Background: Understanding the biological significance of feedback loops requires interrogation at multiple scales.
Results: A nuclear factor κB (NF-κB) negative feedback reporter revealed stimulus-specific dynamics in cells and animals in vivo.
Conclusion: Circulating tumor necrosis factor α (TNFα) doses are perceived by the liver as pulses.
Significance: Bioluminescent imaging of live single cells and cell populations revealed reproducible behaviors that informed interpretation of in vivo data.
Keywords: Bioluminescence, In Vivo Imaging, NF-κB Transcription Factor, Signaling, Tumor Necrosis Factor (TNF)
Abstract
Full understanding of the biological significance of negative feedback processes requires interrogation at multiple scales as follows: in single cells, cell populations, and live animals in vivo. The transcriptionally coupled IκBα/NF-κB negative feedback loop, a pivotal regulatory node of innate immunity and inflammation, represents a model system for multiscalar reporters. Using a κB5→IκBα-FLuc bioluminescent reporter, we rigorously evaluated the dynamics of ΙκBα degradation and subsequent NF-κB transcriptional activity in response to diverse modes of TNFα stimulation. Modulating TNFα concentration or pulse duration yielded complex, reproducible, and differential ΙκBα dynamics in both cell populations and live single cells. Tremendous heterogeneity in the transcriptional amplitudes of individual responding cells was observed, which was greater than the heterogeneity in the transcriptional kinetics of responsive cells. Furthermore, administration of various TNFα doses in vivo generated ΙκBα dynamic profiles in the liver resembling those observed in single cells and populations of cells stimulated with TNFα pulses. This suggested that dose modulation of circulating TNFα was perceived by hepatocytes in vivo as pulses of increasing duration. Thus, a robust bioluminescent reporter strategy enabled rigorous quantitation of NF-κB/ΙκBα dynamics in both live single cells and cell populations and furthermore, revealed reproducible behaviors that informed interpretation of in vivo studies.
Introduction
Cells have evolved complex molecular networks to sense cues from the environment and transmit them throughout the cell to elicit appropriate biological responses. These signaling pathways require certain elemental properties, such as sensitivity, reversibility, a capacity to be regulated, and robustness, that are crucial to reliably maintaining the organization and function of cells within organisms. In addition, these networks equip cells with the ability to distinguish persistently weak signals from background noise with high precision and selectivity (1, 2). These molecular networks include sets of recurring regulation patterns, manifesting as network motifs, that link together in a variety of combinations to create a web of connectivity within a given signaling pathway or between multiple cascades (3). Feedback loops, processes that connect output signals back to their input, represent one of the most frequently observed biological network motifs and are now appreciated as a useful framework for understanding how signaling networks elicit specific cellular responses. In particular, negative feedback loops, defined as sequential regulatory steps that feed the output signal (inverted) back to the input, represent a single motif that is capable of generating many distinct signaling functions, including stabilizing basal signaling levels, limiting maximal signaling output, enabling adaptive response, and creating transient signal responses (2).
Transcriptionally coupled negative feedback loops often serve as critical regulatory nodes within signaling pathways. For example, p532 is negatively regulated by several transcriptional targets, the most well known being MDM2 (4). MDM2 is an E3 ubiquitin ligase that targets p53 for degradation via the proteasome, thus decreasing the amount of p53 present in the cell and dampening/turning off the p53 transcriptional response. The NF-κB transcription factors are similarly regulated by a transcriptionally coupled negative feedback loop; in this case, an ΙκB inhibitor protein (such as ΙκBα) sequesters NF-κB in the cytoplasm until pathway activation through the IKK kinase complex results in ΙκBα phosphorylation and subsequent ubiquitination and degradation. This frees NF-κB to translocate to the nucleus where it transcribes additional ΙκBα, thus preventing chronic activation of NF-κB transcription factors.
In the past, elucidation of these complex molecular networks focused on identifying the key molecules within the network and biochemically defining their individual interactions (1, 5). Conventional techniques typically employed to define these networks were static biochemical methodologies in vitro that were destructive, only semi-quantitative, lacked spatiotemporal resolution, and averaged information from a large number of cells. New developments in optical imaging and biophysical methods have enabled significant advances in the ability to capture spatiotemporal signaling information in a single cell, leading to the development and refinement of mathematical and dynamic models of molecular networks (6). However, to fully understand the biological significance of negative feedback processes, it is critical to study them at multiple scales as follows: in single cells, in cell populations, and in live animals. Multiscalar studies may assist the dissection of which properties of single cells on a coverslip are relevant to how individual cells (or cell populations) actually behave in the context of a tissue in vivo. Bioluminescent reporters are ideally suited for multiscalar studies, given their exceptionally high signal-to-noise levels, ability to continuously monitor dynamic processes, and applicability to in vivo imaging (7, 8). The NF-κB signaling pathway represents a model system for the use of multiscalar bioluminescent reporters to study a complex transcriptionally coupled negative feedback loop.
NF-κB is a pivotal regulator of innate immunity and inflammation and is active in both immune cells and nonimmune tissues (9, 10). Responding to a large number of different stimuli (11), recent work has focused on NF-κB pathway reactivity to the mode of stimulation (i.e. stimulus concentration, stimulus duration (pulse versus continuous), and pulse interval), which may be particularly relevant during cellular responses to inflammatory cytokines, such as TNFα. Cytokines are likely perceived as transient pulses or waves occurring over a wide range of concentrations (12–17). Thus, the NF-κB pathway must rapidly decode different types of signal inputs and integrate intracellular information to control individual cell fate decisions (proliferation, apoptosis, differentiation, etc.) and regulate the production and secretion of cytokines that can amplify, propagate, and terminate the inflammatory response (18, 19). Recently, single cell imaging has been widely utilized to characterize NF-κB signaling in response to different modes of stimulation. These studies have revealed the presence of oscillations in NF-κB nuclear translocation that are dependent upon cycles of degradation and resynthesis of IκB proteins (i.e. negative feedback loops) (20). The frequency of these NF-κB protein oscillations can encode distinct gene expression profiles, as determined with cell population studies (15, 21–24). Furthermore, single cell studies have revealed heterogeneous and asynchronous NF-κB responses in single cells (18, 21, 25), especially in response to low concentrations of TNFα (17). However, the physiological relevance of these findings has yet to be assessed in vivo. Most single cell studies are carried out with fluorescent protein fusions that are not amenable to use in vivo due to the high degree of tissue autofluorescence. Therefore, we have developed a dynamic bioluminescent reporter strategy that enables correlative quantitation of the NF-κB/ΙκBα negative feedback loop in single cells, cell populations, and at the tissue level in live animals.
Previously, we demonstrated that fusing ΙκBα to the firefly luciferase gene (ΙκBα-FLuc) enabled quantitative monitoring of ΙκBα degradation (which directly correlates with IKK activity) in vitro and in vivo (26). We then placed the fusion reporter under the control of an NF-κB-responsive promoter (κB5→ΙκBα-FLuc) and showed that it recapitulated the endogenous ΙκBα negative feedback loop (Fig. 1A) (12, 13, 27). The κB5→ΙκBα-FLuc reporter offers the distinct advantage of monitoring protein activity within different subcellular compartments as opposed to simply measuring changes in total protein content or localization. Thus, κB5→ΙκBα-FLuc temporally reports both TNFα-induced degradation of ΙκBα (which is dependent on the activity of IKK, β-TrCP, and the proteasome) and subsequent NF-κB-dependent transcriptional up-regulation of ΙκBα (which is dependent upon NF-κB nuclear translocation as well as additional post-translational modifications and co-activator associations in the nucleus). Furthermore, the synthetic κB5 promoter has enhanced sensitivity that enables measurement of subtle changes in transcriptional dynamics. This reporter strategy provides a real time dynamic link between fluorescence-based reporters that measure NF-κB nuclear shuttling in single cells and conventional destructive techniques (quantitative-PCR of target genes, transcriptional profiling, EMSA, etc.) that measure downstream NF-κB transcriptional activity in cell populations. In this study, we have exploited the unique characteristics of the κB5→ΙκBα-FLuc reporter for multiscale interrogation of the negative feedback loop in single cells, cell populations, and in vivo. We performed rigorous quantitative analysis of stimulus-specific NF-κB/ΙκBα dynamics in both single cells and populations of cells, and we discovered reproducible behaviors that informed interpretation of in vivo studies.
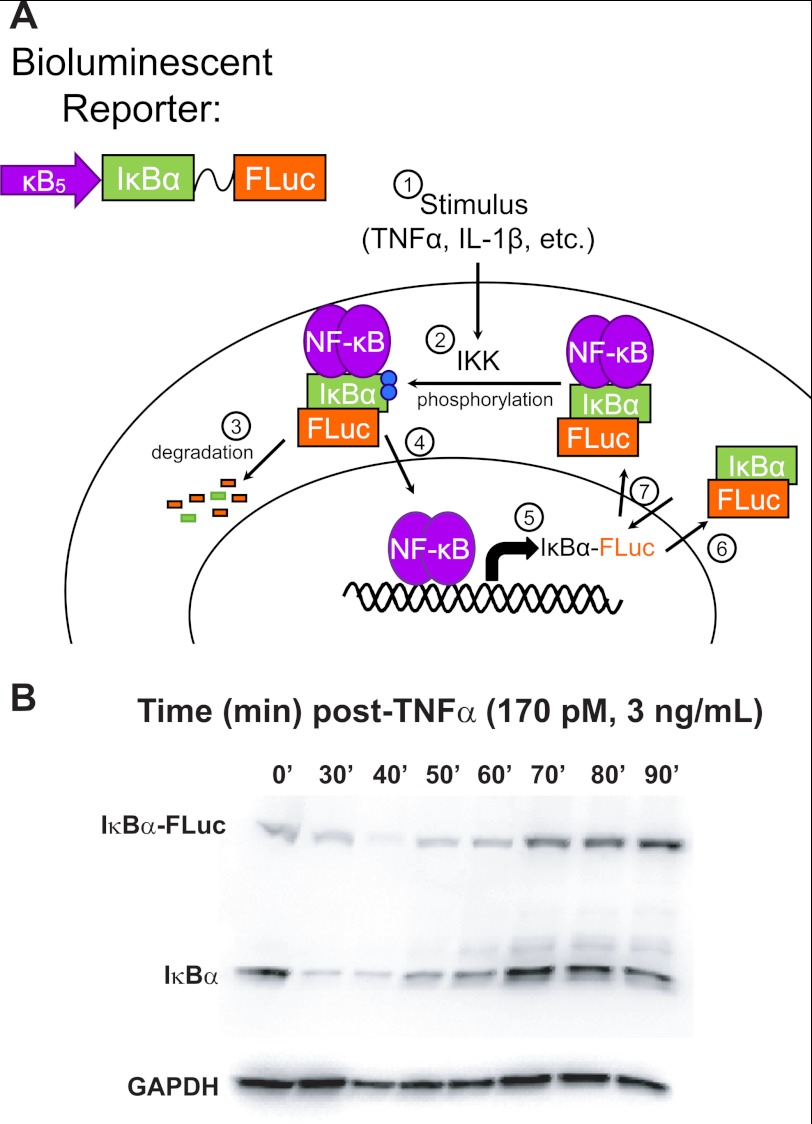
FIGURE 1.
κB5→ΙκBα-FLuc bioluminescent reporter system. A, κB5→ΙκBα-FLuc bioluminescent reporter utilizes the ΙκBα gene fused by a flexible linker to the firefly luciferase (FLuc) gene under the control of a synthetic NF-κB promoter (κB5), thus recapitulating the endogenous negative feedback loop (1→7). B, HepG2 cells were transiently transfected with κB5→ΙκBα-FLuc and stimulated continuously with TNFα (170 pm; 3 ng/ml). Lysates were collected at the indicated time points, resolved on a 7.5% SDS-polyacrylamide gel, and blotted for ΙκBα and GAPDH (loading control).
EXPERIMENTAL PROCEDURES
Dynamic Bioluminescence Imaging in Live Cell Populations
HepG2 cells were transiently transfected with κB5→ΙκBα-FLuc and plated in black-coated 24-well plates. After a 48-h recovery, cells were transferred into fresh clear media containing d-luciferin (150 μg/ml) and exposed to TNFα or vehicle (PBS) for the specified durations and concentrations. Bioluminescence time course measurements were acquired in an IVIS 100 imaging system. Detailed descriptions of cell culture conditions, experimental treatment regimens, image acquisition parameters, and data analysis are provided in the supplemental Experimental Procedures.
Single Cell Bioluminescence Imaging
HepG2 cells were transfected as described above with either the κB5→ΙκBα-FLuc plasmid or the FUW-FLG construct. At 36 h post-transfection, cells were trypsinized, counted, diluted, and plated at 60 cells/well onto pre-plated untransfected HepG2 cells (3 × 105 cells/well plated at the same time as initial transfection) in a black 24-well plate. At 48 h post-transfection, cells were stimulated as indicated with TNFα or vehicle, and bioluminescence images were acquired on an IVIS50 or IVIS100. GFP expression was analyzed on an InCell Analyzer 1000. A monoclonal HCT116 cell line stably expressing the κB5→ΙκBα-FLuc reporter was generated by standard techniques and imaged identically. Details of the image acquisition parameters and data analysis are provided in the supplemental Experimental Procedures.
Hydrodynamic Injections and in Vivo Imaging
In vivo transfection of mouse hepatocytes with the κB5→ΙκBα-FLuc reporter was performed using the hydrodynamic somatic gene transfer method as described (28, 29). Cohorts of four mice were injected with d-luciferin, anesthetized, imaged for basal luciferase activity, administered vehicle or TNFα by tail vein i.v., and imaged every 5 min for 3 h under anesthesia in an IVIS100 system. Expanded descriptions of the injection procedures, image acquisition, and analysis can be found in the supplemental Experimental Procedures.
RESULTS
Characterizing TNFα-induced Regulation of the ΙκBα/NF-κB Negative Feedback Loop in Cell Populations
The ΙκBα/NF-κB negative feedback loop represents a major regulatory node within the NF-κB pathway and is a critical determinant of NF-κB oscillatory behaviors that can encode stimulus-specific gene expression programs (15, 21–24, 30). Bioluminescent reporters are ideally suited for medium to high throughput studies of stimulus-induced cellular signaling in real time in populations of cells. Therefore, we utilized our previously validated κB5→ΙκBα-FLuc reporter expressed at sub-endogenous levels in HepG2 (human hepatocellular carcinoma) cells to systematically evaluate the impact of short duration TNFα pulses on negative feedback loop dynamics within populations of cells in culture (Fig. 1, A and B) (12, 26).
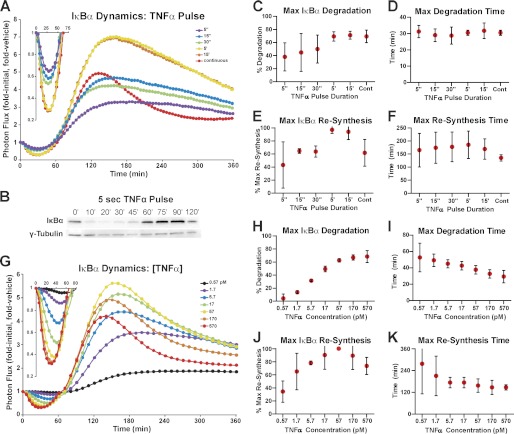
HepG2 cells transfected with κB5→ΙκBα-FLuc were stimulated with the pro-inflammatory cytokine TNFα (1.2 nm, 20 ng/ml) either continuously or as a pulse (5, 15, and 30 s and 5 or 15 min), and images of cells were captured sequentially every 5 min for 6 h. Generally, the normalized ΙκBα-FLuc photon flux (Fig. 2A) rapidly decreased to a transient minimum (due to TNFα-induced degradation of ΙκBα) and then strongly rebounded above initial levels (due to NF-κB-induced resynthesis of ΙκBα). This rebound was previously shown to be consistent with de novo transcription and translation of ΙκBα (12) and with the previously reported ligand-induced stabilization of newly synthesized ΙκBα (31, 32). A TNFα pulse as short as 5 s in duration was capable of inducing substantial ΙκBα degradation (35 ± 9%, mean ± S.E. unless noted otherwise), suggesting that extremely brief exposure can induce significant IKK-dependent activation of canonical NF-κB signaling (Fig. 2A). This was confirmed by Western blot analysis, which revealed substantial ΙκBα degradation in response to a 5-s TNFα pulse (Fig. 2B). As TNFα pulse duration was lengthened from 5 s to 15 min, the degree of ΙκBα degradation increased, and when pulsed for 5 min or longer, ΙκBα degradation saturated at levels equivalent to continuous TNFα stimulation (∼70% degradation) (Fig. 2, A, inset, and C). The time at which maximal degradation occurred did not significantly change as TNFα pulse duration was modulated (Fig. 2D).
FIGURE 2.
ΙκBα dynamics as a function of TNFα pulse duration and concentration. A, HepG2 cells transiently expressing κB5→ΙκBα-FLuc were pulsed with a saturating concentration of TNFα (1.2 nm; 20 ng/ml) or vehicle for the indicated durations with data acquisition over a period of 360 min. Data were normalized as fold initial and fold vehicle (cells pulsed with vehicle for the same duration) and represent the mean of four independent TNFα exposure experiments, each performed in duplicate and averaged. B, HepG2 cells were stimulated with a 5-s pulse of TNFα (1.2 nm; 20 ng/ml), and lysates were collected to capture ΙκBα degradation and resynthesis. Lysates were resolved together on a 4–15% gradient gel and blotted for endogenous ΙκBα and γ-tubulin (loading control). C and D, plots representing the extent of maximal ΙκBα degradation (C), and the time at which maximal ΙκBα degradation occurred (D), as functions of TNFα pulse duration. E and F, plots representing the extent of maximal ΙκBα resynthesis (expressed as a percentage of the maximum level of resynthesis achieved in a given experiment) (E), and the time at which maximal ΙκBα resynthesis occurred (F), as functions of TNFα pulse duration. G, HepG2 cells expressing κB5→ΙκBα-FLuc were continuously treated with TNFα or vehicle at the indicated concentrations, and bioluminescent data were acquired for 360 min. Data were normalized as before and represent three independent experiments, performed in triplicate and averaged. H and I, plots representing the extent of maximal ΙκBα degradation (H), and the time at which maximal ΙκBα degradation occurred (I), as functions of TNFα concentration. J and K, plots representing the extent of maximal ΙκBα resynthesis (J), and the time at which maximal ΙκBα resynthesis occurred (K), as functions of TNFα concentration. Error bars represent 95% confidence interval of the mean. See also supplemental Fig. S3.
Examination of the degree of ΙκBα resynthesis (measured as percent of maximum resynthesis) in response to TNFα pulse duration revealed increasing levels of ΙκBα resynthesis that eventually peaked and leveled off when pulsed for 5 min or longer (Fig. 2E). Interestingly, TNFα pulses elicited a broader ΙκBα resynthesis phase with a less defined peak when compared with continuous TNFα stimulation. Furthermore, maximal ΙκBα resynthesis in response to a 15-min TNFα pulse was higher (97 ± 3% of maximum) than observed for continuous TNFα stimulation (65 ± 8% of maximum). As had been observed for ΙκBα degradation, the timing of the resynthesis peak did not significantly change with increasing pulse duration (Fig. 2F). Additionally, peak ΙκBα resynthesis was later for a 15-min TNFα pulse than for continuous TNFα (164 ± 16 min versus 137 ± 5 min).
We next investigated the impact of TNFα concentration on the dynamic regulation of the ΙκBα/NF-κB negative feedback loop by treating HepG2 cells with a range of TNFα concentrations (0.1–10 ng/ml and 0.57–570 pm) under continuous exposure conditions (Fig. 2G). The degree of ΙκBα degradation increased with increasing TNFα concentration, eventually saturating (68 ± 2%) at the highest concentrations tested (Fig. 2, G, inset, and H), yielding a degradation EC50 value of 6.7 pm TNFα (5.7–7.9 pm, 95% confidence interval). Moreover, examination of ΙκBα degradation kinetics (Fig. 2I) showed that increasing TNFα concentration resulted in faster degradation, with the time of maximal degradation shifting from 53 ± 4 min to 29 ± 2 min.
The relationship between TNFα concentration and ΙκBα resynthesis was more complex than observed for degradation. Increasing the TNFα concentration elicited increasing levels of ΙκBα resynthesis up to a maximum (corresponding to 57 pm; 1 ng/ml TNFα) beyond which higher amounts of TNFα actually elicited lower levels of resynthesis (i.e. a “rollover” back down to 74 ± 3% of maximum levels; Fig. 2, G and J). The lowest TNFα concentrations produced broadly shaped resynthesis profiles with poorly defined peaks, making accurate determination of peak resynthesis time challenging, as evidenced by the wide confidence intervals in Fig. 2K. However, the overall trend showed resynthesis kinetics speeding up, with less variance, as TNFα concentration increased (Fig. 2K).
Thus, we found that the κB5→ΙκBα-FLuc reporter enabled quantitative comparison of the effects of modulating TNFα pulse duration versus concentration in real time in live cells. This systematic analysis revealed that ΙκBα degradation was highly sensitive to both modes of stimulation tested and in each case eventually saturated at ∼70% degradation. Modulation of TNFα pulse duration had little effect on the kinetics of ΙκBα degradation, although increasing TNFα concentration resulted in faster degradation. Both stimulation regimens elicited biphasic patterns in the degree of ΙκBα resynthesis, although modulation of TNFα concentration had a moderate effect on ΙκBα resynthesis kinetics.
Characterizing TNFα-induced Regulation of the ΙκBα/NF-κB Negative Feedback Loop in Single Cells
Having characterized complex and reproducible patterns of ΙκBα dynamics in live cultured cell populations, we sought to determine whether these behaviors could be measured within single cells. Of particular interest was determining whether the broad ΙκBα reporter peaks observed in response to low concentrations of TNFα resulted from the summation of heterogeneous single cell transcriptional responses or from the synchronized responses of all cells within the population. To address these questions, we imaged single HepG2 cells expressing the κB5→ΙκBα-FLuc reporter.
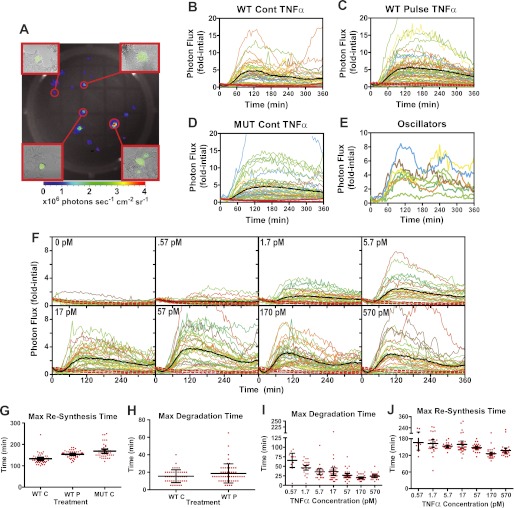
Most single-cell studies utilizing bioluminescent reporters rely upon expensive low light microscopy imaging systems that allow measurement of light emitted from several cells plated on a coverslip or in an imaging chamber. We sought to develop a more accessible, inexpensive, and high throughput means to image single cells expressing the κB5→ΙκBα-FLuc without the use of microscopy. We first verified that we could image single bioluminescent cells in an IVIS100 imaging system by transiently transfecting HepG2 cells with a dual bioluminescent/fluorescent reporter construct, FUW-FLG, comprising pGL3 firefly luciferase fused through a flexible linker to enhanced GFP, driven by a constitutive ubiquitin promoter (33). HepG2 cells were transfected with this plasmid as described above for the κB5→ΙκBα-FLuc reporter; however, 36 h after transfection, cells were trypsinized into a single cell suspension, counted, diluted, and plated at a density of 60 cells/well on top of pre-plated, untransfected HepG2 cells in a black 24-well plate (to best simulate the same conditions used in prior cell population studies). After a 12-h recovery period, cells were imaged sequentially for bioluminescence (IVIS100) and then fluorescence (InCell 1000 imager) (Fig. 3A). We found an excellent correlation between single bioluminescent foci and single cell fluorescence; only a small number of foci corresponded to a cluster of two or three fluorescent cells. Thus, single HepG2 cells expressing a dual-imaging reporter could be imaged in a monolayer of otherwise isogenic cells, without the use of low throughput, expensive low light microscopy equipment.
FIGURE 3.
Characterization of ΙκBα dynamics in single cells. A, image of a single well of a 24-well plate with bioluminescent foci representing sparsely plated HepG2 cells transiently expressing the FUW-FLG luciferase-enhanced GFP fusion reporter. Insets represent 10× fluorescent micrographs of the indicated bioluminescent foci, demonstrating that most foci represent a single transfected cell and occasionally a small group of 2–3 cells. B–E, single HepG2 cells transiently expressing κB5→ΙκBα-FLuc (B and C) or κB5→ΙκBα(S32A,S36A)-Fluc (D) were given continuous (Cont) (B and D) or a 30-s pulse (C) of TNFα (1.2 nm; 20 ng/ml). Data were normalized as fold initial, and data from two independent experiments are plotted together. Black lines represent the experimental mean and red dashed lines represent the 95% confidence interval of the vehicle-treated controls. Mut, mutant. E, select examples of ΙκBα-FLuc oscillations observed in a minor subset of the cells from B. F, single HepG2 cells transiently expressing κB5→ΙκBα-FLuc were continuously treated with TNFα or vehicle at the indicated concentrations. G–J, scatter plots representing the times of maximal ΙκBα-FLuc resynthesis (G) and degradation (H) calculated after smoothing data in B–D, and the times of maximal ΙκBα-FLuc degradation (I) and resynthesis (J) calculated after smoothing data in F. Error bars represent 95% confidence interval of the mean. See also supplemental Fig. S1.
This facile method now enabled hundreds of single HepG2 cells expressing κB5→ΙκBα-FLuc to be monitored simultaneously over time following stimulation with either continuous or 30-s pulses of high TNFα (supplemental Movie 1). Under continuous stimulation, the ΙκBα-FLuc profiles of individual cells (Fig. 3B) remarkably resembled those observed for cell populations (Fig. 2). Interestingly, although single cells exhibited substantial variation in the amplitude of degradation and resynthesis, a plot of the mean photon flux of all individual cells (Fig. 3B, black line) strongly resembled the ΙκBα-FLuc profiles observed in cell populations (Fig. 2A, red line). Maximal resynthesis for all but one of the single cells peaked between 110 and 165 min, with a mean of 133 ± 24 min (± S.D.) (Fig. 3G), identical to the population mean of 137 ± 5 min (Fig. 2F). Similar to cell population studies, a 30-s TNFα pulse elicited broad ΙκBα-FLuc resynthesis profiles in single cells (Fig. 3C), with all of the cells peaking between 115 and 185 min (154 ± 15 min; mean ± S.D.). Variation in the amplitude of ΙκBα-FLuc degradation and resynthesis was observed (Fig. 3C), although the combined mean ΙκBα-FLuc profile for all individual cells (Fig. 3C, black line) and the ΙκBα-FLuc profile observed for a population of cells (Fig. 2A, green line) were nearly identical. The mean percent ΙκBα degradation observed in TNFα pulsed cells (63 ± 3%) was less than that observed under continuous TNFα treatment (78 ± 2%), with similar maximal times (Fig. 3H), recapitulating trends noted in the population studies (Fig. 2, A, C, and D). Interestingly, 18% of continuously stimulated cells exhibited ΙκBα-FLuc oscillatory behavior, peaking once at 109 ± 2 min and again at 244 ± 7 min (Fig. 3E, a periodicity of ∼130 min). This phenomenon was never observed in cells given a 30-s TNFα pulse, again in good agreement with published reports that NF-κB nuclear translocation oscillations are observed only under continuous TNFα stimulation (21, 24).
We next investigated individual cell responses to a range of TNFα concentrations under continuous stimulation as described previously for cell population studies (Fig. 2, G–K). Again, although heterogeneous ΙκBα-FLuc amplitudes were observed, the ΙκBα-FLuc kinetic profiles within individual cells (Fig. 3F) resembled those observed for cell populations (Fig. 2). The coefficient of variation (CV) ranged from 0.36 to 0.77 for the magnitude of maximum resynthesis, greater than the CV range of 0.07 to 0.24 for the kinetics, thus indicating relative synchronicity of kinetics at each corresponding concentration of TNFα. Interestingly, whereas the population average amplitude increased with increasing concentrations of TNFα, substantial overlap was observed in the distribution of individual responses across concentrations such that increasing ligand resulted in both a reduction in the number of cells below the mean and an increase in cells above, with little change in the number of intermediate cells (Fig. 3F). At the lowest concentrations tested, 0.57 pm, most HepG2 cells (62%) did not respond to TNFα stimulation (as defined by falling within the 95% confidence interval of vehicle-stimulated cells and not exhibiting a local maximum; Fig. 3F, 0 pm panel), similar to previous studies that found fewer cells respond to low concentrations of TNF (16). For all other TNFα concentrations examined, less than 7% of cells were nonresponders. When all responding cells were considered, increasing TNFα concentrations resulted in faster ΙκBα-FLuc degradation (Fig. 3I), with the time of maximal degradation shifting from 65 ± 8 to 20 ± 1 min, a trend similar to that seen in our population studies (Fig. 2I) and single cell work by others (16), thus indicating this phenomenon is a property of single cells. Increasing TNFα concentration resulted in higher levels of ΙκBα-FLuc resynthesis (Fig. 3F) and exhibited the same resynthesis rollover observed in cell populations (Fig. 2G). Furthermore, examination of ΙκBα-FLuc resynthesis kinetics demonstrated a high degree of variance at the lowest doses (Fig. 3J) and a moderate trend toward faster kinetics, with lower variance, as TNFα concentration increased, reproducing the trends observed in cell populations. The heterogeneity in resynthesis times at low doses could account for the broad peaks and variability in peak timing observed at 0.57 and 1.7 pm in cell populations (Fig. 2, G and K). However, even at the lowest TNFα concentration tested, we did not observe any cells that exhibited degradation any later than 100 min and resynthesis any later than 240 min.
To determine whether the heterogeneity observed in stimulated cells was simply due to differences in transfection efficiency between cells, we generated a monoclonal HCT116 cell line stably expressing the identical reporter. Importantly, although HCT116 cells stably expressing the reporter presented a different response profile, the cells showed highly comparable heterogeneity in the amplitude of the transcriptional response indicating that amplitude variance was an inherent property of the NF-κB pathway and not an artifact of transient transfection of the reporter construct (HepG2 versus HCT116; 17 pm TNFα, peak amplitude CV 0.64 versus 0.56; and 170 pm TNFα, CV 0.78 versus 0.72) (supplemental Fig. S1).
Thus, a new method for imaging single bioluminescent cells demonstrated the ability to acquire quantitative single cell data in a high throughput manner without the use of low light microscopy. Furthermore, it revealed that whereas heterogeneity in the amplitude, and to a lesser degree the kinetics, was observed in single cells, the different ΙκBα-FLuc profile shapes and dynamic trends observed under various TNFα stimulation conditions in cell populations were recapitulated in individual cells.
Characterization of TNFα-induced Regulation of the ΙκBα/NF-κB Negative Feedback Loop in Live Animals in Vivo
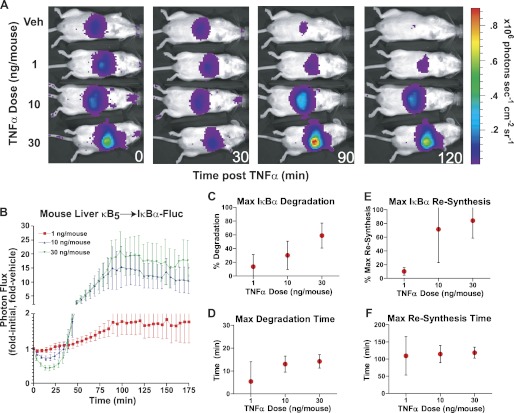
Although studies of single cells and cell populations in culture have proven invaluable in understanding the intricacies underlying the wiring of cellular signaling pathways, full evaluation of signaling events in their native context in vivo is more challenging. Having utilized the κB5→ΙκBα-FLuc reporter successfully in single cells and cell populations to interrogate complex patterns of ΙκBα dynamics in response to modulating TNFα pulse duration and concentration, we subsequently investigated ΙκBα/NF-κB negative feedback loop dynamics in vivo in response to varying TNFα doses. Somatic gene transfer by hydrodynamic transfection was employed to rapidly and efficiently express the κB5→ΙκBα-FLuc plasmid in murine livers (34). Three to 12 weeks post-plasmid injection, sufficient time for hepatocellular recovery and stable integration of reporter plasmids into a subpopulation of hepatocytes, animals were administered vehicle (PBS) or TNFα (1, 10, or 30 ng/mouse) by bolus tail vein injection and imaged at 5-min intervals for 3 h to capture full ΙκBα-FLuc dynamic profiles (Fig. 4, A and B). Each TNFα dose was repeated in five independent experiments, and data were combined for analysis. Of the three TNFα doses used, the lowest (1 ng/mouse) appeared to induce little or no ΙκBα-FLuc reporter degradation, whereas the two higher doses showed increasing amounts of degradation (Fig. 4C; 10 ng/mouse, 30 ± 7%; 30 ng/mouse, 59 ± 7%). Interestingly, the time of maximal degradation appeared to occur slightly earlier in vivo (no later than 20 min, Fig. 4D) than was seen in cellulo (no earlier than 20 min for the highest TNFα concentrations, Figs. 2I and 3I). Increasing the TNFα dose resulted in higher levels of maximal resynthesis (Fig. 4E) that peaked at nearly 20-fold over vehicle-treated animals. The resynthesis phase was broad in shape (similar to the in cellulo ΙκBα profiles in response to TNFα pulses), and it peaked and leveled off at ∼100 min for both the 10 and 30 ng/mouse doses (Fig. 4F). This is in contrast to the highest TNFα concentrations used in cellulo that did not achieve maximal resynthesis until 125 min (Figs. 2K and 3J). Thus, even though TNFα was administered at varying doses in vivo, the resultant ΙκBα dynamic profiles closely resembled those observed upon modulating TNFα pulse duration in cellulo (Figs. 2 and 3), having broad peaks and kinetics that do not significantly change as the dose is modulated. Furthermore, the general profile of κB5→ΙκBα-FLuc activity in vivo closely resembled the profiles observed for individual cells (Fig. 3), suggesting kinetic synchronicity of responding hepatocytes in vivo.
FIGURE 4.
ΙκBα dynamics as a function of TNFα dose in vivo. A, in vivo transfection of mouse hepatocytes was performed using the hydrodynamic somatic gene transfer method. Mice were imaged in an IVIS100 to obtain a pre-stimulation reading, followed by tail vein injection of 100 μl of vehicle (Veh) (sterile PBS) or TNFα (at the indicated doses), and then imaged at 5-min intervals for 3 h. B, data from five independent experiments are plotted normalized to the pre-TNFα stimulation levels (fold initial) and to a vehicle-treated animal (fold untreated); error bars represent mean ± S.E. C–F, quantitative analysis of in vivo measurements representing the extent of maximal ΙκBα degradation (C) and resynthesis (E) and the time of maximal ΙκBα degradation (D) and resynthesis (F), as functions of TNFα dose. All data are presented as mean ± 95% confidence interval. The 1 ng/mouse data point in D represents n = 3 because two animals showed no degradation at that dose and thus no degradation time could be calculated.
Experimental Investigation of Complex ΙκBα Resynthesis Patterns, in Cellulo
Having observed novel and complex patterns in the dynamics of ΙκBα degradation and resynthesis in single cells, cell populations, and in vivo in response to modulation of TNFα pulse duration and concentration, we next sought to investigate potential mechanisms behind these highly reproducible behaviors. Of particular interest were the stimulus-dependent differences in peak shape, amplitude, and kinetics of ΙκBα-Fluc resynthesis. We hypothesized that many of these complex patterns were a consequence of the continuous presence of TNFα driving subsequent rounds of IKK activation and ΙκBα degradation during the resynthesis phase. This hypothesis was supported by our previous finding that HepG2 cells given a 30-s pulse of TNFα regain the capacity to fully re-initiate a second TNFα-induced ΙκBα degradation only after a 60–120-min refractory period, the approximate time frame during which maximal ΙκBα resynthesis and rollover occur (12).
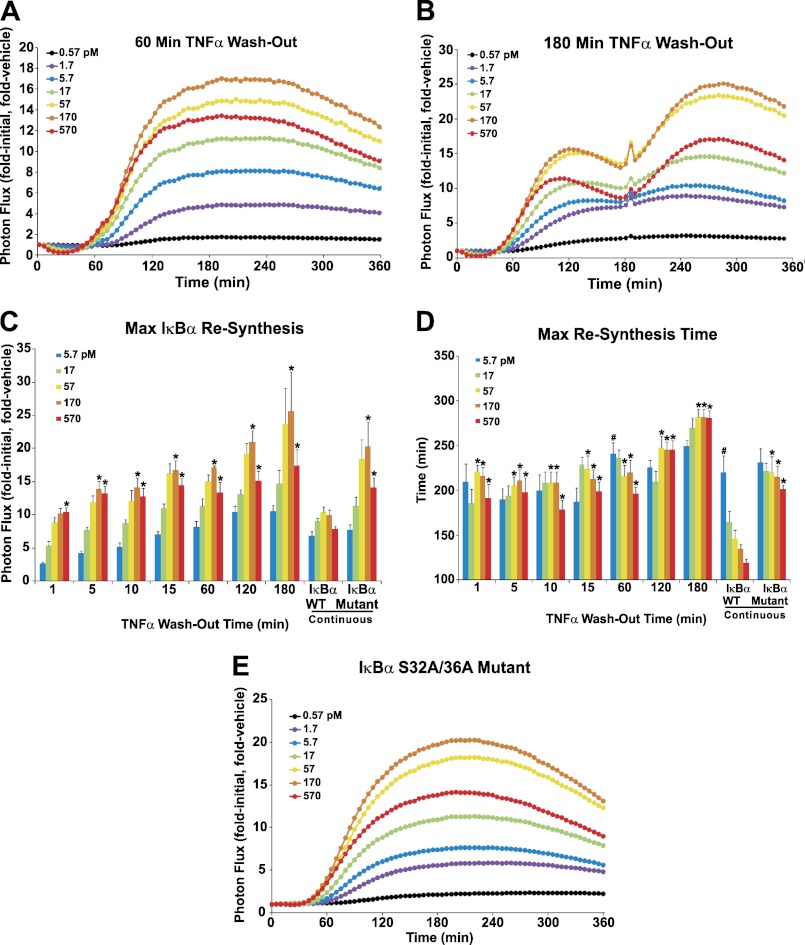
To assess the impact of TNFα presence at various time points before and during ΙκBα resynthesis, HepG2 cells were treated with increasing concentrations of TNFα that was then washed out after 1, 5, 10, 15, 30, 60, 90, 120, and 180 min to remove the effect of continuous TNFα driving subsequent rounds of IKK-mediated ΙκBα degradation. Two representative ΙκBα plots are shown in Fig. 5, A and B (un-normalized photon flux data and additional TNFα and mock washout plots are shown in supplemental Fig. S2). The removal of TNFα at any time before ΙκBα resynthesis had peaked (i.e. up to 120 min), resulted in broadly shaped ΙκBα resynthesis profiles (Fig. 5A and supplemental Fig. S2), rather than the narrower peaks seen under continuous TNFα (Figs. 2 and 3) or mock TNFα washout stimulation (supplemental Fig. S2F). TNFα washouts performed at 120 min (supplemental Fig. S2E) and 180 min (Fig. 5B) exhibited the expected primary ΙκBα resynthesis peak observed at 120 min under continuous TNFα, followed by a second ΙκBα peak (occurring at ∼240 min and 300 min, respectively), more similar to the peaks observed for earlier TNFα washout times (Fig. 5A and supplemental Fig. S2). When high concentrations of TNFα (170–570 pm) were washed out, cells exhibited significantly higher levels of ΙκBα resynthesis compared with continuous TNFα stimulation (Fig. 5C). Furthermore, TNFα washout resulted in ΙκBα resynthesis peaking later than continuously stimulated cells and nearly abolished the pattern of faster ΙκBα resynthesis observed in response to increasing TNFα concentrations (Fig. 5D). Interestingly, ΙκBα resynthesis rollover was still observed when TNFα was washed out (Fig. 5C).
FIGURE 5.
Experimental investigation of complex ΙκBα resynthesis patterns. A and B, HepG2 cells expressing κB5→ΙκBα-FLuc were treated with the indicated TNFα concentrations or vehicle at t = 0 min. At 60 or 180 min, the cells were washed and replenished with fresh TNFα-free media (washout conditions) or media containing TNFα at the initial concentration (a mock washout). Data were acquired every 5 min for 360 min and normalized as before to represent the mean of three or four independent TNFα exposure experiments, each performed in triplicate and averaged. C, plot representing the effect of washout time and TNFα concentration (5.7 pm or higher) on maximum ΙκBα resynthesis magnitude. All 180 min data represent parameters calculated from the second ΙκBα resynthesis peak. Data are mean ± S.E. * indicates p < 0.05 for TNFα (170 or 570 pm) washout or mutant ΙκBα(S32A,S36A)-FLuc versus continuous TNFα wild-type ΙκBα-FLuc. The 30-min data were n = 2 and thus were excluded from statistical analysis. D, plot representing the effect of washout time and TNFα concentration on the timing of maximum ΙκBα resynthesis. # indicates p < 0.05 for lowest versus highest TNFα concentration within a given TNFα treatment. E, HepG2 cells expressing mutant κB5→ΙκBα(S32A,S36A)-FLuc were treated continuously with TNFα or vehicle at the indicated concentrations; data were normalized as described previously and represent three independent experiments, each performed in triplicate and averaged. See also supplemental Figs. S2 and S3.
To further address the role of secondary (i.e. later time point) TNFα-induced ΙκBα degradation in governing ΙκBα-FLuc resynthesis phase dynamics, we utilized a mutant bioluminescent reporter, κB5→ΙκBα(S32A,S36A)-FLuc (35). The serine-to-alanine substitutions render ΙκBα unresponsive to IKK-directed phosphorylation and subsequent proteasomal degradation; however, the reporter is still responsive to the NF-κB transcriptional activity elicited once endogenous ΙκBα is degraded and NF-κB translocates into the nucleus. If re-initiation of ΙκBα degradation is critical in governing the timing, magnitude, and overall shape of ΙκBα-FLuc resynthesis, or the resynthesis rollover effect, then we would not expect to observe these phenomena with the κB5→ΙκBα(S32A,S36A)-FLuc reporter under continuous TNFα stimulation. As anticipated, TNFα stimulation of a population of cells expressing the mutant reporter did not cause any ΙκBα degradation (Fig. 5E) but did exhibit subsequent NF-κB-directed resynthesis of the reporter. Strikingly, these ΙκBα profiles strongly resembled the TNFα washout experiments (Fig. 5, A and B, supplemental Fig. S2) as follows: the ΙκBα resynthesis phases were broad (regardless of TNFα concentration), the peak magnitude increased (Fig. 5C), and the resynthesis kinetics were more synchronized and delayed (Fig. 5D) compared with wild-type reporter, indicating that these patterns were indeed affected by secondary IKK-driven degradation of wild-type ΙκBα. Similarly, continuous TNFα stimulation in single HepG2 cells transfected with the ΙκBα(S32A,S36A)-FLuc mutant reporter also exhibited broad IκBα resynthesis peaks with synchronized and delayed kinetics compared with wild-type reporter (Fig. 3, D and G), further highlighting the cell-autonomous nature of IκBα dynamics. Interestingly, single cells expressing the mutant reporter showed no evidence of ΙκBα-FLuc oscillations (similar to single cells pulsed with TNFα; Fig. 3C), confirming that oscillations in NF-κB activity are a consequence of additional rounds of degradation driven by continuous TNFα exposure.
The observation that ΙκBα resynthesis rollover was still observed even when removing TNFα during the resynthesis phase or when using a nondegradable ΙκBα mutant (Fig. 5C) indicated that this trend was likely caused by an input transcriptional regulation event as opposed to post-translational modification of newly synthesized ΙκBα. To identify potential mechanisms driving this behavior, we generated a set of hypothetical IKK activity profiles (supplemental Fig. S3A) based on an experimentally determined profile (12) and introduced them as input functions in an existing computational model of NF-κB signaling (13, 27). We were able to recapitulate ΙκBα resynthesis rollover computationally (supplemental Fig. S3C) by increasing IKK peak magnitude, holding it constant, and subsequently shortening the IKK deactivation duration (supplemental Fig. S3B), predicting that the temporal relationship of these two IKK parameters is critical in dictating downstream responses to variations in TNFα concentration.
DISCUSSION
Transcriptionally coupled negative feedback loops often serve as critical regulatory nodes within cellular signaling pathways. To fully understand the biological functions of negative feedback processes in their proper context, it would be critical to interrogate feedback loops at multiple scales as follows: in single cells, in populations of cells, and in live animals. To this end, we have employed a bioluminescent reporter of the NF-κB/ΙκBα negative feedback loop for multiscale studies aimed at understanding how diverse stimulation modes impact the dynamics of a critical cellular signaling pathway.
The κB5→ΙκBα-FLuc construct reports not only the rapid IKK-induced degradation of ΙκBα (the mediator of NF-κB nuclear translocation) but also the subsequent NF-κB-dependent transcriptional up-regulation of ΙκBα. Thus, the reporter offers the unique ability to measure both rapid post-translational events and coupled downstream transcriptional activity in real time in live cells, thereby enabling evaluation of the functional consequences (i.e. NF-κB transcriptional activity) of stimulus-specific changes in ΙκBα degradation/NF-κB nuclear translocation. These nondestructive assays are based on luciferase reporters and as such have high temporal resolution, do not rely on antibodies, are amenable to high throughput platforms, are readily translatable to in vivo systems, and have potential for low light microscopic analysis of single cell and subcellular compartments (6, 7, 36–38). Furthermore, although most single cell studies utilizing bioluminescent reporters rely upon costly low light microscopy systems, we sought to develop a more accessible, inexpensive, and high throughput strategy to image single cells expressing the κB5→ΙκBα-FLuc without the use of microscopy. As applied in this study, our method enabled simultaneous imaging of hundreds of single cells under a variety of stimulation regimens.
We employed the unique capabilities of the NF-κB/ΙκBα negative feedback loop reporter to systematically interrogate the impact of modulating TNFα pulse duration and concentration, first focusing on cell populations and then examining single cell responses. We demonstrated that cells are sensitive to pulses of TNFα stimulation as short as 5 s, highlighting that the NF-κB network is remarkably sensitive and tuned to elicit responses to very short bursts of ligand (14). Modulating TNFα pulse duration yielded ΙκBα dynamic profiles (Fig. 2A) that were much broader in shape than observed under continuous TNFα stimulation. As pulse duration increased, so did the amplitude of both ΙκBα degradation and resynthesis, without significantly impacting the kinetics. At the single cell level, continuous and 30-s TNFα pulses yielded ΙκBα-FLuc dynamic profiles (Fig. 3, B and C) that remarkably resembled the qualitative shape and quantitative kinetics of cell population profiles (Fig. 2). Thus, the broad peaks and synchronized resynthesis kinetics observed in cell populations upon stimulation with pulsatile TNFα were not a result of heterogeneous responses from individual cells generating a broad average signal but were intrinsic properties of single cells. A similar trend in invariant temporal NF-κB nuclear localization was observed by Werner et al. (14) in response to TNFα pulses; however, they did not observe changes in the amplitude of NF-κB activity (as measured by EMSA and computational prediction), whereas our reporter measured definitive pulse-dependent changes in the amplitude of ΙκBα resynthesis, a process that is directly dependent upon NF-κB transcriptional activity.
Real time measurements indicated that the ΙκBα/NF-κB negative feedback loop is responsive to a wide range of TNFα concentrations (Figs. 2G and 3F), even as low as 0.57 pm (0.01 ng/ml), affirming what has been observed previously by NF-κB EMSA (39) and single cell microscopy (16, 17). Additionally, we found that upon 0.57 pm TNFα stimulation, only 40% of cells showed evidence of ΙκBα-FLuc degradation and resynthesis, whereas nearly 90% of cells responded at all other concentrations examined; this is in close agreement with previous studies measuring NF-κB nuclear translocation in response to low TNFα concentrations (16, 17). The ΙκBα-FLuc resynthesis peaks became more defined as concentration increased, offering further evidence that this trend is cell autonomous and suggesting that residual TNFα in the media might drive subsequent secondary rounds of ΙκBα-FLuc degradation that impact the shape of the ΙκBα-FLuc resynthesis profile. Furthermore, in both single cells and cell populations, ΙκBα-FLuc degradation kinetics more closely clustered around the mean and sped up as TNFα concentration increased, although the timing of resynthesis also more closely clustered around the mean but exhibited only a moderate trend toward faster kinetics. Both Tay et al. (17) and Turner et al. (16) similarly noted that the time to peak nuclear NF-κB localization in individual cells tended to decrease and became less variable at higher TNFα concentrations. Thus, even though increasing TNFα concentrations result in both faster ΙκBα degradation and faster NF-κB nuclear translocation (16, 17), it does not appear to strongly affect the kinetics (timing) of the subsequent NF-κB transcriptional response. Furthermore, although several other groups have described kinetic heterogeneity in NF-κB translocation, we did not observe substantial heterogeneity in the transcriptional kinetics of responsive cells. We did, however, observe tremendous variability in the transcriptional amplitudes of responding cells, suggesting that the kinetic heterogeneity in NF-κB translocation documented by others could result in heterogeneity in the amplitude of downstream transcriptional responses at NF-κB target genes.
Upon modulating TNFα concentration, we also observed a highly reproducible ΙκBα resynthesis rollover pattern (Figs. 2J and 5C) not previously described. The physiological significance of this phenomenon remains to be characterized, but the fact that it could be modeled in silico (supplemental Fig. S3) by coordinated regulation of IKK activation amplitude and IKK deactivation period suggests these factors as key regulators of downstream responses to levels of TNFα.
Interestingly, the single cell imaging experiments also revealed that 18% of cells continuously stimulated with TNFα exhibited ΙκBα-FLuc oscillatory behavior, with an approximate period of 130 min (Fig. 3, B and E). This correlates with the NF-κB nuclear/cytoplasmic oscillations observed by others with a period of ∼100 min (15–17, 21–24). The slightly longer duty cycle in this study may relate to cell type-specific differences in the pathway or to the time required to transcribe and translate the larger ΙκBα-FLuc chimeric reporter protein. This oscillation phenomenon was never observed in cells given a 30-s pulse of TNFα or in cells expressing the ΙκBα (S32A,S36A)-FLuc mutant reporter, highlighting the critical role that secondary ΙκBα degradation plays in the oscillation phenotype. That we only observed ∼18% of cells oscillating may be due to inherent cell-to-cell variability in initial and total NF-κB concentration and/or ΙκBα translation and degradation rates, as was recently identified by Kalita et al. (40) or may result from the κB5 promoter that has been noted to shift the kinetics of resynthesis somewhat later when compared with the endogenous ΙκBα promoter (12, 21).
After rigorous characterization of the TNFα-induced response patterns of the κB5→ΙκBα-FLuc reporter in single cells and cell populations in culture, we interrogated TNFα-induced activation of the ΙκBα/NF-κB negative feedback loop within mouse livers in vivo (Fig. 4). Somatic gene transfer by hydrodynamic injection is a rapid and convenient strategy to generate mouse models for analysis of cell signaling in vivo (41), and it is a valuable approach to precede or complement development of expensive time-consuming transgenic or knock-in mouse strains.
Our data indicated that circulating TNFα, administered at varying doses, produced ΙκBα dynamic behaviors in vivo with synchronized kinetics and very high levels of ΙκBα resynthesis, patterns that were consistent with in cellulo experiments in which TNFα pulse duration was varied (Fig. 4). This strongly suggested that increasing concentrations of circulating TNFα were perceived by liver cells as increases in pulse duration, also plausible given the dual re-circulation physiology of the liver (hepatic arterial and portal venous) as well as bioavailability and hemodilution effects. Moreover, our findings underscore the importance of studying cytokine signaling pathways under conditions of pulsatile exposure (rather than just continuously bathing cells in ligand), which may better reproduce physiological cytokine stimulation paradigms. If pulsatile stimulation paradigms best recapitulate normal NF-κB stimulation conditions in vivo, this may place reservations on the physiological relevance of oscillatory NF-κB behaviors observed during continuous TNFα stimulation of single cells. However, our data lend support to the relevancy of the synchronous NF-κB oscillatory behaviors that are observed upon sequential TNFα pulse stimulations that drive frequency-encoded transcriptional programs (15, 17). In the future, it will be interesting to utilize low light microscopy techniques to investigate single cell responses within native tissues and further assess the physiological relevance of oscillations within the NF-κB pathway in vivo.
Having discovered novel and complex patterns in ΙκBα dynamics in response to modulation of TNFα pulse duration and concentration in single cells and cell populations, we next sought to investigate potential mechanisms behind these highly reproducible behaviors (Fig. 5). When TNFα was removed from the media any time before the peak of ΙκBα resynthesis or when ΙκBα was rendered insensitive to TNFα-induced degradation (i.e. mutant ΙκBα(S32A,S36A)-FLuc), broad ΙκBα resynthesis peaks with synchronized kinetics were observed, indicating the critical role that secondary TNFα-induced degradation of ΙκBα can play in regulating the ΙκBα/NF-κB negative feedback loop. Previously, we and others discovered a TNFα-induced transient refractory period during which TNFα-preconditioned cells are unable to fully respond (i.e. degrade ΙκBα) upon a second TNFα challenge until 60–120 min post-preconditioning (12, 15, 42). This refractory period is likely governed by the rate of ΙκBα/NF-κB nuclear export that repopulates the cytoplasm with IKK-degradable complexes (12, 38, 43, 44). Thus, the observed patterns in ΙκBα resynthesis dynamics may be a manifestation of this transient refractory period, whereby continuous TNFα is unable to induce subsequent round(s) of ΙκBα degradation until the passage of this refractory period.
The multiscalar approaches described in this study to image signaling dynamics in single cells, cell populations, and in vivo represent comparatively low cost, high throughput means to rapidly study nearly any cellular signaling pathway. Although the use of synthetic promoters such as κB5 can enhance sensitivity and enable measurement of subtle changes in transcriptional dynamics, artificial promoters may not be subject to the same set of regulatory processes encoded in the endogenous locus. Similarly, use of reporters comprising small transgenic coding sequences may fail to account for regulatory elements encoded within intronic and nearby enhancer regions of the endogenous gene. Future use of endogenous promoter regions driving properly engineered bioluminescent fusion reporters may provide a more specific understanding of ΙκBα transcriptional regulation under a variety of stimulation regimens at a variety of scales. In this way it will be possible to most accurately measure the true dynamics of the NF-κB/ΙκBα negative feedback loop in single cells, populations, and tissues in vivo.
In conclusion, this study revealed that the transcriptionally coupled NF-κB/ΙκBα negative feedback loop exhibits complex but highly reproducible dynamic patterns in response to modulating TNFα concentration or pulse duration. Interestingly, administration of TNFα at varying doses in vivo resulted in hepatocellular responses that were most consistent with perception of TNFα in vivo as a single concentration administered with increasing pulse duration. Thus, a single bioluminescent reporter strategy enabled rigorous quantitation of NF-κB/ΙκBα dynamics in both live single cells and cell populations, and it revealed reproducible behaviors that informed interpretation of studies in vivo.
Acknowledgments
We thank Prof. Alexander Hoffmann, Shannon Werner, and Derren Barken (University of California, San Diego) for providing the computational model software. We also thank Kelly Flentie for the HCT116 reporter cell line, Julie Prior for technical assistance with animal imaging, Reece Goiffon for assistance with statistical analysis, and Jayne Marasa for assistance with the InCell Analyzer 1000.
This work was supported, in whole or in part, by National Institutes of Health Grant P50 CA94056. This work was also supported by a predoctoral fellowship Award from the National Science Foundation (to B. L. M.).

This article contains supplemental Figs. 1–3, Movie 1, Experimental Procedures, and additional references.
- p53
- tumor protein 53
- NF-κB
- nuclear factor-κB
- IκBα
- inhibitor of NF-κB
- IKK
- IκB kinase
- FLuc
- firefly luciferase
- MDM2
- murine double minute 2
- CV
- coefficient of variation.
REFERENCES
- 1. Cebecauer M., Spitaler M., Sergé A., Magee A. I. (2010) Signaling complexes and clusters. Functional advantages and methodological hurdles. J. Cell Sci. 123, 309–320 [DOI] [PubMed] [Google Scholar]
- 2. Brandman O., Meyer T. (2008) Feedback loops shape cellular signals in space and time. Science 322, 390–395 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Alon U. (2007) Simplicity in biology. Nature 446, 497. [DOI] [PubMed] [Google Scholar]
- 4. Lahav G. (2008) Oscillations by the p53-Mdm2 feedback loop. Adv. Exp. Med. Biol. 641, 28–38 [DOI] [PubMed] [Google Scholar]
- 5. Sung M. H., McNally J. G. (2011) Live cell imaging and systems biology. Wiley Interdiscip. Rev. Syst. Biol. Med. 3, 167–182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Spiller D. G., Wood C. D., Rand D. A., White M. R. (2010) Measurement of single-cell dynamics. Nature 465, 736–745 [DOI] [PubMed] [Google Scholar]
- 7. Dothager R. S., Flentie K., Moss B., Pan M. H., Kesarwala A., Piwnica-Worms D. (2009) Advances in bioluminescence imaging of live animal models. Curr. Opin. Biotechnol. 20, 45–53 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Prescher J. A., Contag C. H. (2010) Guided by the light. Visualizing biomolecular processes in living animals with bioluminescence. Curr. Opin. Chem. Biol. 14, 80–89 [DOI] [PubMed] [Google Scholar]
- 9. Hayden M. S., West A. P., Ghosh S. (2006) NF-κB and the immune response. Oncogene 25, 6758–6780 [DOI] [PubMed] [Google Scholar]
- 10. Pasparakis M. (2009) Regulation of tissue homeostasis by NF-κB signaling. Implications for inflammatory diseases. Nat. Rev. Immunol. 9, 778–788 [DOI] [PubMed] [Google Scholar]
- 11. Perkins N. D. (2007) Integrating cell-signaling pathways with NF-κB and IKK function. Nat. Rev. Mol. Cell Biol. 8, 49–62 [DOI] [PubMed] [Google Scholar]
- 12. Moss B. L., Gross S., Gammon S. T., Vinjamoori A., Piwnica-Worms D. (2008) Identification of a ligand-induced transient refractory period in nuclear factor-κB signaling. J. Biol. Chem. 283, 8687–8698 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Werner S. L., Barken D., Hoffmann A. (2005) Stimulus specificity of gene expression programs determined by temporal control of IKK activity. Science 309, 1857–1861 [DOI] [PubMed] [Google Scholar]
- 14. Werner S. L., Kearns J. D., Zadorozhnaya V., Lynch C., O'Dea E., Boldin M. P., Ma A., Baltimore D., Hoffmann A. (2008) Encoding NF-κB temporal control in response to TNF. Distinct roles for the negative regulators IκBα and A20. Genes Dev. 22, 2093–2101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Ashall L., Horton C. A., Nelson D. E., Paszek P., Harper C. V., Sillitoe K., Ryan S., Spiller D. G., Unitt J. F., Broomhead D. S., Kell D. B., Rand D. A., Sée V., White M. R. (2009) Pulsatile stimulation determines timing and specificity of NF-κB-dependent transcription. Science 324, 242–246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Turner D. A., Paszek P., Woodcock D. J., Nelson D. E., Horton C. A., Wang Y., Spiller D. G., Rand D. A., White M. R., Harper C. V. (2010) Physiological levels of TNFα stimulation induce stochastic dynamics of NF-κB responses in single living cells. J. Cell Sci. 123, 2834–2843 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Tay S., Hughey J. J., Lee T. K., Lipniacki T., Quake S. R., Covert M. W. (2010) Single-cell NF-κB dynamics reveal digital activation and analogue information processing. Nature 466, 267–271 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lee T. K., Denny E. M., Sanghvi J. C., Gaston J. E., Maynard N. D., Hughey J. J., Covert M. W. (2009) A noisy paracrine signal determines the cellular NF-κB response to lipopolysaccharide. Sci. Signal. 2, ra65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Janes K. A., Gaudet S., Albeck J. G., Nielsen U. B., Lauffenburger D. A., Sorger P. K. (2006) The response of human epithelial cells to TNF involves an inducible autocrine cascade. Cell 124, 1225–1239 [DOI] [PubMed] [Google Scholar]
- 20. David G. L., Yegnasubramanian S., Kumar A., Marchi V. L., De Marzo A. M., Lin X., Nelson W. G. (2004) MDR1 promoter hypermethylation in MCF-7 human breast cancer cells. Changes in chromatin structure induced by treatment with 5-aza-cytidine. Cancer Biol. Ther. 3, 540–548 [DOI] [PubMed] [Google Scholar]
- 21. Nelson D. E., Ihekwaba A. E., Elliott M., Johnson J. R., Gibney C. A., Foreman B. E., Nelson G., See V., Horton C. A., Spiller D. G., Edwards S. W., McDowell H. P., Unitt J. F., Sullivan E., Grimley R., Benson N., Broomhead D., Kell D. B., White M. R. (2004) Oscillations in NF-κB signaling control the dynamics of gene expression. Science 306, 704–708 [DOI] [PubMed] [Google Scholar]
- 22. Kearns J. D., Basak S., Werner S. L., Huang C. S., Hoffmann A. (2006) IκBϵ provides negative feedback to control NF-κB oscillations, signaling dynamics, and inflammatory gene expression. J. Cell Biol. 173, 659–664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Sung M. H., Salvatore L., De Lorenzi R., Indrawan A., Pasparakis M., Hager G. L., Bianchi M. E., Agresti A. (2009) Sustained oscillations of NF-κB produce distinct genome scanning and gene expression profiles. PLoS One 4, e7163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Tian B., Nowak D. E., Brasier A. R. (2005) A TNF-induced gene expression program under oscillatory NF-κB control. BMC Genomics 6, 137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Friedrichsen B. N., Neubauer N., Lee Y. C., Gram V. K., Blume N., Petersen J. S., Nielsen J. H., Møldrup A. (2006) Stimulation of pancreatic beta-cell replication by incretins involves transcriptional induction of cyclin D1 via multiple signaling pathways. J. Endocrinol. 188, 481–492 [DOI] [PubMed] [Google Scholar]
- 26. Gross S., Piwnica-Worms D. (2005) Real time imaging of ligand-induced IKK activation in intact cells and in living mice. Nat. Methods 2, 607–614 [DOI] [PubMed] [Google Scholar]
- 27. Hoffmann A., Levchenko A., Scott M. L., Baltimore D. (2002) The IκB-NF-κB signaling module. Temporal control and selective gene activation. Science 298, 1241–1245 [DOI] [PubMed] [Google Scholar]
- 28. Liu F., Song Y., Liu D. (1999) Hydrodynamics-based transfection in animals by systemic administration of plasmid DNA. Gene Ther. 6, 1258–1266 [DOI] [PubMed] [Google Scholar]
- 29. Pichler A., Zelcer N., Prior J. L., Kuil A. J., Piwnica-Worms D. (2005) In vivo RNA interference-mediated ablation of MDR1 P-glycoprotein. Clin. Cancer Res. 11, 4487–4494 [DOI] [PubMed] [Google Scholar]
- 30. Paszek P., Ryan S., Ashall L., Sillitoe K., Harper C. V., Spiller D. G., Rand D. A., White M. R. (2010) Population robustness arising from cellular heterogeneity. Proc. Natl. Acad. Sci. U.S.A. 107, 11644–11649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Place R. F., Haspeslagh D., Giardina C. (2003) Induced stabilization of IκBα can facilitate its resynthesis and prevent sequential degradation. J. Cell. Physiol. 195, 470–478 [DOI] [PubMed] [Google Scholar]
- 32. Place R. F., Haspeslagh D., Hubbard A. K., Giardina C. (2001) Cytokine-induced stabilization of newly synthesized IκBα. Biochem. Biophys. Res. Commun. 283, 813–820 [DOI] [PubMed] [Google Scholar]
- 33. Warrington N. M., Gianino S. M., Jackson E., Goldhoff P., Garbow J. R., Piwnica-Worms D., Gutmann D. H., Rubin J. B. (2010) Cyclic AMP suppression is sufficient to induce gliomagenesis in a mouse model of neurofibromatosis-1. Cancer Res. 70, 5717–5727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Pichler A., Prior J. L., Luker G. D., Piwnica-Worms D. (2008) Generation of a highly inducible Gal4→Fluc universal reporter mouse for in vivo bioluminescence imaging. Proc. Natl. Acad. Sci. U.S.A. 105, 15932–15937 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Villalobos V., Naik S., Bruinsma M., Dothager R. S., Pan M. H., Samrakandi M., Moss B., Elhammali A., Piwnica-Worms D. (2010) Dual-color click beetle luciferase heteroprotein fragment complementation assays. Chem. Biol. 17, 1018–1029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Harper C. V., Featherstone K., Semprini S., Friedrichsen S., McNeilly J., Paszek P., Spiller D. G., McNeilly A. S., Mullins J. J., Davis J. R., White M. R. (2010) Dynamic organization of prolactin gene expression in living pituitary tissue. J. Cell Sci. 123, 424–430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Webb A. B., Angelo N., Huettner J. E., Herzog E. D. (2009) Intrinsic, nondeterministic circadian rhythm generation in identified mammalian neurons. Proc. Natl. Acad. Sci. U.S.A. 106, 16493–16498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Nelson G., Paraoan L., Spiller D. G., Wilde G. J., Browne M. A., Djali P. K., Unitt J. F., Sullivan E., Floettmann E., White M. R. (2002) Multiparameter analysis of the kinetics of NF-κB signaling and transcription in single living cells. J. Cell Sci. 115, 1137–1148 [DOI] [PubMed] [Google Scholar]
- 39. Cheong R., Bergmann A., Werner S. L., Regal J., Hoffmann A., Levchenko A. (2006) Transient IκB kinase activity mediates temporal NF-κB dynamics in response to a wide range of tumor necrosis factor-α doses. J. Biol. Chem. 281, 2945–2950 [DOI] [PubMed] [Google Scholar]
- 40. Kalita M. K., Sargsyan K., Tian B., Paulucci-Holthauzen A., Najm H. N., Debusschere B. J., Brasier A. R. (2011) Sources of cell-to-cell variability in canonical nuclear factor-κB (NF-κB) signaling pathway inferred from single cell dynamic images. J. Biol. Chem. 286, 37741–37757 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Suda T., Liu D. (2007) Hydrodynamic gene delivery. Its principles and applications. Mol. Ther. 15, 2063–2069 [DOI] [PubMed] [Google Scholar]
- 42. Shih V. F., Kearns J. D., Basak S., Savinova O. V., Ghosh G., Hoffmann A. (2009) Kinetic control of negative feedback regulators of NF-κB/RelA determines their pathogen- and cytokine-receptor signaling specificity. Proc. Natl. Acad. Sci. U.S.A. 106, 9619–9624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Zandi E., Chen Y., Karin M. (1998) Direct phosphorylation of IκB by IKKα and IKKβ. Discrimination between free and NF-κB-bound substrate. Science 281, 1360–1363 [DOI] [PubMed] [Google Scholar]
- 44. O'Dea E. L., Barken D., Peralta R. Q., Tran K. T., Werner S. L., Kearns J. D., Levchenko A., Hoffmann A. (2007) A homeostatic model of IκB metabolism to control constitutive NF-κB activity. Mol. Syst. Biol. 3, 111. [DOI] [PMC free article] [PubMed] [Google Scholar]