Background: Recent results raised doubts regarding the earlier proposed C-23 hydroxylase function of CYP90A1/CPD in brassinosteroid biosynthesis.
Results: The enzymatic role of CYP90A1/CPD is determined by analytical, genetic, and direct biochemical approaches.
Conclusion: CYP90A1 catalyzes C-3 oxidation of early brassinosteroid intermediates.
Significance: Our results highlight the primary role of the campestanol-independent pathway in brassinosteroid biosynthesis.
Keywords: Cytochrome P450, Enzyme Kinetics, Plant Biochemistry, Plant Hormones, Plant Molecular Biology, CPD, CYP90A, Brassinosteroid, Campestanol-independent Pathway, Steroid C-3 Oxidation
Abstract
Brassinosteroids (BRs) are steroidal phytohormones that regulate plant growth and development. Whereas in Arabidopsis the network-like routes of BR biosynthesis have been elucidated in considerable detail, the roles of some of the biosynthetic enzymes and their participation in the different subpathways remained to be clarified. We investigated the function of the cytochrome P450 monooxygenase CYP90A1/CPD, which earlier had been proposed to act as a BR C-23 hydroxylase. Our GC-MS and genetic analyses demonstrated that the cpd mutation arrests BR synthesis upstream of the DET2-mediated 5α reduction step and that overexpression of the C-23 hydroxylase CYP90C1 does not alleviate BR deficiency in the cpd mutant. In line with these results, we found that CYP90A1/CPD heterologously expressed in a baculovirus-insect cell system catalyzes C-3 oxidation of the early BR intermediates (22S)-22-hydroxycampesterol and (22R,23R)-22,23-dihydroxycampesterol, as well as of 6-deoxocathasterone and 6-deoxoteasterone. Enzyme kinetic data of CYP90A1/CPD and DET2, together with those of the earlier studied CYP90B1, CYP90C1, and CYP90D1, suggest that BR biosynthesis proceeds mainly via the campestanol-independent pathway.
Introduction
Brassinosteroids (BRs)4 are steroidal phytohormones that show structural similarity to ecdysone, the molting hormone of insects (1). More than 70 naturally occurring BRs have been identified in a wide range of plant species (2–4). BRs regulate various important physiological and developmental processes, such as cell elongation and division, pollen fertility, photomorphogenesis, differentiation of vascular elements, and stress resistance (5, 6).
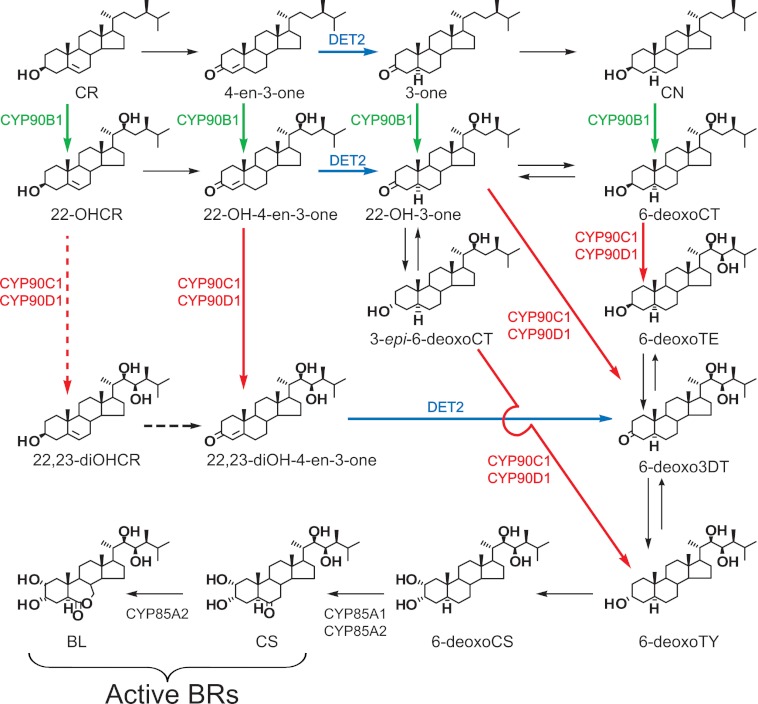
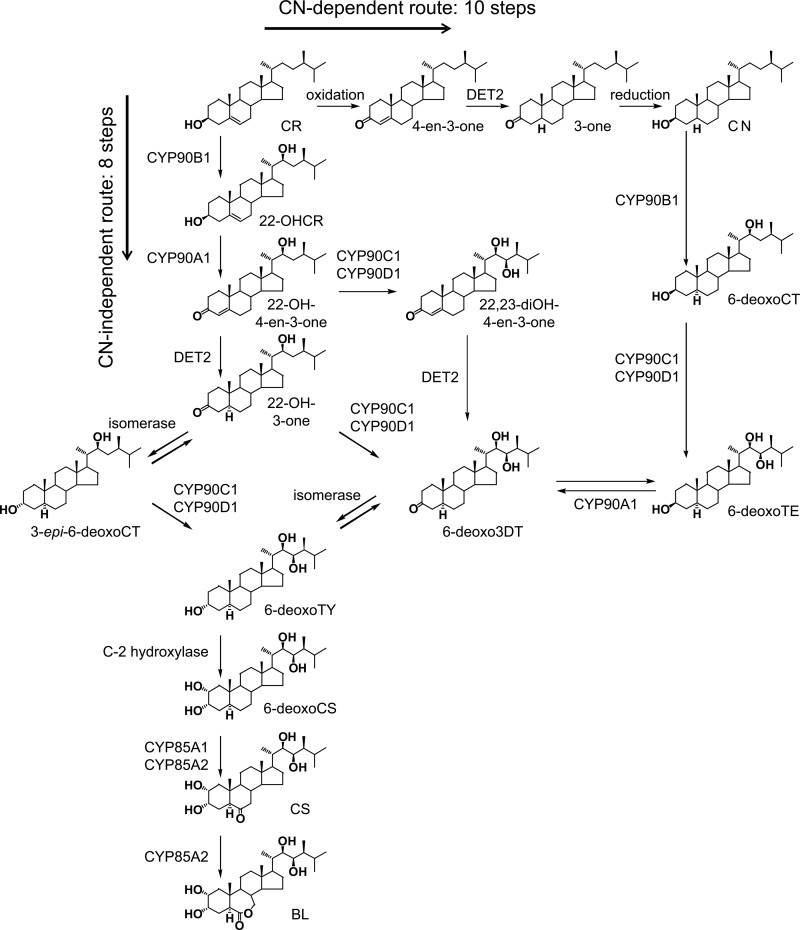
Recent molecular genetic studies of BR-deficient mutants resulted in the detailed characterization of several genes of BR biosynthesis that proceeds from campesterol (CR) to brassinolide (BL), the biologically most active BR (Fig. 1). The structural versatility of the physiologically important C28 BRs, carrying α-methyl at the C-24 side-chain position, results from oxygen moieties at C-3 and/or C-2 and C-6 of the steroid A/B-rings and at the C-22 and C-23 positions of the side chain (4).
FIGURE 1.
BR biosynthetic pathways, including the late C-6, early C-22, and C-23 oxidation routes. The subpathway indicated by dotted lines has not been verified in planta. BL, brassinolide; CN, campestanol; CR, campesterol; CS, castasterone; CT, cathasterone; TE, teasterone; TY, typhasterol; 22-OHCR, (22S)-22-hydroxycampesterol; 22-OH-3-one, (22S)-22-hydroxy-5α-campestan-3-one; 22-OH-4-en-3-one, (22S)-22-hydroxycampest-4-en-3-one; 22,23-diOHCR, (22R,23R)-22,23-dihydroxycampesterol; 22,23-diOH-4-en-3-one, (22R,23R)-22,23-dihydroxycampest-4-en-3-one; 3-epi-6-deoxoCT, 3-epi-6-deoxocathasterone; 3-one, 5α-campestan-3-one; 4-en-3-one, campest-4-en-3-one; 6-deoxoCS, 6-deoxocastasterone; 6-deoxoCT, 6-deoxocathasterone; 6-deoxoTE, 6-deoxoteasterone; 6-deoxoTY, 6-deoxotyphasterol; 6-deoxo3DT, 3-dehydro-6-deoxoteasterone.
So far, numerous BR-deficient dwarf mutants have been identified and characterized in Arabidopsis, pea, tomato, rice, and a number of other species. The de-etiolated 2 (det2) mutant of Arabidopsis has dwarf phenotype with de-etiolation and is defective in the steroid 5α-reduction step (7, 8) early in the BR pathway (Fig. 1). Several other mutants impaired in hydroxylation and oxidation reactions of BR biosynthesis carry lesions in genes that encode cytochromes P450 (P450s). In Arabidopsis the constitutive photomorphogenesis and dwarfism (cpd), dwarf 4 (dwf4), and rotundifolia 3 (rot3) mutants were shown to be deficient in CYP90A1/CPD, CYP90B1, and CYP90C1, respectively (9–12).
In most cases, the functions of these P450s were studied using indirect methods, such as gas chromatography-mass spectrometry (GC-MS) analysis of the endogenous BR content or phenotypic rescue of the mutants by feeding with BR intermediates. For instance, the cpd mutant, lacking CYP90A1/CPD, was rescued by the 22,23-dihydroxylated BRs teasterone, 3-dehydroteasterone, typhasterol, castasterone (CS), and BL but not by the 22-hydroxylated cathasterone (CT), which led to the suggestion that the mutation impairs C-23 hydroxylation (9). Recently, however, we obtained biochemical and genetic evidence that in Arabidopsis this hydroxylation reaction is catalyzed by the redundantly functioning CYP90C1 and CYP90D1 enzymes (13). As cyp90c1/cyp90d1 double null mutants, carrying a fully functional CYP90A1/CPD gene, show severe BR-deficient dwarf phenotypes (13) similar to that of cpd, this was a clear indication that the role of CYP90A1/CPD should be different from those of CYP90C1 and CYP90D1 and other than C-23 hydroxylation.
BR synthesis occurs along a metabolic grid (Fig. 1) where broad substrate specificities of the participating enzymes allow metabolite flow via multiple pathways (13–15). Additionally, most BR biosynthetic genes are up-regulated in BR-deficient mutants (16, 17), thereby increasing the role of subsidiary reaction routes. Because of these intricacies, the use of indirect methods for the functional characterization of BR biosynthetic genes can be difficult and misleading, as has been the case with the cyp90c1 and cyp90d1 mutants (12, 13).
We carried out biochemical and genetic analyses to clarify the enzymatic and physiological functions of Arabidopsis CYP90A1/CPD. Our results show that the cpd mutant accumulates high amounts of 22-OHCR, indicating a lesion early in the pathway. Furthermore, the metabolic consequences of the cpd/det2 double mutation are very similar to those of cpd, demonstrating that CYP90A1/CPD acts upstream of DET2 in the synthesis route. The biochemical characterization of heterologously expressed CYP90A1/CPD revealed that this enzyme does not catalyze C-23 hydroxylation of any C-22-hydroxylated BR forms, but it can oxidize multiple 3β-hydroxylated intermediates to their corresponding 3-dehydro derivatives. It preferentially mediates the conversion of (22S)-22-hydroxycampest-4-en-3-one (22-OH-4-en-3-one), which is an important substrate of the DET2 reaction. CYP90A1/CPD also catalyzes the 6-deoxoCT → (22S)-22-hydroxy-5α-campestan-3-one (22-OH-3-one), 6-deoxoTE → 3-dehydro-6-deoxoteasterone (6-deoxo3DT), and to a lesser extent the 22,23-diOHCR → (22R,23R)-22,23-dihydroxycampest-4-en-3-one (22,23-diOH-4-en-3-one) reactions, but it does not convert 3α-hydroxylated BRs to 3-dehydro intermediates. Our reaction kinetic data suggest that CYP90A1/CPD-driven BR synthesis proceeds mainly via the early C-22 hydroxylation pathway, facilitating the use of the campestanol (CN)-independent route, which allows direct conversion of early 22-hydroxylated intermediates to 6-deoxo3DT and 6-deoxotyphasterol (6-deoxoTY) via C-23 hydroxylation (13).
EXPERIMENTAL PROCEDURES
Chemicals
CR was obtained from Tama Biochemical Co. (Tokyo, Japan); BL and CS were purchased from Fuji Chemical Industries, Ltd. (Toyama, Japan). CN, 22-OHCR, 22-OH-4-en-3-one, 22-OH-3-one, 3-epi-6-deoxocathasterone (3-epi-6-deoxoCT), 22,23-diOHCR, and 22,23-diOH-4-en-3-one were chemically synthesized (13). All other chemicals, analytical grade, were purchased from Sigma, Nacalai Tesque (Kyoto, Japan), and Wako Pure Chemical Industries (Osaka, Japan).
Plant Material and Growth Conditions
Arabidopsis thaliana ecotype Columbia (Col-0) and all mutant lines were grown from surface-sterilized seeds on MS medium (Duchefa, Haarlem, The Netherlands) supplemented with 1% (w/v) sucrose and 0.2% (w/v) Phytagel (Sigma). The plants were raised in controlled environment chambers (SANYO Electronic Co., Osaka, Japan) under alternating 12-h white light (60 μmol m−2 s−1) and 12-h dark photoperiods at 22 °C. For genetic crossing and seed production, plants were grown in soil under controlled greenhouse conditions, at 22 ± 2 °C air temperature, and under 8-h light/16-h dark periods for the first 3 weeks and then 16-h light/8-h dark photoperiods during flower induction and reproductive development. During generation of the cpd/det2 double mutant, the presence of cpd mutation was checked by selection with 15 mg liter−1 hygromycin (Calbiochem), whereas that of the det2-1 allele was verified by restriction fragment length polymorphism analysis (18).
Overexpression of CYP90C1 in cpd Background
For CPD-specific expression of CYP90C1 in cpd mutant plants, the promoter + 5′-UTR region of the CPD gene (−968 to −5 relative to the translational start) was fused to the coding sequence + 3′-UTR part of the CYP90C1 cDNA and then cloned in a glufosinate resistance-conferring version of the pPCV812 T-DNA-based binary vector (19). Transgenic lines expressing the CPD-promoter:CYP90C1 fusion were generated by Agrobacterium-mediated flower dip transformation (20) of cpd/+ heterozygous plants. Progeny lines homozygous for both cpd and the transgene were obtained by self-pollination. Transcript levels of CYP90C1, CPD, and the internal control UBQ10 (At4g05320) were detected by RT-PCRs as described earlier (13).
Determination of the Endogenous BR Content
All plant material, ranging from 15 to 70 g fresh weight, was grown under sterile conditions for 1 month, then collected in liquid nitrogen, and subjected to lyophilization. Quantitative BR analyses of cpd and Col-0 plants, as well as of the cpd, det2, and cpd/det2 mutants, were performed using procedures described by Nomura et al. (21) and Fujioka et al. (14). Methanol extracts were spiked with [2H6]BR internal standards before solvent evaporation. The resulting aqueous residues were partitioned between ethyl acetate and 0.5 m dipotassium phosphate solution. Following purification on a charcoal column (chromatography grade, Wako Pure Chemicals, Wako, Japan), biologically active fractions were subjected to Sephadex LH-20 column chromatography using methanol/chloroform (4:1) as mobile phase. Fractions 37–40 were combined and subjected to reversed phase HPLC using an octadecyl column (8 × 250 mm, 5 μm particles). Elution was conducted at 39 °C and 2.5 ml min−1 flow rate with the following gradient program: 0–15 (min) (45% MeCN), 15–40 (min) (45–100% MeCN), and 40 (min) (100% MeCN). Typical elution profiles were as follows: fractions 14–16 (BL), fractions 21–23 (CS), fractions 36–38 (6-deoxoCS), fractions 40 and 41 (22,23-diOHCR), fraction 42 (22,23-OH-4-en-3-one), fractions 43–45 (6-deoxoTE and 6-deoxo3DT), fractions 46–48 (6-deoxoTY, 22-OHCR, and 22-OH-4-en-3-one), fractions 49 and 50 (6-deoxoCT and 22-OH-3-one), and fractions 51–53 (3-epi-6-deoxoCT).
GC-MS analyses were carried out using a Shimadzu QP2010 instrument fitted with a DB-5 column (0.25-mm inner diameter × 15 m; 0.25-μm film thickness; Agilent Technologies Inc., Palo Alto, CA). Prior to GC-MS analysis, BRs were converted to either methaneboronates (6-deoxo3DT, 6-deoxoCS, CS, and BL), methaneboronate-trimethylsilyl ethers (22,23-diOHCR, 6-deoxoTE, and 6-deoxoTY), or trimethylsilyl ethers (CR, CN, 22-OHCR, 22-OH-3-one, 6-deoxoCT, and 3-epi-6-deoxoCT). GC-MS/MS analyses were conducted using Agilent 7000A triple quad GC/MS to quantify 22-hydroxysteroids by monitoring the m/z 187 to 97 transition for nondeuterated steroids and the m/z 193 to 103 shift for deuterated steroids. LC-MS/MS analyses were carried out using Agilent 6460 Triple Quad LC/MS to quantify 22-OH-4-en-3-one (monitoring m/z 415 to 109 transition) and 22,23-diOH-4-en-3-one (monitoring m/z 431 to 109 transition).
Heterologous Expression of CYP90A1/CPD and DET2 Using Baculovirus-Insect Cell System
CYP90A1/CPD and DET2 open reading frames were amplified by PCR from Arabidopsis cDNA preparations and cloned as BamHI-XhoI fragments in the pCR2.1-TOPO vector (Invitrogen). Following sequence verification, these coding sequences were inserted as BamHI-XhoI fragments in the baculovirus expression vector pFastBac1 (Invitrogen). The pFastBac1-CYP90A1/CPD and pFastBac1-DET2 constructs were then used for preparing the corresponding recombinant bacmid DNAs by transformation of the E. coli strain DH10Bac. Preparation of the recombinant baculovirus DNA and transfection of Sf9 (Spodoptera frugiperda 9) cells was carried out according to the manufacturer's instructions (Invitrogen). Sf9 cells were maintained as a monolayer culture at 27 °C in Grace's insect cell medium supplemented with 10% (v/v) fetal bovine serum. For large scale expression, the Sf9 cells were propagated as suspension cultures in Grace's insect cell medium containing 0.1% (v/v) Pluronic F-68 and incubated at 27 °C in a rotary shaker at 150 rpm. For expression of the recombinant CYP90A1/CPD protein, the culturing medium was supplemented with 200 μm 5-aminolevulinic acid and 200 μm ferrous citrate to compensate for the low heme synthetic capacity of the insect cells. The heterologously expressed CYP90A1/CPD and DET2 proteins were subjected to 10% (w/v) SDS-PAGE and detected by Coomassie Brilliant Blue R-250 staining (Nacalai Tesque).
Preparation of Recombinant CYP90A1/CPD and DET2 Proteins
Preparations containing CYP90A1/CPD and DET2 activities were obtained from infected insect cells grown in 500-ml suspension cultures. The cells were washed with PBS buffer and suspended in buffer A containing 20 mm potassium phosphate (pH 7.25), 20% (v/v) glycerol, 1 mm EDTA, and 1 mm DTT. Following ultrasonic disruption, cell debris was removed by centrifugation at 10,000 × g for 15 min at 4 °C. For DET2 enzyme assays, the supernatant fraction was used. In the case of CYP90A1/CPD, the supernatant was further centrifuged at 100,000 × g for 1 h, and the pellet was homogenized with buffer A to provide the microsomal fractions. These were frozen in liquid nitrogen and stored in aliquots at −80 °C until use in the enzyme assays.
Assays of CYP90A1/CPD and DET2 Enzyme Activities
CYP90A1/CPD activity was reconstituted by mixing the microsomal fraction containing CYP90A1/CPD with NADPH-cytochrome P450 reductase. The reaction mixture consisted of 50 μg of the recombinant microsomes, Arabidopsis NADPH-cytochrome P450 reductase (22), 100 mm potassium phosphate (pH 7.25), 1 mm NADPH, and 20 μm of BR intermediates. DET2 reaction mixtures contained 50 μg of the soluble fraction of DET2, 100 mm potassium phosphate (pH 7.25), 1 mm NADPH, and 20 μm of BR intermediates. Reactions of CPD and DET2 were initiated by the addition of NADPH and were carried out at 30 °C. Incubation time was 2 h for the identification of enzymatic activities and 15 min for the determination of relative activities. Reaction products were extracted three times with a half-volume of ethyl acetate. The organic phase was collected and concentrated by evaporation.
GC-MS Analysis of CYP90A1/CPD and DET2 Reaction Products
Prior to analysis, concentrated products of the CYP90A1/CPD assays were derivatized using methaneboronate-trimethylsilyl ether (13). Concentrated DET2 assay products were either converted to methaneboronate (22,23-diOH-4-en-3-one) or analyzed directly without derivatization (4-en-3-one and 22-OH-4-en-3-one). The derivatized or nonderivatized products were subjected to GC-MS analysis performed on a Autosystem XL gas chromatograph (PerkinElmer Life Sciences) directly coupled to a turbomass mass spectrometer (PerkinElmer Life Sciences). A DB-5 column (15 m × 0.25-mm inner diameter, 0.25-μm film thickness; Agilent Technologies) was used. The carrier gas was helium at a flow rate of 0.5 ml min−1; the injection port temperature was 280 °C, and samples were introduced by splitless injection. The oven temperature program was as follows: 180 °C for 1 min, 180–300 °C at 20 °C/min, and 300 °C for 25 min. The ion source was run in EI mode (70 eV) at 280 °C.
Analyses of Enzyme Kinetics
Kinetic analyses of CYP90A1/CPD were carried out as described at the activity measurements, except that reaction time was 15 min. CYP90A1/CPD-containing microsomes were assayed using BR concentrations ranging from 1 to 30 μm. Kinetic studies of DET2 were carried out in 50 μg ml−1 of soluble DET2 fraction, 100 mm potassium phosphate (pH 7.25), 0.1 mm NADPH, and 1–30 μm of BR intermediates. Reactions were monitored spectrophotometrically at 30 °C for 10 min. The kinetic constants were determined from triplicate data sets. Km and either kcat (for CYP90A1/CPD) or Vmax (for DET2) values were calculated according to the Michaelis-Menten equation using Kaleidagraph (Synergy Software, Reading, PE).
RESULTS
CYP90A1/CPD and CYP90C1 Have Distinct Functions
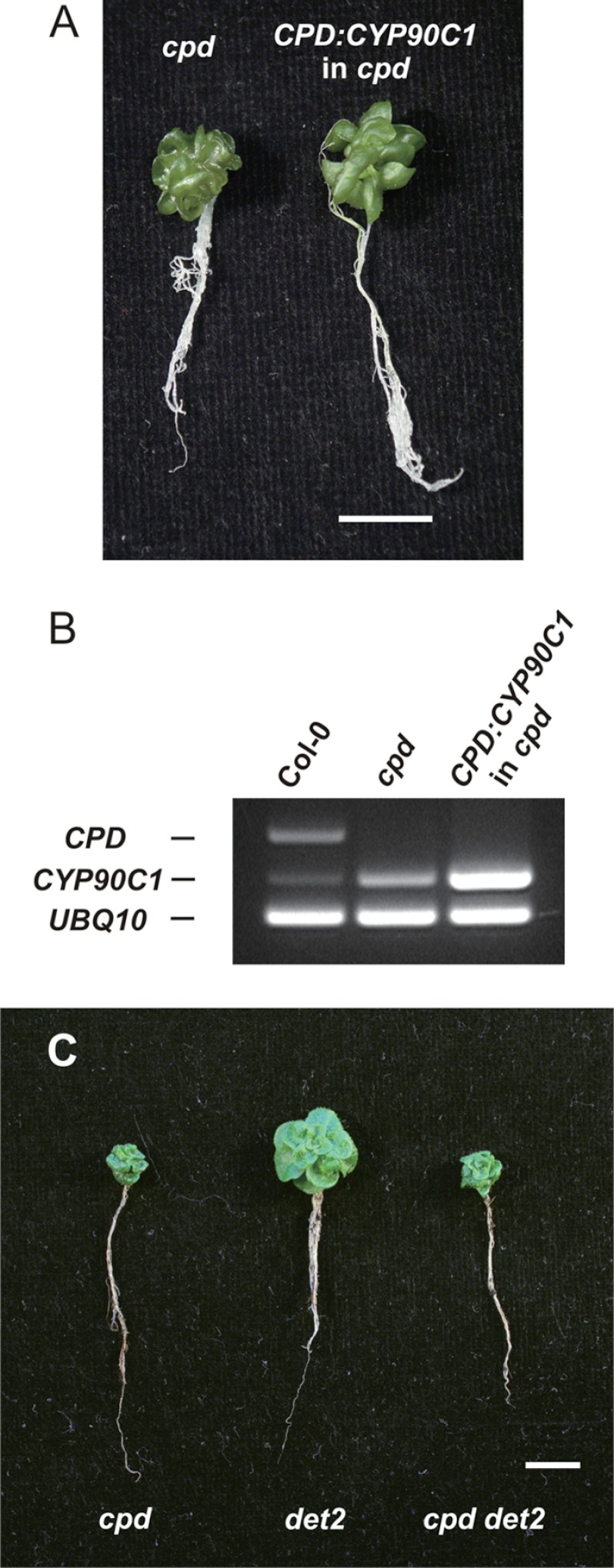
The similar severe dwarf phenotypes of the cpd mutant and the cyp90c1/cyp90d1 double mutant suggested that the biosynthetic role of CYP90A1/CPD differs from the redundantly functioning CYP90C1 and CYP90D1 C-23 hydroxylases. To verify that CYP90C1 cannot alleviate the BR deficiency caused by the loss of CYP90A1/CPD activity, we tested the phenotypic effect of CPD promoter-driven CYP90C1 overexpression in cpd background. Although the transcript level of CYP90C1 was 4-fold higher than in the nontransformed cpd mutant, and it was expressed primarily with the same temporal and spatial pattern as CPD, this did not cause any visible change in the dwarf phenotype (Fig. 2, A and B). Because CYP90C1 and CYP90D1 have redundant enzymatic roles (13), the lack of complementing effect by overexpressed CYP90C1 confirms that the CYP90A1/CPD function does not overlap with those of the CYP90C1 and CYP90D1 C-23 hydroxylases.
FIGURE 2.
Phenotypic consequences of CYP90A1/CPD deficiency are not affected by CYP90C1 gene activity or the det2 mutation. A, CPD promoter-driven overexpression of CYP90C1 in cpd mutant background. One-month-old aseptically grown cpd mutant and CPD:CYP90C1-expressing transgenic plant are shown. Bar, 10 mm. B, semiquantitative RT-PCR detection of the CPD, CYP90C1, and the standard UBQ10 transcripts in wild type Arabidopsis (Col-0), the cpd mutant, and the CPD:CYP90C1 transgenic line with cpd background. C, phenotypes of the cpd, det2, and cpd/det2 mutants. One-month-old soil-grown plantlets. Bar, 10 mm.
Endogenous BR Levels of the cpd Mutant
We quantified the endogenous BR levels of cpd mutant and Col-0 wild type plants from at least two biological replicate samples. In accordance with the BR-deficient dwarf phenotype of the mutant, the amounts of bioactive CS and BL were severely reduced or undetectable in the cpd line. Although the amounts of CR and CN in the Col-0 (1.96 ± 0.28 × 107 and 3.80 ± 0.88 × 105 ng/kg, respectively) and cpd plants (2.36 ± 0.18 × 107 and 4.65 ± 0.88 × 105 ng/kg, respectively) were nearly identical, a decreasing trend was observed in the levels of downstream BR intermediates. The amounts of 6-deoxo3DT and 6-deoxoCS were reduced to 43–69% and ≤1.2% of the respective wild type values, whereas 6-deoxoTE and 6-deoxoTY decreased below the detection level. By contrast, the cpd mutant accumulated high amounts of 22-OHCR, which was found in 85–121-fold excess compared with the wild type level (Table 1). These results suggested that the loss of CYP90A1/CPD function caused BR depletion by impeding C-3 oxidation of 22-OHCR to 22-OH-4-en-3-one or C-23 hydroxylation of 22-OHCR to 22,23-diOHCR. However, 22,23-diOHCR has never been detected in any plant sample, including BR biosynthetic mutants (3), and overexpression of the CYP90C1/ROT3 gene that encodes a BR C-23 hydroxylase could not alleviate the dwarf phenotype of cpd (Fig. 2A). Therefore it seemed more likely that the cpd mutation prevents the C-3 oxidative conversion of 22-OHCR to 22-OH-4-en-3-one (Fig. 1).
TABLE 1.
Endogenous BR content of the Col-0, cpd, det2, and cpd/det2 plants (ng kg-1 fresh weight)
Data correspond to two independent analyses with biological replicates. ND means not detected (below detection limit).
| BRs | Col-0 plant (38.6 g FW) (20.0 g FW) | cpd mutant (16.9 g FW) (20.1 g FW) | det2 mutant (18.1 g FW) (15.0 g FW) | cpd/det2 mutant (16.5 g FW) (21.8 g FW) |
|---|---|---|---|---|
| 22-OHCR | 180 | 16,500 | 7 | 23,800 |
| 193 | 21,800 | 8 | 6070 | |
| 22-OH-4-en-3-one | ND | 158 | 20,200 | 1730 |
| ND | 143 | 25,000 | 2580 | |
| 22-OH-3-one | 950 | 212 | 21 | 31 |
| 805 | 1110 | 47 | 85 | |
| 6-DeoxoCT | 772 | 987 | 39 | 70 |
| 1040 | 827 | 58 | 97 | |
| 3-epi-6-DeoxoCT | 697 | 117 | 10 | 9 |
| 857 | 312 | 28 | 29 | |
| 22,23-diOHCR | ND | ND | ND | ND |
| ND | ND | ND | ND | |
| 22,23-diOH-4-en-3-one | ND | 370 | 1720 | 415 |
| ND | 94 | 1780 | 443 | |
| 6-DeoxoTE | 172 | ND | ND | ND |
| 271 | ND | ND | ND | |
| 6-Deoxo3DT | 255 | 110 | 77 | 40 |
| 206 | 142 | 95 | 47 | |
| 6-DeoxoTY | 988 | ND | ND | ND |
| 729 | ND | ND | ND | |
| 6-DeoxoCS | 1320 | ND | 8 | ND |
| 1020 | ND | 14 | ND | |
| CS | 141 | ND | ND | ND |
| 126 | ND | ND | ND | |
| BL | ND | ND | ND | ND |
| ND | ND | ND | ND |
CYP90A1/CPD Acts Upstream of the C-5α Reduction Step
The analysis of endogenous BRs indicated inefficient conversion of 22-OHCR to 22-OH-4-en-3-one in the cpd mutant (Table 1). To find out if indeed CYP90A1/CPD acts upstream of the DET2-catalyzed reaction in the BR pathway, we determined the amounts of BR intermediates in the cpd and det2 mutants, as well as in the cpd/det2 double mutant (Table 1 and Fig. 2C). While the cpd mutant contained high amounts of 22-OHCR, det2 showed strong accumulation of 22-OH-4-en-3-one, the main substrate of the DET2 enzyme, and to a lesser extent 22,23-diOH-4-en-3-one. In the double mutant, the level of 22-OHCR was as high as in cpd, whereas the 22-OH-4-en-3-one content remained much lower than that of the det2 mutant. The amounts of the intermediates downstream of the DET2 reaction were somewhat additively decreased by the two mutations. These data show that in the cpd/det2 double mutant the lack of CYP90A1/CPD function can hinder accumulation of the DET2 substrate, indicating that the cpd mutation is epistatic to det2. This places the CYP90A1/CPD reaction upstream of the DET2-mediated C-5α reduction step in the BR biosynthetic pathway (Fig. 1).
Catalytic Properties of CYP90A1/CPD
To characterize the biochemical properties of CYP90A1/CPD, the enzyme was expressed using the baculovirus/insect cell system. Expression of the protein and correct targeting to the microsomal fraction were confirmed by SDS-PAGE analysis (supplemental Fig. 1A). Solubilized insect cell microsome preparations showed a cytochrome P450-specific reduced CO difference spectrum with a clear absorption peak at 450 nm (supplemental Fig. 1B). The expression level of the CYP90A1/CPD protein was 28 nmol of P450 liter−1. By contrast, the microsomes of mock-infected cells did not have the characteristic 450 nm peak. These data indicated that the insect cell-expressed CYP90A1/CPD protein is an active P450 enzyme.
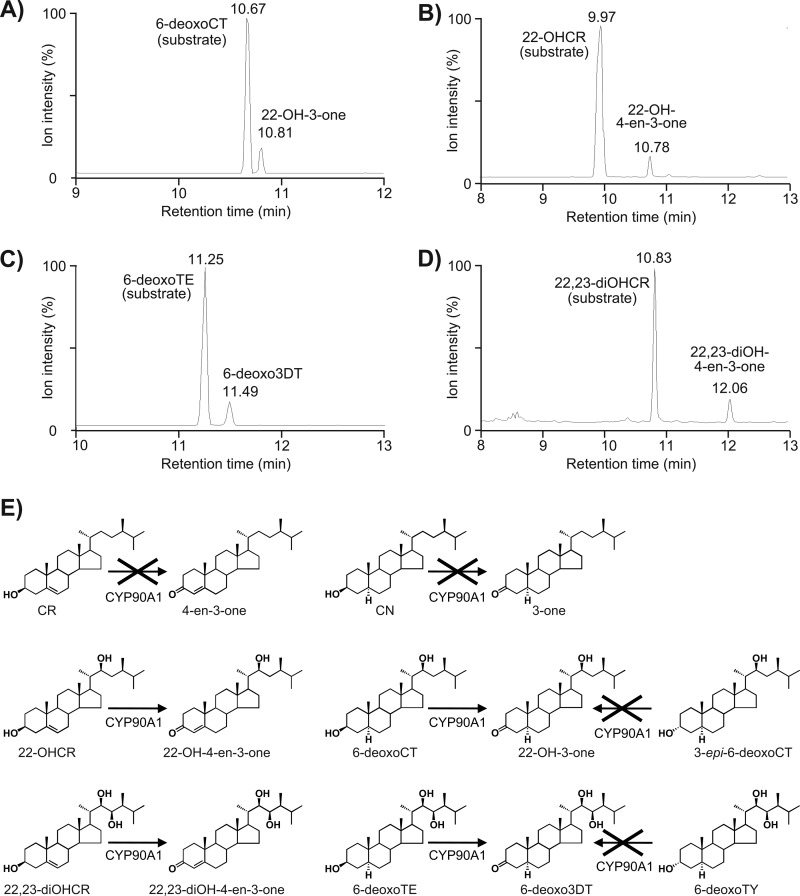
The substrate specificity of CYP90A1/CPD was investigated using 15 intermediates involved in the early or late C-6 oxidation pathways or in the early C-22 hydroxylation routes (Table 2). Because CYP90A1 had initially been regarded as C-23 hydroxylase (9), we tested whether CYP90A1 possesses C-23 hydroxylase activity. With the aid of Arabidopsis NADPH-P450 reductase, CYP90A1-containing microsomal fractions were incubated with the 22-hydroxy BRs CT, 6-deoxoCT, 3-epi-6-deoxoCT, 22-OH-3-one, 22-OH-4-en-3-one, and 22-OHCR as substrates. These assays yielded no detectable C-23 hydroxylated BRs (Table 2). However, the reactions carried out with 6-deoxoCT and 22-OHCR gave products that could be identified as 22-OH-3-one and 22-OH-4-en-3-one, respectively (Fig. 3, A and B), suggesting dehydrogenation of the 3β-hydroxy group.
TABLE 2.
GC-MS data of products obtained in CYP90A1/CPD enzymatic assays
| Substrate | Product | RT | Characteristic ions, m/z (relative intensity percentage) |
|---|---|---|---|
| min | |||
| CR | Not detected | ||
| CN | Not detected | ||
| CT | Not detected | ||
| 6-DeoxoCT | 22-OH-3-one | 10.81 | 374 (2.7), 271 (1.1), 187 (68), 97 (100) |
| Synthesized | 10.83 | 374 (3.6), 271 (2.5), 187 (71), 97 (100) | |
| 22-OHCR | 22-OH-4-en-3-one | 10.78 | 401 (14), 311(18), 269 (35), 196 (51), 187 (50), 97 (100) |
| Synthesized | 10.79 | 401 (10), 311(17), 269 (36), 196 (51), 187 (54), 97 (100) | |
| 22-OH-4-en-3-one | Not detected | ||
| 22-OH-3-one | Not detected | ||
| 3-epi-6-DeoxoCT | Not detected | ||
| 6-DeoxoTE | 6-Deoxo3DT | 11.51 | 456 (0.5), 283 (1.4), 231 (100), 155 (59) |
| Synthesized | 11.50 | 456 (6.3), 283 (7.4), 231(100), 155 (55) | |
| 22,23-diOHCR | 22,23-diOH-4-en-3-one | 12.06 | 454 (6.1), 412 (3.0), 330 (4.0), 229 (28), 124 (100) |
| Synthesized | 12.06 | 454 (6.3), 412 (3.1), 330 (3.8), 229 (30), 124 (100) | |
| 6-Deoxo3DT | Not detected | ||
| 6-DeoxoTY | Not detected | ||
| 6-DeoxoCS | Not detected | ||
| CS | Not detected | ||
| BL | Not detected |
FIGURE 3.
GC-MS analysis of BR metabolites produced by heterologously expressed CYP90A1/CPD. Selected ion chromatograms of the reaction products were obtained with 6-deoxoCT (fragment ion for detection at m/z 97 and 187) (A), 22-OHCR (fragment ion for detection at m/z 97 and 187) (B), 6-deoxoTE (fragment ion for detection at m/z 155 and 231) (C), and 22,23-OHCR (fragment ion for detection at m/z 124 and 229) (D) as substrates. C-3 oxidation of BR biosynthetic intermediates by CYP90A1/CPD (E).
We also assayed CYP90A1/CPD with BRs carrying a 22,23-dihydroxylated side chain. Among them, 6-deoxoTE and 22,23-diOHCR were converted to the respective 3-dehydro compounds, 6-deoxo3DT and 22,23-diOH-4-en-3-one (Fig. 3, C and D). However, CR and CN, devoid of side-chain hydroxylation, as well as 3-epi-6-deoxoCT and 6-deoxoTY, carrying 3α-hydroxy groups, yielded no products in our in vitro assays (Table 2). These observations demonstrated that CYP90A1/CPD catalyzes C-3 oxidation of all 3β-hydroxy BR intermediates with 22-hydroxylated and 22,23-dihydroxylated side chains. Conversely, the metabolites without side-chain hydroxylation or with 3α-hydroxy groups were not substrates of CYP90A1/CPD (Fig. 3E).
Kinetic Studies with CYP90A1/CPD
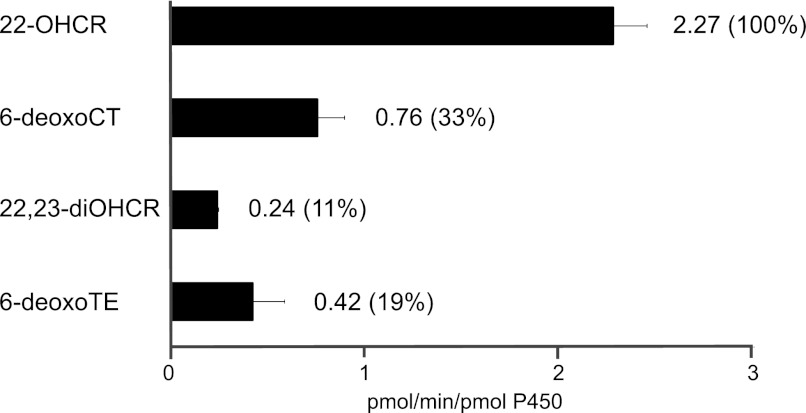
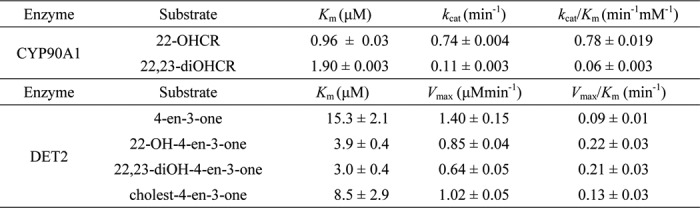
Substrate specificity of CYP90A1/CPD was examined with 22-OHCR, 22,23-diOHCR, 6-deoxoCT, and 6-deoxoTE at 20 μm. The highest specificity was observed for 22-OHCR (relative activity 100%). The lowest specificity (33.2%) was detected in the case of 6-deoxoCT, indicating a preference of the enzyme for side-chain-hydroxylated BRs with Δ5-double bond. Substrate specificities for 6-deoxoTE (18.6%) and 22,23-diOHCR (10.6%) were found lower than those of their respective 22-hydroxy BRs 6-deoxoCT (33.2%) and 22-OHCR (100%) (Fig. 4), indicating that for CYP90A1/CPD 22-hydroxylated substrates are preferred to 22,23-dihydroxylated ones. For further characterization of substrate specificity, we also determined the kinetic parameters of CYP90A1/CPD with 22-OHCR and 22,23-diOHCR. In the case of 22-OHCR, the dissociation constant (Km) was 5.3-fold lower, and the kcat value was 22.6-fold higher than those for 22,23-diOHCR, resulting in a 13-fold higher catalytic efficiency (kcat/Km) than that obtained with 22,23-diOHCR (Table 3). Altogether, these data indicate that CYP90A1/CPD favors substrates possessing Δ5-double bond and 22-hydroxylated side chain in the steroid skeleton.
FIGURE 4.
Relative activities of CYP90A1/CPD with 6-deoxoCT, 22-OHCR, 6-deoxoTE, and 22,23-diOHCR substrates. CYP90A1/CPD assay mixtures were supplemented with BRs (to 20 μm), incubated at 30 °C for 20 min, and the reaction products were then analyzed by GC-MS. The activity obtained with 22-OHCR substrate (100%) corresponds to 2.27 units (defined as pmol of product/min/pmol of P450). Values were determined in four replicate measurements; error bars indicate standard deviation.
TABLE 3.
Kinetic parameters of CYP90A1/CPD and DET2
Substrate Specificity of DET2
Previous in vitro assays demonstrated that DET2 catalyzes 5α reduction of 4-en-3-one to 3-one (7). In vivo feeding of labeled CR has also suggested a role for this enzyme in the conversion of 22-OH-4-en-3-one to 22-OH-3-one, a reaction in the early C-22 oxidation branch of BR biosynthesis (14). To elucidate how the hydroxylation status of the steroid side chain influences DET2-mediated 5α reduction, we characterized the enzymatic properties of heterologously expressed DET2 using the CYP90A1/CPD products 4-en-3-one, 22-OH-4-en-3-one, and 22,23-diOH-4-en-3-one as substrates.
All three substrates were metabolized to give new products that, based on their GC retention times and mass spectra, proved identical to authentic standards of 3-one, 22-OH-3-one, and 6-deoxo3DT, respectively (Table 4). These results reveal that DET2 catalyzes 5α reduction of all the three steroids. We also determined the kinetic parameters of DET2 with these substrates and cholest-4-en-3-one, the cholesterol congener of 4-en-3-one. The catalytic efficiencies (kcat/Km) of these compounds indicate that for DET2 22-OH-4-en-3-one is as favorable a substrate as 22,23-diOH-4-en-3-one, whereas only about half that level of efficiency was recorded for 4-en-3-one and cholest-4-en-3-one. This seems to indicate that the enzyme preferentially uses substrates with monohydroxy or dihydroxy side chains (Table 3).
TABLE 4.
GC-MS data of products obtained in DET2 enzymatic assays
| Substrate | Product | RT | Characteristic ions, m/z (relative intensity percentage) |
|---|---|---|---|
| min | |||
| 4-en-3-one | 3-one | 9.16 | 400 (12), 385 (7), 317 (2), 231 (100) |
| Synthesized | 9.20 | 400 (11), 385 (7), 317 (1), 231 (100) | |
| 22-OH-4-en-3-one | 22-OH-3-one | 11.70 | 398 (6), 302 (35), 271 (40), 231 (100) |
| Synthesized | 11.67 | 398 (6), 302 (30), 271 (43), 231 (100) | |
| 22,23-diOH-4-en-3-one | 6-Deoxo3DT | 11.05 | 456 (7), 283 (6), 231 (100), 155 (59) |
| Synthesized | 11.03 | 456 (10), 283 (8), 231 (100), 155 (56) |
DISCUSSION
CYP90A1/CPD Catalyzes C-3 Oxidation
Earlier characterization of the CYP90A1/CPD-deficient cpd mutant (9) showed that only C-23-hydroxylated BRs were capable of rescuing its dwarf phenotype. BRs of the early C-6 oxidation pathway, such as TE, 3-dehydroteasterone, TY, CS, and BL, restored cell elongation in the hypocotyl and petioles of cpd seedlings both in dark and light. By contrast, treatments with CR and CT, lacking C-23 hydroxylation, did not have a similar rescuing effect. Based on these data, CYP90A1/CPD was proposed to function as BR C-23 hydroxylase (9).
Our recent study, however, showed that in Arabidopsis BR C-23 hydroxylation is catalyzed by the redundantly functioning CYP90C1 and CYP90D1 enzymes and that their inactivation causes BR deficiency even in the presence of fully active CYP90A1/CPD (13). This clearly indicates that the role of CYP90A1/CPD in BR synthesis is different from those of CYP90C1 and CYP90D1, and therefore it should not function as C-23 hydroxylase.
Consistently, our in vitro assays using heterologously expressed CYP90A1/CPD and C-22-hydroxylated BR substrates did not yield any 22,23-dihydroxylated product (Table 2). Instead, we detected C-3 oxidation of 22-OHCR and 22,23-diOHCR, as well as 6-deoxoCT and 6-deoxoTE, to their respective 3-dehydro derivatives (Table 2 and Fig. 3E). Whereas CYP90A1/CPD, similarly to other CYP90 enzymes, was found to have broad substrate specificity, our reaction kinetic studies revealed that its preferred substrate is 22-OHCR (Table 3 and Fig. 4). Accordingly, GC-MS analyses detected high accumulation of 22-OHCR in the cpd mutant, indicating inefficient in planta metabolization of this intermediate in the absence of CYP90A1/CPD. Moreover, we also found that intermediate pool sizes in the cpd/det2 double mutant were determined primarily by the cpd mutation, revealing that it affects BR synthesis upstream of the DET2-mediated reaction. Despite the occasional large variability of parallel BR analysis data, likely resulting from uneven physiological conditions of the biological replicates, these results clearly support our conclusion that CYP90A1/CPD is a C-3 oxidase, which is required for the conversion of 22-OHCR to 22-OH-4-en-3-one, a major DET2 substrate.
In Planta Function of CYP90A1/CPD
The in vitro biochemical function of heterologously expressed CYP90A1/CPD and specific accumulation of 22-OHCR, an early BR intermediate, is consistent with a major role for this enzyme in the initial part of the pathway. Our in vitro enzyme assays imply that CYP90A1/CPD also participates in the 6-deoxoCT → 22-OH-3-one and 6-deoxoTE → 6-deoxo3DT conversions (Fig. 3E). However, compared with 22-OHCR, 6-deoxoCT and 6-deoxoTE are less favored substrates, and they do not accumulate in the cpd mutant (Table 1 and Fig. 4). Therefore, it is not clear how important these reactions are for the functioning of BR synthesis.
Previous intermediate feeding assays revealed that rescuing the dwarf phenotype of cpd plants requires C-23-hydroxylated BRs (9). Surprisingly, the endogenous intermediate pools of cpd (Table 1) are very similar to those determined in the C-23 hydroxylation-deficient cyp90c1/cyp90d1 double mutant (13), except that cyp90c1/cyp90d1 plants do not accumulate 22-OHCR. Whereas both the cpd and cyp90c1/cyp90d1 plants have severe dwarf phenotypes that clearly indicate impairment of distinct biosynthetic functions, BR biosynthesis is similarly affected in the two mutant lines. The physiological background of this unexpected phenomenon has yet to be elucidated. However, one possible explanation can be a functional inter-dependence between CYP90A1/CPD and the CYP90C1 or CYP90D1 enzymes, which may be necessary for the efficient channeling of the C-23 hydroxylation substrates.
A line of evidence shows that CYP90A1/CPD is not the only C-3 oxidase enzyme utilized for BR synthesis in Arabidopsis. CYP90A1/CPD does not catalyze C-3 oxidation of CR, and unlike the det2 mutation (14), cpd does not decrease the endogenous level of CN. These results point out that the synthesis of 3-one from CR requires other probably non-P450-type C-3 oxidase enzymes(s). Analogous reactions in the synthesis of animal steroid hormones are carried out by NAD+- or NADP+-dependent cytosolic 3β-hydroxysteroid dehydrogenase enzymes (23). Homologs of mammalian 3β-hydroxysteroid dehydrogenase genes (e.g. At1g47290) occur in the Arabidopsis genome,5 and a functional 3β-hydroxysteroid dehydrogenase was isolated and characterized from Digitalis lanata (24). It was also shown that Arabidopsis and tomato contain cytosolic C-3 oxidase/isomerase enzymes that are capable of converting 24-epi-TE to 24-epi-3DT and 24-epi-TY in an NAD-dependent manner (25). Nevertheless, it still remains to be clarified which other C-3 oxidases and to what extent contribute to BR biosynthesis, and whether they convert only BR intermediates or have broader specificities.
Main Route of BR Biosynthesis
The first model of BR biosynthesis, leading from CR to the synthesis of BL, was established on the basis of conversion products identified in Catharanthus roseus cell cultures and Arabidopsis seedlings upon feeding with isotope-labeled precursors (26–28). Later in vitro and in vivo studies of BR metabolism (10, 14, 15, 30) revealed that several biosynthetic enzymes have broad specificities for various substrates, resulting in the establishment of a grid-like BR synthesis network (Fig. 1). The originally proposed BR synthesis route is initiated by C-3 oxidation of CR to 4-en-3-one, followed by its DET2-catalyzed hydrogenation to 3-one, which is then converted to CN (Fig. 5). Our results, however, show that C-3 oxidation of CR is not catalyzed by CYP90A1/CPD (Table 2) and that, unlike DET2 (14), this enzyme is not involved in the synthesis of CN. Accordingly, the level of CN in the severely BR-deficient cpd mutant (4.65 ± 0.88 × 105 ng/kg) is comparable with that of the wild type (3.80 ± 0.88 × 105 ng/kg). Considering the severe BR deficiency of cpd plants, these data clearly indicate that the reactions converting CR to CN are not essential steps of BR synthesis.
FIGURE 5.
CN-independent and CN-dependent routes of BR biosynthesis. The CN-independent pathway, via CR → 22-OHCR → 22-OH-4-en-3-one → 22-OH-3-one → 3-epi-6-deoxoCT → 6-deoxoTY → 6-deoxoCS → CS → BL, is two reactions shorter than the CN-dependent route, due to shortcutting the 3-one → CN and 6-deoxoTE → 6-deoxo3DT reactions.
The identification of CN as an important intermediate of BR biosynthesis may have resulted from inherent limitations of in vivo feeding experiments. This method can be used efficiently for tracing metabolic reactions, but high doses of precursors can result in the detection of minor metabolites or induce detoxification reactions.
This work demonstrates the importance of the C-22 hydroxylation pathway in BR biosynthesis, which initiates with the formation of 22-OHCR. This will then be oxidized to 22-OH-4-en-3-one by CYP90A1/CPD. Low affinity of the CYP90C1 and CYP90D1 enzymes to 22-OHCR suggests that 23-hydroxylation of this intermediate to 22,23-diOHCR is not a favored reaction, which is corroborated by the finding that this compound occurs below the detection limit in Col-0 and cpd plants, and even in the det2 mutant, in which BR synthesis is arrested at the immediate downstream intermediate 22,23-diOH-4-en-3-one (Table 1). This also indicates that 22,23-diOH-4-en-3-one, which was now first detected in plant samples, is not synthesized or is only inefficiently synthesized from the putative intermediate 22,23-diOHCR, but it is produced primarily from 22-OH-4-en-3-one in one of the enzymologically favored 23-hydroxylation reactions. As we demonstrated earlier, 22,23-diOH-4-en-3-one, 22-OH-3-one, and 3-epi-6-deoxoCT are also good substrates of CYP90C1 and CYP90D1 (13). Taken together, these results suggest that the initial reactions of BR synthesis proceed primarily via the enzymologically preferred 22-hydroxylated substrates, rather than the less favored intermediates with non- or dihydroxylated side chains.
Based on substrate preferences of the CYP90B1, CYP90C1, CYP90D1, and DET2 enzymes, we proposed a CN-independent BR pathway (Fig. 5) consisting of eight metabolic steps from CR to BL (13, 29, 30). This reaction route is two steps shorter than the originally proposed CN-dependent pathway (27, 28), because it shortcuts the 3-one → CN and 6-deoxoTE → 6-deoxo3DT reactions. Our enzyme kinetic results imply that in Arabidopsis the CR → 22-OHCR → 22-OH-4-en-3-one → 22-OH-3-one → 3-epi-6-deoxoCT/6-deoxo3DT → 6-deoxoTY → 6-deoxoCS → CS → BL route of BR synthesis ensures the most efficient flow of intermediates (Fig. 5). Special physiological conditions altering the availability of particular biosynthetic enzymes and/or their potential substrates can, however, substantially influence the functional hierarchy of subpathways. For instance, seed maturation in garden pea (Pisum sativum) was shown to be accompanied by massive accumulation of 6-deoxoCT, which serves as a storage intermediate for the synthesis of bioactive BRs during germination (31).
Our recent biochemical analyses revealed 22-OHCR as the predominant product of CYP90B1-catalyzed C-22 hydroxylation (29). The data of the reaction kinetic and feeding assays suggested a major role for 22-OHCR in the BR pathway. This is supported by our findings that this compound is the favored substrate of CYP90A1/CPD and that it strongly accumulates in the CYP90A1/CPD-deficient cpd mutant (Table 1). Together these results reveal 22-OHCR as a key early intermediate of BR synthesis.
Supplementary Material
Acknowledgment
We thank Dr. Suguru Takatsuto (Joetsu University of Education, Japan) for supplying deuterium-labeled internal standards for the GC-MS analyses.
This work was supported in part by the Hungarian Scientific Research Fund Grant T68201 (to M. S.), “BRAVISSIMO” Marie Curie Initial Training Grant of the European Union (to B. G.), Ministry of Education, Culture, Sports, Science, and Technology of Japan Grants 19380069 and 23380066 (to S. F.) and Grant 18580091 (to M. M.), Collaborative Research Program of the Institute for Chemical Research, Kyoto University, Grants 2010-16 and 2011-17 (to T. O.), and MEXT-supported Program for the Strategic Research Foundation at Private Universities Grant 2008-2012 (to T. Y.).

This article contains supplemental Fig. 1.
T. Ohnishi and M. Mizutani, unpublished results.
- BR
- brassinosteroid
- BL
- brassinolide
- CN
- campestanol
- CR
- campesterol
- CS
- castasterone
- CT
- cathasterone
- 22-OHCR
- (22S)-22-hydroxycampesterol
- 22-OH-3-one
- (22S)-22-hydroxy-5α-campestan-3-one
- 22-OH-4-en-3-one
- (22S)-22-hydroxycampest-4-en-3-one
- 22,23-diOHCR
- (22R,23R)-22,23-dihydroxycampesterol
- 22,23-diOH-4-en-3-one
- (22R,23R)-22,23-dihydroxycampest-4-en-3-one
- 3-epi-6-deoxoCT
- 3-epi-6-deoxocathasterone
- 3-one
- 5α-campestan-3-one
- 4-en-3-one
- campest-4-en-3-one
- 6-deoxoCS
- 6-deoxocastasterone
- 6-deoxoCT
- 6-deoxocathasterone
- 6-deoxoTE
- 6-deoxoteasterone
- 6-deoxoTY
- 6-deoxotyphasterol
- 6-deoxo3DT
- 3-dehydro-6-deoxoteasterone.
REFERENCES
- 1. Bishop G. J., Yokota T. (2001) Plants steroid hormones, brassinosteroids. Current highlights of molecular aspects on their synthesis/metabolism, transport, perception, and response. Plant Cell Physiol. 42, 114–120 [DOI] [PubMed] [Google Scholar]
- 2. Bajguz A. (2007) Metabolism of brassinosteroids in plants. Plant Physiol. Biochem. 45, 95–107 [DOI] [PubMed] [Google Scholar]
- 3. Bajguz A., Tretyn A. (2003) The chemical characteristic and distribution of brassinosteroids in plants. Phytochemistry 62, 1027–1046 [DOI] [PubMed] [Google Scholar]
- 4. Fujioka S., Yokota T. (2003) Biosynthesis and metabolism of brassinosteroids. Annu. Rev. Plant Biol. 54, 137–164 [DOI] [PubMed] [Google Scholar]
- 5. Clouse S. D., Sasse J. M. (1998) Brassinosteroids. Essential regulators of plant growth and development. Annu. Rev. Plant Physiol. Plant Mol. Biol. 49, 427–451 [DOI] [PubMed] [Google Scholar]
- 6. Mandava B. N. (1988) Plant growth-promoting brassinosteroids. Annu. Rev. Plant Physiol. Plant Mol. Biol. 39, 23–52 [Google Scholar]
- 7. Li J., Biswas M. G., Chao A., Russell D. W., Chory J. (1997) Conservation of function between mammalian and plant steroid 5α-reductases. Proc. Natl. Acad. Sci. U.S.A. 94, 3554–3559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li J., Nagpal P., Vitart V., McMorris T. C., Chory J. (1996) A role for brassinosteroids in light-dependent development of Arabidopsis. Science 272, 398–401 [DOI] [PubMed] [Google Scholar]
- 9. Szekeres M., Németh K., Koncz-Kálmán Z., Mathur J., Kauschmann A., Altmann T., Rédei G. P., Nagy F., Schell J., Koncz C. (1996) Brassinosteroids rescue the deficiency of CYP90, a cytochrome P450, controlling cell elongation and de-etiolation in Arabidopsis. Cell 85, 171–182 [DOI] [PubMed] [Google Scholar]
- 10. Choe S., Dilkes B. P., Fujioka S., Takatsuto S., Sakurai A., Feldmann K. A. (1998) The DWF4 gene of Arabidopsis encodes a cytochrome P450 that mediates multiple 22α-hydroxylation steps in brassinosteroid biosynthesis. Plant Cell 10, 231–243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Kim G. T., Tsukaya H., Uchimiya H. (1998) The ROTUNDIFOLIA3 gene of Arabidopsis thaliana encodes a new member of the cytochrome P-450 family that is required for the regulated polar elongation of leaf cells. Genes Dev. 12, 2381–2391 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kim G. T., Fujioka S., Kozuka T., Tax F. E., Takatsuto S., Yoshida S., Tsukaya H. (2005) CYP90C1 and CYP90D1 are involved in different steps in the brassinosteroid biosynthesis pathway in Arabidopsis thaliana. Plant J. 41, 710–721 [DOI] [PubMed] [Google Scholar]
- 13. Ohnishi T., Szatmari A. M., Watanabe B., Fujita S., Bancos S., Koncz C., Lafos M., Shibata K., Yokota T., Sakata K., Szekeres M., Mizutani M. (2006) C-23 hydroxylation by Arabidopsis CYP90C1 and CYP90D1 reveals a novel shortcut in brassinosteroid biosynthesis. Plant Cell 18, 3275–3288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Fujioka S., Takatsuto S., Yoshida S. (2002) An early C-22 oxidation branch in the brassinosteroid biosynthetic pathway. Plant Physiol. 130, 930–939 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Shimada Y., Fujioka S., Miyauchi N., Kushiro M., Takatsuto S., Nomura T., Yokota T., Kamiya Y., Bishop G. J., Yoshida S. (2001) Brassinosteroid-6-oxidases from Arabidopsis and tomato catalyze multiple C-6 oxidations in brassinosteroid biosynthesis. Plant Physiol. 126, 770–779 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Bancoş S., Nomura T., Sato T., Molnár G., Bishop G. J., Koncz C., Yokota T., Nagy F., Szekeres M. (2002) Regulation of transcript levels of the Arabidopsis cytochrome p450 genes involved in brassinosteroid biosynthesis. Plant Physiol. 130, 504–513 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Goda H., Shimada Y., Asami T., Fujioka S., Yoshida S. (2002) Microarray analysis of brassinosteroid-regulated genes in Arabidopsis. Plant Physiol. 130, 1319–1334 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Wang H., Zhu Y., Fujioka S., Asami T., Li J., Li J. (2009) Regulation of Arabidopsis brassinosteroid signaling by atypical basic helix-loop-helix proteins. Plant Cell 21, 3781–3791 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Koncz C., Martini N., Szabados L., Hrouda M., Brachmair A., Schell J. (1994) in Plant Molecular Biology Manual (Gelvin S. B., Schilperoot A. R., eds) Vol. B2, pp. 1–22, Kluwer Academic Publishers, Dordrecht, The Netherlands [Google Scholar]
- 20. Clough S. J., Bent A. F. (1998) Floral dip. A simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 16, 735–743 [DOI] [PubMed] [Google Scholar]
- 21. Nomura T., Sato T., Bishop G. J., Kamiya Y., Takatsuto S., Yokota T. (2001) Accumulation of 6-deoxocathasterone and 6-deoxocastasterone in Arabidopsis, pea, and tomato is suggestive of common rate-limiting steps in brassinosteroid biosynthesis. Phytochemistry 57, 171–178 [DOI] [PubMed] [Google Scholar]
- 22. Mizutani M., Ohta D. (1998) Two isoforms of NADPH:cytochrome P450 reductase in Arabidopsis thaliana. Gene structure, heterologous expression in insect cells, and differential regulation. Plant Physiol. 116, 357–367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Simard J., Ricketts M. L., Gingras S., Soucy P., Feltus F. A., Melner M. H. (2005) Molecular biology of the 3β-hydroxysteroid dehydrogenase/Δ5-Δ4 isomerase gene family. Endocr. Rev. 26, 525–582 [DOI] [PubMed] [Google Scholar]
- 24. Finsterbusch A., Lindemann P., Grimm R., Eckerskorn C., Luckner M. (1999) Δ(5)-3β-hydroxysteroid dehydrogenase from Digitalis lanata Ehrh. A multifunctional enzyme in steroid metabolism? Planta 209, 478–486 [DOI] [PubMed] [Google Scholar]
- 25. Stündl U., Schneider B. (2001) 3β-Brassinosteroid dehydrogenase activity in Arabidopsis and tomato. Phytochemistry 58, 989–994 [DOI] [PubMed] [Google Scholar]
- 26. Yokota T., Ogino Y., Takahashi N., Saimoto H., Fujioka S., Sakurai A. (1990) Brassinolide is biosynthesized from castasterone in Catharanthus roseus crown gall cells. Agric. Biol. Chem. 54, 1107–1108 [Google Scholar]
- 27. Fujioka S., Noguchi T., Watanabe T., Takatsuto S., Yoshida S. (2000) Biosynthesis of brassinosteroids in cultured cells of Catharanthus roseus. Phytochemistry 53, 549–553 [DOI] [PubMed] [Google Scholar]
- 28. Noguchi T., Fujioka S., Choe S., Takatsuto S., Tax F. E., Yoshida S., Feldmann K. A. (2000) Biosynthetic pathways of brassinolide in Arabidopsis. Plant Physiol. 124, 201–209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Fujita S., Ohnishi T., Watanabe B., Yokota T., Takatsuto S., Fujioka S., Yoshida S., Sakata K., Mizutani M. (2006) Arabidopsis CYP90B1 catalyses the early C-22 hydroxylation of C27, C28, and C29 sterols. Plant J. 45, 765–774 [DOI] [PubMed] [Google Scholar]
- 30. Ohnishi T., Yokota T., Mizutani M. (2009) Insights into the function and evolution of P450s in plant steroid metabolism. Phytochemistry 70, 1918–1929 [DOI] [PubMed] [Google Scholar]
- 31. Nomura T., Ueno M., Yamada Y., Takatsuto S., Takeuchi Y., Yokota T. (2007) Roles of brassinosteroids and related mRNAs in pea seed growth and germination. Plant Physiol. 143, 1680–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.