Background: During epithelial-mesenchymal transition (EMT), cancer cells employ processes to gain migratory and invasive properties that involve dramatic changes in cytoskeletal component organization.
Results: Zyxin silencing increases integrin α5β1 expression, accompanied by enhanced cell motility.
Conclusion: Zyxin is implicated in two diverse activities, regulation of cell adhesion and motility.
Significance: Understanding of cytoskeletal proteins may shed new light on the process of EMT in cancer.
Keywords: Cancer Biology, Cell Motility, Epithelial-to-Mesenchymal Transition, Integrins, Lung Cancer, Transforming Growth Factor beta (TGFbeta)
Abstract
Although TGF-β acts as a tumor suppressor in normal tissues and in early carcinogenesis, these tumor suppressor effects are lost in advanced malignancies. Single cell migration and epithelial-mesenchymal transition (EMT), both of which are regulated by TGF-β, are critical steps in mediating cancer progression. Here, we sought to identify novel direct targets of TGF-β signaling in lung cancer cells and have indentified the zyxin gene as a target of Smad3-mediated TGF-β1 signaling. Zyxin concentrates at focal adhesions and along the actin cytoskeleton; as such, we hypothesized that cytoskeletal organization, motility, and EMT in response to TGF-β1 might be regulated by zyxin expression. We show that TGF-β1 treatment of lung cancer cells caused rapid phospho-Smad3-dependent expression of zyxin. Zyxin expression was critical for the formation and integrity of cell adherens junctions. Silencing of zyxin decreased expression of the focal adhesion protein vasodilator-activated phospho-protein (VASP), although the formation and morphology of focal adhesions remained unchanged. Zyxin-depleted cells displayed significantly increased integrin α5β1 levels, accompanied by enhanced adhesion to fibronectin and acquisition of a mesenchymal phenotype in response to TGF-β1. Zyxin silencing led to elevated integrin α5β1-dependent single cell motility. Importantly, these features are mirrored in the K-ras-driven mouse model of lung cancer. Here, lung tumors revealed decreased levels of both zyxin and phospho-Smad3 when compared with normal tissues. Our data thus demonstrate that zyxin is a novel functional target and effector of TGF-β signaling in lung cancer. By regulating cell-cell junctions, integrin α5β1 expression, and cell-extracellular matrix adhesion, zyxin may regulate cancer cell motility and EMT during lung cancer development and progression.
Introduction
Lung cancer is the leading cause of cancer-related mortality in both men and women worldwide. Aggressive cancer progression has been correlated with disrupted epithelial cell homeostasis that leads to the loss of cell epithelial characteristics and the acquisition of a migratory phenotype (1). Epithelial-mesenchymal transition (EMT)3 plays an important role in the regulation of embryonic development, as well as in various pathological conditions including fibrosis and cancer (2). Different growth factors have been involved in the regulation of EMT, but transforming growth factor-β (TGF-β) has been clearly shown as the major inducer of this process (3–6).
Downstream TGF-β signaling can activate distinctive signal transduction mechanisms, which can be either Smad-mediated or non-Smad-mediated, depending on the precise cellular context. In the Smad-mediated pathway, upon TGF-β binding, the type II receptor kinase activates type I receptor kinase, leading to the phosphorylation and activation of Smad2 and Smad3 proteins. Phosphorylation induces their association with Smad4 and subsequent translocation to the nucleus, where they control the expression of TGF-β-responsive genes (7, 8). Interestingly, at early stages of tumor development, TGF-β acts as a tumor suppressor. In contrast, sustained expression of TGF-β may enhance malignant properties of tumor cells by increasing their motility and promoting EMT. Epithelial cells undergoing EMT lose their apico-basolateral polarity through remodeling of the actin cytoskeleton. Polymerization of the actin cytoskeleton plays a central role in the regulation of cell apoptosis, proliferation, anchorage-independent growth, and invasion (3, 9). Several studies have reported TGF-β effects on cell morphology and actin rearrangement. The formation of actin stress fibers in response to TGF-β requires de novo protein synthesis through Smad-mediated transcription and activation of the Rho family of GTPases, which in turn control cell motility and invasion (10–12). Numerous other actin regulatory proteins including the LIM domain proteins have been described as regulators of cell adhesion and migration.
Zyxin is a LIM domain protein localized to the nucleus, cell-cell contacts, and focal adhesions as well as along the actin stress fibers that harbors distinct actin polymerization activity independent of the Arp2/3 complex (13). Zyxin contains four proline-rich repeats at the N terminus followed by a nuclear export signal, and at the C terminus, three copies of a cysteine- and histidine-rich motif called the LIM domain. With its proline-rich repeats, zyxin directly interacts with α-actinin and Enabled/vasodilator-activated phospho-protein (Ena/VASP) and docks them to the actin filaments (14–18). Furthermore, intact zyxin proline-rich domains are required for efficient VASP binding and strengthening of cell-cell adhesions (17, 19). Zyxin also co-localizes with integrins at focal adhesions where it serves as a docking protein during the multimeric protein complex formation, involved in the regulation of cell-extracellular matrix adhesion (20). When cancer cells become more metastatic, they are able to develop an altered affinity for the extracellular matrix, mostly due to the changed expression of cell-surface receptor integrins. Altered integrin expression has been shown to be important for different cell activities including cell survival, differentiation, proliferation, and motility (21).
Because zyxin appears to have distinct functions in association with cell-cell junctions and cell-extracellular matrix adhesions, we sought to investigate its role in TGF-β-triggered EMT and motility in lung cancer cells. We show that zyxin controls lung cancer cell motility through modulation of cell adhesion and expression of integrins.
EXPERIMENTAL PROCEDURES
Materials
Protein A-agarose beads were purchased from Thermo Scientific (Pierce, Bonn, Germany). Recombinant human TGF-β1 was obtained from R&D Systems (Wiesbaden, Germany), and fibronectin was from Biochrom (Berlin, Germany). Poly-l-lysine was purchased from Sigma-Aldrich (Deisenhofen, Germany). Permanent AP-Red kit and 3,3′-diaminobenzidine substrate kit were from Zytomed (Berlin, Germany). Smad3 inhibitor SIS3 was purchased from Calbiochem (Darmstadt, Germany). All siRNA oligonucleotides were purchased from Ambion (Darmstadt, Germany). Plasmids pEGFP-N2 and zyxin-EGFP were kindly provided by Dr. Gerald Burgstaller (Helmholtz Zentrum Muenchen). Alexa Fluor-labeled secondary antibodies, Alexa Fluor 568-labeled phalloidin, Lipofectamine 2000, and Lipofectamine RNAiMAX were purchased from Invitrogen (Karlsruhe, Germany). The following antibodies were purchased from Abcam (Cambridge, UK): anti-zyxin, anti-VASP (5C6), and anti-Smad3 (ChIP grade). Anti-E-cadherin, anti-p120, anti-paxillin, anti-integrin α5, anti-integrin β1, phycoerythrin anti-human CD49e, and phycoerythrin IgG1 isotype control were purchased from BD Biosciences (Heidelberg, Germany). The antibodies against phospho-paxillin (Tyr118), phospho-Src, Src, and Smad3 were from Cell Signaling (New England Biolabs, Frankfurt, Germany). Anti-actin antibody was purchased from Sigma-Aldrich, and anti-fibronectin was from Santa Cruz Biotechnology (Heidelberg, Germany). Anti-zyxin (clone 2D1) antibody was purchased from Abnova (Heidelberg, Germany), and phospho-Smad3 (pS423/425) was from Epitomics (Burlingame, CA).
Cell Culture and Treatment
The human non-small cell lung cancer (NSCLC) cell lines (A549, H2030, H1299, and H441) were purchased from the American Type Culture Collection (ATCC), and human SCLC cell lines (DMS273 and H82) were from the German Collection of Microorganisms and Cell Cultures (DSMZ). All cell lines were maintained according to the instructions of the manufacturer. The cell culture medium was supplemented with 10% fetal bovine serum (FBS), and cells were maintained in a humidified incubator in an atmosphere of 5% CO2 at 37 °C. Cells were routinely passaged every 3–4 days. For experimental use, cell cultures were made quiescent by growing them to 80% confluence and overnight incubation in serum-free medium.
Mice
K-rasLA2 mice (22) and control mice (C57BL/6 background) were maintained under pathogen-free conditions and handled in accordance with the European Communities recommendations for experimentation. 5-month-old mice were sacrificed for lung tumor analysis, immunohistochemistry, and gene expression profile.
Reverse Transcription and Quantitative Real-time PCR
Total RNA was extracted from frozen mouse lung tissues and cells using the TRIzol (Invitrogen) or peqGOLD total RNA kit (peqlab, Erlangen, Germany) according to the manufacturer's instructions. RNA (1 μg) was reverse-transcribed in a 40-μl reaction using the M-MLV reverse transcriptase (Promega, Mannheim, Germany). Quantitative real-time PCR (qRT-PCR) was performed using SYBR Green PCR master mix (Roche Applied Science, Mannheim, Germany). All qRT-PCR assays were performed in triplicate. For standardization of relative mRNA expression, human hypoxanthine-guanine phosphoribosyltransferase (HPRT) or mouse glyceraldehyde-3-phosphate dehydrogenase(GAPDH) primers were used. Relative transcript abundance of a gene is expressed in ΔCp values (ΔCp = Cpreference − Cptarget). Relative changes of mRNA levels are presented as 2−ΔΔCp calculations (ΔΔCp = ΔCptreated − ΔCpcontrol). PCR amplification was performed with the gene-specific primers listed in Table 1.
TABLE 1.
Primer list
HPRT, hypoxanthine-guanine phosphoribosyltransferase.
| Gene | Forward primer (5′-3′) | Reverse primer (5′-3′) |
|---|---|---|
| Human zyxin | GCAGAATGTGGCTGTCAACGAAC | TGAAGCAGGCGATGTGGAACAG |
| Human ITGα5 | TCAGTGGAGTTTTACCGGCCGGG | TGGCTGGTATTAGCCTTGGGTGC |
| Human ITGβ1 | ACCAAATGATGGACAATGTCACCTGGA | ACAGGCTGAAATTCTTCAGTAACTGCA |
| Human HPRT | AAGGACCCCACGAAGTGTTG | GGCTTTGTATTTTGCTTTTCCA |
| Human zyx-1 (ChIP) | TTACCCAGGTCTGGCTTCTTTCGT | TACTGGCCAGACTGGATTTCTC |
| Human zyx-2 (ChIP) | TGAGAAATCCAGTCTGGCCAGTAG | TGCTCGTCTTTTACGGCCCTTCAA |
| Human zyx-ctr (ChIP) | TGGCAACCCAGAAAGGGACATA | TTCCAGTTCTGCCCAGAATG |
| Human Smad7 (ChIP) | GAAACCCGATCTGTTGTTTGC | CTCTGCTCGGCTGGTTCCAC |
| Human Smad3 | TGGCTACCTGAGTGAAGATGGA | ATTATGTGCTGGGGACATCGGA |
| Mouse zyxin | GGCTGCTACACCGACACTTTG | CTCAGCATGCGGTCAGTGAT |
| Mouse TGFbRII | AACGTGGAGTCGTTCAAGCAGA | ATTGCAGCGGGACGTCATTT |
| Mouse GAPDH | TGTGTCCGTCGTGGATCTGA | CCTGCTTCACCACCTTCTTGA |
Western Blot Analysis
Cultures were lysed for 30 min at 4 °C with lysis buffer (10 mm Tris-HCl, pH 7.5, 5 mm EDTA, pH 8, 150 mm NaCl, 30 mm sodium pyrophosphate, 50 mm NaF, 10%, glycerol, 0.5% Triton X-100, supplemented with CompleteTM proteinase inhibitor mixture (Merck Biosciences, Darmstadt, Germany)). Cellular debris were removed by centrifugation at 13,000 × g at 4 °C for 15 min, and protein concentration was quantified using the BCA assay (Pierce). Lysates were extracted in sample buffer (260 mm Tris-HCl, 40% glycerol, 8% SDS, 0.004% bromphenol blue, 5% β-mercaptoethanol), resolved by SDS-PAGE, and transferred to polyvinylidene difluoride (PVDF) membrane (Amersham Biosciences, Bucks, UK). After blocking with Tris-buffered saline (TBS) containing 5% nonfat milk and 0.1% Tween, membranes were incubated with primary antibody either for 3 h at room temperature or overnight at 4 °C. Immunoreactive proteins were visualized by autoradiography following incubation with horseradish peroxidase (HRP)-coupled secondary antibody and enhanced chemiluminescence (ECL) reagent. Prior to reprobing, PVDF membranes were incubated with ReBlot Plus buffer (Millipore, Darmstadt, Germany) according to the manufacturer's protocol.
Cell Adhesion Assay
96-well plates were coated with 10 μg/ml fibronectin and incubated at 4 °C overnight. Coated wells were blocked with 0.5% bovine serum albumin (BSA) in phosphate-buffered saline (PBS) for 1 h at 37 °C. After siRNA transfection, cells were serum-starved and then incubated in the absence or presence of TGF-β1 (5 ng/ml) for 24 h. 2 × 104 cells were added to each well and incubated for 30 min at 37 °C under an atmosphere of 5% CO2. Plates were shaken for 10 s on a plate shaker followed by three washing steps in washing buffer (0.1% BSA in PBS) to remove nonadherent cells. The bound cells were fixed in 3.7% paraformaldehyde and stained with crystal violet for 20 min. Cell-associated crystal violet was eluted with 10% acetic acid, and optical density was measured at a wavelength of 570 nm.
Chromatin Immunoprecipitation Assay
Cells were serum-starved overnight and then treated with TGF-β1 (5 ng/ml) for 30 min. Cross-linking was performed by the direct addition of formaldehyde (1.42% final concentration) for 10 min at room temperature followed by glycine (125 mm final concentration) addition to quench the formaldehyde. After washing with ice-cold PBS, cells were collected by centrifugation. Collected cells were washed twice in 1 ml of cell lysis buffer (150 mm NaCl, 50 mm Tris-HCl, pH 7.5, 5 mm EDTA, 0.5% v/v Nonidet P-40, 1% v/v Triton X-100, supplemented with CompleteTM proteinase inhibitor mixture). Nuclei were pelleted, resuspended in lysis buffer, and incubated 30 min at 4 °C. After incubation, samples were sonicated to yield sheared DNA fragments between 200 and 500 bp and clarified by centrifugation at 12,000 × g for 10 min at 4 °C. Immunoprecipitation was carried out with 5 μg of antibody against Smad3 or control IgG at 4 °C overnight followed by a 2 h incubation with protein A-agarose beads. Immunoprecipitates were washed as follows: three times with lysis buffer, two times with TE buffer (10 mm Tris-HCl, pH 8.0, 1 mm EDTA, pH 8.0). Elutions were performed at room temperature by adding elution buffer (50 mm NaHCO3, 1% SDS, 10 mm DTT) to the beads. Cross-linking was reversed by incubation at 65 °C overnight. The samples were treated with RNase and proteinase K for 2 h at 45 °C. DNA was extracted with phenol/chloroform and ethanol precipitation. The purified DNA was dissolved in 100 μl water. 5 μl of purified DNA was amplified and analyzed by qRT-PCR using primers specific for the zyxin promoter region (primers are given in Table 1).
Immunohistochemical Analysis
For immunohistochemistry, mouse lungs were perfused with PBS, fixed in 4% paraformaldehyde (pH 7.0), and embedded in paraffin. Paraffin sections (2 μm) were deparaffinized, rehydrated, and pressure-cooked (30 s at 125 °C and then for 10 s at 90 °C) in citrate buffer (10 mm, pH 6.0) for an antigen retrieval followed by blocking of endogenous peroxidase (3% H2O2/dH2O; 15 min). Immunostaining employed the streptavidin-alkaline phosphatase technique (Permanent AP-Red kit) for zyxin (clone 2D1) and streptavidin-horseradish-peroxidase technique (3,3′-diaminobenzidine substrate kit) for pSmad3. Stained slides were counterstained for 1 min in Mayer's Haemalaun solution (Carl Roth, Karlsruhe, Germany). Images were taken with an Axio Imager (Zeiss) using a 40× oil immersion objective. All immunohistochemical slides were evaluated by two independent observers.
Immunofluorescence Microscopy
A549 cells were treated as described under “Results” and plated onto poly l-lysine-coated glass coverslips. After a 24-h incubation, cultures were serum-starved and treated in the absence or presence of TGF-β1 (5 ng/ml) for the indicated times. Cells were washed in PBS, fixed in 3.7% paraformaldehyde, and permeabilized with PBS containing 0.1% Triton X-100. After blocking with PBS containing 5% BSA at room temperature, cells were incubated with the primary antibody for 1 h, washed, and incubated for an additional 1 h with either Alexa Fluor 488- or Alexa Fluor 568-labeled secondary antibody. Afterward, cells were washed and incubated with 4′,6-diamidino-2-phenylindole (DAPI) and phalloidin for 30 min at room temperature.
Coverslips were mounted in fluorescent mounting medium (Dako, Hamburg, Germany) and fluorescent microscopy performed using the laser-scanning microscope LSM710 (Carl Zeiss, Munich, Germany). Images were analyzed using the ZEN 2010 software (Carl Zeiss).
Small Interfering RNA Transfection
Reverse transfection of siRNA into cells was performed with Lipofectamine RNAiMAX according to the manufacturer's instructions. The final siRNA concentration of 10 nm was used in all experiments. Cells transfected with siRNA were incubated 72 h before harvest.
Flow Cytometry
Treated cells were washed with PBS and harvested. The cells were washed once with FACS buffer containing 2% FBS in PBS. One million cells were incubated with the primary antibody (conjugated with phycoerythrin (PE)) for 20 min on ice. The cells were washed in FACS buffer after the incubation, resuspended in 700 μl of the same buffer, and acquired in a BD Biosciences LSR II flow cytometer. The results were analyzed using the software Cell Quest. Unstained cells and cells incubated with a suitable isotype control antibody were used as a negative control.
Plasmid Transfection
A549 cells were transfected with 3 μg of plasmid DNA using Lipofectamine 2000 according to manufacturer's instructions and used within 72 h.
Time-lapse Microscopy
Cells were transfected with siRNA and plated into 24-well plates. Cultures were incubated for 24 h and serum-starved overnight. Starved cultures were incubated in either the absence or the presence of TGF-β1 (5 ng/ml). Time-lapse movies were acquired over a period of 24 h using the Axio Observer microscope equipped with an AxioCam camera (Carl Zeiss). Images were captured at 10-min intervals and analyzed by AxioVision 4.0 software (Carl Zeiss). Cells were tracked, and migration velocity (μm/min) and persistence (displacement/track length) were determined by ImageJ analysis software (rsbweb.nih.gov/ij/).
Aggregation Assay
siRNA-transfected cells were trypsinized, washed in PBS, and resuspended in normal growth medium. 5 × 103 cells were suspended in each hanging drop from the lid of a 24-well plate and incubated for 24 h at 37 °C under an atmosphere of 5% CO2. Cells were subjected to the shear force (passaged five times through a 200-μl pipette tip) and imaged using a 10× phase contrast objective. For quantification, individual fields of cells were counted after the shear stress, and aggregation index was measured according to the formula (Nt/N0), where N0 is the total number of particles at the start of incubation and Nt is the total particle number after the incubation.
Calcium Switch Assay
Cells were transfected with specific siRNA and allowed to form junctions. Transfected cells were serum-starved and then treated with 4 mm EGTA for 30 min followed by replacement of the fresh medium containing Ca2+ (1.8 mm) to initiate junction reassembly. At the indicated times, cells were fixed in 3.7% paraformaldehyde and processed for immunofluorescent staining.
Wound Healing Assay
For wound healing migration assay, cells were seeded on 24-well plates. Serum-starved confluent monolayers were scratched with a fine pipette tip, and cell migration in the presence or absence of TGF-β1 or inhibitory antibody was visualized by phase contrast microscopy. Photographed wounds were analyzed by ImageJ software, and wound closure was represented as a percentage of the initial wound area.
Statistical Analysis
Data were presented as mean ± S.D. or mean ± S.E., and analysis was performed using the Student's t test. A value of p < 0.05 was considered significant. All values represent at least three independent experiments.
RESULTS
TGF-β1-induced Zyxin Expression Is Smad3-dependent
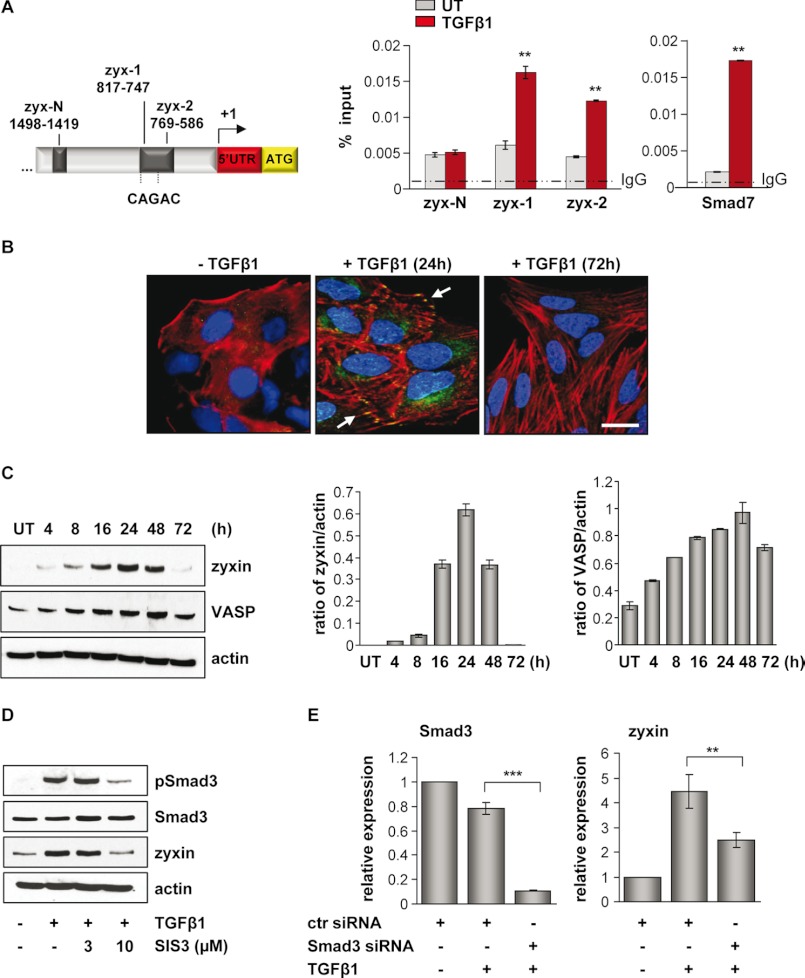
The EMT process in A549 cells upon TGF-β1 stimulation was characterized by changes in cell morphology, loss of the epithelial marker E-cadherin, and expression of the mesenchymal marker fibronectin (supplemental Fig. S1). Recently, with the aim of identifying novel target genes of TGF-β1/Smad3 signaling in lung epithelial cancer cells, we have performed a ChIP-on-chip screen (23). Using this approach, the zyxin gene was revealed as a novel target of Smad3-mediated TGF-β1 signaling. To validate the binding of Smad3 to the zyxin promoter region, the ChIP-qRT-PCR analysis was performed by using primers specific to the zyxin promoter region (Table 1). The strong Smad3 binding to the zyxin (zyx-1, zyx-2) promoter regions was observed 30 min after TGF-β1 stimulation when compared with untreated controls (Fig. 1A). Amplified promoter region contained two Smad-binding elements, CAGAC sequences. Primers for the Smad7 promoter region were used as a positive control. Amplification of zyx-N promoter region (negative control at about 1000 bp downstream of the Smad3 binding site) was the same in TGF-β1-treated and untreated cells, indicating the specificity of Smad3 binding. We next determined the effect of TGF-β1 stimulation on endogenous zyxin expression levels in A549 cells. Increased levels of zyxin and its binding partner VASP were detected at 24 and 48 h, respectively (Fig. 1C). At 72 h after TGF-β1 stimulation, the level of zyxin was barely detectable. Immunofluorescent staining of A549 cells also revealed TGF-β1-dependent increase in zyxin expression and showed sustained actin stress fiber formation and its reorganization (Fig. 1B). The most apparent observation was that zyxin expression levels correlated with an increase in Smad3 phosphorylation and its translocation to the nucleus (supplemental Fig. S1). Therefore, to further demonstrate the Smad3-dependent zyxin expression, we investigated zyxin expression levels in the presence or absence of a specific inhibitor of Smad3 (SIS3) or Smad3 siRNA. As shown in Fig. 1D, the treatment of cells with 10 μm SIS3 inhibitor markedly reduced the TGF-β1-induced phosphorylation of Smad3 and affected the endogenous expression of zyxin protein. Likewise, siRNA-mediated silencing of Smad3 resulted in decreased zyxin mRNA levels even in the presence of TGF-β1 (Fig. 1E).
FIGURE 1.
Zyxin is a direct target of TGF-β/Smad3-dependent signaling. A, schematic presentation of the zyxin promoter area. Positions of potential Smad3 binding are indicated relative to the transcription start site. Arrow indicates the transcription start site. ChIP analysis revealed Smad3 binding to the zyxin promoter region after 30 min of TGF-β1 stimulation, as analyzed by the qRT-PCR method (zyx-1 and zyx-2 represent two different primer sets in zyxin promoter region). The ChIP-qRT-PCR primers amplifying a fragment of Smad7 promoter were used as a positive control. Gray bars indicate Smad3 base-line binding to the zyxin promoter in the untreated (UT) condition, whereas red bars indicate Smad3 binding after TGF-β1 stimulation. Data are presented as mean ± S.D. **, p < 0.01. B, A549 cells were either untreated or treated with TGF-β1, fixed, and immunostained with anti-zyxin antibody (green). Actin cytoskeleton was visualized by Alexa Fluor 568 phalloidin staining (red), and nuclei were visualized with DAPI (blue). White arrows indicate zyxin localization. Scale bar: 10 μm. C, immunoblot analysis of zyxin and VASP in untreated or TGF-β1-treated A549 cells. Densitometric analysis was used to determine the zyxin/actin and VASP/actin ratios (ImageJ software). Data are shown as mean ± S.E. D, A549 cells were cultured in either the presence or the absence of SIS3 inhibitor and TGF-β1 for 24 h. Protein lysates were prepared and Western blotted using anti-phospho-Smad3, Smad3, or zyxin antibodies. β-Actin was used as a loading control. E, A549 cells were transfected with control (ctr) or Smad3 siRNA, serum-starved, and then incubated with or without TGF-β1 for 24 h. The mRNA levels of Smad3 and zyxin were quantified by qRT-PCR. Data are presented as mean ± S.D. of three independent experiments; **, p < 0.01, ***, p < 0.001.
Zyxin Depletion Alters the Stability and Dynamics of Adherens Junctions
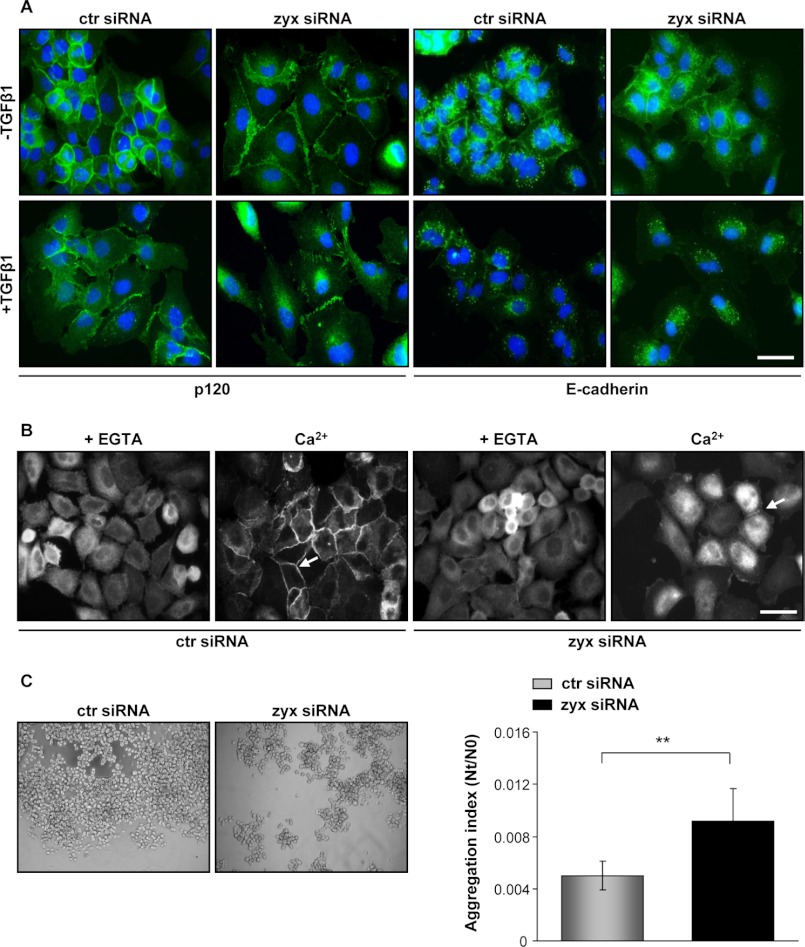
To determine the precise role of zyxin in lung cancer cells, we have used RNA interference to knock down endogenous levels of zyxin. Because TGF-β1 induced changes in cell morphology but did not promote a complete cell scattering, the expression and distribution of junctional markers were studied by immunofluorescence microscopy. Cancer A549 cells treated with TGF-β1 for 24 h showed decreased protein levels of E-cadherin and its internalization from cell-cell junctions in both control and zyxin-silenced cells. In contrast, junctional protein p120 persisted at cell-cell contacts upon TGF-β1 stimulation in control siRNA-transfected cells. Interestingly, the p120-positive junctions observed in zyxin-depleted cells were “zipper-like” when compared with scrambled siRNA-transfected cells that exhibited typical “sealed” cell-cell contact phenotype (Fig. 2A). Moreover, zyxin-depleted cells showed spindle shape-like morphology (data not shown) and increased scattering upon TGF-β1 stimulation.
FIGURE 2.
Silencing of zyxin affects cell-cell junction integrity. A, A549 cells were transfected with the indicated siRNAs, starved, and then incubated in either the absence or the presence of TGF-β1 for 24 h. Fixed and permeabilized cells were processed for fluorescence with antibodies recognizing p120 or E-cadherin (both green). Nuclei were visualized with DAPI (blue). Scale bar: 20 μm. ctr siRNA, control siRNA; zyx siRNA, zyxin-specific siRNA. B, confluent siRNA-transfected A549 cells were treated with EGTA-containing medium and then incubated with calcium supplemented medium for 1 h. Cell layers were fixed and stained for p120. White arrows indicate formation of adherens junctions. Scale bar: 20 μm. C, cell aggregates were subjected to the phase contrast microscopy. The extent of cell aggregation is presented by the ratio of the total particle number at time of incubation (Nt) to the initial particle number (N0). Lower levels represent higher degrees of aggregation; Nt/N0 = 1 no cell-cell adhesion; Nt/N0 < 1 specific cell-cell adhesion. Results are presented as mean ± S.D. of three independent experiments; **, p < 0.01.
To determine whether zyxin is localized at adherens cell-cell junctions, we have performed immunofluorescent staining. As shown in supplemental Fig. S2, after TGF-β1 stimulation, zyxin was mostly found at places of focal adhesion but also at cell-cell contacts co-localizing with junctional protein p120. Adherens junctions are dynamic structures that undergo constant remodeling. To test the involvement of zyxin in the formation of adherens junctions, we next performed a Ca+2 switch assay. A549 cell monolayers were placed in low calcium low medium to disrupt cell-cell junctions and were switched to high calcium medium to trigger junction reassembly. Most of the cells transfected with scrambled control siRNA formed sealed cell-cell contacts after 1 h in high calcium medium. In contrast, zyxin-depleted cells were able to form just a few discrete adherens junctions during the same period of time, suggesting zyxin involvement in junction dynamics (Fig. 2B). The influence of zyxin on adherens junctions was further confirmed by a cell aggregation assay. Zyxin-depleted A549 cells exhibited reduced aggregation capacity by 45% when compared with control siRNA-transfected cells (Fig. 2C).
Formation of Focal Adhesions in Lung Cancer Cells Is Zyxin-independent
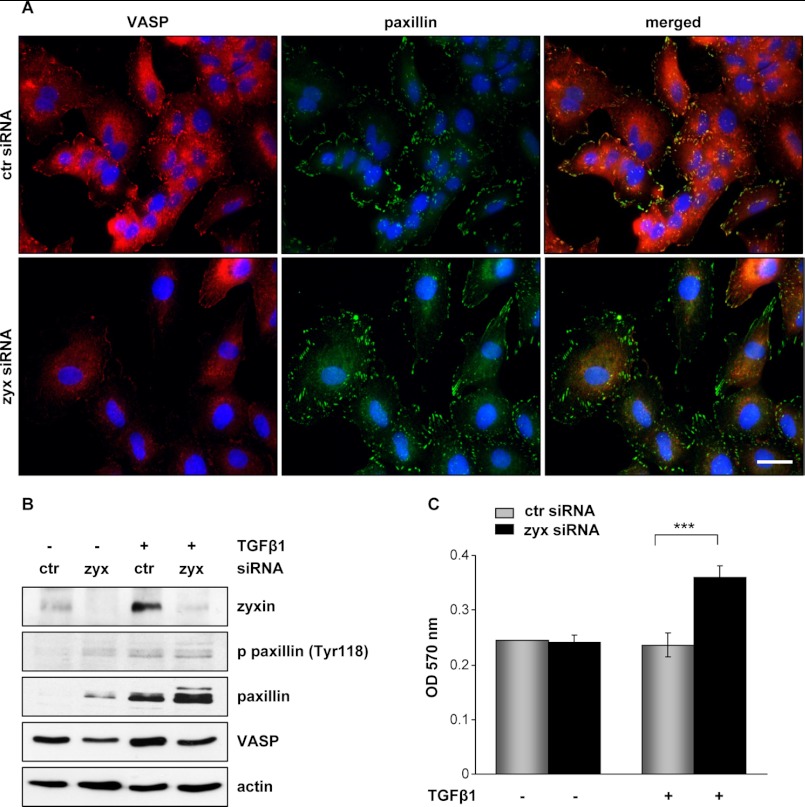
Zyxin is present at places of mature focal adhesion, crucial signaling components involved in cytoskeleton organization and cell migration. To explore the contribution of zyxin to lung cancer cell adhesion and motility, we examined both the formation of focal adhesions and the localization of zyxin-interacting partner VASP. Silencing of zyxin in A549 cells followed by TGF-β1 stimulation resulted in marked cell shape changes, increased cell scattering, and pronounced diminution in VASP expression (Fig. 3, A and B). Of notice, zyxin depletion and VASP down-regulation did not influence the formation and morphology of focal adhesions as assessed by immunofluorescent staining and Western blot analysis of focal adhesion protein paxillin. Likewise, TGF-β1 was shown to up-regulate paxillin levels in control cells, whereas zyxin knockdown alone induced a significant increase in paxillin expression.
FIGURE 3.
Effect of zyxin depletion on cancer cell adhesion. A, A549 cells transfected with the indicated siRNAs were stimulated for 24 h with TGF-β1. Fixed cells were stained with anti-VASP (red) or anti-paxillin (green) antibodies. Images were acquired by confocal microscopy. Scale bar: 20 μm. ctr siRNA, control siRNA; zyx siRNA, zyxin-specific siRNA. B, cells were transfected with either control or zyxin-specific siRNA, serum-starved, and incubated in the medium with or without TGF-β1 for 24 h. Cleared cell lysates were immunoblotted with anti-phospho-paxillin (Tyr118) (p-paxillin (Tyr118)), anti-paxillin, and anti-VASP antibodies. Equal protein loading was confirmed by reprobing the stripped blots with an anti-β-actin antibody. Representative blots are shown. C, cells transfected with either control or zyxin siRNA were grown in the absence or presence of TGF-β1 for 24 h. Transfected cells were plated onto fibronectin-coated wells. After 30 min, attached cells were stained with 0.1% crystal violet, and optical density was measured at 570 nm. Data represent mean ± S.D. of three independent experiments; ***, p < 0.001.
The addition of TGF-β further elevated this effect (Fig. 3B). Next, we examined the adhesion capacity of cells to fibronectin in TGF-β1-free or supplemented cell culture medium. As shown in Fig. 3C, a remarkable increase of cell adhesion to the fibronectin (∼35%) was observed in zyxin-depleted cells when compared with control cells. Both zyxin and control siRNA-transfected cells showed no significant differences in adhesion capacity to fibronectin when grown without TGF-β1.
Zyxin Expression Modulates Integrin α5β1 Levels in Lung Cancer Cells
To explore the functional specificity of TGF-β1-induced zyxin expression, we examined protein levels of zyxin and pSmad3 in the panels of NSCLC (A549, H2030, H441, and H1299) and SCLC (DMS273 and H82) cell lines. All examined cell lines responded well to TGF-β1 treatment and exhibited prominent Smad3 phosphorylation. The expression profile of zyxin correlated positively with the Smad3 phosphorylation in most of the NSCLC and SCLC lines. Interestingly, TGF-β1 treatment did not markedly affect zyxin expression levels in p53-null H1299 cells (supplemental Fig. S3). After identifying TGF-β1-dependent zyxin expression in different lung cancer cell subtypes and significantly higher adhesion of zyxin-depleted cells to fibronectin, we sought to determine their integrin expression profile. We observed striking differences in integrin α5 and β1 expression levels when comparing zyxin-depleted and control cancer cells, particularly upon TGF-β1 treatment (Fig. 4, A–C). Elevated integrin α5 cell surface expression pattern in zyxin-depleted A549 cells was further confirmed by flow cytometry (supplemental Fig. S4A). On the other hand, zyxin overexpression did not remarkably influence integrin α5 and integrin β1 mRNA levels in both TGF-β1-treated and untreated conditions (supplemental Fig. S4B).
FIGURE 4.
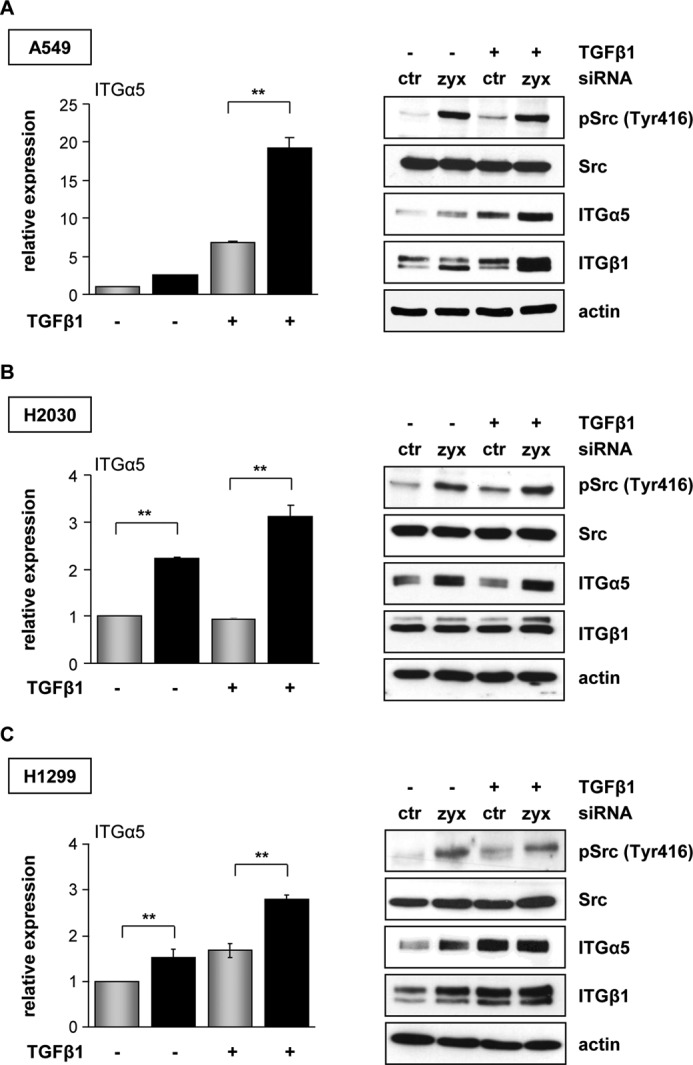
Zyxin mediates integrin α5β1 expression. A–C, A549, H2030, and H1299 cancer cells were transfected with control (ctr) (gray bars) or zyxin (zyx) (black bars) siRNA, serum-starved, and incubated in the absence or presence of TGF-β1 for 24 h. The mRNA levels of integrin α5 were quantified by real-time qRT-PCR. Data are presented as mean ± S.D. of three independent experiments performed in duplicate; **, p < 0.01. Control and zyxin-depleted A549, H2030, and H1299 cancer cells were grown in the absence or presence of TGF-β1. Cells were lysed, and cleared lysates were immunoblotted with anti-phospho-Src (Tyr416) (pSrc (Tyr416)), anti-Src, anti-integrin α5, or anti-integrin β1 antibodies. Equal protein loading was confirmed by reprobing the stripped blots with an anti-β-actin antibody. Representative blots are shown.
An important molecular hallmark of the integrin activation state is the autophosphorylation of focal adhesion kinase (FAK), which in turn phosphorylates Src protein. Because Src catalytic activity is required for integrin-regulated events, we next addressed the phosphorylation state of Src in zyxin-depleted cancer cells. The phospho-Tyr-416 signal of Src protein was higher in all zyxin-silenced cancer cells when compared with control cells (Fig. 4, A–C). Control siRNA-transfected cells were able to display only a slight increase in Src phosphorylation at position Tyr-416 after TGF-β1 stimulation.
Increased Cell Motility of Zyxin-depleted Cells Is Integrin α5β1-dependent
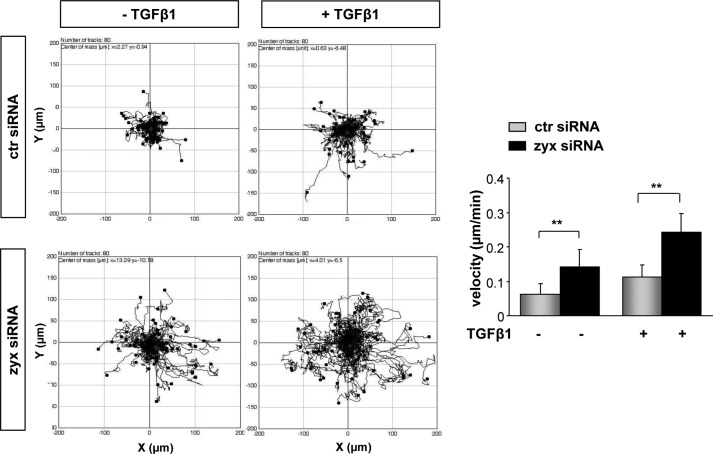
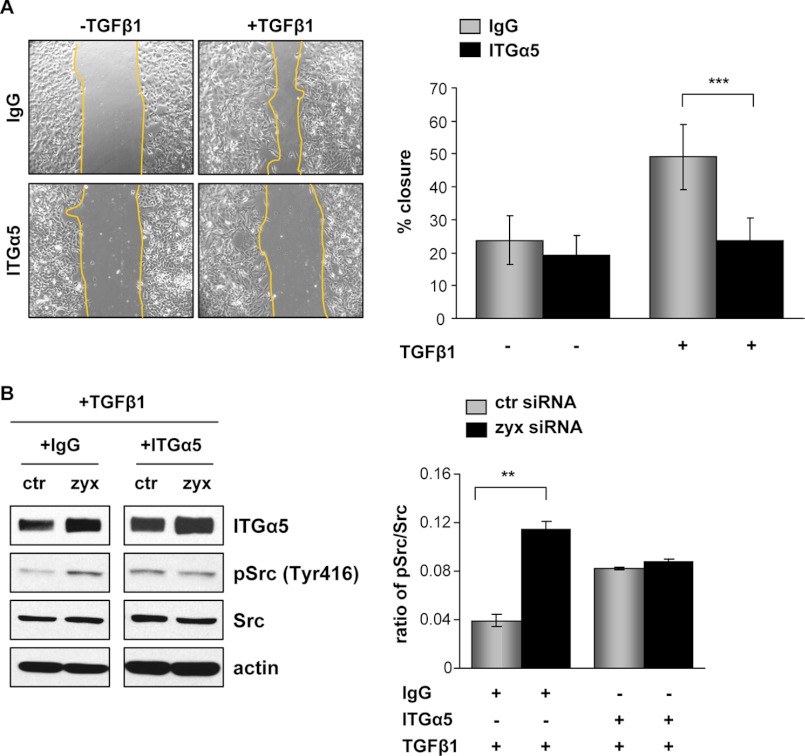
To establish a role of zyxin in lung cancer cell motility, zyxin-silenced and control A549 cells were monitored by time-lapse microscopy over 24 h and tracked to determine their migration distance and velocity. The single cell tracks of 80 cells assayed in three independent experiments demonstrated a significant increase in migratory distance traveled by zyxin-depleted A549 cells when compared with control siRNA-transfected cells (Fig. 5), both under base-line condition and after TGF-β1 addition. Analysis of cell velocity rates revealed clear differences in the motility of zyxin-depleted cells when compared with control cells. TGF-β1 treatment of zyxin-silenced A549 cells increased their velocity by ∼40% when compared with control siRNA-transfected cells. Scratch wound healing assay of confluent cell monolayers confirmed elevated migratory ability of zyxin-depleted cells, notably after TGF-β1 addition (data not shown). Because cell proliferation over this period was negligible (data not shown), wound closure under these conditions primarily reflected cell spreading and migration ability. To assess the functional importance of fibronectin-specific receptor integrin α5β1 in the regulation of A549 cell migration, we performed a scratch wound healing assay in the presence of blocking antibody to integrin α5 or isotype control IgG. An inhibitory antibody directed against α5 subunit was used to block the function of α5β1-integrin complexes because α5 subunit has only been found in α5β1-integrin complexes. As shown in Fig. 6A, the capacity of wound closing of zyxin-depleted cells after TGF-β1 treatment was markedly inhibited by the blocking antibody to α5 integrin when compared with isotype IgG antibody-treated cells that exhibited almost 50% of wound closure after 24 h. In contrast, no significant difference in wound closing was observed between α5- and IgG-treated zyxin-depleted cells under nonstimulated conditions. Given that integrin engagement and activation induces Src tyrosine kinase activity by enhancing phosphorylation at Tyr-416, we examined the effect of α5 inhibitory antibody on the Src phosphorylation levels in both zyxin-depleted and control scrambled siRNA-transfected cells under TGF-β1-stimulated condition. Depletion of zyxin led to an increase in Src phosphorylation levels in isotype IgG (+IgG)-treated A549 cells. In addition, zyxin-depleted cells did not exhibit an increase in Src phosphorylation when compared with control cells when treated with the α5 inhibitory antibody (+ITGα5) (Fig. 6B). Taken together, these observations suggest that zyxin-mediated cell motility is highly dependent on integrin α5β1 expression levels and their engagement.
FIGURE 5.
Zyxin regulates cancer cell motility. A549 cells transfected with the indicated siRNAs were incubated in the presence or absence of TGF-β1. Cells were monitored by live cell microscopy acquiring an image every 10 min over a period of 24 h. Migration tracks of 80 cells from three independent experiments are shown. Cell migration velocity was determined using ImageJ software. Data are presented as mean ± S.D. of three independent experiments; **, p < 0.01. ctr siRNA, control siRNA; zyx siRNA, zyxin-specific siRNA.
FIGURE 6.
Zyxin-mediated cell motility is integrin α5β1-dependent. A, a scratch wound assay was carried out in confluent cultures of zyxin siRNA-transfected A549 cells. An isotype control anti-IgG or anti-integrin α5 antibody (1:100) was added to the culture medium, and wound closure was measured in the presence and absence of TGF-β1. The percentage of wound closure was determined 24 h after injury. Representative phase contrast images of wounded areas are shown. Data are mean ± S.D. from three independent experiments; ***, p < 0.001. B, A549 cells transfected with either control siRNA (ctr siRNA) or zyxin siRNA (zyx siRNA) were grown for 24 h in TGF-β1-containing medium in the presence or absence of anti-integrin α5 inhibitory antibody. Anti-IgG was used as an isotype control. Cells were lysed, and Western blotting was performed with whole cell lysates by using anti-integrin α5, anti-phospho-Src (Tyr416) (pSrc (Tyr416)), or anti-Src antibody. Equal protein loading was confirmed by reprobing the stripped blots with an anti-β-actin antibody. Representative blots are shown. Densitometric analysis was used to determine the phospho-Src/Src ratio (ImageJ software). Data are shown as mean ± S.E.; **, p < 0.01.
Zyxin Expression Strongly Associates with pSmad3 Levels in Mouse Model of NSCLC
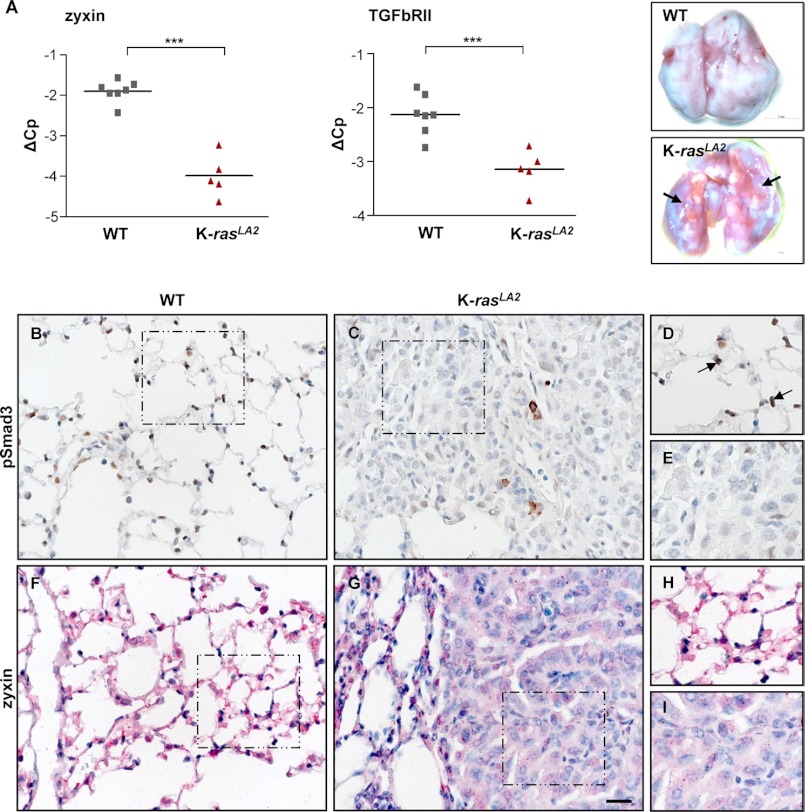
To explore the expression levels of zyxin and pSmad3 in the pathogenesis of NSCLC, we utilized the 5-month-old K-rasLA2 mouse lung cancer model, which harbors a targeted, latent K-ras G12D allele. These mice develop multifocal lung tumors (Fig. 7A) in 100% of mice. We found that all K-rasLA2 mice had significantly decreased zyxin mRNA levels when compared with control wild type (WT) mice. Reduced TGFbRII expression has been previously reported in NSCLC samples and is associated with more aggressive tumor behavior (48). Therefore, we sought to examine the TGFbRII mRNA levels in control and K-rasLA2 mice. As expected, TGFbRII mRNA levels were greatly decreased in lung homogenates from the K-rasLA2 mice (Fig. 7A). In addition, we have found the lowest zyxin expression exclusively in the tumor areas when compared with a strong expression in healthy lung tissue (Fig. 7, F–I). We also examined the levels of phosphorylated Smad3 proteins in the lungs of WT and K-rasLA2 mice. A striking difference was observed not only in levels, but also in localization of pSmad3, whereby high levels of nuclear pSmad3 were detected in healthy WT mouse lungs when compared with remarkable lower levels in K-rasLA2-derived tumors (Fig. 7, B–E). Interestingly, only a few cells within the tumor displayed increased pSmad3 that mostly appeared as cytoplasmic staining.
FIGURE 7.
Decreased levels of zyxin and pSmad3 in K-rasLA2 model of lung cancer. A, qRT-PCR analysis for zyxin and TGFbRII mRNA levels in the lungs from WT (n = 7) and K-rasLA2 (n = 5) mice; ***, p < 0.001. Photographs of WT and K-rasLA2 mouse lungs are shown. Arrows indicate tumor nodules in the K-rasLA2 mouse lungs. B–I, formalin-fixed sections of lung tissues from K-rasLA2 and WT mice were used for immunohistochemical analysis. Representative examples of pSmad3 (brown) and zyxin (red) immunostaining are presented. Sections were counterstained using hematoxylin (blue). D, E, H and I, magnified images for the boxed areas. The arrows indicate cells that show nuclear pSmad3 immunoreactivity (D). Scale bar: 20 μm.
DISCUSSION
In this study, we have demonstrated that the zyxin promoter is a direct target of Smad3 after TGF-β1 treatment in NSCLC cell line A549. The essential role of TGF-β1-mediated Smad3 activation has been well demonstrated in the EMT process associated with tumor progression (4, 24, 25). However, depending on the cell type and the local activity of TGF-β1, TGF-β1 can have both disease-preventing and disease-promoting effects (26–28). Although the TGF-β-mediated Smad signaling has received much attention in the past years, non-Smad signaling pathways take an important place in a context-specific fashion and can result in activation of different targets such as PI3K, ERK1/ERK2, p38 MAPK, JNK, or small GTPases (7, 29). In the Smad-mediated pathway, phosphorylation of Smad2 and Smad3 after TGF-β1 stimulation induces their translocation to the nucleus, where they interact with other transcription factors that regulate transcription of TGF-β-responsive genes (30, 31). Here, we showed that activation of Smad3 upon TGF-β1 treatment in K-ras mutated NSCLC cells (A549, H2030, and H441), resulted in prominent zyxin expression. Importantly, TGF-β1-dependent zyxin regulation has also been observed in two small cell lung cancer cell lines, indicating that similar underlying mechanisms might regulate zyxin expression in two primary types of lung cancer. In addition, blockade of Smad3 phosphorylation or its expression abrogated zyxin mRNA and protein levels, confirming TGF-β1/Smad3-dependent zyxin regulation.
It is known that zyxin does not interact directly with actin filaments, but can dock the proteins involved in cytoskeletal organization and therefore regulate actin polymerization and dynamics (13, 32, 33). Furthermore, TGF-β1 has been shown to induce stress fiber formation in epithelial cells by Smad/p38 MAPK-dependent signaling, and thus controls cell motility (10). We have shown here that the depletion of zyxin plays an important role in regulating the integrity of cell-cell contacts, which in turn affects TGF-β1-mediated cell motility. Importantly, several studies have described zyxin co-localization with the VASP protein at cell-cell junctions and their importance in actin filament formation and binding (33). Zyxin connection with the VASP protein and actin filaments generates places of relatively strong intercellular adhesions and prevents their dissociation (17, 34). In contrast, zyxin localization at cell-cell contacts was not observed in NMuMG cells (35), which suggests the context- and cell-specific role of zyxin at cell junctions.
We further revealed that although TGF-β1 stimulation induced down-regulation of E-cadherin and its internalization, it had no significant effect on A549 cell junction morphology and single cell scattering. This cell phenotype corresponds to a partial EMT process, which is mostly characterized by the maintenance of cell-cell adhesions and implicates an important, but not crucial, role of E-cadherin in the regulation of cellular contacts and induction of the mesenchymal phenotype. Furthermore, we showed that silencing of zyxin accompanied by simultaneous TGF-β1 treatment led to p120 dissociation and disassembly of adherens junctions. This effect correlated with increased E-cadherin internalization; however, E-cadherin internalization itself was not sufficient to induce single cell scattering. We also demonstrated that TGF-β1 stimulation elevated zyxin expression, but also enhanced its localization to focal adhesions and cell junctions. Interestingly, zyxin depletion in A549 cells did not lead to changes in the formation of focal adhesions or their morphology, although VASP accumulation was reduced. This finding further indicates that the presence of VASP is not critical for the formation of focal adhesions in lung cancer A549 cells. Consistent with this notion, several studies have reported that zyxin depletion causes the mislocalization of VASP, but does not interfere with the capacity of Ena/VASP localization at the lamellipodial protrusions and leading edge of migrating cell (36, 37). In fact, recruitment of Ena/VASP proteins to the focal adhesions requires interactions between their N-terminal EVH1 domain and proteins containing the FPPPP motifs such as zyxin and vinculin (38). Despite these findings, more work will be required to identify the precise molecular mechanisms by which these molecules regulate formation of focal adhesions.
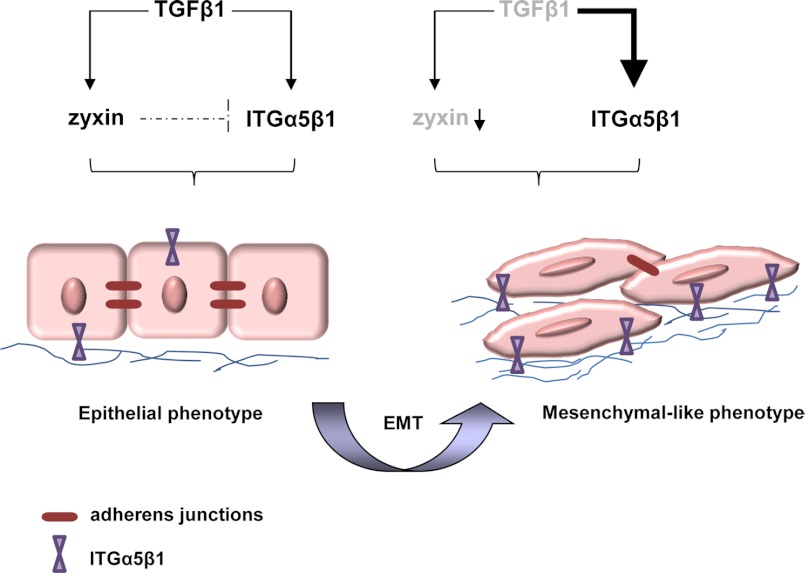
Moreover, our study is the first to demonstrate that cytoskeletal protein zyxin can regulate integrin α5β1 levels. Knockdown of endogenous zyxin induced significant expression and activation of integrin α5β1, which resulted not only in increased adhesion to the fibronectin, but also elevated cell motility. The observed effect was even more prominent when cells were treated with TGF-β1, suggesting a negative feedback mechanism that involves a functional pathway linking zyxin to the ability of TGF-β1 to control the integrin α5β1 expression (Fig. 8).
FIGURE 8.
Proposed model of zyxin-mediated lung cancer cell EMT. TGF-β1 increases zyxin expression, which balances integrin α5β1 (ITGα5β1) expression and stabilizes cell-cell adherens junctions. In cancer epithelial cells during the EMT process, both zyxin expression levels and TGF-β signaling might be deregulated. These processes contribute to the formation of loose cell-cell junctions, increased ITGα5β1 expression, and elevated cell motility.
Several hypothetical mechanisms have been proposed by which zyxin could affect gene expression in different cell types because it is also found in the cell nucleus (20, 39, 40). Thus, it is tempting to speculate that zyxin might act as an important regulator of TGF-β-induced integrin α5β1 expression in lung cancer cells. Most of the studies supported a role of TGF-β1 in inducing the expression of several integrins, such as α5β1, αVβ6, αVβ3, and αVβ5, thereby enhancing the migratory phenotype of cells.
Intriguingly, it has been described that TGF-β1 alone in many cases is not sufficient to drive single cell motility and that other factors might be involved (41). TGF-β, in addition, can influence the A549 cell phenotype by stimulating fibronectin production. In fact, remodeling of the extracellular matrix is a cell-mediated process that involves integrins, such as α5β1, which can promote formation of multimeric fibrils from dimeric fibronectin (42). In line with other studies, we showed that TGF-β1 not only regulated expression of EMT markers in A549 cells, but also stimulated the expression of focal adhesion-associated protein paxillin (43). Nonetheless, depletion of zyxin in those cells caused elevated paxillin levels independently of TGF-β stimuli. Strikingly, increased activation of Src protein (upstream paxillin effector) in zyxin-silenced cells is likely due to increased integrin α5β1 expression and its clustering on the cell surface. These observations could at least partly explain the undisturbed formation of focal adhesions and enhanced cell adhesion to the fibronectin. Application of integrin α5 inhibitory antibody prevented Src phosphorylation in zyxin-silenced cells and decreased their migratory capacity.
Some previous studies have linked low zyxin expression levels to bladder cancer progression and identified zyxin protein as potential tumor suppressor in Ewing tumor cells and prostate carcinomas (44–46). Our data show that lung tumors derived form the K-rasLA2 mouse models exhibit dramatically reduced levels of zyxin and nuclear pSmad3. Consistent with this finding, it has been reported that Smad3 level decreases during carcinogenesis in some tissues and that Smad3 protein levels can be a crucial determinant of TGF-β responsiveness during tumor progression (47). Analysis of K-rasLA2 mouse lungs also revealed decreased levels of TGFbRII mRNA. Thus, it is interesting to note that in the large number of human NSCLC samples, reduced TGFbRII expression is associated with more aggressive tumor behavior (48).
In summary, our findings implicate zyxin in two widely diverse activities, the regulation of cell-cell adhesion and expression of integrin α5β1. These results provide novel, interesting insights into the process underlying TGF-β-mediated lung cancer cell adhesion and motility. In that view, it would be of interest to know whether zyxin plays a part in development of lung cancer metastasis. We believe that further examination of zyxin levels and its function in metastatic lung cancers will answer these key questions.
Supplementary Material
Acknowledgments
We thank Ann-Christin Beitel and Daniela Dietel for excellent technical assistance and Dr. Thomas Hofer for assistance with the flow cytometric analysis.
This work was supported by the Helmholtz Association.

This article contains supplemental Figs. S1–S4.
- EMT
- epithelial-to-mesenchymal transition
- LIM
- Lin11, Isl-1, and Mec-3 domain
- SIS3
- specific inhibitor of Smad3
- pSmad3
- phosphorylated Smad3
- VASP
- vasodilator-stimulated phosphoprotein
- SCLC
- small cell lung cancer
- NCSLC
- non-small cell lung cancer
- ITG
- integrin
- qRT-PCR
- quantitative real-time PCR.
REFERENCES
- 1. Xiao D., He J. (2010) Epithelial-mesenchymal transition and lung cancer. J. Thorac. Dis. 2, 154–159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Thiery J. P., Acloque H., Huang R. Y., Nieto M. A. (2009) Epithelial-mesenchymal transitions in development and disease. Cell 139, 871–890 [DOI] [PubMed] [Google Scholar]
- 3. Miettinen P. J., Ebner R., Lopez A. R., Derynck R. (1994) TGF-β-induced transdifferentiation of mammary epithelial cells to mesenchymal cells: involvement of type I receptors. J. Cell Biol. 127, 2021–2036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Piek E., Moustakas A., Kurisaki A., Heldin C. H., ten Dijke P. (1999) TGF-β type I receptor/ALK-5 and Smad proteins mediate epithelial-to-mesenchymal transdifferentiation in NMuMG breast epithelial cells. J. Cell Sci. 112, 4557–4568 [DOI] [PubMed] [Google Scholar]
- 5. Portella G., Cumming S. A., Liddell J., Cui W., Ireland H., Akhurst R. J., Balmain A. (1998) Transforming growth factor-β is essential for spindle cell conversion of mouse skin carcinoma in vivo: implications for tumor invasion. Cell Growth Differ. 9, 393–404 [PubMed] [Google Scholar]
- 6. Willis B. C., Borok Z. (2007) TGF-β-induced EMT: mechanisms and implications for fibrotic lung disease. Am. J. Physiol. Lung Cell Mol. Physiol. 293, L525–L534 [DOI] [PubMed] [Google Scholar]
- 7. Derynck R., Zhang Y. E. (2003) Smad-dependent and Smad-independent pathways in TGF-β family signaling. Nature 425, 577–584 [DOI] [PubMed] [Google Scholar]
- 8. ten Dijke P., Hill C. S. (2004) New insights into TGF-β-Smad signaling. Trends Biochem. Sci. 29, 265–273 [DOI] [PubMed] [Google Scholar]
- 9. Thiery J. P., Sleeman J. P. (2006) Complex networks orchestrate epithelial-mesenchymal transitions. Nat. Rev. Mol. Cell Biol. 7, 131–142 [DOI] [PubMed] [Google Scholar]
- 10. Bakin A. V., Safina A., Rinehart C., Daroqui C., Darbary H., Helfman D. M. (2004) A critical role of tropomyosins in TGF-β regulation of the actin cytoskeleton and cell motility in epithelial cells. Mol. Biol. Cell 15, 4682–4694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bhowmick N. A., Ghiassi M., Bakin A., Aakre M., Lundquist C. A., Engel M. E., Arteaga C. L., Moses H. L. (2001) Transforming growth factor-β1 mediates epithelial-to-mesenchymal transdifferentiation through a RhoA-dependent mechanism. Mol. Biol. Cell 12, 27–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Ridley A. J., Hall A. (1992) The small GTP-binding protein rho regulates the assembly of focal adhesions and actin stress fibers in response to growth factors. Cell 70, 389–399 [DOI] [PubMed] [Google Scholar]
- 13. Fradelizi J., Noireaux V., Plastino J., Menichi B., Louvard D., Sykes C., Golsteyn R. M., Friederich E. (2001) ActA and human zyxin harbor Arp2/3-independent actin polymerization activity. Nat. Cell Biol. 3, 699–707 [DOI] [PubMed] [Google Scholar]
- 14. Beckerle M. C. (1998) Spatial control of actin filament assembly: lessons from Listeria. Cell 95, 741–748 [DOI] [PubMed] [Google Scholar]
- 15. Hoffman L. M., Nix D. A., Benson B., Boot-Hanford R., Gustafsson E., Jamora C., Menzies A. S., Goh K. L., Jensen C. C., Gertler F. B., Fuchs E., Fässler R., Beckerle M. C. (2003) Targeted disruption of the murine zyxin gene. Mol. Cell Biol. 23, 70–79 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Scott J. A., Shewan A. M., den Elzen N. R., Loureiro J. J., Gertler F. B., Yap A. S. (2006) Ena/VASP proteins can regulate distinct modes of actin organization at cadherin-adhesive contacts. Mol. Biol. Cell 17, 1085–1095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Vasioukhin V., Bauer C., Yin M., Fuchs E. (2000) Directed actin polymerization is the driving force for epithelial cell-cell adhesion. Cell 100, 209–219 [DOI] [PubMed] [Google Scholar]
- 18. Crawford A. W., Michelsen J. W., Beckerle M. C. (1992) An interaction between zyxin and α-actinin. J. Cell Biol. 116, 1381–1393 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hansen M. D., Beckerle M. C. (2006) Opposing roles of zyxin/LPP ACTA repeats and the LIM domain region in cell-cell adhesion. J. Biol. Chem. 281, 16178–16188 [DOI] [PubMed] [Google Scholar]
- 20. Nix D. A., Fradelizi J., Bockholt S., Menichi B., Louvard D., Friederich E., Beckerle M. C. (2001) Targeting of zyxin to sites of actin membrane interaction and to the nucleus. J. Biol. Chem. 276, 34759–34767 [DOI] [PubMed] [Google Scholar]
- 21. Desgrosellier J. S., Cheresh D. A. (2010) Integrins in cancer: biological implications and therapeutic opportunities. Nat. Rev. Cancer 10, 9–22 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Johnson L., Mercer K., Greenbaum D., Bronson R. T., Crowley D., Tuveson D. A., Jacks T. (2001) Somatic activation of the K-ras oncogene causes early onset lung cancer in mice. Nature 410, 1111–1116 [DOI] [PubMed] [Google Scholar]
- 23. Zhang Y., Handley D., Kaplan T., Yu H., Bais A. S., Richards T., Pandit K. V., Zeng Q., Benos P. V., Friedman N., Eickelberg O., Kaminski N. (2011) High throughput determination of TGF-β1/SMAD3 targets in A549 lung epithelial cells. PLoS One 6, e20319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Oft M., Akhurst R. J., Balmain A. (2002) Metastasis is driven by sequential elevation of H-ras and Smad2 levels. Nat. Cell Biol. 4, 487–494 [DOI] [PubMed] [Google Scholar]
- 25. Valcourt U., Kowanetz M., Niimi H., Heldin C. H., Moustakas A. (2005) TGF-β and the Smad signaling pathway support transcriptomic reprogramming during epithelial-mesenchymal cell transition. Mol. Biol. Cell 16, 1987–2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Böttinger E. P., Jakubczak J. L., Roberts I. S., Mumy M., Hemmati P., Bagnall K., Merlino G., Wakefield L. M. (1997) Expression of a dominant-negative mutant TGF-β type II receptor in transgenic mice reveals essential roles for TGF-β in regulation of growth and differentiation in the exocrine pancreas. EMBO J. 16, 2621–2633 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Ikushima H., Miyazono K. (2010) TGF-β signaling: a complex web in cancer progression. Nat. Rev. Cancer 10, 415–424 [DOI] [PubMed] [Google Scholar]
- 28. Pardali E., ten Dijke P. (2009) Transforming growth factor-β signaling and tumor angiogenesis. Front Biosci. 14, 4848–4861 [DOI] [PubMed] [Google Scholar]
- 29. Tian M., Neil J. R., Schiemann W. P. (2011) Transforming growth factor-β and the hallmarks of cancer. Cell Signal 23, 951–962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Feng X. H., Zhang Y., Wu R. Y., Derynck R. (1998) The tumor suppressor Smad4/DPC4 and transcriptional adaptor CBP/p300 are coactivators for smad3 in TGF-β-induced transcriptional activation. Genes Dev. 12, 2153–2163 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang Y., Feng X. H., Derynck R. (1998) Smad3 and Smad4 cooperate with c-Jun/c-Fos to mediate TGF-β-induced transcription. Nature 394, 909–913 [DOI] [PubMed] [Google Scholar]
- 32. Reinhard M., Zumbrunn J., Jaquemar D., Kuhn M., Walter U., Trueb B. (1999) An α-actinin binding site of zyxin is essential for subcellular zyxin localization and α-actinin recruitment. J. Biol. Chem. 274, 13410–13418 [DOI] [PubMed] [Google Scholar]
- 33. Drees B., Friederich E., Fradelizi J., Louvard D., Beckerle M. C., Golsteyn R. M. (2000) Characterization of the interaction between zyxin and members of the Ena/vasodilator-stimulated phosphoprotein family of proteins. J. Biol. Chem. 275, 22503–22511 [DOI] [PubMed] [Google Scholar]
- 34. Sperry R. B., Bishop N. H., Bramwell J. J., Brodeur M. N., Carter M. J., Fowler B. T., Lewis Z. B., Maxfield S. D., Staley D. M., Vellinga R. M., Hansen M. D. (2010) Zyxin controls migration in epithelial-mesenchymal transition by mediating actin-membrane linkages at cell-cell junctions. J. Cell Physiol. 222, 612–624 [DOI] [PubMed] [Google Scholar]
- 35. Mori M., Nakagami H., Koibuchi N., Miura K., Takami Y., Koriyama H., Hayashi H., Sabe H., Mochizuki N., Morishita R., Kaneda Y. (2009) Zyxin mediates actin fiber reorganization in epithelial-mesenchymal transition and contributes to endocardial morphogenesis. Mol. Biol. Cell 20, 3115–3124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hoffman L. M., Jensen C. C., Kloeker S., Wang C. L., Yoshigi M., Beckerle M. C. (2006) Genetic ablation of zyxin causes Mena/VASP mislocalization, increased motility, and deficits in actin remodeling. J. Cell Biol. 172, 771–782 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Krause M., Leslie J. D., Stewart M., Lafuente E. M., Valderrama F., Jagannathan R., Strasser G. A., Rubinson D. A., Liu H., Way M., Yaffe M. B., Boussiotis V. A., Gertler F. B. (2004) Lamellipodin, an Ena/VASP ligand, is implicated in the regulation of lamellipodial dynamics. Dev. Cell 7, 571–583 [DOI] [PubMed] [Google Scholar]
- 38. Ball L. J., Kühne R., Hoffmann B., Häfner A., Schmieder P., Volkmer-Engert R., Hof M., Wahl M., Schneider-Mergener J., Walter U., Oschkinat H., Jarchau T. (2000) Dual epitope recognition by the VASP EVH1 domain modulates polyproline ligand specificity and binding affinity. EMBO J. 19, 4903–4914 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Nix D. A., Beckerle M. C. (1997) Nuclear-cytoplasmic shuttling of the focal contact protein, zyxin: a potential mechanism for communication between sites of cell adhesion and the nucleus. J. Cell Biol. 138, 1139–1147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Chan C. B., Liu X., Tang X., Fu H., Ye K. (2007) Akt phosphorylation of zyxin mediates its interaction with acinus-S and prevents acinus-triggered chromatin condensation. Cell Death Differ. 14, 1688–1699 [DOI] [PubMed] [Google Scholar]
- 41. Giampieri S., Manning C., Hooper S., Jones L., Hill C. S., Sahai E. (2009) Localized and reversible TGF-β signaling switches breast cancer cells from cohesive to single cell motility. Nat. Cell Biol. 11, 1287–1296 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Hynes R. O. (1992) Integrins: versatility, modulation, and signaling in cell adhesion. Cell 69, 11–25 [DOI] [PubMed] [Google Scholar]
- 43. Han X., Stewart J. E., Jr., Bellis S. L., Benveniste E. N., Ding Q., Tachibana K., Grammer J. R., Gladson C. L. (2001) TGF-β1 up-regulates paxillin protein expression in malignant astrocytoma cells: requirement for a fibronectin substrate. Oncogene 20, 7976–7986 [DOI] [PubMed] [Google Scholar]
- 44. Sanchez-Carbayo M., Socci N. D., Charytonowicz E., Lu M., Prystowsky M., Childs G., Cordon-Cardo C. (2002) Molecular profiling of bladder cancer using cDNA microarrays: defining histogenesis and biological phenotypes. Cancer Research 62, 6973–6980 [PubMed] [Google Scholar]
- 45. Yu Y. P., Luo J. H. (2011) Phosphorylation and interaction of myopodin by integrin-link kinase lead to suppression of cell growth and motility in prostate cancer cells. Oncogene 30, 4855–4863 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Amsellem V., Kryszke M. H., Hervy M., Subra F., Athman R., Leh H., Brachet-Ducos C., Auclair C. (2005) The actin cytoskeleton-associated protein zyxin acts as a tumor suppressor in Ewing tumor cells. Exp. Cell Res. 304, 443–456 [DOI] [PubMed] [Google Scholar]
- 47. Daly A. C., Vizán P., Hill C. S. (2010) Smad3 protein levels are modulated by Ras activity and during the cell cycle to dictate transforming growth factor-β responses. J. Biol. Chem. 285, 6489–6497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Malkoski S. P., Haeger S. M., Cleaver T. G., Rodriguez K. J., Li H., Lu S. L., Feser W. J., Barón A. E., Merrick D., Lighthall J. G., Ijichi H., Franklin W., Wang X. J. (2012) Loss of transforming growth factor-β type II receptor increases aggressive tumor behavior and reduces survival in lung adenocarcinoma and squamous cell carcinoma. Clin. Cancer Res. 18, 2173–2183 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.