Background: Nikkomycins are potent antibiotics. NikO is a key enzyme in their biosynthesis.
Results: NikO is structurally closely related to other enolpyruvyl transferases and is inhibited by fosfomycin.
Conclusion: Conservation of important active site residues indicates mechanistic similarities to MurA.
Significance: The structure of NikO is the first of an enzyme participating in the formation of the aminohexuronic acid moiety in nikkomycin biosynthesis.
Keywords: Antibiotics, Crystal Structure, Docking, Enzyme Inhibitors, X-ray Crystallography, Phosphoenol Pyruvate
Abstract
Nikkomycins are peptide-nucleoside compounds with fungicidal, acaricidal, and insecticidal properties because of their strong inhibition of chitin synthase. Thus, they are potential antibiotics especially for the treatment of immunosuppressed patients, for those undergoing chemotherapy, or after organ transplants. Although their chemical structure has been known for more than 30 years, only little is known about their complex biosynthesis. The genes encoding for proteins involved in the biosynthesis of the nucleoside moiety of nikkomycins are co-transcribed in the same operon, comprising the genes nikIJKLMNO. The gene product NikO was shown to belong to the family of enolpyruvyl transferases and to catalyze the transfer of an enolpyruvyl moiety from phosphoenolpyruvate to the 3′-hydroxyl group of UMP. Here, we report activity and inhibition studies of the wild-type enzyme and the variants C130A and D342A. The x-ray crystal structure revealed differences between NikO and its homologs. Furthermore, our studies led to conclusions concerning substrate binding and preference as well as to conclusions about inhibition/alkylation by the antibiotic fosfomycin.
Introduction
The microbial strains of Streptomyces sp. have been one of the richest sources for antibiotic drugs throughout the last 50 years, with every strain presenting its own, specific set of medically useful compounds. Among them, several structurally related peptide nucleosides, isolated from cell filtrates of Streptomyces tendae and Streptomyces ansochromogenes (1, 2), have been found and termed nikkomycins. Structurally these compounds show similarities to UDP-N-acetylglucosamine, the substrate of chitin synthase, and thus are potent inhibitors of this enzyme (3, 4). As chitin is an essential component in fungal cell walls as well as an integrated compound in the exoskeleton of insects and invertebrates, nikkomycins exhibit fungicidal, insecticidal, and acaricidal properties. Moreover, nikkomycins were shown to be nontoxic to mammals and easily degradable in nature (5, 6). This makes them ideal compounds for the treatment of immunosuppressed patients, for those undergoing chemotherapy, or after organ transplantation. Moreover, the rapid biodegradation of nikkomycins in nature suggests that they may have potential as fungicides in agriculture.
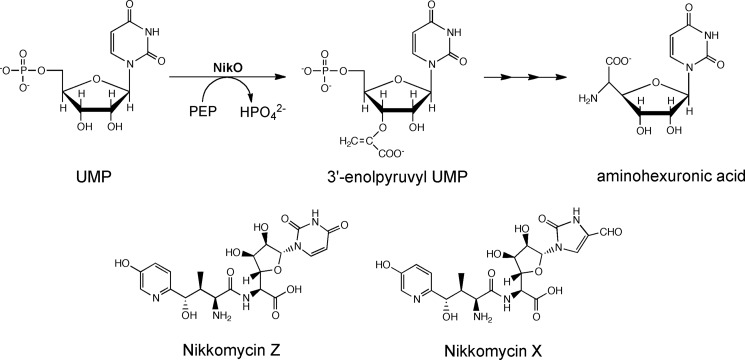
As part of the gene operon encoding proteins required for nikkomycin biosynthesis, a putative new member of the family of phosphoenolpyruvate (PEP)3-utilizing enzymes has been described in S. tendae and termed NikO (7). NikO was annotated as an enolpyruvyl transferase showing highest sequence identity to bacterial UDP-N-acetylglucosamine enolpyruvyl transferases (MurA) from various species (∼30%). We could demonstrate that NikO is, indeed, an enolpyruvyl transferase catalyzing the transfer of the enolpyruvyl moiety from PEP to the 3′-hydroxyl group of UMP (Scheme 1) (8). It is also assumed that NikO catalyzes the corresponding reaction with 5′-phosphoribofuranosyl-4-formyl-4-imidazolin-2-one because disruption of the nikO gene leads to the accumulation of ribofuranosyl-4-formyl-4-imidazolone (7). In any case, the transfer of the enolpyruvyl moiety to either substrate constitutes the first step toward the biosynthesis of the aminohexuronic acid moiety, which is one of two building blocks required for nikkomycin biosynthesis (7).
SCHEME 1.
The reaction catalyzed by NikO, the structure of the synthesized compound, aminohexuronic acid, and the two most abundantly found nikkomycins are shown. The enolpyruvyl group is transferred onto 3′-OH of UMP.
PEP is a commonly used metabolite in diverse biosynthetic pathways. In the reactions found in nature, the phosphate group can be released either by P–O or by C–O bond cleavage (for review, see Walsh et al. (9)). The latter type of cleavage reaction leads either to a net aldol condensation with the substrate, as exemplified by the 3-deoxy-d-arabino-2-heptulosonate-7-phosphate synthase reaction, or to the transfer of the intact enolpyruvyl moiety, as seen in the MurA and 5-enolpyruvylshikimate 3-phosphate synthase reactions. The former enzyme transfers the enolpyruvyl moiety to UDP-N-acetylglucosamine, which constitutes the committing step in the biosynthesis of murein, an essential bacterial cell wall component (10), whereas the latter enzyme catalyzes the sixth and penultimate step in the shikimate pathway (9).
These two PEP-utilizing enzymes exhibit a unique three-dimensional structure termed an “inside-out α/β-barrel” (11–13). Several x-ray crystallographic structures of MurA and 5-enolpyruvylshikimate 3-phosphate synthase have been determined to date, providing structural insight into the function of enolpyruvyl transferases (11–13). Characteristically, these structures consist of two domains, connected by loops forming a hinge region as well as a large cleft between the two domains. The active site is set up by residues from both domains and situated in the center of the cleft. In MurA, a conformational change in the hinge region is supposed to be triggered by a lysine-arginine residue pair that forms repulsive interactions in the apo-state of the enzyme and thus drives the domains farther apart (12). Upon substrate binding, a conformational change in the two side chains abolishes the repulsive charge-charge interaction and thereby induces a movement of the domains toward each other.
Concomitant with this domain movement, the reorientation of a long loop from the upper domain has been observed in MurA (14–17). This flexible loop contains a cysteine residue, which reacts with the naturally occurring antibiotic fosfomycin ((1R,2S)-1,2-epoxypropylphosphonic acid) via a nucleophilic attack of the thiolate at the epoxypropyl ring (12, 18), leading to the inactivation of the enzyme (9, 18). From sequence alignments, it is known that NikO has a cysteine residue present in a similar position as in MurA, suggesting that NikO might also be inhibited by fosfomycin (7, 19).
In the present study, we demonstrate that fosfomycin indeed inactivates NikO via alkylation at residue Cys-130 but that this inactivation is not promoted (as in MurA), but rather impaired by the presence of substrate. We also determined the three-dimensional structure of NikO to confirm its structural relationship to the family of enolpyruvyl transferases and to gain functional insights. Furthermore, two variants of NikO were constructed (C130A and D342A) and characterized. Moreover, molecular docking experiments were performed to elucidate the mode of substrate binding and to identify other, potentially important active site residues.
EXPERIMENTAL PROCEDURES
Reagents
All chemicals were of highest grade commercially available (Sigma-Aldrich; Fluka, Buchs, Switzerland; or Merck, Darmstadt, Germany). Nickel-nitrilotriacetic acid-agarose was from Qiagen (Hilden, Germany), and Sephadex was from GE Healthcare (Uppsala, Sweden). Crystallization equipment and materials were from Hampton Research (Aliso Viejo, CA), Molecular Dimensions (Newmarket, UK), Qiagen (Hilden, Germany), and Douglas Instruments (Hungerford, UK).
Expression and Purification
Chemically competent Escherichia coli BL21 (DE3) cells were transformed with the expression plasmid pET21d(nikO). The gene was cloned such that the protein is generated with a hexahistidine tag at the C terminus. Typically, 750 ml of LB medium were inoculated with an overnight culture (10 ml) and then incubated at 37 °C in the presence of 100 μg/ml ampicillin until an absorbance at 600 nm of 0.5 was reached. Adding isopropyl-1-thio-β-d-galactopyranoside (IPTG) to a final concentration of 1 mm then induced expression of the target gene. The culture was incubated for 3 h at 37 °C, and cells were harvested by centrifugation. The pellet was washed with 0.9% NaCl solution and stored at −20 °C.
For purification of NikO, the pellet was thawed and resuspended in lysis buffer (100 mm Tris/HCl buffer, pH 8.0, containing 300 mm NaCl and 10 mm imidazole), using 2 ml of buffer/g of wet cells. Cells were disrupted by incubation with lysozyme (30 min) and sonicated (0.5-s pulses for 10 min on ice). Cell debris was removed by centrifugation at 18,000 × g for 30 min at 4 °C.
NikO was then purified by nickel-nitrilotriacetic acid affinity chromatography following the manufacturer's instructions. The purity of the protein was determined by SDS-PAGE. Pooled fractions were concentrated by Amicon Ultra centrifugal filter units (Millipore, Billerica, MA). The elution buffer was exchanged to 100 mm Tris buffer, pH 7.5, using PD-10 desalting columns (GE Healthcare). The protein was then stored at −20 °C. For crystallization trials, the protein was further purified by molecular sieve chromatography (HiLoadTM 16/70 Superdex 75 prep grade column, GE Healthcare) exchanging the buffer to 50 mm Tris/HCl, pH 7.5.
Generation of NikO C130A and D342A Variants by Site-specific Mutagenesis
Site-directed mutagenesis was carried out according to the QuikChange® XL site-directed mutagenesis kit from Stratagene (Santa Clara, CA). The pET21d(nikO) plasmid served as template. The following primers and their complementary counterparts were used: 5′-GCG AGC GGC GGC GCG CCG ATC GGC GAA GG-3′ for the NikO C130A variant and 5′-CGC GGC GTG TTC AGC GCG AGC CAG CCC TTC CTG-3′ for the NikO D342A variant. The underlined nucleotides denote the mutated codons. After mutagenesis, the sequence of the transformation construct was verified by DNA sequence analysis (VBC-Biotech, Vienna, Austria). Transformation and expression were carried out as described for wild-type NikO.
Activity Assays
NikO activity was measured with the EnzChek phosphate assay kit (Molecular Probes). This assay is based on a method originally described in Ref. 20. Briefly, the substrate 2-amino-6-mercapto-7-methylpurine riboside (MESG) is converted to ribose-1-phosphate and 2-amino-6-mercapto-7-methylpurine by purine nucleoside phosphorylase in the presence of phosphate (Pi), resulting in a shift of the absorption maximum from 330 to 360 nm. The sensitivity of the assay is in the range of 2–150 μm Pi. All reagents provided in the kit were prepared according to the manufacturer's instructions except for the enolpyruvyl test buffer (50 mm Tris/HCl, 2 mm DTT, pH 7.5). A calibration curve was established in the range of 5–50 μm Pi. The final concentrations in the assay mixture were 200 μm MESG, 1 mm PEP, and 0.5–1.0 μm NikO. Three μl of the purine nucleoside phosphorylase solution were used (containing 0.3 units of activity). The reaction mixture was preincubated for 5 min at 30 °C and transferred to a cuvette to record a base line at 360 nm. After 2–3 min, the reaction was started by adding UMP.
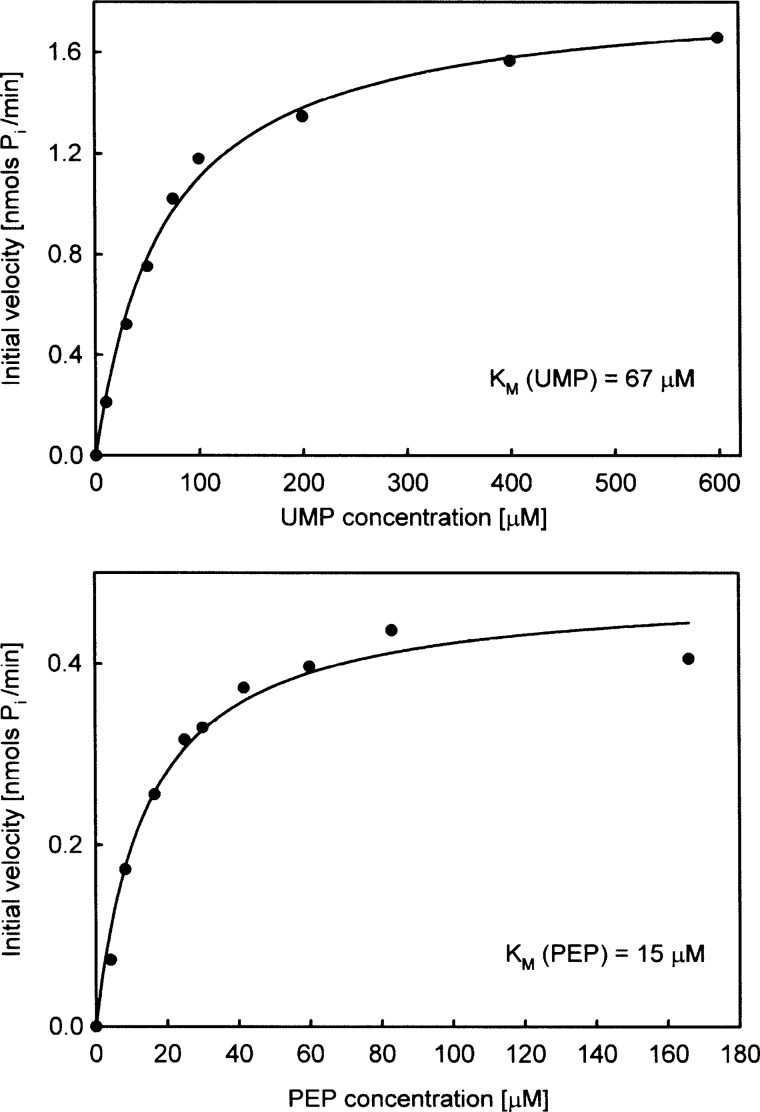
Km values were determined using the continuous assay method described in the previous paragraph. For the determination of Km (UMP), the sample cuvette contained 0.2 mm MESG, 1 mm PEP, 0.1 units/μl purine nucleoside phosphorylase, and 0.9 μm NikO. Initial velocities were measured by adding variable amounts (10–600 μm) of UMP to the mixture. The reference cuvette contained all the components except UMP. All measurements were performed in triplicate, and the average was taken and plotted in nmol of Pi min−1 against the UMP concentration (see Fig. 3). Km and Vmax were obtained by fitting the experimental curve to the Michaelis-Menten equation (i.e. a hyperbolic function: y = a x/(b + x) using the SigmaPlot software (version 8.0). For the determination of Km (PEP), the sample cuvette contained 0.2 mm MESG, 0.66 mm UMP, 0.1 units/μl purine nucleoside phosphorylase, and 0.45 μm NikO. Initial velocities were measured by adding variable amounts of PEP in the range of 4.1–166 μm to the mixture. The reference cuvette contained all the components except PEP. Experimental data were plotted and treated as described for UMP.
FIGURE 3.
Michaelis-Menten plots for UMP (upper panel) and PEP (lower panel).
Inhibition by Fosfomycin
Time- and concentration-dependent inhibition of NikO by fosfomycin was determined spectrophotometrically, employing the continuous assay described above. NikO (23 μm) was incubated with fosfomycin (50, 100, 250, 500, or 1000 μm) in 50 mm Tris/HCl buffer, containing 2 mm DTT at pH 7.5 and 30 °C. At various time intervals (2–20 min), the mix was diluted by adding 0.1 mm MESG, 1 mm PEP, and 1 mm UMP. The final concentration of NikO was 1.5 μm, and that of fosfomycin was between 3.3 and 66 μm. Measurements were performed in thermostated cuvettes at 30 °C using a reference without fosfomycin and NikO. Residual activities were plotted against the incubation time and fitted to a single exponential function (y = y0 + exp(−b x) using SigmaPlot v8.0). For studying the effect of UMP on the rate of inhibition by fosfomycin, NikO was preincubated with 6 mm UMP for 5 min at 30 °C and then incubated with fosfomycin as before (final concentration of UMP was 0.5 mm).
Fluorescent Phospho- and Coomassie Blue Staining
10–20 μg of NikO, NikO C130A, NikO D342A, or MurA were incubated with 1–10 mm fosfomycin for 5–10 min at 30 °C, in the absence or presence of the cognate substrate (UMP or UDPNAG, 5 mm). After incubation with the antibiotic, the protein was mixed with loading buffer, denatured for 3 min at 95 °C, and analyzed on a 10% acrylamide gel. A low range protein marker (Bio-Rad) was loaded on the gel as a standard (ovalbumin with a molecular mass of 45 kDa has two phosphate residues bound and hence gives rise to a single band in the phospho-stain). For fluorescent photo staining, the gel was treated with ProQ® Diamond phosphoprotein gel stain according to the manufacturer's recommendations (Molecular Probes) and scanned with a Molecular Imager FX from Bio-Rad (excitation at 535 nm). After destaining, the gel was treated with Coomassie Blue and scanned again with a standard flatbed device.
Isothermal Titration Calorimetry
Binding of fosfomycin was determined by isothermal titration calorimetry with a solution of 200–350 μm NikO in the cell and 5–10 mm fosfomycin in the syringe. Both solutions were prepared in 50 mm Hepes, 2 mm DTT, 0.5 mm EDTA, and 10% v/v glycerol, pH 7.5 at 25 °C, freshly degassed, and filtered before the experiment. All measurements were performed with an MCS titration calorimeter from MicroCal Inc. (Northampton, MA). The data were analyzed using MicroCal Origin v2.9 following the guidelines provided by the manufacturer.
Crystallization
A purified sample of native NikO in 10 or 50 mm Tris-HCl, pH 7.5, at a concentration of 5.75 mg/ml was treated with 10 mm fosfomycin. Crystallization drops were set up with commercial crystallization screens using the microbatch method employing an Oryx 7 crystallization robot (Douglas Instruments Ltd.) and incubated at 289 K. The drop volume was 1 μl with equal amounts of protein and precipitant solution. These setups gave initial crystals in several conditions, of which one (0.2 m lithium sulfate, 0.1 m Bis-Tris, pH 6.5, 25% w/v PEG 3350: Hampton Research Index Screen, condition 75) could be optimized through matrix seeding (21). The crystals grew to full size within 3–4 days. All measured crystals were harvested from their mother liquor with CryoLoopsTM (Hampton Research) and cryo-protected prior to flash-freezing in liquid nitrogen and data collection by soaking in a 20% (v/v) glycerol solution for a few seconds.
Structure Determination and Refinement
Diffraction quality crystals were taken to the Swiss Light Source (Paul Scherrer Institute, Villigen, Switzerland) and measured at beamline X06DA-PXIII (λ = 1.00Å). A full dataset (180°) was collected to 2.5 Å resolution from an orthorhombic crystal (space group C2221). The dataset was processed with the programs MOSFLM (22) and SCALA (23) as well as with programs of the CCP4 suite (24). The calculated Matthews coefficient (24) showed a 98% probability for the presence of two molecules in the asymmetric unit. Blast searches against the Protein Data Bank (PDB) and homology detection with HHPred (25, 26) identified the structure of UDP-N-acetylglucosamine 1-carboxyvinyltransferase (MurA) from Aquifex aeolicus VF5 (28% sequence identity, PDB entry 2yvw) as a suitable search model for molecular replacement. Structure determination was conducted by following a recently developed protocol by DiMaio et al. (27). The best solution of this approach (based on log-likelihood-gain) was further used as input for the automated chain-tracing/model-building program PHENIX AutoBuild (28). The resulting model showed R/Rfree values of 31 and 32%. Rfree values (29) were computed from 5% randomly chosen reflections, which were not used during refinement.
The model resulting from the AutoBuild procedure was further refined against the reflection data by alternating real-space fitting steps against σA-weighted 2Fo − Fc and Fo − Fc electron density maps and least squares optimizations using the programs PHENIX (28) and Coot (30, 31). Water molecules were placed automatically into difference electron maps and accepted or rejected according to geometry criteria and their refined B-factors defined in the refinement protocol. Noncrystallographic symmetry restraints were applied during all refinement steps. The final model could be refined to R = 19.53% and Rfree = 23.28%.
No electron density was observed for the first 10 and the last 15 residues in both chains. Moreover, density for residues 127–138 in chain A as well as residues 127–139 in chain B, corresponding to the typical mobile loop in enolpyruvyl transferases, was missing. Three sulfate ions were included into the model. Details of the data collection, processing, and refinement are summarized in Table 1. Validation of the structure was carried out with the web-based program MOLPROBITY (32), giving a Ramachandran plot with 97% of the residues in favored, 3% in allowed, and 0% in disallowed regions.
TABLE 1.
Statistics on data collection and refinement of NikO co-crystallized with fosfomycin
Values in parentheses account for highest resolution shell.
| NikO + Fosfomycin | |
|---|---|
| Data collection | |
| X-ray source | Swiss Light Source PX-III |
| Wavelength (Å) | 1.00 |
| Temperature | 100 K |
| Space group | C2221 |
| Cell dimensions | |
| a, b, c (Å) | 96.21, 120.05, 153.21 |
| Resolution (Å) | 51.07-2.50 (2.64-2.50) |
| Total no. of reflections | 139,398 |
| Unique no. of reflections | 28,405 |
| Multiplicity | 4.9 (5.0) |
| Completeness (%) | 91.3 (100) |
| Rp.i.m. | 0.051 (0.139) |
| Rlinear | 0.099 (0.270) |
| 〈I/σI〉 | 13.0 (6.6) |
| Refinement | |
| Rwork/Rfree | 0.1953/0.2328 |
| No. of atoms | |
| Protein | 6948 |
| Water | 339 |
| B-factors (total) | |
| Protein (Å2) | 22.85 |
| Water (Å2) | 21.76 |
| All atoms (Å2) | 22.80 |
| r.m.s. deviations | |
| Bond lengths (Å) | 0.0024 |
| Bond angles (°) | 0.6323 |
Loop Modeling
The missing loop (11 amino acids from residues 127–138 in chain A) was built using a protocol adapted from DiMaio et al. (27). Loop remodeling employed full atom refinement in conjunction with the improved, density-guided energy function of Rosetta (33) and by taking noncrystallographic symmetry into account. First electron density maps were calculated from the final experimentally obtained NikO model. Based on an energy cutoff of 5%, 51 structures were chosen and rescored against the reflection data using the program PHASER in MR_RNP mode (34). The structure with the highest Log-Likelihood-Gain was taken as the final loop model.
Docking
The reaction product enolpyruvyl-UMP (EPUMP) was docked into the active site of NikO using AutoDock v4.2 (35). Coordinates of an idealized structure of UMP were downloaded from the PDB (www.rcsb.org/pdb), the missing enolpyruvyl moiety was added and the geometry was optimized in Maestro (48) using the OPLS force field (36, 37). Partial charges for the substrate were computed using AutoDock python scripts. The NikO structure was prepared according to requirements of AutoDock v4.2, and the simulations were performed employing the implemented Lamarckian genetic algorithm with a population size of 350 individuals and an average number of generations of about 6000. The cubic energy grid was centered 2 Å away from Oδ2 of Asp-342 and had an extension of 22.5 Å in each direction. Although the ligands were treated as fully flexible, all protein residues were kept rigid. 50 independent docking runs were performed, and the resulting binding modes were clustered based on an r.m.s. deviation cut-off of 2.0 Å.
RESULTS
Overall Structure
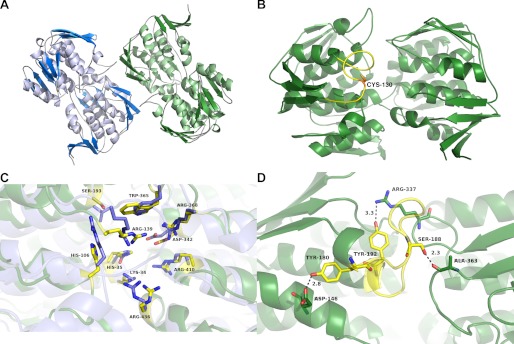
We determined the structure of NikO at a crystallographic resolution of 2.5 Å and refined it to final R-values of R = 19.53% and Rfree = 23.28 (Table 1). The structure contains two NikO molecules in the asymmetric unit connected by a noncrystallographic two-fold symmetry axis. A PDBe-PISA (Protein Interfaces, Surfaces, and Assemblies) (38) analysis predicts that the interactions between the monomers are too weak to form a stable dimer in solution. This is not in line with NikO eluting as a dimer from a size exclusion column (results not shown). Every NikO monomer is composed of two domains with an inside-out α/β-barrel structure characteristic for enolpyruvyl transferases (12, 13) (Fig. 1A). A superposition of the two chains resulted in an r.m.s. deviation of 0.335Å for 436 aligned Cα atoms. Several deviations between the deposited amino acid sequence (UniProt accession code: Q712I1) and the electron density were found upon model building (P52R, D96E, V97L, and V141D). In the homologous enzyme MurA, which had been used as a search template for molecular replacement, residue 67 was found to be an isoaspartic acid (39). This aspartic acid residue is conserved between MurA and NikO (Asp-79), but the electron density clearly showed no isoaspartate formation.
FIGURE 1.
Crystal structure of NikO. A, the final refined NikO structure (comprising two molecules in the asymmetric unit) is shown in a graphic representation. The molecules exhibit a rarely found inverse α,β-barrel fold and are colored according to their secondary structure elements (α-helices, light blue or light green; β-strands, dark blue or dark green; loops, gray). B, modeled loop structure of residues 127–139 in NikO. The best scoring loop model was chosen, exhibiting an open loop conformation. The cysteine residue (Cys-130) of this loop is shown as sticks. C, active site of NikO with catalytic residues shown as sticks. NikO is colored in dark green, and MurA is in blue. Active site residues of NikO are colored yellow. Residues are labeled according to NikO numbering. D, structure of the loop region (residues 180–192) in NikO. The structure is shown in a graphic representation with the loop colored in yellow. The figure was prepared with PyMOL (49).
Unexpectedly, no interpretable density was observed for the co-crystallized inhibitor. Continuous density was also missing for residues 127–138 in chain A as well as for residues 127–139 in chain B. The corresponding loop in homologous MurAs changes its conformation upon substrate binding and is involved in product release (15, 40). This loop also contains the residue Cys-130, which corresponds to the reactive cysteine residue in MurA (e.g. Cys-115 in the enzyme from E. coli) and was therefore supposed to be modified by fosfomycin in NikO as well. Therefore, we modeled this loop (see “Experimental Procedures”), which resulted in a twisted conformation that is tilted three times due to the presence of three prolines (Pro-131, Pro-135, and Pro-140; Fig. 1B). In a superposition with the structures of substrate bound MurA enzymes (i.e. with a closed loop conformation), Cys-130 is between 8 and 11 Å away from the corresponding residue in MurA.
The active site of NikO is located between the two domains and is built up from residues of both domains. It is lined mostly with polar and aromatic amino acids and exhibits high similarities to the active site of MurA enzymes (Fig. 1C). A loop from residues 180–192 forms one edge of the active site cavity. It starts and ends with a tyrosine residue (Tyr-180 and Tyr-192) and is anchored between both domains via an H-bond of Ser-188 to the main chain carbonyl of Ala-363 (Fig. 1D).
Docking
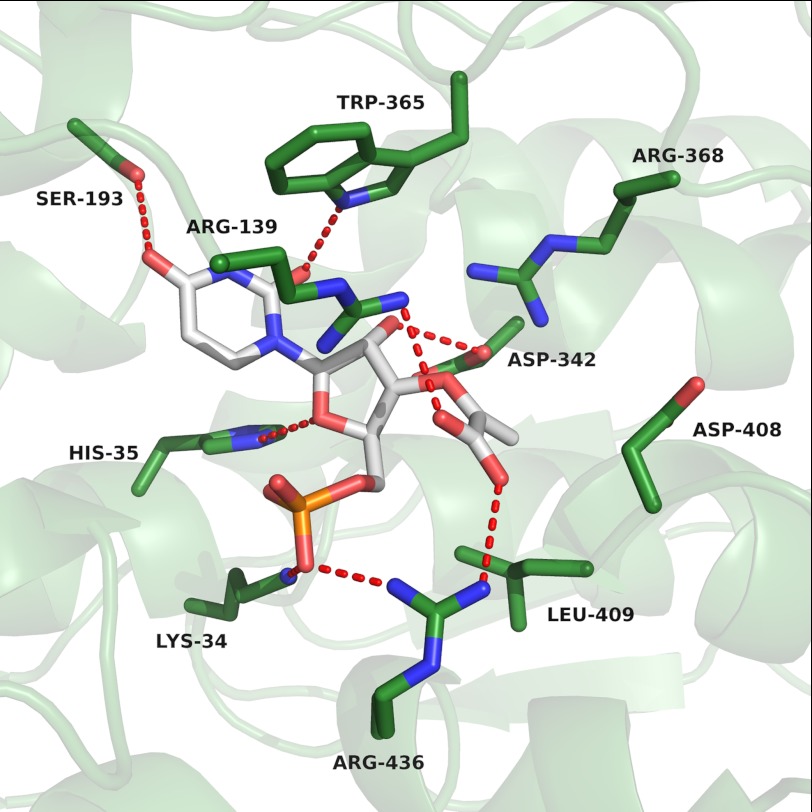
Docking of the reaction product EPUMP to the active site of NikO resulted in the lowest energy binding mode with the phosphate moiety interacting with Lys-34 and Arg-436. The ribose moiety is within H-bonding distances to Asp-342 as well as His-35. The carboxyl group of the enolpyruvyl side chain is forming a salt bridge with Arg-436, the 3′-oxygen atom is H-bonded to Arg-139, and the rest of the enolpyruvyl moiety is forming hydrophobic interactions with Leu-409. The uracil ring is bound between the side chains Ser-193 and Trp-365, which form H-bonds with the two carbonyl groups. In addition, the tryptophan residue forms a T-shaped π-stacking interaction with the uracil ring (41)(Fig. 2).
FIGURE 2.
Close-up view of the lowest energy binding mode obtained upon docking of EPUMP into the active site of NikO (EPUMP, white; NikO-residues, green). The figure was prepared with PyMOL (49).
Kinetic Properties of NikO and Inhibition by Fosfomycin
A plot of the initial velocity as a function of UMP and PEP concentration (at saturating concentrations of the respective other substrate) shows Michaelis-Menten behavior for both substrates (Fig. 3, upper and lower panels). A fit of the experimental data to the Michaelis-Menten equation yielded Km values of 67 ± 5.7 and 15 ± 2.9 μm for UMP and PEP, respectively. At saturating substrate concentrations (0.66 mm UMP and 1 mm PEP), the kcat of the reaction was 6.8 min−1. The specific activity of NikO was 132 nmol Pi min−1 mg−1. For comparison, MurA from Enterobacter cloacae has a specific activity of 1300 nmol Pi min−1 mg−1 and a kcat of 60 min−1 (42). Activity measurements with both NikO variants (C130A and D342A) showed that both amino acid exchanges lead to inactive enzyme (supplemental Fig. 4).
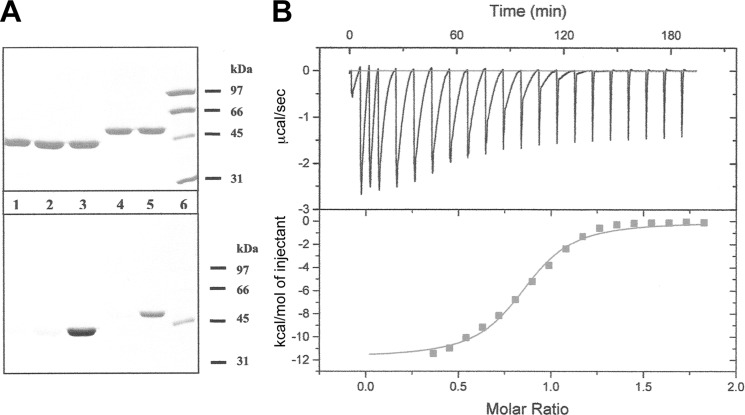
To test for fosfomycin binding, we incubated wild-type NikO as well as NikO C130A with the antibiotic and then qualitatively analyzed the phosphate content of the protein by phospho-staining using MurA as control. As shown in Fig. 4A (bottom panel, lanes 4 and 5), the test was clearly positive, indicating that NikO binds fosfomycin even in the absence of UMP. This finding is in contrast to the behavior of MurA whose reactivity toward fosfomycin is enhanced in the presence of UDPNAG (15, 42) (Fig. 4A, bottom panel, lane 3). Repeating the experiment with the C130A variant of NikO showed that the sole site of alkylation by fosfomycin is Cys-130 (Fig. 5, lanes 1 and 3).
FIGURE 4.
Fosfomycin binding. A, NikO binds to fosfomycin in the absence of UMP. Upper panel, gel stained with Coomassie Blue; lower panel, gel stained with ProQ Diamond. The lanes were loaded with the following samples: lane 1, MurA; lane 2, MurA and UDPNAG; lane 3, MurA, UDPNAG and fosfomycin; lane 4, NikO; lane 5, NikO and fosfomycin; lane 6, protein marker. B, binding of fosfomycin to NikO determined by isothermal titration microcalorimetry. 346 μm NikO was titrated with 5 mm fosfomycin.
FIGURE 5.

The C130A variant of NikO does not bind fosfomycin. A gel stained with ProQ Diamond is shown. The lanes were loaded with the following samples: lane 1, NikO C130A and fosfomycin; lane 2, NikO C130A; lane 3, NikO and fosfomycin; lane 4, NikO; lane 5, protein marker. +ffm, fosfomycin added; −ffm, no fosfomycin added; MW-Std., molecular weight standard.
Binding of fosfomycin to NikO was further investigated by isothermal microcalorimetry (Fig. 4B). The apparent KD for binding of fosfomycin to NikO was 17.1 μm and only slightly lower when the enzyme was preincubated with UMP (KD = 13.8 μm). The stoichiometry for fosfomycin binding was n = 0.82 and 0.85 in the absence and presence of UMP, indicating that NikO has one binding site per protein molecule.
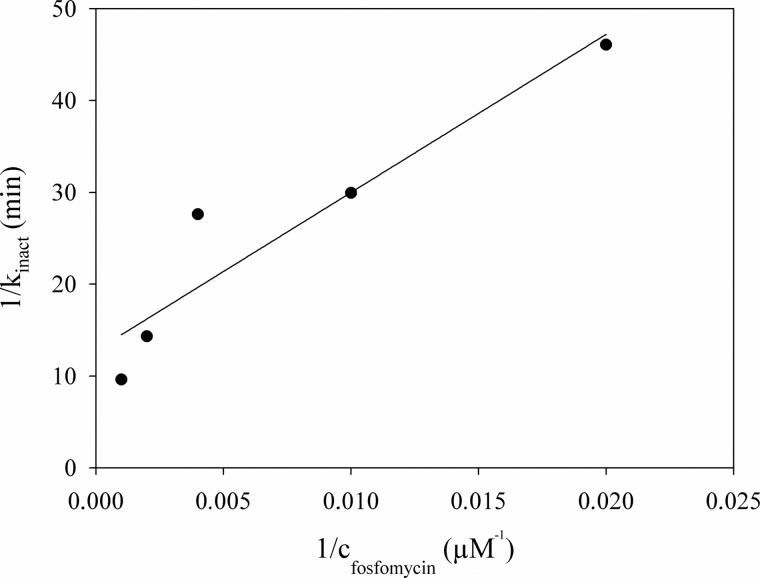
Incubation of NikO with fosfomycin resulted in a time-dependent inactivation of the enzyme (Fig. 6). UMP was not required for inactivation and, in contrast to MurA, the substrate slowed down the rate of inactivation (supplemental Fig. 5). The first-order inactivation rate constants were obtained by fitting the experimental data to a single exponential function. A double reciprocal plot of inactivation rates versus fosfomycin concentrations and extrapolation to find the y axis intercept yielded a kinact of 0.08 min−1 (Fig. 6).
FIGURE 6.
Determination of the limiting rate of kinact. In the figure, the reciprocal value of kinact was plotted against the reciprocal value of the fosfomycin concentration. The y-intercept is 12.8 min, so the limiting rate of kinact was determined to be 0.08 min−1. To determine kinact, time-dependent inhibition of NikO by fosfomycin was measured, and the residual enzymatic activity was plotted against the incubation time. For each concentration of fosfomycin in the incubation mix (50 μm, 100 μm, 250 μm, 500 μm, 1 mm, respectively), kinact was determined by fitting the data to the equation y = y0 + exp(−kinact × x).
DISCUSSION
Nikkomycins are promising new antibiotics for the use against human pathogens as well as in agriculture. Their biosynthesis is complex and proceeds via two separate pathways producing hydroxypyridyl-homothreonine and amino hexuronic acid, which are joined by a peptide bond in a final step (43, 44). The enzyme catalyzing the first step in the formation of the aminohexuronic acid moiety, NikO, belongs to the Structural Classification of Proteins (SCOP) superfamily of enolpyruvyl transferases (SCOP number: 55209). The only other known members of this family are MurA from bacteria involved in murein biosynthesis and 5-enolpyruvylshikimate 3-phosphate synthase, found in bacteria, fungi, and plants, catalyzing the penultimate step in the shikimate pathway. Structures of MurA and 5-enolpyruvylshikimate 3-phosphate synthase have been determined for enzymes from various species. These enzymes have been shown to occur in an ”open“ and a ”closed“ conformation defined by domain motions and loop conformations (12, 17). The NikO structure is more similar to the closed conformation of MurA (Cα, r.m.s. deviation of 1.68 Å) and 5-enolpyruvylshikimate 3-phosphate synthase (2.91 Å, supplemental Table 1 and supplemental Fig. 1). Because of the closer structural resemblance to MurAs, we focus the further discussion on the comparison of NikO and MurA.
Active Site
In addition to sharing similar architectures, several active site residues are conserved between NikO and MurA. The active site residue Asp-342 is equivalent to Asp-305 in MurA, which acts as a base for proton abstraction at C-3 in the elimination part of the enolpyruvyl transferase reaction (45) (Fig. 1C). Lys-34 corresponds to Lys-22 in MurA, where it is involved in de- and reprotonation of the substrate during the reaction (45). Interestingly, Arg-91 from MurA, a residue described to be important for the stabilization of the closed loop conformation by H-bonding to the backbone carbonyl group of Pro-121 (46), is exchanged to a histidine (His-106) in NikO. This indicates that the equivalent loop region in NikO, which is not visible in the current structure, either is not stabilized, if it occurs in a closed conformation, or has to fold down farther into the cleft to get stabilized after substrate binding. Similarly, the conserved Asn-23 in MurA is replaced by a histidine (His-35) in NikO. Our docking results indicate that this histidine may play a role in binding the ribose and/or the phosphate moiety of UMP (Fig. 2). Another interesting difference is the exchange of Ser-193 in NikO versus Val-163 in MurA. The docking results suggest that this serine residue acts as H-bond donor for the uracil moiety (Fig. 2).
Conformational Changes
Substrate binding has been investigated in greater detail (45) for MurA enzymes. These studies showed that substrate binding triggers a conformational change in the hinge, as well as the loop region covering the active site (15, 17). Concomitant with this structural change, the active site of these enzymes is completed (15). This conformational change in the hinge region is supposed to be triggered by a lysine-arginine residue pair that forms repulsive interactions in the apo-state of the enzyme and thus drives the domains farther apart (12). Upon substrate binding, a conformational change in the two side chains abolishes the repulsive charge-charge interaction and thereby induces a movement of the domains toward each other. Additionally, it has been shown that the naturally occurring antibiotic fosfomycin covalently binds to the Cys-115 residue in MurA and thereby inhibits the enzyme through occupation of the PEP binding site (12, 13). Although this effect is enhanced in the presence of the substrate (UDPNAG), binding of the inhibitor was also observed without preincubation with substrate but with a greatly reduced affinity (15). Previously, a function in product release was ascribed to Cys-115 in MurA rather than protonation during the enolpyruvyl transfer reaction (40). Very recently, however, a covalent tetrahedral reaction intermediate between Cys-115 in MurA and PEP has been reported. This intermediate is supposedly attacked by the deprotonated substrate, whereas Cys-115 gets reprotonated by a close-by Arg-120 (47). Both of these residues are conserved in NikO (Cys-130 and Arg-139 in NikO) and very likely act in a similar fashion.
From structural superpositions, it became clear that the structure of NikO more closely resembles a closed MurA structure. A superposition of NikO with the structure of MurA from E. cloacae in its ligated and unligated states also showed that the lysine residue that stabilizes the open conformation in MurA is exchanged to a valine (Val-60) residue in NikO, preventing any repulsive charge-charge interactions (supplemental Fig. 2). Therefore, we propose that NikO does not exhibit the large domain movement seen in MurA and rather prefers a closed conformation.
Substrate Specificity
The structure of NikO provides insights into the determinants of the substrate specificity of the enzyme and its inhibition by the naturally occurring antibiotic fosfomycin. Initially, it was proposed that NikO uses uridine as a substrate (3, 7). However, attempts to demonstrate the transfer of an enolpyruvyl moiety to uridine failed and led to the discovery that UMP is the cognate substrate of the enzyme (8). To get a better understanding of the differences in substrate specificity between NikO and MurA, a comparison of the active sites was conducted by superimposing experimentally determined complex structures of MurA and the modeled product (EPUMP) complex of NikO. The comparison revealed that the loop region of residues 180–192 in NikO would be blocking the binding site for the uridine moiety in a substrate like UDPNAG (supplemental Fig. 3A). This loop is anchored between two tyrosine residues, which are engaged in multiple hydrogen-bonding interactions to an aspartic acid residue (Tyr-180 to Asp-146) as well as a backbone carbonyl (Tyr-192 to Pro-215). In addition, an H-bond was found between Ser-188 and the carbonyl group of Ala-363 (Fig. 1D). Sequence alignments show that NikO has an insertion of about seven residues within this region and therefore exhibits the longest loop of all enolpyruvyl transferase structures currently available in the PDB, leading to a more restricted binding site. The docked complex structure of NikO and EPUMP was superimposed onto the experimentally determined complex structure of MurA co-crystallized with the tetrahedral reaction intermediate. This showed the ribose moiety of EPUMP and the N-acetylglucosamine of enolpyruvoyl-UDPNAG to be coinciding (supplemental Fig. 3B), thereby validating the docking results.
The lowest energy docking pose showed that the phosphate moiety of EPUMP is positioned in interaction distance to Lys-34 and Arg-436 and that the 2′-OH of the sugar is H-bonding to Asp-342. One of the few differences in this region of the structure is the exchange of a hydrophobic valine residue to a hydrophilic serine (Val-163 in MurA to Ser-193 in NikO), which facilitates binding of one of the uracil carbonyl groups (Fig. 2). Additional T-shaped π-stacking interactions of Trp-365 with the uracil moiety provide further stabilization of this part of the ligand. Because the obtained docking pose closely resembles a part of an experimentally observed substrate binding mode from MurA, it provides further evidence that the identified active site residues can be considered catalytically important and that the catalytic mechanism of enolpyruvyl transfer is similar in the two enzymes.
Construction of two active site residue variants, C130A and D342A, showed complete inactivation of the enzyme as determined by activity measurements. This further corroborates the mechanistic importance of these residues. In the C130A variant, the enolpyruvyl transfer is presumably impaired through the inability of forming a tetrahedral intermediate with PEP, which was very recently found to be essential in the reaction (47). In the case of D342A, the proton abstraction from the substrate UMP may be impeded, and thus, the nucleophilic attack on the PEP enzyme intermediate is abolished.
Enzyme Kinetics
Detailed biochemical data on NikO were obtained through steady-state and isothermal titration calorimetry experiments (Figs. 3 and 6) showing that the specific activity of enolpyruvyl transfer to UMP is about 10-fold lower than the specific activity found for enolpyruvyl transfer onto UDPNAG in MurA. Likewise, catalytic turnover is about 10-fold slower than for E. cloacae MurA. Apart from that, our results show that NikO is specifically inhibited by the antibiotic fosfomycin, through a covalent linkage to a cysteine residue of the enzyme (Cys-130), which is conserved in many members of the family of enolpyruvyl transferases. In MurA, this cysteine residue is part of a loop that closes around the active site upon substrate binding (12, 13). The corresponding loop in NikO was not visible in the electron density and was modeled. In this model, the loop is twisted due to the presence of three proline residues, and Cys-130 is completely solvent-accessible (Fig. 1B).
Time-dependent inhibition studies showed that inhibition of NikO is slowed down rather than enhanced in the presence of substrate. This is contrasting the observation for MurA, in which inhibition of the enzyme is faster when preincubated with substrate (15). Conclusively, the mode of inhibition by fosfomycin is different in NikO when compared with MurA. Considering our docking results, it can be speculated that the flexible loop is closing down on the interdomain cleft after substrate binding and thereby ”buries“ the reactive cysteine residue, thereby preventing reaction with the antibiotic. Alternatively, fosfomycin binding might be limited due to steric hindrances. This assumption is supported by the modeled binding mode of EPUMP (Fig. 2), in which the 5′-phosphate group occupies the analogous region of the active site as the phosphate group of fosfomycin in the structure of the substrate-inhibitor complex of MurA (13) (PDB code: 1uae). In the absence of UMP, the flexible loop is most probably in an arbitrary orientation (consistent with the lack of electron density for this region) and hence is accessible for reaction with fosfomycin.
Conclusion
The structure of NikO is the first of an enzyme participating in the formation of the aminohexuronic acid moiety in nikkomycin biosynthesis. The structure underlines the close relationship to other enolpyruvyl transferases and corresponds to a closed conformation. The structure and accompanying modeling studies as well as mutagenesis studies have revealed important active site residues, showing mechanistic similarities to MurA. Activity assays and inhibition studies confirm the previously identified enolpyruvyl transferase activity of the enzyme as well as the susceptibility of inactivation by the antibiotic fosfomycin. This inactivation is slowed down in the presence of substrate, probably due to burying of the reactive cysteine triggered by a substrate-induced loop movement.
Supplementary Material
Acknowledgments
We appreciate the support of the beamline staff at the Swiss Light Source (SLS), Villigen, Switzerland during data collection.
This work was supported by the Fonds zur Förderung der wissenschaftlichen Forschung (FWF) through grant P19858 and the Ph. D. program “Molecular Enzymology” (FWF W901 to K. G. and P. M.).

This article contains supplemental Table 1 and Figs. 1–5.
The atomic coordinates and structure factors (code 4FQD) have been deposited in the Protein Data Bank, Research Collaboratory for Structural Bioinformatics, Rutgers University, New Brunswick, NJ (http://www.rcsb.org/).
- PEP
- phosphoenolpyruvate
- MESG
- 2-amino-6-mercapto-7-methylpurine riboside
- Bis-Tris
- 2-(bis(2-hydroxyethyl)amino)-2-(hydroxymethyl)propane-1,3-diol
- EPUMP
- enolpyruvyl-UMP
- UDPNAG
- UDP-N-acetylglucosamine
- r.m.s.
- root mean square.
REFERENCES
- 1. Bormann C., Mattern S., Schrempf H., Fiedler H. P., Zähner H. (1989) Isolation of Streptomyces tendae mutants with an altered nikkomycin spectrum. J. Antibiot. 42, 913–918 [DOI] [PubMed] [Google Scholar]
- 2. Chen H., Hubbard B. K., O'Connor S. E., Walsh C. T. (2002) Formation of β-hydroxy histidine in the biosynthesis of nikkomycin antibiotics. Chem. Biol. 9, 103–112 [DOI] [PubMed] [Google Scholar]
- 3. Dähn U., Hagenmaier H., Höhne H., König W. A., Wolf G., Zähner H. (1976) Stoffwechselprodukte von mikroorganismen. 154. Mitteilung. Nikkomycin, ein neuer hemmstoff der chitinsynthese bei pilzen. Arch. Microbiol. 107, 143–160 [DOI] [PubMed] [Google Scholar]
- 4. Kim M. K., Park H. S., Kim C. H., Park H. M., Choi W. (2002) Inhibitory effect of nikkomycin Z on chitin synthases in Candida albicans. Yeast 19, 341–349 [DOI] [PubMed] [Google Scholar]
- 5. Brillinger G. U. (1979) Metabolic products of microorganisms. 181. Chitin synthase from fungi, a test model for substances with insecticidal properties. Arch. Microbiol. 121, 71–74 [DOI] [PubMed] [Google Scholar]
- 6. Hector R. F. (1993) Compounds active against cell walls of medically important fungi. Clin. Microbiol. Rev. 6, 1–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Lauer B., Süssmuth R., Kaiser D., Jung G., Bormann C. (2000) A putative enolpyruvyl transferase gene involved in nikkomycin biosynthesis. J. Antibiot. 53, 385–392 [DOI] [PubMed] [Google Scholar]
- 8. Ginj C., Rüegger H., Amrhein N., Macheroux P. (2005) 3′-Enolpyruvyl-UMP, a novel and unexpected metabolite in nikkomycin biosynthesis. ChemBioChem 6, 1974–1976 [DOI] [PubMed] [Google Scholar]
- 9. Walsh C. T., Benson T. E., Kim D. H., Lees W. J. (1996) The versatility of phosphoenolpyruvate and its vinyl ether products in biosynthesis. Chem. Biol. 3, 83–91 [DOI] [PubMed] [Google Scholar]
- 10. Bugg T. D., Walsh C. T. (1992) Intracellular steps of bacterial cell wall peptidoglycan biosynthesis: enzymology, antibiotics, and antibiotic resistance. Nat. Prod. Rep. 9, 199–215 [DOI] [PubMed] [Google Scholar]
- 11. Schönbrunn E., Eschenburg S., Shuttleworth W. A., Schloss J. V., Amrhein N., Evans J. N., Kabsch W. (2001) Interaction of the herbicide glyphosate with its target enzyme 5-enolpyruvylshikimate 3-phosphate synthase in atomic detail. Proc. Natl. Acad. Sci. U.S.A. 98, 1376–1380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Schönbrunn E., Sack S., Eschenburg S., Perrakis A., Krekel F., Amrhein N., Mandelkow E. (1996) Crystal structure of UDP-N-acetylglucosamine enolpyruvyl transferase, the target of the antibiotic fosfomycin. Structure 4, 1065–1075 [DOI] [PubMed] [Google Scholar]
- 13. Skarzynski T., Mistry A., Wonacott A., Hutchinson S. E., Kelly V. A., Duncan K. (1996) Structure of UDP-N-acetylglucosamine enolpyruvyl transferase, an enzyme essential for the synthesis of bacterial peptidoglycan, complexed with substrate UDP-N-acetylglucosamine and the drug fosfomycin. Structure 4, 1465–1474 [DOI] [PubMed] [Google Scholar]
- 14. Eschenburg S., Priestman M. A., Abdul-Latif F. A., Delachaume C., Fassy F., Schönbrunn E. (2005) A novel inhibitor that suspends the induced fit mechanism of UDP-N-acetylglucosamine enolpyruvyl transferase (MurA). J. Biol. Chem. 280, 14070–14075 [DOI] [PubMed] [Google Scholar]
- 15. Samland A. K., Jelesarov I., Kuhn R., Amrhein N., Macheroux P. (2001) Thermodynamic characterization of ligand-induced conformational changes in UDP-N-acetylglucosamine enolpyruvyl transferase. Biochemistry 40, 9950–9956 [DOI] [PubMed] [Google Scholar]
- 16. Schönbrunn E., Eschenburg S., Krekel F., Luger K., Amrhein N. (2000) Role of the loop containing residue 115 in the induced-fit mechanism of the bacterial cell wall biosynthetic enzyme MurA. Biochemistry 39, 2164–2173 [DOI] [PubMed] [Google Scholar]
- 17. Schönbrunn E., Svergun D. I., Amrhein N., Koch M. H. (1998) Studies on the conformational changes in the bacterial cell wall biosynthetic enzyme UDP-N-acetylglucosamine enolpyruvyl transferase (MurA). Eur. J. Biochem. 253, 406–412 [DOI] [PubMed] [Google Scholar]
- 18. Kim D. H., Lees W. J., Kempsell K. E., Lane W. S., Duncan K., Walsh C. T. (1996) Characterization of a Cys-115 to Asp substitution in the Escherichia coli cell wall biosynthetic enzyme UDP-GlcNAc enolpyruvyl transferase (MurA) that confers resistance to inactivation by the antibiotic fosfomycin. Biochemistry 35, 4923–4928 [DOI] [PubMed] [Google Scholar]
- 19. Marquardt J. L., Siegele D. A., Kolter R., Walsh C. T. (1992) Cloning and sequencing of Escherichia coli murZ and purification of its product, a UDP-N-acetylglucosamine enolpyruvyl transferase. J. Bacteriol. 174, 5748–5752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Webb M. R., Hunter J. L. (1992) Interaction of GTPase-activating protein with p21ras, measured using a continuous assay for inorganic phosphate release. Biochem. J. 287, 555–559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. D'Arcy A., Villard F., Marsh M. (2007) An automated microseed matrix-screening method for protein crystallization. Acta Crystallogr. D 63, 550–554 [DOI] [PubMed] [Google Scholar]
- 22. Leslie A. G. (2006) The integration of macromolecular diffraction data. Acta Crystallogr. D 62, 48–57 [DOI] [PubMed] [Google Scholar]
- 23. Evans P. (2006) Scaling and assessment of data quality. Acta Crystallogr. D 62, 72–82 [DOI] [PubMed] [Google Scholar]
- 24. Collaborative Computational Project, Number 4 (1994) The CCP4 suite: programs for protein crystallography. Acta Crystallogr. D 50, 760–763 [DOI] [PubMed] [Google Scholar]
- 25. Söding J. (2005) Protein homology detection by HMM-HMM comparison. Bioinformatics 21, 951–960 [DOI] [PubMed] [Google Scholar]
- 26. Söding J., Biegert A., Lupas A. (2005) The HHpred interactive server for protein homology detection and structure prediction. Nucleic Acids Res. 33, W244–W248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. DiMaio F., Terwilliger T. C., Read R. J., Wlodawer A., Oberdorfer G., Wagner U., Valkov E., Alon A., Fass D., Axelrod H. L., Das D., Vorobiev S. M., Iwaï H., Pokkuluri P. R., Baker D. (2011) Improved molecular replacement by density- and energy-guided protein structure optimization. Nature 473, 540–543 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Adams P. D., Grosse-Kunstleve R. W., Hung L. W., Ioerger T. R., McCoy A. J., Moriarty N. W., Read R. J., Sacchettini J. C., Sauter N. K., Terwilliger T. C. (2002) PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr. D 58, 1948–1954 [DOI] [PubMed] [Google Scholar]
- 29. Kleywegt G. J., Brünger A. T. (1996) Checking your imagination: applications of the free R-value. Structure 4, 897–904 [DOI] [PubMed] [Google Scholar]
- 30. Emsley P., Cowtan K. (2004) Coot: model-building tools for molecular graphics. Acta Crystallogr. D 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 31. Emsley P., Lohkamp B., Scott W. G., Cowtan K. (2010) Features and development of Coot. Acta Crystallogr. D 66, 486–501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Chen V. B., Arendall W. B., 3rd, Headd J. J., Keedy D. A., Immormino R. M., Kapral G. J., Murray L. W., Richardson J. S., Richardson D. C. (2010) MolProbity: all-atom structure validation for macromolecular crystallography. Acta Crystallogr. D Biol. Crystallogr. 66, 12–21 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Leaver-Fay A., Tyka M., Lewis S. M., Lange O. F., Thompson J., Jacak R., Kaufman K., Renfrew P. D., Smith C. A., Sheffler W., Davis I. W., Cooper S., Treuille A., Mandell D. J., Richter F., Ban Y. E., Fleishman S. J., Corn J. E., Kim D. E., Lyskov S., Berrondo M., Mentzer S., Popović Z., Havranek J. J., Karanicolas J., Das R., Meiler J., Kortemme T., Gray J. J., Kuhlman B., Baker D., Bradley P. (2011) ROSETTA3: an object-oriented software suite for the simulation and design of macromolecules. Methods Enzymol. 487, 545–574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Morris G. M., Goodsell D. S., Halliday R. S., Huey R., Hart W. E., Belew R. K., Olson A. J. (1998) Automated docking using a Lamarckian genetic algorithm and empirical binding free energy function. J. Comput. Chem. 19, 1639–1662 [Google Scholar]
- 36. Jorgensen W. L., Maxwell D. S., Tirado-Rives J. (1996) Development and testing of the OPLS all-atom force field on conformational energetics and properties of organic liquids. J. Am. Chem. Soc. 118, 11225–11236 [Google Scholar]
- 37. Jorgensen W. L., Tirado-Rives J. (1988) The OPLS force field for proteins: energy minimizations for crystals of cyclic peptides and Crambin. J. Am. Chem. Soc. 110, 1657–1666 [DOI] [PubMed] [Google Scholar]
- 38. Krissinel E., Henrick K. (2007) Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 372, 774–797 [DOI] [PubMed] [Google Scholar]
- 39. Eschenburg S., Schönbrunn E. (2000) Comparative x-ray analysis of the unliganded fosfomycin-target murA. Proteins 40, 290–298 [DOI] [PubMed] [Google Scholar]
- 40. Eschenburg S., Priestman M., Schönbrunn E. (2005) Evidence that the fosfomycin target Cys-115 in UDP-N-acetylglucosamine enolpyruvyl transferase (MurA) is essential for product release. J. Biol. Chem. 280, 3757–3763 [DOI] [PubMed] [Google Scholar]
- 41. Meyer E. A., Castellano R. K., Diederich F. (2003) Interactions with aromatic rings in chemical and biological recognition. Angew. Chem. Int. Ed. Engl. 42, 1210–1250 [DOI] [PubMed] [Google Scholar]
- 42. Samland A. K., Amrhein N., Macheroux P. (1999) Lysine 22 in UDP-N-acetylglucosamine enolpyruvyl transferase from Enterobacter cloacae is crucial for enzymatic activity and the formation of covalent adducts with the substrate phosphoenolpyruvate and the antibiotic fosfomycin. Biochemistry 38, 13162–13169 [DOI] [PubMed] [Google Scholar]
- 43. Lauer B., Russwurm R., Schwarz W., Kálmánczhelyi A., Bruntner C., Rosemeier A., Bormann C. (2001) Molecular characterization of co-transcribed genes from Streptomyces tendae Tü901 involved in the biosynthesis of the peptidyl moiety and assembly of the peptidyl nucleoside antibiotic nikkomycin. Mol. Gen. Genet. 264, 662–673 [DOI] [PubMed] [Google Scholar]
- 44. Bruntner C., Lauer B., Schwarz W., Möhrle V., Bormann C. (1999) Molecular characterization of co-transcribed genes from Streptomyces tendae Tü901 involved in the biosynthesis of the peptidyl moiety of the peptidyl nucleoside antibiotic nikkomycin. Mol. Gen. Genet. 262, 102–114 [DOI] [PubMed] [Google Scholar]
- 45. Eschenburg S., Kabsch W., Healy M. L., Schonbrunn E. (2003) A new view of the mechanisms of UDP-N-acetylglucosamine enolpyruvyl transferase (MurA) and 5-enolpyruvylshikimate-3-phosphate synthase (AroA) derived from x-ray structures of their tetrahedral reaction intermediate states. J. Biol. Chem. 278, 49215–49222 [DOI] [PubMed] [Google Scholar]
- 46. Schonbrunn E., Eschenburg S., Luger K., Kabsch W., Amrhein N. (2000) Structural basis for the interaction of the fluorescence probe 8-anilino-1-naphthalene sulfonate (ANS) with the antibiotic target MurA. Proc. Natl. Acad. Sci. U.S.A. 97, 6345–6349 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Zhu J. Y., Yang Y., Han H., Betzi S., Olesen S. H., Marsilio F., Schönbrunn E. (2012) Functional consequence of covalent reaction of phosphoenolpyruvate with UDP-N-acetylglucosamine 1-carboxyvinyltransferase (MurA). J. Biol. Chem. 287, 12657–12667 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Schrödinger, LLC (2011) Maestro, v 9.1, Schrödinger, LLC, New York [Google Scholar]
- 49. Schrödinger, LLC (2011) The PyMOL Molecular Graphics System, version 1.4, Schrödinger, LLC, New York [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.