Abstract
We have completed the total chemical synthesis of cytochrome b562 and an axial ligand analogue, [SeMet7]cyt b562, by thioester-mediated chemical ligation of unprotected peptide segments. A novel auxiliary-mediated native chemical ligation that enables peptide ligation to be applied to protein sequences lacking cysteine was used. A cleavable thiol-containing auxiliary group, 1-phenyl-2-mercaptoethyl, was added to the α-amino group of one peptide segment to facilitate amide bond-forming ligation. The amine-linked 1-phenyl-2-mercaptoethyl auxiliary was stable to anhydrous hydrogen fluoride used to cleave and deprotect peptides after solid-phase peptide synthesis. Following native chemical ligation with a thioester-containing segment, the auxiliary group was cleanly removed from the newly formed amide bond by treatment with anhydrous hydrogen fluoride, yielding a full-length unmodified polypeptide product. The resulting polypeptide was reconstituted with heme and folded to form the functional protein molecule. Synthetic wild-type cyt b562 exhibited spectroscopic and electrochemical properties identical to the recombinant protein, whereas the engineered [SeMet7]cyt b562 analogue protein was spectroscopically and functionally distinct, with a reduction potential shifted by ≈45 mV. The use of the 1-phenyl-2-mercaptoethyl removable auxiliary reported here will greatly expand the applicability of total protein synthesis by native chemical ligation of unprotected peptide segments.
Because of its unmatched flexibility, total chemical synthesis is emerging as a powerful tool for protein engineering (1, 2). Chemical access to proteins inherently provides the ability to incorporate unnatural amino acids into proteins in a completely general fashion and has opened many new avenues for the study of protein function. The large number and variety of unnatural amino acids available enables researchers to systematically probe size, shape, acidity, hydrogen bonding, and electronic structure effects on the reactivity properties of the protein molecule. Studies of bioinorganic systems in particular can benefit greatly from the ability to place unnatural metal-chelating residues into metalloprotein structures in a site-specific manner.
Chemical ligation reactions (3) have been used in the total synthesis of myriad engineered protein targets with unusual backbone and side chain groups. Several ligation chemistries have been used (4–7). In particular, the native chemical ligation (NCL) reaction (8) has enabled the synthesis of many wild-type and engineered protein targets by allowing full-length polypeptide sequences to be built by linking readily obtained smaller unprotected peptide segments with native amide bonds. In the original NCL scheme (9), a thioester-containing peptide reacts in a chemoselective manner with a second peptide that has a cysteine as its N-terminal residue. The side chain thiol of that cysteine undergoes thiol exchange with the thioester moiety of the first peptide. A thioester-linked intermediate is formed, which, because of the presence of an amino group beta to the thiol moiety, spontaneously rearranges to form an amide bond. The result of this thioester-mediated reaction is a product polypeptide containing a native amide bond at the ligation site. However, the requirement for cysteine at the site of peptide ligation is an intrinsic limitation of the NCL strategy. In proteins, Cys occurs with a relatively low frequency (only Trp, Met, and His appear less frequently). Many protein sequences lack Cys residues entirely; others do not have suitably disposed cysteines for the NCL synthetic strategy and are therefore inaccessible. As was pointed out by Canne and coworkers (10), a potential solution to this sequence restriction would involve the use of an Nα-linked removable thiol moiety to fulfill the same function as the side chain of Cys. Such an approach would enable ligation at a variety of amino acid sequences and would greatly increase the number of proteins accessible to synthesis by thioester-mediated native chemical ligation.
Here, we demonstrate the utility of a new removable thiol-containing auxiliary group, 1-phenyl-2-mercaptoethyl, in the chemical synthesis of a protein, cytochrome b562 (cyt b562), and an engineered variant featuring a noncoded selenomethionine in place of the native axial methionine ligand to the heme iron ([SeMet7]cyt b562). The 1-phenyl-2-mercaptoethyl auxiliary facilitates ligation in sequences that are devoid of Cys residues and is easily removed to give unmodified polypeptides, which can subsequently be folded to form the corresponding functional proteins.
Materials and Methods
Materials.
All reagents used were of the highest quality available and used as received. Standard Boc amino acids were obtained from Midwest Biotech Inc. (Fishers, IN) Trifluoroacetic acid was purchased from Halocarbon Products (Hackensack, NJ). Solvents were HPLC grade or higher. Dimethylformamide, acetonitrile, and water for HPLC purifications were from Burdick and Jackson. Water for buffer preparation was obtained from NERL Diagnostics (East Providence, RI). Heme (protoporphyrin IX) and Tris⋅HCl were purchased from Sigma. Diethyl ether was purchased from VWR Scientific. Anhydrous hydrogen fluoride was obtained from Matheson, and 2-mercaptoethanol, tris-carboxyethyl phosphine, thiophenol, and p-cresol were obtained from Fluka.
Methods.
Automated solid-phase peptide synthesis was carried out as described previously (11). The 1-(4-methoxyphenyl)-2-mercaptoethyl auxiliary was introduced as an on-resin modification of the amino terminus of the peptide cyt b562(65–106), as described in an earlier report (12). Peptides were cleaved from solid supports by treatment with anhydrous hydrogen fluoride (HF) at 0°C for 1 h in a specialized Teflon apparatus (Peptide Institute, Osaka), after which the HF was removed in vacuo (13). Crude peptides were then precipitated with ether and collected on a Teflon membrane by suction filtration and finally dissolved in a 1:1 water:acetonitrile mixture containing 0.1% trifluoroacetic acid. Crude peptides were purified by preparative HPLC (Rainin Dynamax solvent delivery system, Vydac C4 column, water–acetonitrile solvent system containing 0.1% trifluoroacetic acid). HPLC fractions were analyzed by using electrospray mass spectrometry (ES-MS), and fractions containing the desired products were pooled and lyophilized.
Native chemical ligation reactions were performed at ambient temperature. Peptides were dissolved in 6 M guanidine hydrochloride buffered at pH 7 with 0.3 M sodium phosphate. Thiophenol (0.5–2%) was added as a catalyst. Reactions were monitored by analytical HPLC (Vydac C4, water–acetonitrile solvent system). Peptides bearing a 1-phenyl-2-mercaptoethyl auxiliary often elute in two peaks because of the racemic nature of the auxiliary, which gives rise to diastereomeric peptides that may be resolved by HPLC. Ligation reactions were typically carried out with a 1.5-fold excess of thioester peptide and were complete after stirring gently overnight at ambient temperatures. Ligation reactions were treated with β-mercaptoethanol and tris-carboxyethyl phosphine to reduce disulfides, and the products were then isolated by semipreparative HPLC. The 1-(4-methoxyphenyl)-2-mercaptoethyl group was cleaved from the ligation product by treatment with HF (1 h, 0°C), and the resulting native polypeptide product was isolated by HPLC.
The following procedure was used to fold and reconstitute the synthetic proteins. A stock solution of heme was prepared by dissolving solid heme in 0.1 M NaOH. The appropriate apoprotein was then dissolved in phosphate buffer, pH 7. The heme was then slowly added to the apoprotein solution, and the mixture allowed to gently stir at 4°C overnight. The mixture was then purified by ion-exchange FPLC by using an Amersham Pharmacia FPLC system equipped with a Resource-Q column (1 ml). The solvent system used was 20 mM Tris⋅HCl, pH 8, and a 0–200 mM NaCl gradient was used. Both synthetic wild-type and SeMet7 cyt b562 eluted as a single, sharp peak at 10 mM NaCl.
Mass spectra were obtained by using Sciex API 1 and API 3 mass spectrometers. Peptide samples were typically dissolved in aqueous acetonitrile mixtures and infused directly into the electrospray interface by using a syringe pump (Harvard Apparatus) at 5–10 μl/s. Molecular mass spectra were reconstructed from charge state spectra by using the program MacSpec. Circular dichroism spectra were collected on a Jasco J600 spectropolarimeter in a 1 mM circular quartz cuvette. Samples for CD were prepared in 10 mM sodium phosphate buffer, pH 7. Spectra shown are the average of three scans. UV-visible spectra were collected by using a Hewlett-Packard HP8453 spectrophotometer in 1-cm pathlength masked cuvettes. Spectra of reduced proteins were obtained by adding a minimum amount of solid sodium dithionite (Sigma) directly to a solution of the oxidized protein.
All electrochemical experiments were performed by using a Bioanalytical Systems BAS CV-50W electrochemical analyzer. Cyclic voltammetry was carried out in a two-compartment cell consisting of a modified-gold working electrode, a platinum auxiliary electrode, and a saturated calomel reference electrode. The working/auxiliary compartment of the electrochemical cell was separated from the reference compartment by a modified Luggin capillary. Gold electrodes were polished with 0.05 μm alumina, sonicated in water, and electrochemically etched in 1 M H2SO4 immediately before use. They were then modified by cycling the potential between 0 and −1 V (≈30 cycles) in a 1 mM solution of the hexapeptide KCTCCA in 0.1 mM potassium phosphate, pH 7. Plots of ip versus scan rate (ν) (1/2) yield straight lines that pass through the origin, consistent with a diffusion-controlled process.
Experimental Design.
Heme proteins mediate a large number of important biological processes, from small molecule binding and activation (myoglobins and peroxidases) to multielectron redox processes (cytochrome c oxidase, cytochrome P450s) (14). The functional properties of heme proteins can be greatly influenced by the nature of the axial ligands and by the chemical properties of other amino acids near the heme active site (15–17). Hence, the idea of using chemical protein synthesis to investigate these effects is an attractive one. In our previous studies of engineered metalloproteins, we used chemical protein synthesis to explore the effects of polypeptide backbone hydrogen bonding to redox centers (18) and second-shell aromatic effects on metalloprotein redox properties (19). Relatively few of the naturally occurring amino acids are found as axial ligands for the iron atom in heme proteins (histidine, methionine, cysteine) (20). Semisynthesis has been used to study the effects of chemically introduced amino acids on the cytochrome c molecule (21), but this approach is not generally applicable. Chemical access to heme proteins will therefore allow much greater flexibility in designing heme proteins with tailored properties by greatly expanding the number of possible metal-coordinating residues. Thus far, the effects of introduction of unnatural heme ligands by total chemical synthesis remain unexplored. We therefore began a study of heme proteins using an expanded set of amino acids to systematically vary metal–ligand bonding.
A cursory survey of databases reveals that many heme proteins do not have sequences suitable for assembly using native chemical ligation at cysteine residues. We elected to establish synthetic entry into the heme protein family by synthesizing a typical small heme protein, cytochrome b562, and an engineered variant using a new strategy involving amide bond-forming ligation at non-Cys residues by means of a removable auxiliary group. The 106-aa sequence of cyt b562 lacks cysteine and is an ideal size for synthesis by ligation of two peptide segments {the full-length sequences synthesized were: ADLEDNXETLNDNLKVIEKADNAAQVKDALTKMRAAALDAQKATPPKLEDKSPDSPEMKDFRHGFDILVGQIDDALKLANEGKVKEAQAAAEQLKTTRNAYHQKYR, where X = Met (cyt b562) or SeMet ([SeMet7]cyt b562)}. We also anticipated that the well-documented spectroscopic properties of the heme group would serve as a useful reporter of the folded state of our synthetic proteins and that electrochemical measurements would serve as a simple and sensitive assay for functional integrity.
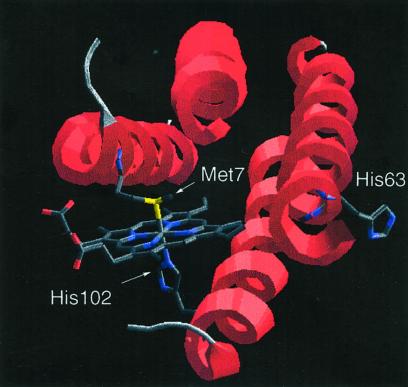
Cyt b562 is representative of a diverse family of four-helical bundle metalloproteins that are currently of great interest as the basis for protein engineering (22–25) and protein-folding studies (26, 27). The folded cyt b562 polypeptide coordinates a single heme cofactor through heteroatoms in the side chains of His-102 and Met-7 as axial ligands for the iron atom (Fig. 1) (28, 29). Inspection of the cyt b562 sequence suggested a convenient native chemical ligation site at the His-63–Gly-64 sequence at the N-terminal end of helix 3 near the heme binding pocket. Synthesis by ligation at this site would involve the reaction of 63- and 43-residue unprotected peptide segments. Although conversion of Gly-64 to a Cys residue would allow the synthesis of full-length mutant polypeptides using NCL reactions, we felt that folding of these mutants would likely be precluded by steric interactions between the introduced Cys side chain and the heme cofactor. In principle, placement of a D-Cys at residue 64 should relieve unfavorable steric interactions with the heme, but in practice we found that the ligated [D-Cys64]-containing cyt b562 polypeptide chain did not fold under the conditions examined, possibly due to the intrinsic helix-destabilizing effect of the D-amino acid (30). The auxiliary-mediated native chemical ligation strategy elegantly circumvents these issues as removal of the auxiliary group will leave a native Gly at position 64. We therefore placed a Gly(N-Aux) residue at position 64 for use in native chemical ligation of the two peptide segments.
Figure 1.
Molecular structure of recombinant wild-type cyt b562 showing the heme coordination environment and the location of the His-63–Gly-64 ligation site. Coordinates were obtained from the Protein Data Bank (K. Hamada, P. H. Bethge, F. S. Mathews, PDB accession no. 256B; see also ref. 28), and the illustration was made using DEEP VIEW in slab mode to provide an unobstructed view of the active site approximately along the four-helical bundle axis.
In designing the NCL removable auxiliary, the following factors were taken into account (12). The auxiliary must include a thiol group suitably disposed to mimic the side chain of the Cys residue that is crucial to thioester-mediated NCL reactions (10), hence the use of the Nα-(2-mercaptoethyl) moiety as the core building block in the auxiliary. Further, the auxiliary must be stable to conditions of peptide synthesis but removable after the ligation under conditions that do not damage the newly formed polypeptide product. For this reason, a suitably substituted 1-phenyl group was introduced into the auxiliary to generate a benzylamine derivative. It was anticipated that the resulting Nα-(1-phenyl-2-mercaptoethyl) moiety would be stable to HF cleavage conditions when linked to the α-amino group of a peptide. We reasoned that protonation of the amino group under strongly acidic conditions would result in a positive charge that would disfavor formation of a nascent carbocation at the benzylic position. However, after participating in the ligation reaction, the former N-terminal amino group becomes integrated into the polypeptide as a backbone amide, and thus the auxiliary can then be cleaved by HF treatment. Indeed, we found this to be the case; peptides containing Nα-linked 1-phenyl-2-mercaptoethyl moieties were readily cleaved from their solid supports with intact auxiliary groups on their amino termini, as shown by ES-MS, and model studies showed facile removal of the auxiliary after ligation (12). In the work reported here, we have applied this auxiliary-mediated native chemical ligation to the total synthesis of a functional protein molecule, cytochrome b562.
Results and Discussion
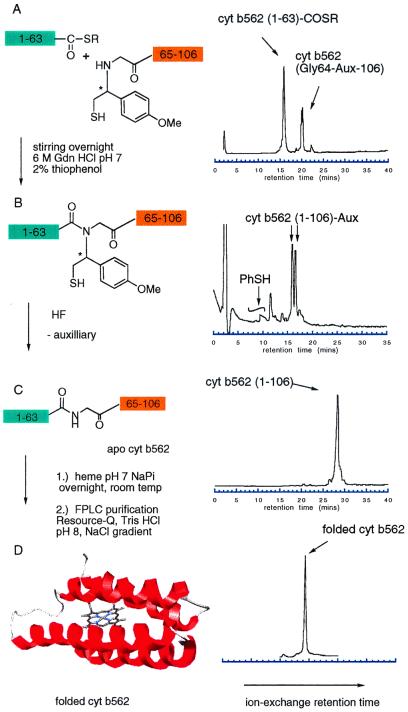
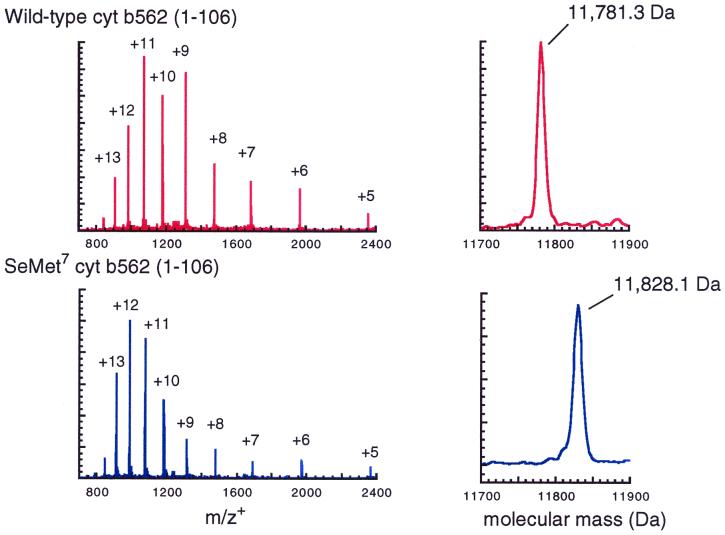
Our two-segment auxiliary-mediated NCL strategy required the preparation of a total of three peptide segments for synthesis of both the wild-type and the [SeMet7]cyt b562 protein molecules. We prepared cyt b562(1–63) with either Met or SeMet at position 7 using highly optimized Boc chemistry solid-phase peptide synthesis (11) on a thioester-generating resin (31, 32). The third peptide segment, cyt b562(64–106), was also prepared by using Boc chemistry solid-phase peptide synthesis, with the Nα-(1-phenyl-2-mercaptoethyl) auxiliary appended to the peptide N terminus by polymer-supported organic chemistry (12, 33). Following deprotection and cleavage from the resin by treatment with HF and HPLC purification, the two cyt b562(1–63) thioester segments and the cyt b562(Gly64(Aux)-106) segment were ready for use in ligation reactions. A 1.5-fold excess of each (1–63) segment was used in separate ligation reactions with the (Gly64(Aux)-106) segment, at 5 mM peptide concentration in pH 7 aqueous buffer with 0.5–2% thiophenol added. All auxiliary-modified C-terminal peptides were consumed after stirring overnight as judged by analytical HPLC (Fig. 2 A and B). Product polypeptides were then isolated by semipreparative HPLC and lyophilized. Full-length polypeptides containing the 1-phenyl-2-mercaptoethyl auxiliary-modified Gly at residue 64 were then treated with anhydrous HF to remove the auxiliary group, yielding the full-length unmodified apoproteins (Fig. 2C). ES-MS measurements were consistent with removal of the 1-phenyl-2-mercaptoethyl group, resulting in the expected native backbone polypeptides (Fig. 3).
Figure 2.
Ligation reactions between cyt b562(1–63) and cyt b562 (Gly64(Aux)-106) immediately after the peptides were dissolved in 6 M guanidine hydrochloride, pH 7 (A), and after stirring at room temperature overnight (B). Peptides bearing 1-phenyl-2-mercaptoethyl auxiliaries were prepared as diastereotopic mixtures due to the presence of the chiral center (denoted by the asterisk). These diasteromers are often seen to elute as two resolved peaks in HPLC chromatograms. A large peak in the chromatogram after overnight reaction due to elution of thiophenol catalyst was truncated for clarity. Following peptide ligation, the auxiliary was removed by treatment with anhydrous HF (C). The resulting holoprotein finally was folded in the presence of heme and purified by ion-exchange chromatography (D).
Figure 3.
Electrospray mass spectra of synthetic cyt 562 (Upper) and [SeMet7]cyt b562 (Lower). Molecular mass data were obtained from the unprocessed charge state spectra shown at left using the hypermass reconstruction function in the MacSpec software package. Measured molecular masses are in agreement with calculated masses (average isotope composition) to within ±1.5 Da.
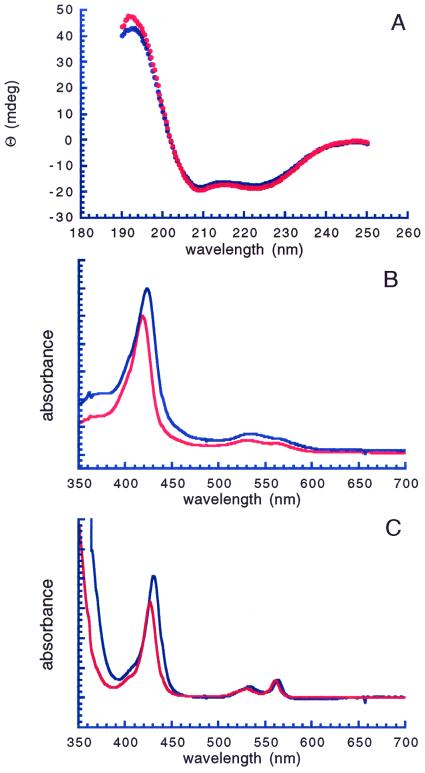
The apoproteins were then folded in the presence of heme (34) and subjected to a final purification by ion exchange FPLC (Fig. 2D). Analytical ion-exchange chromatograms of the crude folding mixture exhibited a single sharp peak corresponding to reconstituted protein, indicating that heme incorporation was essentially quantitative (Fig. 2D). UV circular dichroism spectra of synthetic cyt b562 and [SeMet7]cyt b562 were virtually identical (Fig. 4A), corresponding to the spectra expected for high α-helical content, indicating that the presence of the SeMet residue did not significantly perturb the overall secondary structure of [SeMet7]cyt b562. The optical spectra of oxidized and chemically reduced synthetic cyt b562 were in excellent agreement with literature values for the Escherichia coli-derived protein (35) (oxidized, λmax = 418, 530 nm; reduced λmax = 427, 531, 562 nm, Fig. 4B). In its oxidized Fe3+ form, heme iron has five unpaired electrons, and a low-spin configuration with a single unpaired electron is commonly found in cytochrome-type proteins with strong electron-donating axial ligands. As expected, the optical spectrum of the oxidized Fe3+ [SeMet7]cyt b562 analogue is consistent with a six-coordinate low-spin heme iron, indicating that the selenium is coordinated to the heme (36). The absorption bands for oxidized and reduced [SeMet7]cyt b562 (oxidized, λmax = 423, 533 nm, reduced, λmax = 430, 533, 564 nm, Fig. 4C) were shifted compared with the analogous bands in wild-type cyt b562, consistent with observations of other metalloproteins that have sulfur to selenium substitutions in their ligand spheres (37–39). For instance, the replacement of the native methionine 80 with selenomethionine in cytochrome c by semisynthesis results in a red-shift in Soret absorbance in the oxidized form from 408 to 412 nm (37).
Figure 4.
(A) UV circular dichroism spectra of synthetic wild-type (red) and [SeMet7]cyt b562 (blue). Spectra shown were corrected for baseline absorption but were otherwise unprocessed. (B) UV-visible spectra of oxidized synthetic cyt b562 (red) and [SeMet7]cyt b562. (C) UV-visible spectra of reduced synthetic cyt b562 (red) and [SeMet7]cyt b562.
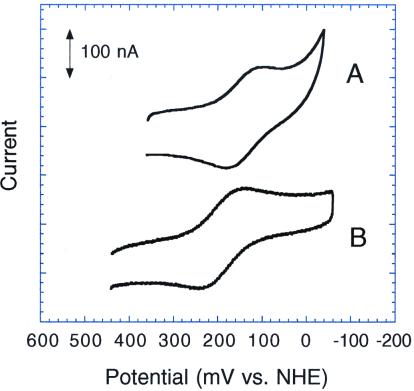
Cyt b562 is believed to be an electron transfer protein (40), and we therefore opted to use electrochemical measurements as a simple and sensitive functional assay for integrity of the protein redox center. We predicted that in [SeMet7]cyt b562, coordination of selenium, with its more extensive 4p electronic configuration, should act as a stronger electron donor to the heme iron compared with sulfur, a lighter group IVB element. More efficient electron donation should serve to stabilize the Fe(III) state relative to the Fe(II) state, resulting in a lowered reduction potential for the [SeMet7]cyt b562 analogue. This prediction was borne out by electrochemical measurements on the synthetic wild-type and [SeMet7]cyt b562 proteins. Both exhibited well behaved, reversible electrochemistry at a gold electrode in buffered aqueous solution in the presence of a small peptide mediator (41). As determined by cyclic voltammetry, synthetic cyt b562 had a reduction potential of −55 mV vs. standard calomel electrode, identical to recombinant cyt b562 (41), whereas we found a potential of −99 mV for SeMet7 cyt b562 (Fig. 5). This ≈45 mV potential shift is in excellent agreement with that observed by Wallace and Clark-Lewis for their semisynthetic SeMet80 cytochrome c mutant (37), whose potential is shifted by −48 mV compared with the methionine-ligated wild-type cytochrome c. This effect is much larger than that observed for the analogous Met ↔ SeMet change in the purple CuA domain from cytochrome ba3; presumably, this difference is a result of the relatively strong bonding between heme iron and its axial ligands compared with the relatively weak copper methionine interaction in the CuA domain (38).
Figure 5.
Cyclic voltammograms of [SeMet7]cyt b562 (A) and synthetic wild-type cyt b562 (B). Data were corrected for background and processed to remove 60-MHz noise. Potentials reported are accurate to within ±10 mV.
Conclusions
In this article, we have described the total chemical synthesis of cytochrome b562, a protein with no Cys residues, using a novel native chemical ligation strategy that employs a thiol-containing removable auxiliary moiety. The protein folded properly and exhibited the expected spectroscopic and functional properties. We further demonstrated the utility of our auxiliary-mediated ligation approach to site-specific incorporation of unnatural amino acids for protein engineering by preparing a cytochrome b562 protein analogue featuring a noncoded selenomethionine as axial ligand for the heme iron atom. Whereas the relatively conservative change of sulfur to selenium resulted in only modest changes in functional and spectroscopic properties between wild-type and [SeMet7]cyt b562, approaches based on total chemical synthesis will facilitate further engineering of heme protein active sites with unprecedented flexibility.
With straightforward synthetic access to cyt b562 established, we can now undertake a number of interesting experiments. These include a systematic examination of ligand properties on heme reactivity (i.e., classic coordination chemistry in a protein matrix) (16, 17); the synthesis of heme proteins tailored to dissect the complex array of factors that determine the kinetics of protein folding (27); and the design of novel active sites for small molecule binding and activation (24). We are now able to uniquely control every aspect of the covalent chemical structure of this heme protein, in an atom-by-atom fashion, at one or more specific sites anywhere in the protein molecule. Such experiments are made possible only through the use of chemical protein synthesis and potentially could be of great significance for elucidating the molecular basis of protein function.
These syntheses represent the first successful applications of the removable auxiliary native chemical ligation strategy to protein targets. The method overcomes the sequence restrictions imposed on previous peptide ligation strategies and represents a critical step in the expansion of chemical synthesis to any native or engineered protein regardless of sequence. Chemical synthesis of proteins is fast, precise, and versatile; it is inherently not limited to the genetically coded amino acids as building blocks and enables the incorporation of any number and type of substitutions throughout the protein molecule.
Acknowledgments
We thank Jennifer C. Lee for assistance with cyt b562 folding conditions. D.W.L. received funding from National Institutes of Health Postdoctoral Fellowship GM190402. M.G.H. is the recipient of a grant from the Packard Foundation. A preliminary account of this work was presented at the 26th European Peptide Symposium, Montpellier, France, September 9–15, 2000.
Abbreviations
- NCL
native chemical ligation
- ES-MS
electrospray mass spectrometry
References
- 1.Wilken J, Kent S B H. Curr Opin Biotech. 1998;9:412–426. doi: 10.1016/s0958-1669(98)80016-5. [DOI] [PubMed] [Google Scholar]
- 2.Kochendoerfer G G, Kent S B H. Curr Opin Chem Biol. 1999;3:665–671. doi: 10.1016/s1367-5931(99)00024-1. [DOI] [PubMed] [Google Scholar]
- 3.Schnolzer M, Kent S B H. Science. 1992;256:221–225. doi: 10.1126/science.1566069. [DOI] [PubMed] [Google Scholar]
- 4.Camarero J, Muir T W. Curr Protocol Protein Sci. 1999;18:1–21. [Google Scholar]
- 5.Saxon E, Armstrong J I, Bertozzi C R. Org Lett. 2000;2:2141–2143. doi: 10.1021/ol006054v. [DOI] [PubMed] [Google Scholar]
- 6.Tam J P, Yu Q, Miao Z. Biopolymers. 1999;51:311–332. doi: 10.1002/(SICI)1097-0282(1999)51:5<311::AID-BIP2>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- 7.Muir T W, Dawson P E, Kent S B H. Methods Enzymol. 1997;289:266–298. doi: 10.1016/s0076-6879(97)89052-0. [DOI] [PubMed] [Google Scholar]
- 8.Dawson P E, Kent S B H. Annu Rev Biochem. 2000;69:923–960. doi: 10.1146/annurev.biochem.69.1.923. [DOI] [PubMed] [Google Scholar]
- 9.Dawson P E, Muir T W, Clark-Lewis I, Kent S B H. Science. 1994;266:776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- 10.Canne L E, Bark S J, Kent S B H. J Am Chem Soc. 1996;118:5891–5896. [Google Scholar]
- 11.Schnolzer M, Alewood P, Jones A, Alewood D, Kent S B H. Int J Peptide Protein Res. 1992;40:180–193. doi: 10.1111/j.1399-3011.1992.tb00291.x. [DOI] [PubMed] [Google Scholar]
- 12.Botti P, Carrasco M R, Kent S B H. Tetrahedron Lett. 2001;42:1831–1833. [Google Scholar]
- 13.Miranda L P, Jones A, Meutermans W D F, Alewood P F. J Am Chem Soc. 1998;120:1410–1420. [Google Scholar]
- 14.Eichhorn G L, Marzilli L G. Heme Proteins. New York: Elsevier; 1988. [Google Scholar]
- 15.Bren K L, Gray H B. J Am Chem Soc. 1993;115:10382–10383. [Google Scholar]
- 16.De Pillis G, Decatur S M, Barrick D, Boxer S G. J Am Chem Soc. 1994;116:6981–6982. [Google Scholar]
- 17.Hirst J, Wilcox S K, Jingyuan A, Moenne-Loccoz P, Loehr T M, Goodin D B. Biochemistry. 2001;40:1274–1283. doi: 10.1021/bi002090q. [DOI] [PubMed] [Google Scholar]
- 18.Low D W, Hill M G. J Am Chem Soc. 2000;122:11039–11040. [Google Scholar]
- 19.Low D W, Hill M G. J Am Chem Soc. 1998;120:11536–11537. [Google Scholar]
- 20.Lippard S J, Berg J M. Principles of Bioinorganic Chemistry. Mill Valley, CA: Univ. Sci. Books; 1994. [Google Scholar]
- 21.Raphael A L, Gray H B. Proteins. 1989;6:338–340. doi: 10.1002/prot.340060316. [DOI] [PubMed] [Google Scholar]
- 22.Springs S L, Bass S E, McLendon G L. Biochemistry. 2000;39:6075–6082. doi: 10.1021/bi0001675. [DOI] [PubMed] [Google Scholar]
- 23.Hill R B, Raleigh D P, Lombardi A, DeGrado W F. Acc Chem Res. 2000;33:745–754. doi: 10.1021/ar970004h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Uno T, Yukinari A, Moriyama Y, Ishikawa Y, Tomisugi Y, Brannigan J A, Wilkinson A J. J Am Chem Soc. 2001;123:512–513. doi: 10.1021/ja002079d. [DOI] [PubMed] [Google Scholar]
- 25.Shifman J M, Gibney B R, Sharp R E, Dutton P L. Biochemistry. 2000;39:14813–14821. doi: 10.1021/bi000927b. [DOI] [PubMed] [Google Scholar]
- 26.Brunet A P, Huang E S, Huffine M E, Loeb J E, Weltman R J, Hecht M H. Nature (London) 1993;364:355–358. doi: 10.1038/364355a0. [DOI] [PubMed] [Google Scholar]
- 27.Wittung-Stafshede P, Lee J C, Winkler J R, Gray H B. Proc Natl Acad Sci USA. 1999;96:6587–6590. doi: 10.1073/pnas.96.12.6587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hamada K, Bethge P H, Mathews F S. J Mol Biol. 1995;247:947–962. doi: 10.1006/jmbi.1995.0192. [DOI] [PubMed] [Google Scholar]
- 29.Arnesano F, Banci L, Bertini I, Faraone-Mennella J, Rosato A, Barker P D, Fersht A R. Biochemistry. 1999;38:8657–8670. doi: 10.1021/bi982785f. [DOI] [PubMed] [Google Scholar]
- 30.Krause E, Bienert M, Schindler P, Wenschuh H. J Am Chem Soc. 2000;122:4865–4870. [Google Scholar]
- 31.Hackeng T M, Griffin J H, Dawson P E. Proc Natl Acad Sci USA. 1999;96:10068–10073. doi: 10.1073/pnas.96.18.10068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hojo H, Aimoto S. Bull Chem Soc Jpn. 1991;64:111–117. [Google Scholar]
- 33.Zuckermann R, Kerr J M, Kent S B H, Moos W H. J Am Chem Soc. 1992;114:10646–10647. [Google Scholar]
- 34.Itagaki E, Palmer G, Hager L P. J Biol Chem. 1967;242:2272–2277. [PubMed] [Google Scholar]
- 35.Itagaki E, Hager L P. J Biol Chem. 1966;241:3687–3693. [PubMed] [Google Scholar]
- 36.Makinen M W, Churg A K. In: Iron Porphyrins: Part I. Lever A B P, Gray H B, editors. Reading, MA: Addison–Wesley; 1983. [Google Scholar]
- 37.Wallace C J, Clark-Lewis I. J Biol Chem. 1992;267:3852–3861. [PubMed] [Google Scholar]
- 38.Blackburn N J, Ralle M, Gomez E, Hill M G, Pastuszyn A, Sanders D, Fee J A. Biochemistry. 1999;38:7075–7084. doi: 10.1021/bi982500z. [DOI] [PubMed] [Google Scholar]
- 39.Fee J A, Mayhew S G, Palmer G. Biochim Biophys Acta. 1971;245:196–200. doi: 10.1016/0005-2728(71)90021-1. [DOI] [PubMed] [Google Scholar]
- 40.Iwasaki T, Oshima T. FEMS Microbiol Lett. 1996;144:259–266. doi: 10.1111/j.1574-6968.1996.tb08539.x. [DOI] [PubMed] [Google Scholar]
- 41.Barker P D, Butler J L, de Oliveria P, Hill H A O, Hunt N I. Inorg Chim Acta. 1996;252:71–77. [Google Scholar]