Background: To become antigenic, mycobacterial hexamannosylated phosphatidyl-myo-inositol (PIM6) undergo CD1e-assisted α-mannosidase processing.
Results: Measuring membrane-to-membrane PIM transfer, CD1e selectively transferred diacylated PIM species in accordance with the fact that liposome-inserted di-acylated PIM6 were the only PIM species digested into antigenic molecules by α-mannosidase.
Conclusion: CD1e assists PIM processing through its lipid transfer protein property.
Significance: This study reveals the molecular mechanisms by which CD1e contributes to lipid immunoediting.
Keywords: Antigen Presentation, Antigen Processing, Lipid Transport, Liposomes, Mycobacterium tuberculosis, Tuberculosis, Lipid Transfer Protein, Saposin
Abstract
Lipids are important antigens that induce T cell-mediated specific immune responses. They are presented to T lymphocytes by a specific class of MHC-I like proteins, termed CD1. The majority of the described CD1-presented mycobacterial antigens are presented by the CD1b isoform. We previously demonstrated that the stimulation of CD1b-restricted T cells by the hexamannosylated phosphatidyl-myo-inositol (PIM6), a family of mycobacterial antigens, requires a prior partial digestion of the antigen oligomannoside moiety by α-mannosidase and that CD1e is an accessory protein absolutely required for the generation of the lipid immunogenic form. Here, we show that CD1e behaves as a lipid transfer protein influencing lipid immunoediting and membrane transfer of PIM lipids. CD1e selectively assists the α-mannosidase-dependent digestion of PIM6 species according to their degree of acylation. Moreover, CD1e transfers only diacylated PIM from donor to acceptor liposomes and also from membranes to CD1b. This study provides new insight into the molecular mechanisms by which CD1e contributes to lipid immunoediting and CD1-restricted presentation to T cells.
Introduction
CD1 proteins are membrane proteins specialized in the presentation of endogenous and exogenous lipid antigens to T lymphocytes (1). A large fraction of the known exogenous antigens are components of the Mycobacterium tuberculosis envelope and induce specific T cell responses (2). Human antigen-presenting cells express five CD1 isoforms, CD1a, CD1b, CD1c, CD1d, and CD1e, that traffic in different endocytic compartments (3). During recycling, CD1a, -b, -c, and -d molecules bind and form stable complexes with lipid antigens, which are then exposed on the cell surface for T cell stimulation (4). CD1e displays very distinct characteristics. In contrast to other CD1 isoforms, it is expressed only in dendritic cells. CD1e accumulates in the Golgi compartments of immature dendritic cells, and upon dendritic cell maturation, CD1e is transported to late endosomes/lysosomes where it is cleaved and accumulates as a soluble form (sCD1e)5 (5, 6). CD1e is never expressed on the cell surface, thus preventing any direct interaction with T cells, but is implicated in the processing of the complex mycobacterial lipid antigens hexamannosylated phosphatidyl-myo-inositol (PIM6)6 into dimannosylated phosphatidyl-myo-inositol (PIM2) (7). In addition to this role, CD1e may positively or negatively affect lipid presentation by CD1b, CD1c, and CD1d, due to the capacity of CD1e to facilitate rapid formation of CD1-lipid complexes and also to accelerate their turnover (8). Thus, CD1e is an important modulator of both group 1 and group 2 CD1-restricted responses influencing the lipid antigen availability as well as the generation and persistence of CD1-lipid complexes (8). The crystal structure of human recombinant sCD1e (rsCD1e) at 2.90 Å resolution revealed that CD1e displays a main portal wider than that of other CD1 molecules as well as a less intricate groove, allowing accommodation of a variety of lipid antigens and facilitating the rapid exchange of bound lipids (9).
Recent studies have implicated saposins in CD1 lipid antigen loading and presentation. To date, five activator proteins of sphingolipid catabolism have been described, saposin-A to saposin-D (Sap-A to Sap-D) and the GM2 activator protein (GM2AP). These are non-enzymatic cofactors involved in the degradation of sphingolipids by lysosomal hydrolases (10), assisting membrane-inserted substrate digestion. They mediate the interaction between the water-soluble exohydrolase and their membrane-embedded glycolipid substrates at the lipid-water interface. Saposins may act via two distinct mechanisms, including membrane destabilization (11) and direct binding and transferring of monomeric lipids (12). In the context of CD1 proteins, saposins facilitate binding of α-GalCer to CD1d (13), with Sap-B involved in the loading of CD1d with lipid antigens (14, 15), and Sap-C involved in the loading of CD1b with mycobacterial lipoarabinomannan antigen (16).
Similarly to saposins, CD1e participates in lipid antigen editing. Processing of the PIM6 oligosaccharide requires the presence of CD1e and α-mannosidase in lysosomes to produce PIM2, which are the antigenic forms recognized by CD1b-restricted T cells (7). In the absence of CD1e, PIM6 are digested to phosphatidyl-myo-inositol trimannosides (PIM3), which are not able to stimulate the PIM-specific T cells.
Both the multiple effects of CD1e on antigen presentation (8) and its structural characteristics (9) suggest that CD1e might have evolved to mediate lipid exchange/editing processes. Although CD1e/PIM association has been shown by isoelectric focusing experiments (7, 9), the mechanisms by which CD1e protein participates in PIM processing and influences the immune responses remains to be precisely determined. PIM exist naturally in the mycobacterial envelope as a mixture of different acyl forms, including mono-, di-, tri-, and tetra-acylated forms (17). Using liposome-inserted PIM and membrane-to-membrane transfer assays, we find that CD1e is an LTP, transferring only diacylated PIM species. This finding is in accordance with the fact that only liposome-inserted diacylated PIM6 species can be digested by α-mannosidase to generate PIM2.
EXPERIMENTAL PROCEDURES
Preparation of Mycobacterial PIM2 and PIM6 Mixtures
PIM2 and PIM6 mixtures were purified from Mycobacterium smegmatis total lipid extract. Briefly, total lipids were extracted according to the Folch method (18). Acetone was then added to the concentrated organic phase to precipitate acetone-insoluble lipids (overnight at 4 °C). After centrifugation (20 min, 2000 × g, 4 °C), the acetone-insoluble fraction was dried under nitrogen flow and loaded on a silica column (25 × 2.3 cm; KG60, 230–400 mesh, Merck), successively eluted with 300 ml of chloroform, 300 ml of chloroform/methanol/water 9:1:0.1 (by volume), 200 ml of chloroform/methanol/water 8:2:0.1 (by volume), 200 ml of chloroform/methanol/water 7:3:0.2 (by volume), 300 ml of chloroform/methanol/water 7:3:0.4 (by volume), 300 ml of chloroform/methanol/water 60:35:6 (by volume), and 300 ml of chloroform/methanol/water 60:35:8 (by volume). PIM2 were eluted using 7:3:0.2 and 7:3:0.4 proportions and PIM6 using 60:35:6 and 60:35:8.
Purification of PIM2 and PIM6 Acyl Forms
Different PIM2 and PIM6 acyl forms were purified as described previously (19). PIM2- or PIM6-enriched fractions (40 mg) were loaded in equilibrating buffer (0.1 m ammonium acetate solution containing 20% propanol-1), on an octyl-Sepharose CL-4B column (20 × 1.5 cm; Amersham Biosciences). The elution was performed with a linear gradient from 20 to 50% propanol-1 (135-ml each) at 5 ml/h flow rate. The purification process was monitored by TLC and MALDI-TOF mass spectrometry analysis, and each purified acyl form was controlled by 1H NMR spectroscopy as described previously (20, 21).
Liposome Preparations
Liposomes contained POPC (1-palmitoyl-2-oleyl-sn-glycero-3-phosphocholine; Sigma; 4.55 μmol), cholesterol (Sigma; 1.75 μmol), POPS (1-palmitoyl-2-oleyl-sn-glycero-3-phosphoserine; Avanti polar lipids; 0.7 μmol) and optionally, PIM6 mixture (0.28 μmol). PIM and commercial lipids were solubilized in the solvent systems chloroform/methanol/water 60:35:8 (by volume) or chloroform, respectively, mixed in the desired proportions and dried under nitrogen flow. Lipid films were hydrated in appropriate volume of Tris buffer (10 mm Tris, pH 7.4, 150 mm NaCl, 1 mm EDTA), mixed by vortexing and subjected to 10 freeze/thaw cycles by alternative passages in liquid nitrogen and water (25 °C) to obtain large unilamellar vesicles (LUV). For fluorophore-encapsulated LUV, the entire procedure was performed in the presence of 20 mm calcein. LUV size was homogenized by sequential extrusion through 0.2- and 0.1-μm pore diameter polycarbonate membranes. Liposomes were dialyzed against 50 mm phosphate buffer, pH 6.0, or acetate buffer, pH 4.7. LUV size (diameter of 120 ± 10 nm) was assessed by dynamic light scattering and electron microscopy.
Recombinant Soluble Human CD1e Production (rsCD1e)
rsCD1e was expressed in Drosophila S2 cells, previously transduced with human β2 microglobulin, and purified as described (7). Briefly, rsCD1e with the propeptide (amino acids 20–305 of the pre-α chain) was expressed as a fusion protein with the BIP signal peptide and a C-terminal tag containing the V5 epitope followed by a hexahistidine sequence (pMT BIP-V5His, Invitrogen). The recombinant protein was purified by metal chelate chromatography followed by affinity chromatography using 20.6 anti-hCD1e mAb (5).
Recombinant Saposins
Human saposins B and C (Sap-B and Sap-C) cDNA were amplified from human spleen RNA (Clontech), using Platinum TaqDNA polymerase high fidelity (Invitrogen), cloned into pCR2.1-TOPO (Invitrogen) and sequenced (7). After subcloning into the NcoI-BamHI sites of the procaryotic expression vector pQE-60 (Qiagen), the plasmids were used to transform Escherichia coli BL21(DE3) LysS (Novagen). Recombinant saposins were purified from bacterial lysates via their C-terminal His6 tag with the His-select spin columns (Sigma) according to standard procedures. Purity of the preparations was estimated by Coomassie staining on SDS-PAGE.
In Vitro α-Mannosidase Digestion of PIM6
In vitro α-mannosidase digestion assays were carried out as described previously (7). PIM6 (27.7 nmol), Ac1PIM6 (24.1 nmol), or Ac2PIM6 (21.5 nmol) were sonicated in 100 μl of mannosidase buffer (0.1 m sodium acetate, pH 4.5, 1 mm ZnSO4). The enzymatic reaction was performed in the presence of jack bean α-mannosidase (Sigma) (0.44 unit) and rsCD1e (1.05 nmol) for 1 h at 37 °C (total volume of 150 μl). In vitro α-mannosidase digestion of sonicated mixPIM6 (100 μg) in the presence of Sap-B or Sap-C (30 μg; 3 nmol) instead of rsCD1e (120 μg; 3 nmol) were also tested. These assays were also carried out with LUV (18 mm of total lipids) composed by POPC/cholesterol/PIM6 or Ac1PIM6 65:25:10 (by mol): typically, 14 μl of LUV solution (252 nmol of lipids) were incubated with rsCD1e (80 μg; 2.1 nmol) and α-mannosidase (20 μg; 0.44 unit) in a final volume of 150 μl for 8 h at 37 °C.
In all cases, PIM were extracted following the reaction by the Folch method as described previously (7). The organic extracts were dried under nitrogen stream and resuspended in 20 μl of chloroform/methanol/water 60:35:8 (by volume) for MALDI-TOF MS analysis.
Fluorophore Leakage
Calcein-encapsulated LUV were prepared as described above and dialyzed against acetate buffer, pH 4.7, to remove free calcein and purified on a Sephadex G-75 column (10 × 2 cm; Sigma). The elution was performed with acetate buffer at 0.3 ml/min flow rate, and 1-ml fractions were collected. The presence of liposomes within each fraction was determined by spectrophotometry (600-nm wavelength), and the most enriched fractions were used for leakage experiments.
Calcein-encapsulated LUV (40 nmol of lipids) were incubated with jack bean α-mannosidase (0.44 unit) and rsCD1e (2.1 nmol) or Sap-C (0.5 nmol), as a positive control, in 2 ml of acetate buffer (0.1 m sodium acetate, pH 4.5, 1 mm ZnSO4). Fluorophore leakage was measured by spectrofluorometry (SAFAS) at 37 °C under agitation. Excitation wavelength was 480 nm, and emission was recorded at 517 nm with 4-nm slit widths. For each experiment, the maximum fluorophore leakage was obtained by final addition of 10% Triton X-100 (10 μl), which induces disorganization of liposomes.
Dynamic Light Scattering
Dynamic light scattering measurements were performed at 20 °C in a microcuvette (90° scattering angle) with a DynaPro-MS/X instrument (ProteinSolution, Inc.) equipped with a laser diode operating at 830 nm wavelength and a digital correlator with 248 exponentially spaced channels. Diluted LUV solutions (1 μl in 100 μl) were centrifuged for 10 min at 30,000 × g and transferred into a 100-μl microcuvette. The laser intensity was set at 10% of its maximum power, and 30 acquisitions of 10 s were cumulated. Each single experiment resulted from the sum of 30 acquired spectra. Data analysis was performed with the cumulative method using Dynamics V6 software (ProteinSolution, Inc.).
Electron Microscopy
For negative staining, 10 μl of LUV preparations were applied to carbon-coated copper grids. After 30 s, solution in excess was blotted off with filter paper, and samples were stained with 1% (w/v) uranyl acetate for 1 min. Analysis was performed using a Jeol 120 EX transmission electron microscope, and images were acquired with a digital camera (AMT-USA) at 75,000–120,000× magnification.
Lipid Transfer by CD1e
LUV characterized by different lipid compositions were prepared as described above and used at 25 mm lipid concentration. Acceptor liposomes (LUV-A) contained POPC (7.5 μmol) and cholesterol (2.5 μmol), whereas donor liposomes (LUV-D) contained POPC (6.5 μmol), cholesterol (2.5 μmol), and negatively charged phospholipids (1.0 μmol), including POPS, PI (Sigma), or PIM2 acyl forms.
Neutral liposomes (40 μl, LUV-A; 1.0 μmol total lipids) were incubated with an equal amount of negatively charged liposomes (LUV-D), in the presence of rsCD1e (2.1 nmol) in 50 mm phosphate buffer, pH 6.0 (total volume of 160 μl), for 36 h at 37 °C on agitation. Samples were then loaded on a 1.2 ml of DEAE-Sephadex A-25 (GE Healthcare) column pre-equilibrated with phosphate buffer. The elution was performed with phosphate buffer containing increasing concentrations of NaCl (12.5, 60, and 1 m). Three 1-ml fractions were collected for each NaCl concentration, and liposome elution was monitored by measuring absorbance at 600 nm wavelength. A progressive and selective elution of LUV-A (0% of POPS), LUV-I (5% of POPS), and LUV-D (10% of POPS) from the DEAE column was obtained using increasing NaCl concentrations (0, 60, and 1 m, respectively).
The presence of rsCD1e and lipids in the collected fractions was assessed by SDS-PAGE (15% acrylamide) and silver nitrate staining. Control experiments were performed in the absence of rsCD1e and in the presence of denaturated rsCD1e or BSA. The stability of individual liposome populations over incubation time in the presence of rsCD1e was verified by electron microscopy and analyzing their chromatographic behavior.
CD1b PI Loading Assays
LUV-D (3.6 mm of total lipids), containing POPC (86.4 nmol), cholesterol (28.8 nmol), and PI (28.8 nmol) were prepared as described above. PI was supplemented with [3H]PI (PerkinElmer Life Sciences) to obtain a 280 × 103 dpm/nmol specific activity. LUV-D were incubated with rsCD1b (19.6 pmol) in the absence or presence of rsCD1e (6.6 and 13.2 pmol) or N Oct-3 transactivation domain (16.7 pmol) in 50 mm phosphate buffer, pH 6.0, 60 mm NaCl (4 μl of final volume). The reaction was performed at 37 °C for 5 h under agitation. 2 μl of each sample were then loaded on an isoelectric focusing gel (PhastGel 4–6.5, GE Healthcare) to separate proteins and free lipids. Electrophoresis was performed in a PhastGel system (Amersham Biosciences) at 600 accumulated volts/h. After the migration, bands corresponding to rsCD1b were excised from the gel, and the associated [3H] radioactivity was counted. The capacity of rsCD1e to load PI into rsCD1b molecules was analyzed at different time points (5, 30, and 300 min). Control experiments in the absence of rsCD1b were performed to assess the radioactivity diffusion within the pH range corresponding to rsCD1b isoelectric point. The percentage of rsCD1b loaded with PI was determined by comparing the amounts of PI detected by radioactivity with the total amounts of rsCD1b loaded on the gel, assuming that 1 mol of rsCD1b is associated to 1 mol of PI.
Matrix-assisted Laser Desorption/Ionization Mass Spectrometry
MALDI-TOF MS analysis was carried out using a 4700 proteomics analyzer (with TOF/TOF optics, Applied Biosystems, Voyager DE-STR, Framingham, MA) in reflectron mode. Ionization was achieved by irradiation with a Nd:YAG laser (355 nm) operating at 500-ps pulses with a frequency of 200 Hz. PIM were analyzed in negative-ion mode. Spectra from 2000 to 4000 laser shots were summed to obtain the final spectrum. Typically, 2 μl (2–5 μg) of PIM dissolved in the solvent system chloroform/methanol/water 60:35:8 (by volume) were mixed with 2 μl of matrix solution, 2-(4-hydroxyphenylazo)benzoic dissolved at 5 mg/ml in ethanol/water 1:1 (by volume) (Sigma). 0.4 μl of this mixture were spotted on the target.
RESULTS
CD1e Assists Mannosidase in Processing of Specific Acyl Forms of PIM
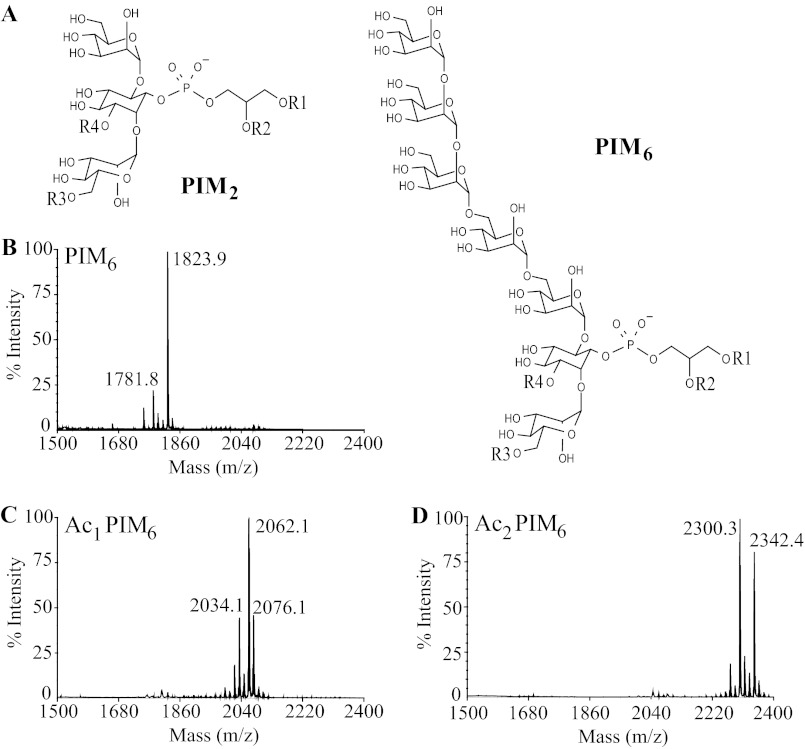
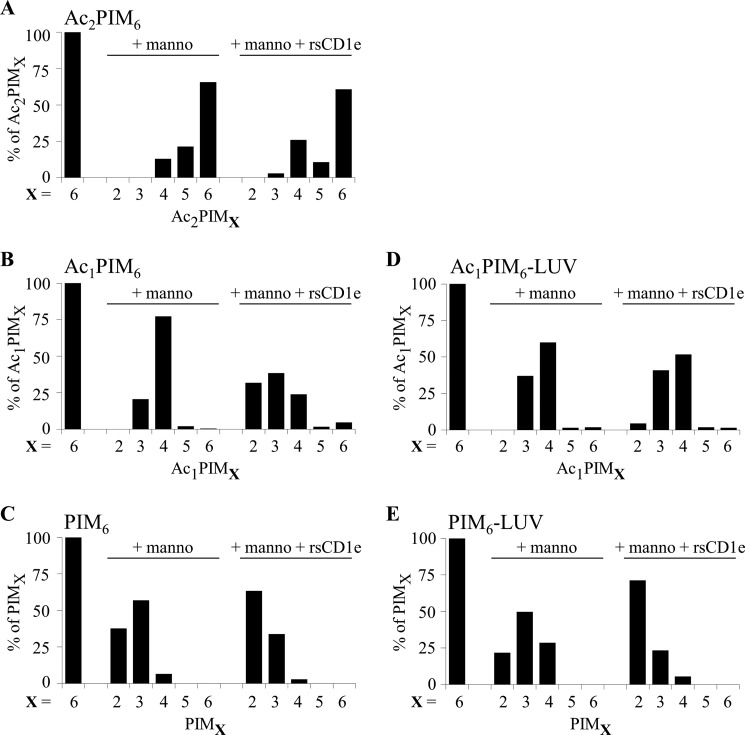
Previously, we showed that CD1e acts as an accessory protein to assist α-mannosidase-mediated processing of PIM6 into PIM2, which is required for CD1b-restricted T cells activation (7). Using the natural pool of PIM6, containing a mixture of different acyl forms (mixPIM6) as substrate for in vitro digestion (Fig. 1A), we noticed that only di-acylated (PIM6) and tri-acylated (Ac1PIM6) forms of PIM6, but not tetra-acylated PIM6 (Ac2PIM6) were digested by α-mannosidase in the presence of rsCD1e to generate the respective PIM2 acyl forms (7). Moreover, insertion of mixPIM6 into liposomes, to better mimic physiological conditions, resulted in a nearly exclusive digestion of diacylated forms (7). These prior observations suggested that the degree of PIM acylation interferes with their glycosidic processing efficiency. To better understand this selective hydrolysis, in vitro digestion assays were performed on individual purified PIM6, Ac1PIM6, and Ac2PIM6 acyl forms (Fig. 1, B–D), in the absence or presence of rsCD1e. The reaction products were analyzed by negative-ion mode MALDI-TOF mass spectrometry (supplemental Fig. S1), and the relative abundances of the generated PIM glycoform were determined (Fig. 2). In the presence of α-mannosidase only (+ manno), a more effective enzymatic digestion was observed for PIM with a lower acylation degree, as Ac2PIM6, Ac1PIM6, and PIM6 acyl forms were digested to Ac2PIM4, Ac1PIM3, and PIM2 glyco-forms, respectively (Fig. 2, A–C). The addition of rsCD1e to the reaction mixture (+ manno + rsCD1e) did not alter the digestion profile of Ac2PIM6 (Fig. 2A), suggesting that CD1e does not assist their digestion by α-mannosidase. In contrast, rsCD1e improved the digestion of both Ac1PIM6 and PIM6, with Ac1PIM6 digested to Ac1PIM2 (Fig. 2B), and the proportion of the PIM2 glycoform resulting from PIM6 was greatly increased (Fig. 2C). To get closer to physiological conditions, purified PIM6 acyl forms were individually inserted into liposomes and used in similar digestion experiments. As previously observed with mixPIM6 (7), liposome inclusion greatly modified the type of Ac1PIM6 digested products generated by α-mannosidase in the presence of rsCD1e + manno + rsCD1e as almost no Ac1PIM2 were obtained (Fig. 2D). However, PIM6 in liposomes +manno + rsCD1e were almost completely degraded into PIM2 (Fig. 2E).
FIGURE 1.
Structure of PIM2 and PIM6 subfamilies (A) and MALDI-TOF MS spectra in negative-ion mode of purified PIM6 (B), Ac1PIM6 (C), and Ac2PIM6 (D) acyl forms. In monoacylated PIM (lyso-PIM), R1 represents acyl group; R2–R4 represents H. In diacylated PIM (PIM), R1 and R2 represent acyl group; R3 and R4 indicate H. In triacylated PIM (Ac1PIM), R1, R2, R3 indicate acyl group; R4 indicates H. In tetraacylated PIM (Ac2PIM), R1 to R4 indicates acyl group.
FIGURE 2.
PIM6 acylation degree determines their glycosidic processing. The different purified PIM6 acyl forms, Ac2PIM6 (A), Ac1PIM6 (B and D) and PIM6 (C and E), were digested by α-mannosidase in the absence (+manno) or in presence (+manno +rsCD1e) of rsCD1e. The relative abundance of the different PIM species generated (PIMX, where X indicates the number of mannosyl units) was determined by negative-ion mode MALDI-TOF MS analysis of the reaction mixture (mass spectra are shown in supplemental Fig. 1). Ac1PIM6 and PIM6 were either sonicated (B and C) or inserted in liposomes (D and E). The results presented here are from one representative experiment of three recorded spectra.
Taken together, these results demonstrate that degree of acylation of PIM6 determines their in vitro digestion by α-mannosidase and that when PIM6 is inserted into liposomes, rsCD1e selectively assists the digestion of diacylated forms. To understand this selectivity, we next attempted to uncover the mode of action of CD1e.
Lipid Transfer Protein Assists α-Mannosidase Digestion of PIM6
CD1e properties seem to be reminiscent to that of saposin assistance of the sphingolipid digestion (10). Saposins have been shown to increase lipid substrate accessibility for enzymatic digestion by either (i) membrane disorganization as for Sap-C or (ii) lipid transfer properties from membrane-to-membrane or from membrane to protein as for Sap-B (12).
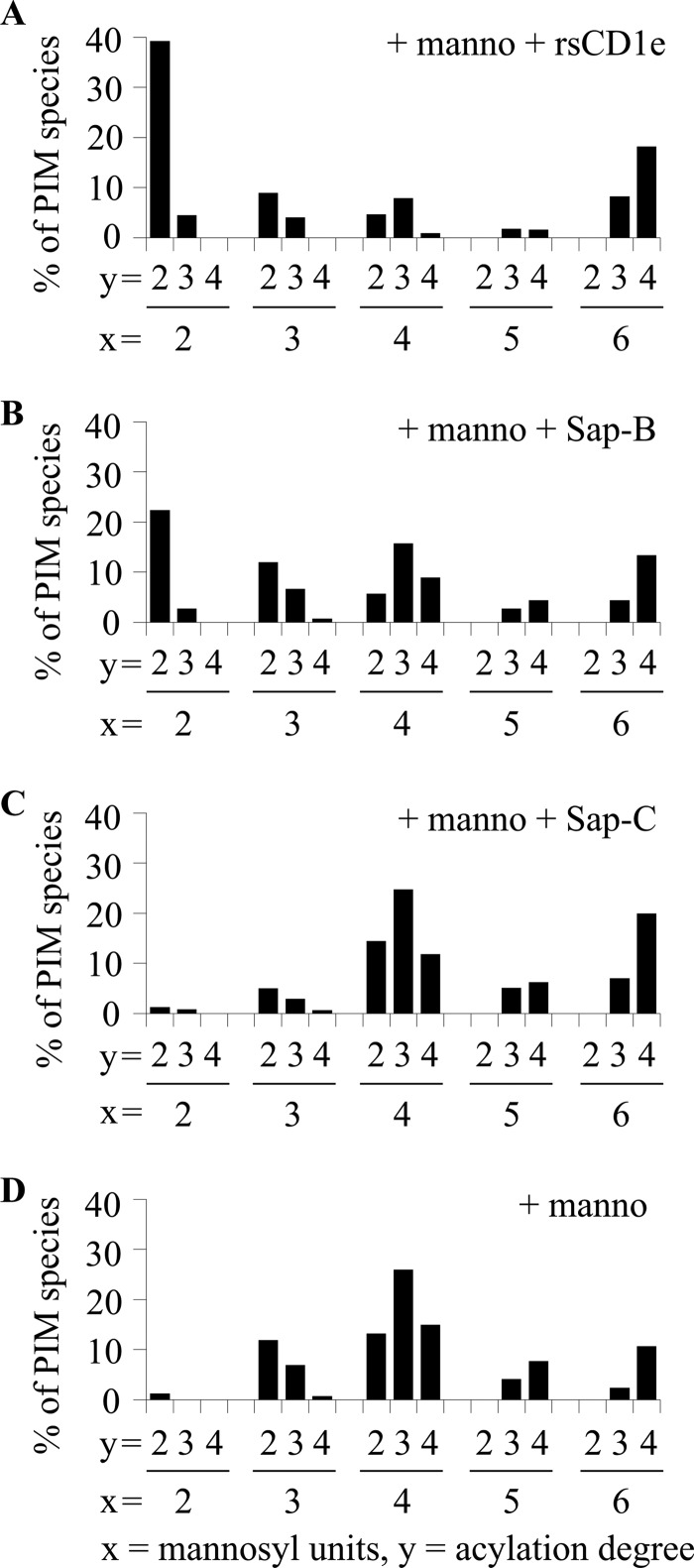
Thus, to get better clues on CD1e activity, we compared it with that of saposins in the in vitro α-mannosidase digestion of mixPIM6. In the presence of rsCD1e or Sap-B (Fig. 3A and B), we observed that mix-PIM6 were degraded up to diacylated PIM2. In contrast, Sap-C had no effect (Fig. 3C and D). These results suggest that the mechanism of CD1e as a lipid transfer protein (LTP) is similar to that of Sap-B but not of Sap-C.
FIGURE 3.
rsCD1e and Sap-B but not Sap-C promote in vitro α-mannosidase digestion of purified mixPIM6. Relative abundance of the different glycoforms deduced from the negative-ion mode MALDI-TOF MS analysis of Ac2PIM6 after hydrolysis by α-mannosidase in the presence of rsCD1e (A), Sap-B (B), Sap-C (C), or in the absence of any protein (D). x and y indicate mannosyl unit and fatty acid numbers in PIM species, respectively. The results presented here are from one representative experiment of three recorded spectra.
CD1e Selectively Transfers Diacylated PIM
To determine whether rsCD1e is able to promote membrane-to-membrane lipid transfer, we studied the transfer of a negatively charged phospholipid, POPS from POPS-containing donor liposomes (LUV-D) to acceptor liposomes devoid of POPS (LUV-A) in the presence of CD1e (22). Dynamic light scattering analysis revealed homogenous population of both liposomes with apparent hydrodynamic radius of ∼60 nm (supplemental Fig. S2). Accordingly, the images from microscopy allowed determining a diameter of ∼120 nm.
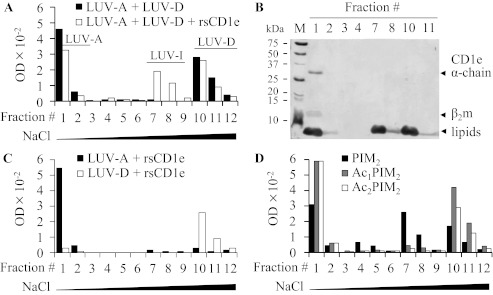
The transfer of POPS from donor to acceptor liposomes was expected to generate a new liposome population with an intermediate POPS content (LUV-I) that can be separated from LUV-A and LUV-D by anionic exchange chromatography (DEAE). The sodium chloride gradient was adjusted to provide an effective separation of LUV-A, LUV-D, and potential new LUV-I liposome populations. LUV-A liposomes, devoid of charged POPS, were not retained on the column and were eluted in fractions 1 and 2 (F1 and F2). However, the elution of LUV-D occurred in the presence of 1 m NaCl in F10, F11, and F12. Importantly, co-incubation of rsCD1e with a LUV-A/LUV-D mixture (ratio 1/1) yielded a new population of liposomes, which eluted with 60 mm NaCl in F7 and F8, indicated as LUV-I (Fig. 4A). This population was not observed when the liposomes were incubated with heat-denaturated rsCD1e or a non-relevant lipid binding protein, BSA (data not shown). The presence of POPS in F7 and F8 was confirmed by TLC analysis (data not shown). Moreover, because the molar ratio between rsCD1e and total phospholipids (POPS and POPC) is 1:714, it is unlikely that LUV-I results from a POPS depletion of LUV-D caused by the formation of CD1e-POPS stable complexes. SDS-PAGE analysis revealed that rsCD1e was present only in the first NaCl-free fractions (Fig. 4B), excluding the possibility that LUV-I resulted from a complex between CD1e and LUV-D. In addition, when LUV-A and LUV-D were individually incubated in the presence of rsCD1e (Fig. 4C), neither a modification of their respective chromatographic behavior nor the appearance of the intermediate population were noticed. Furthermore, dynamic light scattering measurements and transmission electron microscopy showed that the size of the liposomes was not modified during the experiment, excluding the possibility of fusion events (supplemental Fig. S3). Altogether, these data indicated that rsCD1e was directly responsible for the generation of LUV-I, resulting from the transfer of POPS from LUV-D to LUV-A. Similarly, rsCD1e was able to transfer PI (supplemental Fig. S4). However, when LUV-D were prepared using the mixture of PIM2 acyl forms (mixPIM2) instead of POPS or PI, LUV-I were hardly detectable. Because mixPIM2 is composed of ∼80% of tetra- and triacylated molecules (Ac2PIM2 and Ac1PIM2, respectively) (19), which might not be efficiently transferred by rsCD1e (Fig. 2), we tested purified PIM2 acyl forms and found that only diacylated PIM2 were transferred by rsCD1e (Fig. 4D). The presence of PIM2 in LUV-I was confirmed by MALDI-TOF MS and TLC analyses of F7 (data not shown). Moreover, LUV-I were not observed in absence of rsCD1e (supplemental Fig. S5).
FIGURE 4.
rsCD1e transfers diacylated PIM2. A, LUV-A (1 μmol of total lipids) and POPS-containing LUV-D (1 μmol of total lipids) were incubated with (open squares) or without (black filled squares) rsCD1e (2.1 nmol), and final liposomes populations were separated on DEAE chromatography using increasing concentrations of NaCl (fractions 1–3, no NaCl; 4–6, 12.5 mm NaCl; 7–9, 60 mm NaCl; 10–12, 1 m NaCl) and monitored by turbidity measurement at 600 nm of the collected fractions. B, SDS-PAGE analysis of DEAE fractions obtained in A in the presence of rsCD1e. Both proteins and lipids are stained. rsCD1e was eluted in F1 and lipids were present in F1, F2, F7, F8, F10, and F11. C, DEAE chromatography of LUV-A (black filled squares) or LUV-D (open squares) incubated separately with rsCD1e. Fractions 1–12 were eluted using NaCl concentrations as described in A. D, LUV-A and LUV-D containing Ac2PIM2 (open squares), Ac1PIM2 (gray filled squares) or PIM2 (black filled squares) were incubated with rsCD1e and separated on DEAE chromatography as described in A. One representative experiment of three independent ones is shown in A to D.
Altogether, our data support a LTP function for the rsCD1e protein. This activity is specific for diacylated forms of PIM and suggests that in antigen-presenting cells, CD1e might extract PIM6 from the membranes and present them to the α-mannosidase for processing of the glycosidic moiety.
CD1e Does Not Disturb Membrane Organization
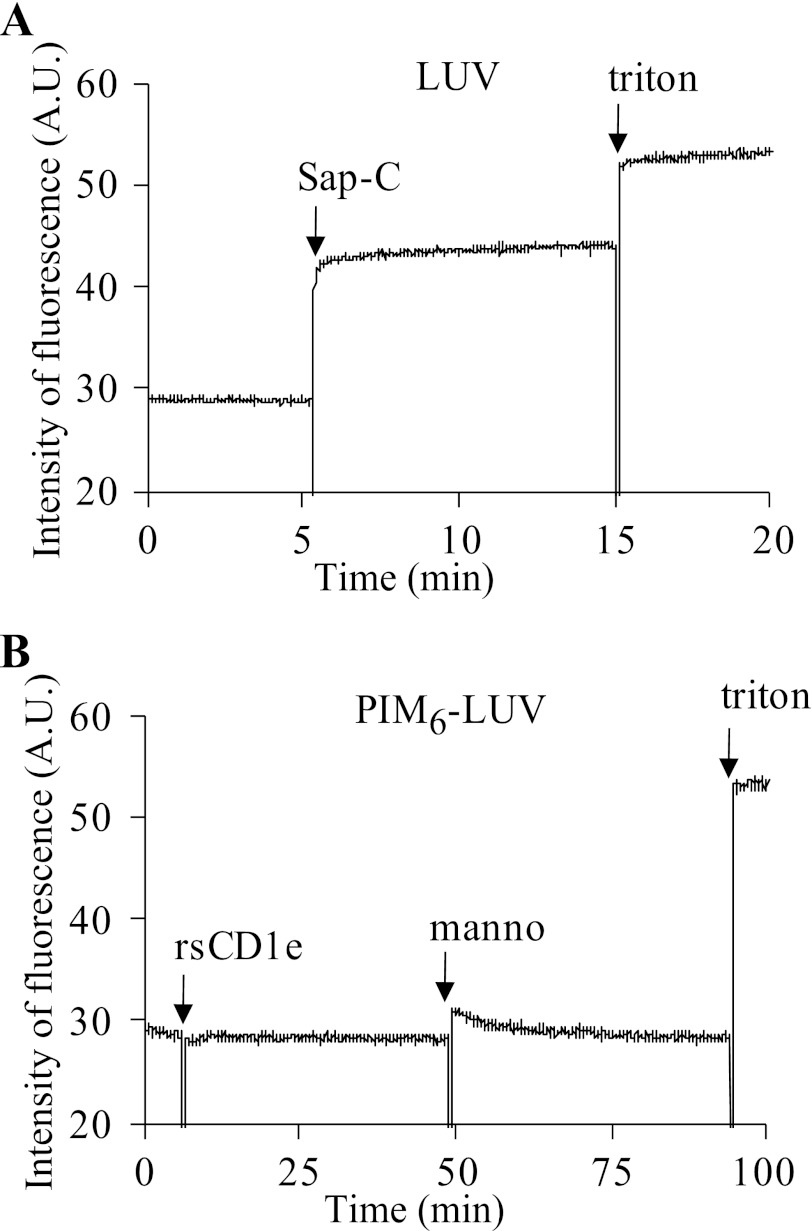
Membrane disorganization properties of rsCD1e were investigated by measuring the leakage of calcein entrapped within liposomes composed of POPC, cholesterol, and POPS in a 65/25/10 ratio, at an acidic pH. As observed by Vaccaro et al. (23), addition of Sap-C (0.5 nmol) disrupted the liposomes and induced calcein leakage, validating our experimental design (Fig. 5A). The addition of 0.21 nmol (data not shown) or 2.1 nmol rsCD1e did not induce any calcein release. We then tested whether PIM, known to bind to CD1e, caused membrane perturbation. Neither the use of liposomes containing 4% of mixPIM6 nor the addition of α-mannosidase promoted calcein release (Fig. 5B). Moreover, varying experimental parameters, such as liposome size (SUV or LUV), pH (acidic or neutral), rsCD1e concentration or incubation time (few minutes to four hours) did not make any difference, strongly suggesting that CD1e does not harbor membrane disruption properties, at least in this in vitro assay.
FIGURE 5.
rsCD1e does not alter model membrane integrity. LUV were loaded with 20 mm of calcein, and calcein release was measured by recording induced fluorescence at 517 nm. A, calcein release from LUV composed of POPC/cholesterol/POPS, 65/25/10 (40 nmol of total lipids) in the presence of Sap-C (0.5 nmol) at pH 4.7. B, calcein release from LUV composed of POPC/cholesterol/POPS/PIM6, 65/25/10/4 (40 nmol of total lipids) in the presence of rsCD1e (2.1 nmol), with or without α-mannosidase (manno; 0.44 unit) at pH 4.7. The maximum fluorescence, indicating complete liposome disruption, was observed after addition of 10 μl of Triton X-100 (triton; 10%). A.U., arbitrary units.
CD1e Loads Liposome-inserted PI onto CD1b
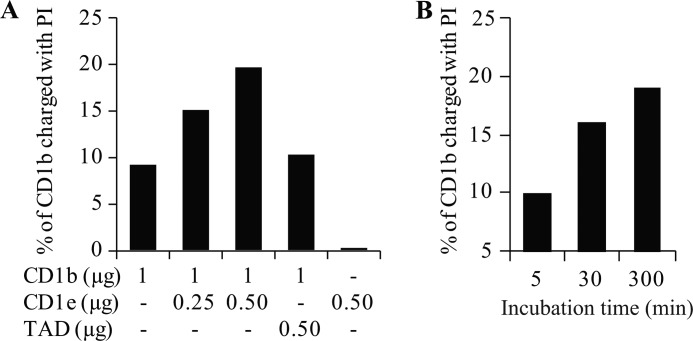
PIM are carried by sCD1e during processing but are ultimately presented to T cells by CD1b molecules. To test whether CD1e was able to transfer liposome-inserted PIM onto CD1b, liposome-inserted radiolabeled diacylated phosphatidyl-inositol ([3H]PI) was used as a lipid antigen model and radiolabeled lipid loading onto rsCD1b was measured in the presence and absence of rsCD1e. After a 5-h incubation, 10% of rsCD1b proteins were loaded by [3H]PI, whereas addition of rsCD1e allowed this proportion to increase up to 20% (Fig. 6A). PI loading onto CD1b by CD1e was both CD1e concentration- and time-dependent (Fig. 6B). In contrast, an irrelevant protein (transactivation domain, N Oct-3 transactivation domain) showing a pI similar to CD1e had no effect (Fig. 6A). These results indicate that CD1e has the capacity to transfer liposome-inserted processed antigen onto CD1b.
FIGURE 6.
rsCD1e promotes loading of liposome-inserted phosphatidyl-inositol onto rsCD1b. A- [3H]-PI-containing liposomes (LUV-[3H]-PI; 3.6 nmol total lipids) were incubated for 5 h with rsCD1b (1 μg; 19.6 pmol) and with or without rsCD1e (0.25 or 0.5 μg; 6.6 and 13.2 pmol, respectively). rsCD1b and rsCD1e were separated on isoelectric focusing gel, and radioactivity associated to rsCD1b was determined. LUV-[3H]-PI were incubated with rsCD1b and 0.5 μg of N Oct-3 transactivation domain (TAD) as control. One of three independent experiments is shown. B, LUV-[3H]-PI were incubated with rsCD1b and rsCD1e (0.5 μg) during different periods of time and rsCD1b-associated radioactivity was measured as described in A.
DISCUSSION
Antigen presentation to T cells is a multistep process, including antigen uptake, editing/processing, and loading onto presenting molecules followed by T cell antigen receptor recognition of the antigenic complex. Although accessory proteins and molecular mechanisms involved in the presentation of peptide antigens have been extensively studied, details on lipid antigen presentation by the CD1 proteins are not yet fully understood. Lipid antigens taken in charge by antigen-presenting cells are embedded in internal vesicles inside lysosomes. Their hydrophobic nature causes a series of biophysical constraints to their presentation because the different steps preceding CD1-lipid complex formation occur at the interface between hydrophobic and hydrosoluble environments. The stimulation of CD1b-restricted T cells by PIM6, a family of mycobacterial antigens, requires a partial digestion of the PIM oligomannoside moiety by an α-mannosidase and CD1e is absolutely required for antigen processing and T cell stimulation (7). We previously found that rsCD1e binds glycolipid antigens and assists in vitro enzymatic oligosaccharide hydrolysis of PIM6. In all of these previous studies, the mechanism by which CD1e mediates these effects remained unknown. Here, we show that CD1e behaves as a LTP capable of facilitating selective transport of unique acylated forms of PIM. Indeed, PIM structure not only contains a variable number of mannosyl residues, but also a variable number (one to four) of acyl chains (19, 21). Thus, this family of antigens represents an ideal model to investigate the functions of CD1e and its eventual specificity for individual PIM species. Using an in vitro assay in which purified PIM6 acyl forms were digested by α-mannosidase in the presence of rsCD1e, we found that the degree of acylation of PIM impacts upon their CD1e-assisted digestion. When the different acyl forms of PIM6 were inserted into liposomes, rsCD1e showed a selective assistance in the processing of diacylated forms only. These findings are in apparent contrast with our previous report showing that rsCD1e may form stable complexes with a wide range of pure two- or three-tailed lipids (9) and with the finding that CD1e also facilitates transfer of triacylated PIM. However, in all these previous experiments, the binding and transfer capacity of CD1e was tested with lipids in solution. Here, the selectivity toward the diacylated forms of PIM instead was observed in the more physiological context of lipids embedded in membranes. These important differences highlight the relevance of membrane localization of lipid antigens on their processing and transfer to other membranes.
Comparing the activity of rsCD1e with that of saposins in the in vitro α-mannosidase digestion assay, we observed that CD1e and Sap-B, a LTP expressed in all cell types, display a similar in vitro activity in terms of assistance of α-mannosidase digestion of PIM. In contrast, Sap-C was completely ineffective in this in vitro digestion assay. Recent studies comparing the mode of action of Sap-B and Sap-C showed that the two investigated saposins use different strategies for structurally diverse lipid antigen presentation (24). Although Sap-B forms soluble saposin-lipid complexes and can directly load CD1 proteins, Sap-C inserts itself directly into the membrane bilayer, thereby disrupting the tightly packed lipid bilayer and thus facilitating antigen loading onto the presenting CD1 protein. Our findings suggest that the activity of CD1e is more similar to that of Sap-B. Indeed, CD1e selectively transfers diacylated PIM from donor to acceptor liposomes, resembling GM2AP and Sap-B (12, 22, 25) without affecting vesicle structure and membrane fusion, which are instead hallmarks of Sap-C and Sap-D (23, 26). However, CD1e and Sap-B do not have completely overlapping functions as suggested by the finding that CD1e is absolutely required for PIM6 antigenicity (7). CD1e can be viewed as an LTP exposing the saccharidic units of PIM to the enzyme for degradation. Thus, in vivo, Sap-B could not replace per se CD1e in the context of α-mannosidase-dependent PIM processing. Winau et al. (16) have previously shown that Sap-C extracts lipid antigens from membranes and binds to CD1b, promoting loading of the mycobacterial antigenic lipids on CD1b. Therefore, we cannot exclude a participation of other saposins such as Sap-C in the transfer of processed PIM onto CD1b in vivo.
An important issue is whether CD1e makes cognate interactions with other CD1 molecules and with hydrolases involved in glycolipid antigen processing. Following a series of experiments, it was not possible to detect contacts between CD1e and other CD1 molecules.7 Whether CD1e directly interacts with α-mannosidase remains a point to further investigate. A precedent is the case of lysosomal degradation of GM2 by β-hexosaminidase A, in which GM2AP was shown to be an essential cofactor (27). GM2AP has an enzyme-binding region (28), which is important for its function, suggesting that GM2AP participates in GM2 degradation by penetrating into the hydrophobic region of the membrane structure, lifting GM2 out of the lipid plane and interacting specifically with the β-hexosaminidase A. Whether CD1e specifically interacts with the α-mannosidase, making the CD1e-PIM complex the true enzyme substrate, will be explored in future studies.
Among all PIM6 acyl forms, CD1e selects only diacylated PIM. This step may act as a checkpoint in CD1e-dependent antigen processing and subsequent presentation to T cells. Among all PIM6 acyl forms, only diacylated PIM might become antigenic and be presented by CD1b as a consequence of being the only ones transferred by CD1e and efficiently processed by α-mannosidase. This possibility raises the question of how tri- and tetra-acylated PIM species, which are the most abundant acyl forms present in the mycobacterial envelope, become antigenic. One possibility is that they are first processed by lipases that generate diacylated forms and then are assisted by CD1e for the trimming of mannose residues. This hypothesis is supported by current literature showing that all the antigenic structures presented by CD1b contain a maximum of two fatty acyl chains (2). Several lysosomal glycosidases have been shown to be involved in glycolipid processing (7, 29, 30), but lipases processing multiacylated glycolipids have never been identified. Whether CD1e also facilitates lipase-mediated processing of multiacylated lipid antigens remains an open possibility and PIM antigen recognition may represent an ideal model to investigate the mechanisms of lipolytic processing.
Supplementary Material
Acknowledgments
We thank S. Mazères (IPBS, Toulouse, France) for fluorescence experiments and N. Benmeradi (IBCG, Toulouse, France) for microscopic analysis of liposomes. We are also grateful to professor E. Clottes for kind gift of transactivation domain protein, Dr. C. Lebrun for discussions, and Drs. J. Nigou and D. Ly for reading the manuscript.
This work was supported by the Centre National de la Recherche Scientifique, the Agence Nationale de la Recherche Emergence (ANR-05-MIIM-006), the Etablissement Français du Sang-Alsace, European Community's Seventh Framework Program Grant 241745, and Swiss National Foundation Grants 3100AO-122464/1 and Sinergia CRS133-124819.

This article contains supplemental Table S1 and Figs. S1–S5.
PIM is used to describe the global family of PIM that carries one to four fatty acids and one to six Manp residues. In AcXPIMY, X refers to the number of acyl groups esterified to available hydroxyls on the Manp or myo-inositol residues, y refers to the number of Manp residues, e.g. Ac1PIM6 corresponds to triacylated PIM6, the phosphatidyl-myo-inositol hexamannoside PIM6 carrying two acyl groups attached to the glycerol (the diacylglycerol substituent) and one acyl group esterified to the Manp residue. Inherent in the abbreviation for PI is the diacylglycerol unit. Accordingly, natural acyl form mixtures of PIM2 or PIM6 will be designated by mixPIM2 and mixPIM6.
G. de Libero, L. Mori, and H. de la Salle, unpublished data.
- sCD1e
- soluble form of CD1e
- rsCD1e
- recombinant soluble form of CD1e
- GM2AP
- GM2-activator protein
- LTP
- lipid transfer protein
- LUV
- large unilamellar vesicle(s)
- myo-Ins
- myo-inositol
- PIM
- phosphatidyl-myo-inositol mannoside(s)
- PIM3
- phosphatidyl-myo-inositol trimannoside(s)
- Sap
- saposin
- SUV
- small unilamellar vesicle(s)
- PI
- phosphatidyl-myo-inositol
- POPS
- 1-palmitoyl-2-oleyl-sn-glycero-3-phosphoserine.
REFERENCES
- 1. Barral D. C., Brenner M. B. (2007) CD1 antigen presentation: How it works. Nat. Rev. Immunol. 7, 929–941 [DOI] [PubMed] [Google Scholar]
- 2. De Libero G., Collmann A., Mori L. (2009) The cellular and biochemical rules of lipid antigen presentation. Eur. J. Immunol. 39, 2648–2656 [DOI] [PubMed] [Google Scholar]
- 3. Dougan S. K., Kaser A., Blumberg R. S. (2007) CD1 expression on antigen-presenting cells. Curr. Top Microbiol. Immunol. 314, 113–141 [DOI] [PubMed] [Google Scholar]
- 4. Sugita M., Grant E. P., van Donselaar E., Hsu V. W., Rogers R. A., Peters P. J., Brenner M. B. (1999) Separate pathways for antigen presentation by CD1 molecules. Immunity 11, 743–752 [DOI] [PubMed] [Google Scholar]
- 5. Angenieux C., Salamero J., Fricker D., Cazenave J. P., Goud B., Hanau D., de La Salle H. (2000) Characterization of CD1e, a third type of CD1 molecule expressed in dendritic cells. J. Biol. Chem. 275, 37757–37764 [DOI] [PubMed] [Google Scholar]
- 6. Angénieux C., Fraisier V., Maître B., Racine V., van der Wel N., Fricker D., Proamer F., Sachse M., Cazenave J. P., Peters P., Goud B., Hanau D., Sibarita J. B., Salamero J., de la Salle H. (2005) The cellular pathway of CD1e in immature and maturing dendritic cells. Traffic 6, 286–302 [DOI] [PubMed] [Google Scholar]
- 7. de la Salle H., Mariotti S., Angenieux C., Gilleron M., Garcia-Alles L. F., Malm D., Berg T., Paoletti S., Maître B., Mourey L., Salamero J., Cazenave J. P., Hanau D., Mori L., Puzo G., De Libero G. (2005) Assistance of microbial glycolipid antigen processing by CD1e. Science 310, 1321–1324 [DOI] [PubMed] [Google Scholar]
- 8. Facciotti F., Cavallari M., Angénieux C., Garcia-Alles L. F., Signorino-Gelo F., Angman L., Gilleron M., Prandi J., Puzo G., Panza L., Xia C., Wang P. G., Dellabona P., Casorati G., Porcelli S. A., de la Salle H., Mori L., De Libero G. (2011) Fine tuning by human CD1e of lipid-specific immune responses. Proc. Natl. Acad. Sci. U.S.A. 108, 14228–14233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Garcia-Alles L. F., Giacometti G., Versluis C., Maveyraud L., de Paepe D., Guiard J., Tranier S., Gilleron M., Prandi J., Hanau D., Heck A. J., Mori L., De Libero G., Puzo G., Mourey L., de la Salle H. (2011) Crystal structure of human CD1e reveals a groove suited for lipid-exchange processes. Proc. Natl. Acad. Sci. U.S.A. 108, 13230–13235 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Schulze H., Kolter T., Sandhoff K. (2009) Principles of lysosomal membrane degradation: Cellular topology and biochemistry of lysosomal lipid degradation. Biochim. Biophys. Acta 1793, 674–683 [DOI] [PubMed] [Google Scholar]
- 11. Vaccaro A. M., Tatti M., Ciaffoni F., Salvioli R., Maras B., Barca A. (1993) Function of saposin C in the reconstitution of glucosylceramidase by phosphatidylserine liposomes. FEBS Lett. 336, 159–162 [DOI] [PubMed] [Google Scholar]
- 12. Ciaffoni F., Tatti M., Boe A., Salvioli R., Fluharty A., Sonnino S., Vaccaro A. M. (2006) Saposin B binds and transfers phospholipids. J. Lipid Res. 47, 1045–1053 [DOI] [PubMed] [Google Scholar]
- 13. Kang S. J., Cresswell P. (2004) Saposins facilitate CD1d-restricted presentation of an exogenous lipid antigen to T cells. Nat. Immunol. 5, 175–181 [DOI] [PubMed] [Google Scholar]
- 14. Zhou D., Cantu C., 3rd, Sagiv Y., Schrantz N., Kulkarni A. B., Qi X., Mahuran D. J., Morales C. R., Grabowski G. A., Benlagha K., Savage P., Bendelac A., Teyton L. (2004) Editing of CD1d-bound lipid antigens by endosomal lipid transfer proteins. Science 303, 523–527 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Yuan W., Qi X., Tsang P., Kang S. J., Illarionov P. A., Besra G. S., Gumperz J., Cresswell P. (2007) Saposin B is the dominant saposin that facilitates lipid binding to human CD1d molecules. Proc. Natl. Acad. Sci. U.S.A. 104, 5551–5556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Winau F., Schwierzeck V., Hurwitz R., Remmel N., Sieling P. A., Modlin R. L., Porcelli S. A., Brinkmann V., Sugita M., Sandhoff K., Kaufmann S. H., Schaible U. E. (2004) Saposin C is required for lipid presentation by human CD1b. Nat. Immunol. 5, 169–174 [DOI] [PubMed] [Google Scholar]
- 17. Gilleron M., Jackson M., Nigou J., Puzo G. (2008) Structure, biosynthesis, and activities of the phosphatidyl-myo-inositol-based lipoglycans in The Mycobacterial Cell Envelope (Daffé M., Reyrat J. M., ed) pp. 75–105, ASM Press, Washington D.C [Google Scholar]
- 18. Folch J., Lees M., Sloane Stanley G. H. (1957) A simple method for the isolation and purification of total lipides from animal tissues. J. Biol. Chem. 226, 497–509 [PubMed] [Google Scholar]
- 19. Gilleron M., Ronet C., Mempel M., Monsarrat B., Gachelin G., Puzo G. (2001) Acylation state of the phosphatidylinositol mannosides from Mycobacterium bovis bacillus Calmette Guerin and ability to induce granuloma and recruit natural killer T cells. J. Biol. Chem. 276, 34896–34904 [DOI] [PubMed] [Google Scholar]
- 20. Gilleron M., Nigou J., Cahuzac B., Puzo G. (1999) Structural study of the lipomannans from Mycobacterium bovis BCG: Characterization of multiacylated forms of the phosphatidyl-myo-inositol anchor. J. Mol. Biol. 285, 2147–2160 [DOI] [PubMed] [Google Scholar]
- 21. Gilleron M., Quesniaux V. F., Puzo G. (2003) Acylation state of the phosphatidylinositol hexamannosides from Mycobacterium bovis bacillus Calmette Guerin and Mycobacterium tuberculosis H37Rv and its implication in Toll-like receptor response. J. Biol. Chem. 278, 29880–29889 [DOI] [PubMed] [Google Scholar]
- 22. Vogel A., Schwarzmann G., Sandhoff K. (1991) Glycosphingolipid specificity of the human sulfatide activator protein. Eur. J. Biochem. 200, 591–597 [DOI] [PubMed] [Google Scholar]
- 23. Vaccaro A. M., Ciaffoni F., Tatti M., Salvioli R., Barca A., Tognozzi D., Scerch C. (1995) pH-dependent conformational properties of saposins and their interactions with phospholipid membranes. J. Biol. Chem. 270, 30576–30580 [DOI] [PubMed] [Google Scholar]
- 24. León L., Tatituri R. V., Grenha R., Sun Y., Barral D. C., Minnaard A. J., Bhowruth V., Veerapen N., Besra G. S., Kasmar A., Peng W., Moody D. B., Grabowski G. A., Brenner M. B. (2012) Saposins utilize two strategies for lipid transfer and CD1 antigen presentation. Proc. Natl. Acad. Sci. U.S.A. 109, 4357–4364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Conzelmann E., Burg J., Stephan G., Sandhoff K. (1982) Complexing of glycolipids and their transfer between membranes by the activator protein for degradation of lysosomal ganglioside GM2. Eur. J. Biochem. 123, 455–464 [DOI] [PubMed] [Google Scholar]
- 26. Ciaffoni F., Salvioli R., Tatti M., Arancia G., Crateri P., Vaccaro A. M. (2001) Saposin D solubilizes anionic phospholipid-containing membranes. J. Biol. Chem. 276, 31583–31589 [DOI] [PubMed] [Google Scholar]
- 27. Conzelmann E., Sandhoff K. (1979) Purification and characterization of an activator protein for the degradation of glycolipids GM2 and GA2 by hexosaminidase A. Hoppe-Seylers Z. Physiol. Chem. 360, 1837–1849 [DOI] [PubMed] [Google Scholar]
- 28. Wendeler M., Werth N., Maier T., Schwarzmann G., Kolter T., Schoeniger M., Hoffmann D., Lemm T., Saenger W., Sandhoff K. (2006) The enzyme-binding region of human GM2-activator protein. FEBS J. 273, 982–991 [DOI] [PubMed] [Google Scholar]
- 29. Prigozy T. I., Naidenko O., Qasba P., Elewaut D., Brossay L., Khurana A., Natori T., Koezuka Y., Kulkarni A., Kronenberg M. (2001) Glycolipid antigen processing for presentation by CD1d molecules. Science 291, 664–667 [DOI] [PubMed] [Google Scholar]
- 30. Zhou D., Mattner J., Cantu C., 3rd, Schrantz N., Yin N., Gao Y., Sagiv Y., Hudspeth K., Wu Y. P., Yamashita T., Teneberg S., Wang D., Proia R. L., Levery S. B., Savage P. B., Teyton L., Bendelac A. (2004) Lysosomal glycosphingolipid recognition by NKT cells. Science 306, 1786–1789 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.