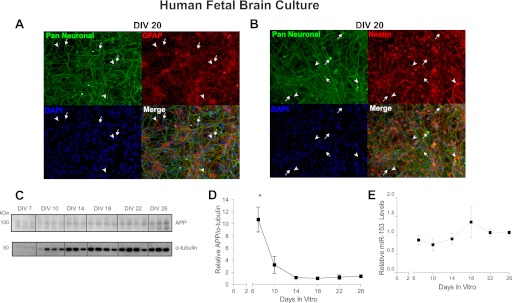
To better characterize the distribution of cellular phenotypes in these cultures, cells were fixed at specific time intervals during culture ranging from DIV 8 to 24. One set of cells was co-labeled with the combination of a pan-neuronal antibody mixture designed to label neurites, cell soma, and nucleus and anti-GFAP. A second set was co-labeled with anti-nestin and anti-GFAP. Early cultures (e.g. DIV 8) consisted almost entirely of cells co-expressing GFAP, nestin, and neuronal markers (data not shown). Later stage cultures (e.g. DIV 20) consisted of a mixture of cells. Some cells co-expressed GFAP and neuronal markers (Fig. 3A) or GFAP and nestin (Fig. 3B). Other cells expressed only neuronal markers, GFAP, or nestin (Fig. 3, A and B). Given that radial glia represent the predominant neural stem cell in the developing human cortex (59) and that nestin is a known marker of neural stem cells (60), we conclude that our late stage culture contains a mixture of immature neural stem cells (GFAP-, nestin-, and neuronal marker-positive) and differentiated neurons and astrocytes (positive for a single marker). The persistence of neural stem cells in the culture might be explained by the maintenance of bFGF in the medium throughout culture. A previous study has also described a similar mixture of cultured cells derived from human fetal brain parenchyma (61). Importantly, our primary HFB culture closely mimics the in vivo fetal brain cellular network.
FIGURE 3.
Time profile of APP and miR-153 levels in an HFB culture. A and B, HFB cultures at DIV20 following continuous bFGF exposure were co-labeled with a pan-neuronal antibody mixture and anti-GFAP (A) or with anti-nestin and anti-GFAP (B). Significant co-labeling with each combination as well as individual labeling with pan-neuronal and anti-GFAP antibodies suggests the presence of immature neural stem cells as well as both differentiated neurons and astrocytes. Arrows point to cells only labeled the pan-neuronal mixture. The arrowheads point to cells only labeled by either anti-GFAP or anti-nestin. C, Western blot analysis of APP and α-tubulin levels across time (DIV 7–26) in a HFB culture. D, densitometric analysis of APP normalized to α-tubulin demonstrated that APP levels rapidly decrease from DIV 7 to 14 and exhibit the lowest expression levels at DIV 18 (*, p < 0.001 versus all time points by Tukey's HSD test). E, RT-qPCR analysis of miR-153 levels across time in the same HFB culture as in B and C. miR-153 expression exhibits an inverse pattern relative to APP, with the highest expression at DIV 18, although there are no statistically significant differences between any of the time points (ANOVA, p = 0.462). Error bars, S.E.
We next assayed levels of APP and miR-153 at DIV 7, 10, 14, 18, 22, and 26 in HFB to better characterize the nature of the culture with respect to these key analytes. Western blot analysis of APP levels revealed very high levels of APP expression at DIV 7 that plateaued over time, with the lowest expression levels observed at DIV 18 (Fig. 3, C and D). Interestingly, miR-153 had a somewhat inverse expression pattern, with the highest expression levels at DIV 18, although no differences in expression were statistically significant (Fig. 3E). These data demonstrate that APP and miR-153 exhibit inverse expression patterns across time in HFB culture and suggest that the inhibitory relationship between miR-153 and APP could underlie these patterns.
Endogenous miR-153 Regulates APP Expression in Human Brain Cultures
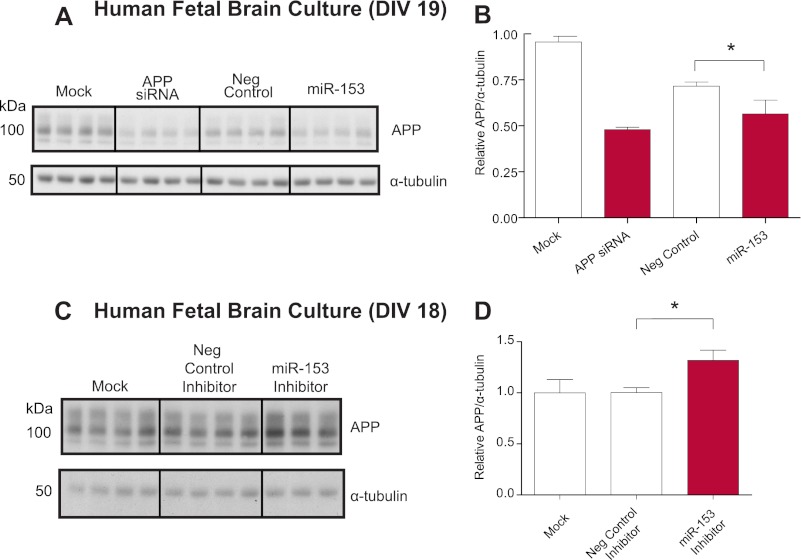
We first sought to establish whether delivery of miR-153 to HFB cultures would reduce APP expression as observed with HeLa. Therefore, we transfected DIV 17 HFB cultures with miR-153 mimic and assayed APP expression by Western blot (Fig. 4A). Transfection of miR-153 significantly reduced APP expression by ∼20% relative to transfection with negative control mimic (Fig. 4B). Therefore, we were able to successfully transfect these primary human neuronal cells and demonstrate down-regulation of APP following miR-153 delivery.
FIGURE 4.
miR-153 endogenously regulates APP expression in HFB cultures. A, Western blot analysis of APP and α-tubulin levels in transfected HFB cultures. HFB cells at DIV 17 were either mock-transfected, transfected with 20 nm APP siRNA, or transfected with 150 nm negative control or miR-153 mimic. Cell lysates were prepared 48 h post-transfection. B, densitometric analysis of APP levels normalized to α-tubulin levels demonstrated that miR-153 significantly reduced APP expression in HFB cells (*, p < 0.05 by Student-Neuman-Keuls test; n = 4). C, Western blot analysis of APP and α-tubulin levels in transfected HFB cultures. HFB cells at DIV 17 were either mock-transfected or transfected with 1000 nm negative control or miR-153 antisense inhibitor. Cell lysates for proteins were prepared 24 h post-transfecton. D, densitometric analysis of APP normalized to α-tubulin demonstrated that miR-153 inhibitor significantly increased APP expression in HFB cells (*, p = 0.018 by post hoc Dunnett's t test; n = 3–4). Error bars, S.E.
To establish whether miR-153 participates in the regulatory network that controls APP expression in human neurons, we utilized miRNA antisense inhibitors to bind to and disrupt the interaction of miR-153 with mRNA targets. We transfected DIV 17 HFB cultures with miR-153 inhibitor and assayed expression of APP by Western blot. Transfection of miR-153 inhibitor significantly increased APP expression relative to negative control inhibitor transfection by 30% (Fig. 3, C and D). Therefore, endogenous miR-153 actively inhibits APP expression in human neurons under physiological culture conditions.
miR-153 Inhibits Production of Secreted Aβ Peptide in Human Neurons
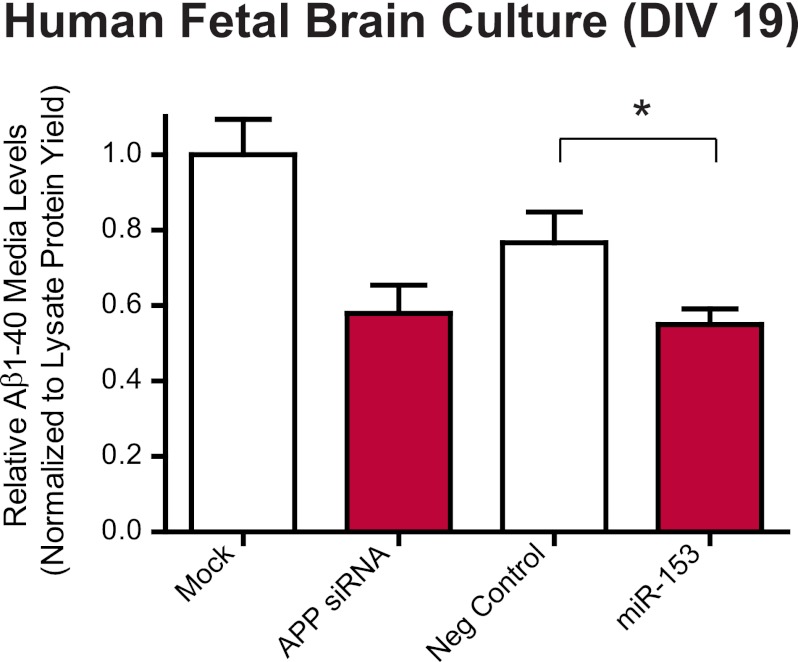
Our hypothesis was that inhibition of APP by miR-153 would lead to reduced production of APP metabolites, including the neurotoxic Aβ peptide. We tested this hypothesis by measuring levels of Aβ(1–40) by a sensitive sandwich ELISA in the CM of HFB cultures transfected with miR-153 at DIV 17 and harvested at DIV 19. To control for any variability associated with differences in cell number, absolute Aβ(1–40) values (pg/ml) obtained by ELISA were normalized to cell lysate total protein yield as a surrogate for absolute cell number and expressed relative to mock-transfected cells. Aβ(1–40) levels were significantly decreased (by ∼30%) in HFB cultures transfected with miR-153 as compared with negative control mimic transfections (Fig. 5). This confirms that miR-153 can also regulate Aβ levels, presumably via its activity against APP.
FIGURE 5.
miR-153 reduces secretion of Aβ(1–40) into the conditioned media of HFB cultures. HFB cells at DIV 17 were either mock-transfected, transfected with 20 nm APP siRNA, or transfected with 150 nm negative control or miR-153 mimic. Conditioned media were collected 48 h post-transfection. Aβ(1–40) levels were measured in CM by ELISA as described under “Experimental Procedures.” Absolute values (pg/ml) were normalized to the total protein yield of crude cell lysates and scaled relative to mock transfection to account for variability associated with differences in cell number and viability as described under “Experimental Procedures.” Transfection of miR-153 significantly reduced levels of Aβ(1–40) released in the CM of HFB cultures as compared with negative control-transfected cultures (*, p = 0.04 by Student's t test; n = 4). Error bars, S.E.
miR-153 Also Down-regulates Expression of APP-like Protein 2 (APLP2) in Human Brain Cultures
APLP2 3′-UTR contains a miR-153 target site predicted by TargetScan (seed sequence located at nucleotides 1264–1270 relative to the 3′-UTR start site). This is significant because APP and APLP2 are highly redundant protein products that probably function in similar biological pathways important for neurodevelopment (62). Dual regulation of APP and APLP2 by a single miRNA may signal a master mechanism for controlling a biological pathway with built-in redundancy.
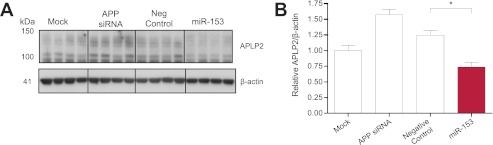
To determine whether delivery of miR-153 may also down-regulate APLP2, we transfected DIV 17 HFB cultures with miR-153 mimic and assayed APLP2 expression by Western blot (Fig. 6A). Transfection of miR-153 significantly reduced APLP2 expression by ∼35% relative to transfection with negative control mimic (Fig. 6B). Therefore, miR-153 is capable of down-regulating of APLP2 expression in human brain cultures.
FIGURE 6.
miR-153 down-regulates expression of APLP2 in primary HFB cultures. A, Western blot analysis of APLP2 and β-actin levels in transfected HFB cultures. HFB cells at DIV 17 were either mock-transfected, transfected with 20 nm APP siRNA, or transfected with 150 nm negative control or miR-153 mimic. Cell lysates were prepared 48 h post-transfection. B, densitometric analysis of APLP2 levels normalized to β-actin levels demonstrated that miR-153 significantly reduced APLP2 expression in HFB cells (*, p < 0.01 by post hoc Tukey's HSD test; n = 4). Error bars, S.E.
APP and miR-153 Are Inversely Dysregulated in AD Brain Specimens
Several miRNA species have been previously reported to be dysregulated in the post-mortem AD brain (63–67). We next investigated whether miR-153 might also be dysregulated. We analyzed APP expression by Western blot and miR-153 expression by RT-qPCR in brain specimens from control and early and late stage AD patients (Braak stages I/II (early), stages III/IV (definite), and stages V/VI (severe)) isolated from the frontal cortex (BA9). We also grouped specimens into two higher level categories for analysis: 1) control and Braak stage I/II specimens and 2) Braak stage III, IV, V, and VI specimens. The rationale for comparing these two supergroups is that neurofibrillary tangle (NFT) pathology (the basis of Braak staging) does not progress into the neocortex until Braak stages III and IV (46). Because specimens analyzed here are from BA9 of the neocortex, the progression from Braak stage II to stage III delineates a distinct transition in the pathogenic environment of this region of the brain. Clinicopathological correlation studies have also demonstrated that the progression of NFT in the neocortex better correlates with cognitive decline than stages of disease where NFT are restricted to the allocortex (68). Grouping also served to enhance the power of analysis by increasing the sample size of each group to 10.
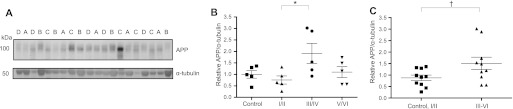
When compared across Braak stages, APP levels were significantly increased in Braak stage III/IV AD samples (Fig. 7, A and B). When compared between the two supergroups, APP expression was also significantly elevated in specimens with the presence of neocortical NFT pathology (stages III–VI) (Fig. 7C).
FIGURE 7.
APP levels are dysregulated in advanced AD brain specimens. A, Western blot analysis of APP and α-tubulin levels in control (A), early (stage I/II) (B), definite (stage III/IV) (C), and severe (stage V/VI) (D) AD brain specimens. B, densitometric analysis of APP levels normalized to α-tubulin levels demonstrated that APP levels were significantly elevated in definite (Braak stages III/IV) AD specimens (*, p = 0.041 by post hoc Tukey's HSD test; n = 5). C, densitometric analysis also revealed that APP levels were significantly increased in specimens with neocortical NFT pathology (Braak stages III–VI) as compared with specimens lacking neocortical NFT pathology (control and stages I/II) (†, p = 0.048 by Student's t test; n = 10). Error bars, S.E.
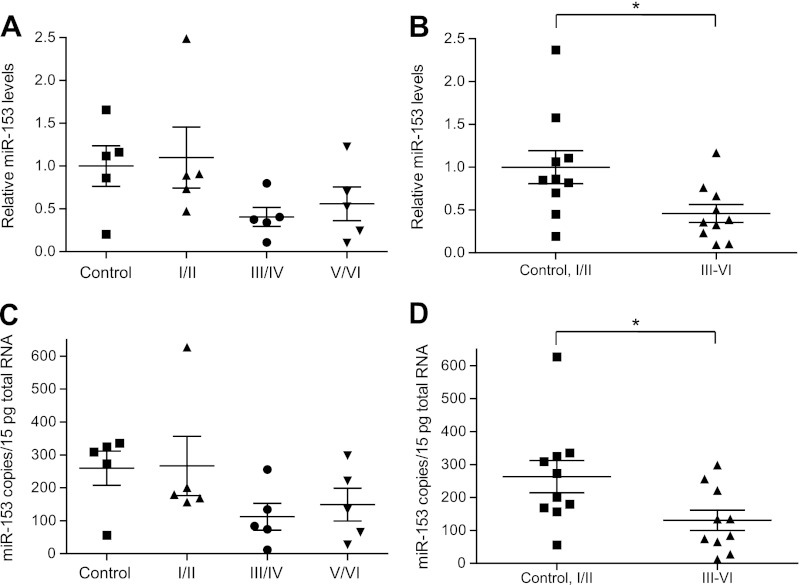
Interestingly, miR-153 levels were significantly decreased in a similar pattern. When compared across Braak stages, miR-153 levels were detectably lower in stage III/IV and stage V/VI specimens, although these trends did not reach statistical significance following corrections for multiple comparisons (Fig. 8, A and C). However, miR-153 levels were significantly decreased in specimens with neocortical NFT pathology (stages III–VI) as compared with earlier stage specimens (control, stage I/II) (Fig. 8, B and D).
FIGURE 8.
miR-153 levels are dysregulated in advanced AD brain specimens. miR-153 levels were quantified by RT-qPCR analysis. In A and B, relative expression levels were normalized to the geometric mean of four endogenous controls: RNU6B, RNU48, RNU49, and miR-16. In C and D, expression levels were quantified in absolute terms as miRNA copy counts/15 pg of total RNA using standard curves prepared from serial dilutions of miRNA oligonucleotide standards with known concentrations. In A and C, expression levels were compared across control and Braak stage I/II, III/IV, and V/VI specimens. In B and D, expression levels were compared across two supergroups either with neocortical NFT pathology (Braak stages III/VI) or without neocortical NFT (control and Braak stages I/II). A, miR-153 levels were lowest in Braak stages III/IV and stages V/VI, but no statistical difference was observed between groups (ANOVA, p = 0.215). B, miR-153 levels were significantly decreased in AD specimens with neocortical NFT (Braak stages III–VI) as compared with specimens lacking neocortical NFT (control, stages I/II) (*, p = 0.024 by Student's t test). C, miR-153 levels were lowest in Braak stages III/IV and stages V/VI, but no statistical difference was observed between groups (ANOVA, p = 0.420). D, as in B, miR-153 levels were significantly decreased in AD specimens with neocortical NFT as compared with specimens without (*, p = 0.035 by Student's t test). Error bars, S.E.
Importantly, decreased miR-153 levels were observed when using two distinct RT-qPCR quantification strategies: 1) relative quantification with normalization to the geometric mean of multiple small RNA reference controls (RNU6B, RNU48, RNU49, and miR-16) (Fig. 8, A and B) and 2) absolute quantification by comparison with standard curves prepared from synthetic miRNA oligonucleotides (Fig. 8, C and D). Therefore, changes in miRNA expression patterns cannot be attributed to normalization bias. The inverse pattern of APP and miR-153 dysregulation in advanced stage AD specimens with neocortical NFT suggests that decreased miR-153 levels may contribute to elevated APP in these specimens.
DISCUSSION
This study outlines a novel inhibitory interaction between miR-153 and the APP transcript, an interaction that participates in the physiological regulatory scheme responsible for the control of APP protein levels in human cultured primary brain cells. This claim is supported by multiple findings described above. First, mutation of the predicted miR-153 target site eliminated inhibition of reporter expression mediated by miR-153, indicating that sequence integrity of this site is essential for a functional regulatory interaction. Second, direct delivery of miR-153 to both HeLa and human brain cells down-regulated expression of endogenous APP. Finally, disruption of miR-153 function by use of an antisense inhibitor elevated APP levels in human brains cells, confirming that miR-153 basally inhibits APP expression in human brain cells under typical culture conditions.
A recent study has shown that direct exogenous delivery of miR-153 into HeLa cells or Neuro2A cells can reduce APP expression (40). This study also tested the effect of an AD-specific SNP in the APP 3′-UTR directly juxtaposed to the miR-153 target site seed sequence. They found, however, that this SNP did not influence the functional effect of miR-153 on reporter expression. Our study still stands, to our knowledge, as the first to demonstrate that miR-153 physiologically regulates APP expression and does so in human primary brain cells.
An additional putative miR-153 target was also examined here. The APP paralog, APLP2, has a predicted miR-153 target site in the 3′-UTR. We found that miR-153 delivery in human fetal brain cultures also reduced endogenous APLP2 expression. Therefore, both APP and APLP2 appear to be bona fide targets of miR-153. Given the redundancy in function between APP and APLP2, as suggested by the subtle phenotype of single gene knockouts but postnatal lethality associated with APP-APLP2 double knockouts (62), it is tempting to speculate that miR-153 may target both of these gene products as an evolutionarily conserved mechanism to regulate certain biological pathways in which they both participate. The predicted APLP2 target site does not appear to be paralogous to the target site in the APP 3′-UTR, suggesting that convergent evolutionary mechanisms may have promoted the appearance and maintenance of miR-153 3′-UTR target sites in APP and APLP2. The functional efficacy of this site must be first validated to confirm that miR-153 mediates its inhibitory effects on APLP2 expression through it.
A third important discovery demonstrated here is that miR-153 levels were significantly decreased in the cohort of advanced AD post-mortem brain specimens with neocortical NFT pathology (Braak stage III–VI) as compared with specimens lacking neocortical NFT pathology (control and Braak stage I/II specimens). Importantly this trend survived multiple normalization schemes, confirming that normalization bias was not responsible for this trend. Interestingly, APP levels were also significantly increased in Braak stage III–VI specimens. This raises the intriguing possibility that the decrease in miR-153 in these specimens may at least partially underlie the elevated APP levels.
One might question why dysregulated expression of miR-153 and APP only appears at Braak stage III and beyond in this analysis and is not represented progressively across Braak stages. One consideration in response is that all of these specimens were derived from the frontal cortex (BA9), a region that only begins to become invested with neurofibrillary pathology in stages III and IV of the disease (46). A second consideration is that changes in miR-153 and APP expression would be expected to contribute to amyloid pathology in the AD brain. The development of amyloid and neurofibrillary pathology do not initially overlap anatomically and do not progress with direct linear correlations between each other in the AD brain (5). Therefore, we might not expect dysregulation of APP and miR-153 to vary linearly with Braak staging.
We are aware of certain caveats in our analysis. First, the sample size here is small (n = 5–10/group), and the reliability of the finding would be greatly aided if it was observed in an independent cohort of larger sample size. Second, RNA integrity of some samples was rather low due to prior processing of specimens, possibly introducing bias into the analysis. However, specimens in Braak stages III–VI were not preferentially represented among the low RNA integrity extracts. The low integrity extracts also did not demonstrate detectably higher Ct values or lower normalized miR-153 expression values (data not shown). Again, replication of these findings in an independent cohort of low post-mortem interval specimens and high RNA quality extracts would address these concerns. It should also be noted that procuring a large sample of high quality, low post-mortem interval AD brain tissue spread across Braak stages is a significant challenge. A final caveat is that we are not able to rule out that the “dysregulation” of miR-153 and APP might be an epiphenomenal manifestation of cell type distribution changes that occurs during the progression of AD. During the course of disease, neurons are lost, and astrogliosis results in increased relative numbers of glia to neurons (69). Therefore, changes at the molecular level may simply reflect changes in cell types. However, both APP (70) and miR-153 (71) are more highly expressed in neurons than astrocytes, suggesting that the changes observed cannot be solely explained by changes in cell type distribution. Clarification could be provided by the analysis of miR-153 by in situ hybridization and APP by immunohistochemistry in sections from specimens across Braak stages. In situ hybridization and immunohistochemistry allows for cellular level resolution of miRNA and protein levels, respectively.
Other studies have profiled miRNA expression in the AD brain (38, 64–67) and in peripheral blood mononuclear cells (72) and identified miRNAs that are dysregulated (e.g. miR-107, miR-29a/b, miR-106b, etc.) (see Ref. 73 for a review). None of these studies to our knowledge have previously identified miR-153 among these. One study specifically measured miR-153 levels in AD brain using a DNA dot blot array but was unable to detect any miR-153 expression in specimens due to low sensitivity (63). Because most of these studies did not segment brain specimens by severity of NFT pathology (Braak staging), one would not necessarily expect that the decrease in miR-153 expression observed here would be replicated in these studies.
miR-153 certainly is functional in other molecular pathways aside from APP. This miRNA was originally reported to be brain-specific (74), although other studies have reported detection outside of the brain (75–77). miR-153 has been proposed to function as a tumor suppressor based upon its low expression in cancer cell lines versus normal tissue (75, 76) and by the effect of miR-153 overexpression to reduce cancer cell line viability (78). This tumor suppressor activity may be mediated by inhibitory targeting of antiapoptotic and prosurvival pathways. Two demonstrated targets of miR-153 in glioblastoma multiforme cell lines include B-cell lymphoma 2 (Bcl-2) and myeloid cell leukemia sequence 1 (Mcl-1), two antiapoptotic proteins (78). miR-153 also down-regulates insulin receptor substrate 2 (Irs2), thereby inhibiting the prosurvival effect of the PI3K/Akt signaling pathway (79). In drug-resistant leukemic cancer cells, miR-153 has been shown to be down-regulated. Restoring miR-153 levels in these cells was shown to sensitize them to As2O3-induced apoptosis (33). Finally, in an experimental model of pulmonary fibrosis, miR-153 was up-regulated, with its proapoptotic activity hypothesized to contribute to disease etiology (77).
miR-153 also targets a gene product especially relevant to AD and Parkinson disease: α-synuclein (SNCA) (71). Doxakis (71) demonstrates that endogenous miR-153 regulates SNCA in rodent neurons. SNCA is an abundant component of the Lewy bodies (LB) found in Parkinson disease and dementia with LB (80). Therefore, miR-153 negatively regulates two gene products implicated in two of the most common forms of neurodegenerative disease. LB are also found on autopsy in a common subtype of AD known as the Lewy body variant (AD-LBV) (81–84). This subtype has been associated with more rapid cognitive decline compared with “pure” AD (85, 86), along with lower levels of presynaptic proteins (87). A recent animal model of AD-LBV created by crossing 3xTg-AD mice with A53T SNCA transgenic mice revealed accelerated amyloid, tau, and Lewy body pathology in the AD-LBV animals as compared with the parental strains, suggesting synergistic effects between SNCA, Aβ, and tau in promoting pathology (88). An interesting question is whether miR-153 expression might be decreased more substantially in the brains of AD-LBV patients relative to AD patients. Measurement of miR-153 levels in these patients is warranted.
miR-153 is not likely to mediate its regulatory effects on APP expression in isolation. Indeed, regulatory interactions between APP and other miRNA have already been reported (as reviewed in Refs. 34 and 89). We and others have recently described the inhibitory effect of miR-101 against APP expression (36, 37). Other miRNAs reported to negatively regulate APP expression include miR-147 (40), the miR-20a family (miR-20a, -17, and -106b) (38, 42), miR-106a, miR-520c (43), and miR-16 (41). Some but not all of these studies have demonstrated physiological regulation of APP expression by these miRNA. The presence of AD-specific SNPs in the APP 3′-UTR has been shown to interfere with the ability of several miRNAs to regulate APP expression (40). Finally, miRNAs have been shown to play vital roles in regulating other aspects of APP biology, including alternative splicing (39).
The full complement of miRNAs that participate in the regulation of APP expression is yet to be fully elucidated. In this study, we have demonstrated that miR-153 is a member of this network in human neurons and appears to be dysregulated in at least a subset of advanced stage AD patients. Therefore, miR-153 makes for an attractive therapeutic target. Enhancing miR-153 levels would be expected to reduce the expression of two gene products (APP and SNCA) and downstream metabolites (e.g. Aβ peptides) that may have synergistic roles in promoting AD pathology and cognitive decline. Two recent studies have demonstrated that treating transformed cell lines with chromatin-modifying drugs can induce miR-153 expression by demethylating the miR-153 promoter and promoting histone acetylation (79, 90). How these manipulations would translate to neurons, where miR-153 expression is relatively high and chromatin structure might already favor transcription, is yet to be determined. Regardless of application to neurons, these studies demonstrate proof of principle for modifying miR-153 therapeutically by small molecules. The next steps will require assessing whether miR-153 manipulations in vivo ameliorate AD pathology and memory deficits in appropriate AD animal models.
Acknowledgments
We sincerely appreciate the assistance of Dr. P. H. Reddy for providing brain specimens, Dr. A. B. Niculescu for the use of real-time PCR instrumentation, Dr. J. T. Rogers for providing the pGALA construct, and Dr. J. A. Bailey for technical assistance.
Note Added in Proof
Since acceptance of this manuscript, the authors have become aware of recent work that independently demonstrates regulation of APP and APLP2 by miR-153 (see Ref. 91).
This work was supported, in whole or in part, by National Institutes of Health Grants AG18379 and AG18884 (to D. K. L.). This work was also supported by the Alzheimer's Association (Zenith award and investigator-initiated research grant).
- AD
- Alzheimer disease
- Aβ
- amyloid-β
- APP
- amyloid-β precursor protein
- miRNA
- microRNA
- qPCR
- quantitative PCR
- HFB
- human fetal brain cell(s)
- DIV
- day(s) in vitro
- GFAP
- glial fibrillary acidic protein
- SNCA
- α-synuclein
- LB
- Lewy bodies
- AD-LBV
- Lewy body variant of Alzheimer disease
- RIN
- RNA integrity number
- bFGF
- basic fibroblast growth factor
- BisTris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- ANOVA
- analysis of variance
- HSD
- honest significant difference
- NFT
- neurofibrillary tangle
- CM
- conditioned media.
REFERENCES
- 1. Alzheimer's Association, Thies W., Bleiler L. (2011) 2011 Alzheimer's disease facts and figures. Alzheimers Dement. 7, 208–244 [DOI] [PubMed] [Google Scholar]
- 2. Hardy J., Selkoe D. J. (2002) The amyloid hypothesis of Alzheimer's disease. Progress and problems on the road to therapeutics. Science 297, 353–356 [DOI] [PubMed] [Google Scholar]
- 3. Karran E., Mercken M., De Strooper B. (2011) The amyloid cascade hypothesis for Alzheimer's disease. An appraisal for the development of therapeutics. Nat. Rev. Drug Discov. 10, 698–712 [DOI] [PubMed] [Google Scholar]
- 4. Thinakaran G., Koo E. H. (2008) Amyloid precursor protein trafficking, processing, and function. J. Biol. Chem. 283, 29615–29619 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Nelson P. T., Braak H., Markesbery W. R. (2009) Neuropathology and cognitive impairment in Alzheimer disease. A complex but coherent relationship. J. Neuropathol. Exp. Neurol. 68, 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Rumble B., Retallack R., Hilbich C., Simms G., Multhaup G., Martins R., Hockey A., Montgomery P., Beyreuther K., Masters C. L. (1989) Amyloid A4 protein and its precursor in Down's syndrome and Alzheimer's disease. N. Engl. J. Med. 320, 1446–1452 [DOI] [PubMed] [Google Scholar]
- 7. Rovelet-Lecrux A., Hannequin D., Raux G., Le Meur N., Laquerrière A., Vital A., Dumanchin C., Feuillette S., Brice A., Vercelletto M., Dubas F., Frebourg T., Campion D. (2006) APP locus duplication causes autosomal dominant early onset Alzheimer disease with cerebral amyloid angiopathy. Nat. Genet. 38, 24–26 [DOI] [PubMed] [Google Scholar]
- 8. Theuns J., Brouwers N., Engelborghs S., Sleegers K., Bogaerts V., Corsmit E., De Pooter T., van Duijn C. M., De Deyn P. P., Van Broeckhoven C. (2006) Promoter mutations that increase amyloid precursor-protein expression are associated with Alzheimer disease. Am. J. Hum. Genet. 78, 936–946 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Lahiri D. K., Ge Y. W., Maloney B., Wavrant-De Vrièze F., Hardy J. (2005) Characterization of two APP gene promoter polymorphisms that appear to influence risk of late onset Alzheimer's disease. Neurobiol. Aging 26, 1329–1341 [DOI] [PubMed] [Google Scholar]
- 10. Song W., Lahiri D. K. (1998) Functional identification of the promoter of the gene encoding the Rhesus monkey β-amyloid precursor protein. Gene 217, 165–176 [DOI] [PubMed] [Google Scholar]
- 11. Lahiri D. K., Robakis N. K. (1991) The promoter activity of the gene encoding Alzheimer β-amyloid precursor protein (APP) is regulated by two blocks of upstream sequences. Brain Res. Mol. Brain Res. 9, 253–257 [DOI] [PubMed] [Google Scholar]
- 12. Pollwein P., Masters C. L., Beyreuther K. (1992) The expression of the amyloid precursor protein (APP) is regulated by two GC-elements in the promoter. Nucleic Acids Res. 20, 63–68 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Quitschke W. W., Goldgaber D. (1992) The amyloid β-protein precursor promoter. A region essential for transcriptional activity contains a nuclear factor binding domain. J. Biol. Chem. 267, 17362–17368 [PubMed] [Google Scholar]
- 14. Maloney B., Ge Y. W., Greig N., Lahiri D. K. (2004) Presence of a “CAGA box” in the APP gene unique to amyloid plaque-forming species and absent in all APLP-1/2 genes. Implications in Alzheimer disease. FASEB J. 18, 1288–1290 [DOI] [PubMed] [Google Scholar]
- 15. Lahiri D. K., Chen D., Vivien D., Ge Y. W., Greig N. H., Rogers J. T. (2003) Role of cytokines in the gene expression of amyloid β-protein precursor. Identification of a 5′-UTR-binding nuclear factor and its implications in Alzheimer's disease. J. Alzheimers Dis. 5, 81–90 [DOI] [PubMed] [Google Scholar]
- 16. Lesné S., Docagne F., Gabriel C., Liot G., Lahiri D. K., Buée L., Plawinski L., Delacourte A., MacKenzie E. T., Buisson A., Vivien D. (2003) Transforming growth factor-β1 potentiates amyloid-β generation in astrocytes and in transgenic mice. J. Biol. Chem. 278, 18408–18418 [DOI] [PubMed] [Google Scholar]
- 17. Bellingham S. A., Lahiri D. K., Maloney B., La Fontaine S., Multhaup G., Camakaris J. (2004) Copper depletion down-regulates expression of the Alzheimer's disease amyloid-β precursor protein gene. J. Biol. Chem. 279, 20378–20386 [DOI] [PubMed] [Google Scholar]
- 18. Lahiri D. K., Ge Y. W., Maloney B. (2005) Characterization of the APP proximal promoter and 5′-untranslated regions. Identification of cell type-specific domains and implications in APP gene expression and Alzheimer's disease. FASEB J. 19, 653–655 [DOI] [PubMed] [Google Scholar]
- 19. Rogers J. T., Leiter L. M., McPhee J., Cahill C. M., Zhan S. S., Potter H., Nilsson L. N. (1999) Translation of the Alzheimer amyloid precursor protein mRNA is up-regulated by interleukin-1 through 5′-untranslated region sequences. J. Biol. Chem. 274, 6421–6431 [DOI] [PubMed] [Google Scholar]
- 20. Rogers J. T., Randall J. D., Cahill C. M., Eder P. S., Huang X., Gunshin H., Leiter L., McPhee J., Sarang S. S., Utsuki T., Greig N. H., Lahiri D. K., Tanzi R. E., Bush A. I., Giordano T., Gullans S. R. (2002) An iron-responsive element type II in the 5′-untranslated region of the Alzheimer's amyloid precursor protein transcript. J. Biol. Chem. 277, 45518–45528 [DOI] [PubMed] [Google Scholar]
- 21. Beaudoin M. E., Poirel V. J., Krushel L. A. (2008) Regulating amyloid precursor protein synthesis through an internal ribosomal entry site. Nucleic Acids Res. 36, 6835–6847 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zaidi S. H., Malter J. S. (1994) Amyloid precursor protein mRNA stability is controlled by a 29-base element in the 3′-untranslated region. J. Biol. Chem. 269, 24007–24013 [PubMed] [Google Scholar]
- 23. Zaidi S. H., Denman R., Malter J. S. (1994) Multiple proteins interact at a unique cis-element in the 3′-untranslated region of amyloid precursor protein mRNA. J. Biol. Chem. 269, 24000–24006 [PubMed] [Google Scholar]
- 24. Westmark P. R., Shin H. C., Westmark C. J., Soltaninassab S. R., Reinke E. K., Malter J. S. (2006) Decoy mRNAs reduce β-amyloid precursor protein mRNA in neuronal cells. Neurobiol. Aging 27, 787–796 [DOI] [PubMed] [Google Scholar]
- 25. Rajagopalan L. E., Westmark C. J., Jarzembowski J. A., Malter J. S. (1998) hnRNP C increases amyloid precursor protein (APP) production by stabilizing APP mRNA. Nucleic Acids Res. 26, 3418–3423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. de Sauvage F., Kruys V., Marinx O., Huez G., Octave J. N. (1992) Alternative polyadenylation of the amyloid protein precursor mRNA regulates translation. EMBO J. 11, 3099–3103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mbella E. G., Bertrand S., Huez G., Octave J. N. (2000) A GG nucleotide sequence of the 3′-untranslated region of amyloid precursor protein mRNA plays a key role in the regulation of translation and the binding of proteins. Mol. Cell Biol. 20, 4572–4579 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Bartel D. P. (2009) MicroRNAs. Target recognition and regulatory functions. Cell 136, 215–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lewis B. P., Shih I. H., Jones-Rhoades M. W., Bartel D. P., Burge C. B. (2003) Prediction of mammalian microRNA targets. Cell 115, 787–798 [DOI] [PubMed] [Google Scholar]
- 30. Lewis B. P., Burge C. B., Bartel D. P. (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120, 15–20 [DOI] [PubMed] [Google Scholar]
- 31. Pratt A. J., MacRae I. J. (2009) The RNA-induced silencing complex. A versatile gene-silencing machine. J. Biol. Chem. 284, 17897–17901 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Filipowicz W., Bhattacharyya S. N., Sonenberg N. (2008) Mechanisms of post-transcriptional regulation by microRNAs. Are the answers in sight? Nat. Rev. Genet. 9, 102–114 [DOI] [PubMed] [Google Scholar]
- 33. Guo H., Ingolia N. T., Weissman J. S., Bartel D. P. (2010) Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature 466, 835–840 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Long J. M., Lahiri D. K. (2011) Current drug targets for modulating Alzheimer's amyloid precursor protein. Role of specific microRNA species. Curr. Med. Chem. 18, 3314–3321 [DOI] [PubMed] [Google Scholar]
- 35. Long J. M., Lahiri D. K. (2012) Advances in microRNA experimental approaches to study physiological regulation of gene products implicated in CNS disorders. Exp. Neurol. 235, 402–418 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Long J. M., Lahiri D. K. (2011) MicroRNA-101 down-regulates Alzheimer's amyloid-β precursor protein levels in human cell cultures and is differentially expressed. Biochem. Biophys. Res. Commun. 404, 889–895 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Vilardo E., Barbato C., Ciotti M., Cogoni C., Ruberti F. (2010) MicroRNA-101 regulates amyloid precursor protein expression in hippocampal neurons. J. Biol. Chem. 285, 18344–18351 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hébert S. S., Horré K., Nicolaï L., Bergmans B., Papadopoulou A. S., Delacourte A., De Strooper B. (2009) MicroRNA regulation of Alzheimer's amyloid precursor protein expression. Neurobiol. Dis. 33, 422–428 [DOI] [PubMed] [Google Scholar]
- 39. Smith P., Al Hashimi A., Girard J., Delay C., Hébert S. S. (2011) In vivo regulation of amyloid precursor protein neuronal splicing by microRNAs. J. Neurochem. 116, 240–247 [DOI] [PubMed] [Google Scholar]
- 40. Delay C., Calon F., Mathews P., Hébert S. S. (2011) Alzheimer-specific variants in the 3′-UTR of amyloid precursor protein affect microRNA function. Mol. Neurodegener. 6, 70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Liu W., Liu C., Zhu J., Shu P., Yin B., Gong Y., Qiang B., Yuan J., Peng X. (2012) MicroRNA-16 targets amyloid precursor protein to potentially modulate Alzheimer's-associated pathogenesis in SAMP8 mice. Neurobiol. Aging 33, 522–534 [DOI] [PubMed] [Google Scholar]
- 42. Fan X., Liu Y., Jiang J., Ma Z., Wu H., Liu T., Liu M., Li X., Tang H. (2010) miR-20a promotes proliferation and invasion by targeting APP in human ovarian cancer cells. Acta Biochim. Biophys. Sin. 42, 318–324 [DOI] [PubMed] [Google Scholar]
- 43. Patel N., Hoang D., Miller N., Ansaloni S., Huang Q., Rogers J. T., Lee J. C., Saunders A. J. (2008) MicroRNAs can regulate human APP levels. Mol. Neurodegener. 3, 10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ray B., Bailey J. A., Sarkar S., Lahiri D. K. (2009) Molecular and immunocytochemical characterization of primary neuronal cultures from adult rat brain. Differential expression of neuronal and glial protein markers. J. Neurosci. Methods 184, 294–302 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Vandesompele J., De Preter K., Pattyn F., Poppe B., Van Roy N., De Paepe A., Speleman F. (2002) Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. 3, research0034.1–0034.11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Braak H., Braak E. (1991) Neuropathological staging of Alzheimer-related changes. Acta Neuropathol. 82, 239–259 [DOI] [PubMed] [Google Scholar]
- 47. Manczak M., Calkins M. J., Reddy P. H. (2011) Impaired mitochondrial dynamics and abnormal interaction of amyloid β with mitochondrial protein Drp1 in neurons from patients with Alzheimer's disease. Implications for neuronal damage. Hum. Mol. Genet. 20, 2495–2509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Hellemans J., Mortier G., De Paepe A., Speleman F., Vandesompele J. (2007) qBase relative quantification framework and software for management and automated analysis of real-time quantitative PCR data. Genome Biol. 8, R19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Garcia D. M., Baek D., Shin C., Bell G. W., Grimson A., Bartel D. P. (2011) Weak seed-pairing stability and high target site abundance decrease the proficiency of lsy-6 and other microRNAs. Nat. Struct. Mol. Biol. 18, 1139–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Grimson A., Farh K. K., Johnston W. K., Garrett-Engele P., Lim L. P., Bartel D. P. (2007) MicroRNA targeting specificity in mammals. Determinants beyond seed pairing. Mol. Cell 27, 91–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Friedman R. C., Farh K. K., Burge C. B., Bartel D. P. (2009) Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 19, 92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Krek A., Grün D., Poy M. N., Wolf R., Rosenberg L., Epstein E. J., MacMenamin P., da Piedade I., Gunsalus K. C., Stoffel M., Rajewsky N. (2005) Combinatorial microRNA target predictions. Nat. Genet. 37, 495–500 [DOI] [PubMed] [Google Scholar]
- 53. Maragkakis M., Vergoulis T., Alexiou P., Reczko M., Plomaritou K., Gousis M., Kourtis K., Koziris N., Dalamagas T., Hatzigeorgiou A. G. (2011) DIANA-microT Web server upgrade supports Fly and Worm miRNA target prediction and bibliographic miRNA to disease association. Nucleic Acids Res. 39, W145–W148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Maragkakis M., Reczko M., Simossis V. A., Alexiou P., Papadopoulos G. L., Dalamagas T., Giannopoulos G., Goumas G., Koukis E., Kourtis K., Vergoulis T., Koziris N., Sellis T., Tsanakas P., Hatzigeorgiou A. G. (2009) DIANA-microT web server. Elucidating microRNA functions through target prediction. Nucleic Acids Res. 37, W273–W276 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. John B., Enright A. J., Aravin A., Tuschl T., Sander C., Marks D. S. (2004) Human MicroRNA targets. PLoS Biol. 2, e363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Betel D., Koppal A., Agius P., Sander C., Leslie C. (2010) Comprehensive modeling of microRNA targets predicts functional non-conserved and non-canonical sites. Genome Biol. 11, R90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Betel D., Wilson M., Gabow A., Marks D. S., Sander C. (2008) The microRNA.org resource. Targets and expression. Nucleic Acids Res. 36, D149–153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Miranda K. C., Huynh T., Tay Y., Ang Y. S., Tam W. L., Thomson A. M., Lim B., Rigoutsos I. (2006) A pattern-based method for the identification of microRNA binding sites and their corresponding heteroduplexes. Cell 126, 1203–1217 [DOI] [PubMed] [Google Scholar]
- 59. Kriegstein A., Alvarez-Buylla A. (2009) The glial nature of embryonic and adult neural stem cells. Annu. Rev. Neurosci. 32, 149–184 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Lendahl U., Zimmerman L. B., McKay R. D. (1990) CNS stem cells express a new class of intermediate filament protein. Cell 60, 585–595 [DOI] [PubMed] [Google Scholar]
- 61. Rieske P., Azizi S. A., Augelli B., Gaughan J., Krynska B. (2007) A population of human brain parenchymal cells express markers of glial, neuronal and early neural cells and differentiate into cells of neuronal and glial lineages. Eur. J. Neurosci. 25, 31–37 [DOI] [PubMed] [Google Scholar]
- 62. Heber S., Herms J., Gajic V., Hainfellner J., Aguzzi A., Rülicke T., von Kretzschmar H., von Koch C., Sisodia S., Tremml P., Lipp H. P., Wolfer D. P., Müller U. (2000) Mice with combined gene knock-outs reveal essential and partially redundant functions of amyloid precursor protein family members. J. Neurosci. 20, 7951–7963 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Lukiw W. J. (2007) MicroRNA speciation in fetal, adult and Alzheimer's disease hippocampus. Neuroreport 18, 297–300 [DOI] [PubMed] [Google Scholar]
- 64. Wang W. X., Rajeev B. W., Stromberg A. J., Ren N., Tang G., Huang Q., Rigoutsos I., Nelson P. T. (2008) The expression of microRNA miR-107 decreases early in Alzheimer's disease and may accelerate disease progression through regulation of β-site amyloid precursor protein-cleaving enzyme 1. J. Neurosci. 28, 1213–1223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Hébert S. S., Horré K., Nicolaï L., Papadopoulou A. S., Mandemakers W., Silahtaroglu A. N., Kauppinen S., Delacourte A., De Strooper B. (2008) Loss of microRNA cluster miR-29a/b-1 in sporadic Alzheimer's disease correlates with increased BACE1/β-secretase expression. Proc. Natl. Acad. Sci. U.S.A. 105, 6415–6420 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Nunez-Iglesias J., Liu C. C., Morgan T. E., Finch C. E., Zhou X. J. (2010) Joint genome-wide profiling of miRNA and mRNA expression in Alzheimer's disease cortex reveals altered miRNA regulation. PLoS ONE 5, e8898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Cogswell J. P., Ward J., Taylor I. A., Waters M., Shi Y., Cannon B., Kelnar K., Kemppainen J., Brown D., Chen C., Prinjha R. K., Richardson J. C., Saunders A. M., Roses A. D., Richards C. A. (2008) Identification of miRNA changes in Alzheimer's disease brain and CSF yields putative biomarkers and insights into disease pathways. J. Alzheimers Dis. 14, 27–41 [DOI] [PubMed] [Google Scholar]
- 68. Nelson P. T., Jicha G. A., Schmitt F. A., Liu H., Davis D. G., Mendiondo M. S., Abner E. L., Markesbery W. R. (2007) Clinicopathologic correlations in a large Alzheimer disease center autopsy cohort. Neuritic plaques and neurofibrillary tangles “do count” when staging disease severity. J. Neuropathol. Exp. Neurol. 66, 1136–1146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Rodríguez J. J., Olabarria M., Chvatal A., Verkhratsky A. (2009) Astroglia in dementia and Alzheimer's disease. Cell Death Differ. 16, 378–385 [DOI] [PubMed] [Google Scholar]
- 70. Guo Q., Li H., Gaddam S. S., Justice N. J., Robertson C. S., Zheng H. (2012) Amyloid precursor protein revisited. Neuron-specific expression and highly stable nature of soluble derivatives. J. Biol. Chem. 287, 2437–2445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Doxakis E. (2010) Post-transcriptional regulation of α-synuclein expression by mir-7 and mir-153. J. Biol. Chem. 285, 12726–12734 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Schipper H. M., Maes O. C., Chertkow H. M., Wang E. (2007) MicroRNA expression in Alzheimer blood mononuclear cells. Gene Regul. Syst. Bio. 1, 263–274 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Delay C., Mandemakers W., Hébert S. S. (2012) MicroRNAs in Alzheimer's disease. Neurobiol. Dis. 46, 285–290 [DOI] [PubMed] [Google Scholar]
- 74. Sempere L. F., Freemantle S., Pitha-Rowe I., Moss E., Dmitrovsky E., Ambros V. (2004) Expression profiling of mammalian microRNAs uncovers a subset of brain-expressed microRNAs with possible roles in murine and human neuronal differentiation. Genome Biol. 5, R13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Gaur A., Jewell D. A., Liang Y., Ridzon D., Moore J. H., Chen C., Ambros V. R., Israel M. A. (2007) Characterization of microRNA expression levels and their biological correlates in human cancer cell lines. Cancer Res. 67, 2456–2468 [DOI] [PubMed] [Google Scholar]
- 76. Kim T. H., Kim Y. K., Kwon Y., Heo J. H., Kang H., Kim G., An H. J. (2010) Deregulation of miR-519a, 153, and 485–5p and its clinicopathological relevance in ovarian epithelial tumors. Histopathology 57, 734–743 [DOI] [PubMed] [Google Scholar]
- 77. Xie T., Liang J., Guo R., Liu N., Noble P. W., Jiang D. (2011) Comprehensive microRNA analysis in bleomycin-induced pulmonary fibrosis identifies multiple sites of molecular regulation. Physiol. Genomics 43, 479–487 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Xu J., Liao X., Wong C. (2010) Down-regulations of B-cell lymphoma 2 and myeloid cell leukemia sequence 1 by microRNA 153 induce apoptosis in a glioblastoma cell line DBTRG-05MG. Int. J. Cancer 126, 1029–1035 [DOI] [PubMed] [Google Scholar]
- 79. Xu J., Liao X., Lu N., Liu W., Wong C. W. (2011) Chromatin-modifying drugs induce miRNA-153 expression to suppress Irs-2 in glioblastoma cell lines. Int. J. Cancer 129, 2527–2531 [DOI] [PubMed] [Google Scholar]
- 80. Baba M., Nakajo S., Tu P. H., Tomita T., Nakaya K., Lee V. M., Trojanowski J. Q., Iwatsubo T. (1998) Aggregation of α-synuclein in Lewy bodies of sporadic Parkinson's disease and dementia with Lewy bodies. Am. J. Pathol. 152, 879–884 [PMC free article] [PubMed] [Google Scholar]
- 81. Förstl H., Burns A., Luthert P., Cairns N., Levy R. (1993) The Lewy-body variant of Alzheimer's disease. Clinical and pathological findings. Br. J. Psychiatry 162, 385–392 [DOI] [PubMed] [Google Scholar]
- 82. Heyman A., Fillenbaum G. G., Gearing M., Mirra S. S., Welsh-Bohmer K. A., Peterson B., Pieper C. (1999) Comparison of Lewy body variant of Alzheimer's disease with pure Alzheimer's disease. Consortium to Establish a Registry for Alzheimer's Disease, Part XIX. Neurology 52, 1839–1844 [DOI] [PubMed] [Google Scholar]
- 83. Hansen L., Salmon D., Galasko D., Masliah E., Katzman R., DeTeresa R., Thal L., Pay M. M., Hofstetter R., Klauber M. (1990) Neurology 40, 1–8 [DOI] [PubMed] [Google Scholar]
- 84. Hamilton R. L. (2000) Lewy bodies in Alzheimer's disease. A neuropathological review of 145 cases using α-synuclein immunohistochemistry. Brain Pathol. 10, 378–384 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Kraybill M. L., Larson E. B., Tsuang D. W., Teri L., McCormick W. C., Bowen J. D., Kukull W. A., Leverenz J. B., Cherrier M. M. (2005) Cognitive differences in dementia patients with autopsy-verified AD, Lewy body pathology, or both. Neurology 64, 2069–2073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Olichney J. M., Galasko D., Salmon D. P., Hofstetter C. R., Hansen L. A., Katzman R., Thal L. J. (1998) Cognitive decline is faster in Lewy body variant than in Alzheimer's disease. Neurology 51, 351–357 [DOI] [PubMed] [Google Scholar]
- 87. Mukaetova-Ladinska E. B., Xuereb J. H., Garcia-Sierra F., Hurt J., Gertz H. J., Hills R., Brayne C., Huppert F. A., Paykel E. S., McGee M. A., Jakes R., Honer W. G., Harrington C. R., Wischik C. M. (2009) Lewy body variant of Alzheimer's disease. Selective neocortical loss of t-SNARE proteins and loss of MAP2 and α-synuclein in medial temporal lobe. Scientific World Journal 9, 1463–1475 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Clinton L. K., Blurton-Jones M., Myczek K., Trojanowski J. Q., LaFerla F. M. (2010) Synergistic Interactions between Aβ, Tau, and α-synuclein. Acceleration of neuropathology and cognitive decline. J. Neurosci. 30, 7281–7289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Schonrock N., Matamales M., Ittner L. M., Götz J. (2012) MicroRNA networks surrounding APP and amyloid-β metabolism. Implications for Alzheimer's disease. Exp. Neurol. 235, 447–454 [DOI] [PubMed] [Google Scholar]
- 90. Bao B., Rodriguez-Melendez R., Zempleni J. (2012) Cytosine methylation in miR-153 gene promoters increases the expression of holocarboxylase synthetase, thereby increasing the abundance of histone H4 biotinylation marks in HEK-293 human kidney cells. J. Nutr. Biochem. 23, 635–639 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Liang C., Zhu H., Xu Y., Huang L., Ma C., Deng W., Liu Y., Qin C. (2012) MicroRNA-153 negatively regulates the expression of amyloid precursor protein and amyloid precursor-like protein 2. Brain Res. 1455, 103–113 [DOI] [PubMed] [Google Scholar]