The causes of vascular diseases are complex and strongly influenced by environmental factors as well as genetic predisposition. Ultimately, these factors alter the structure and/or functional properties of the arterial role disrupting vascular homeostasis. Vascular smooth muscle cells (SMCs) have been implicated in the pathogenesis of vascular diseases including atherosclerosis, systemic and pulmonary hypertension, aortic aneurysm and dissection, post-transplant vasculopathy and restenosis post-PCI.1 In response to arterial injury and the release and activation of growth factors, contractile SMCs populating the tunica media of the arterial wall down-regulate a set of genes encoding SMC-restricted contractile proteins and concomitantly up-regulate genes involved in the secretion of extracellular matrix, cell migration, adhesion and proliferation.1, 2 Comprehensive understanding of the molecular programs regulating modulation of SMC phenotype will be necessary to develop preventive and targeted therapeutic strategies for vascular disease.
The MADS box transcription factor, serum response factor (SRF), and the SMC lineage-restricted transcriptional co-activator, myocardin, lie at the center of a molecular program regulating SMC differentiation and the contractile SMC phenotype.2–4 Most, but not all, genes encoding SMC contractile proteins contain CArG boxes, or SRF binding sites, in their promoter and/or transcriptional enhancers.3, 4 In response to a variety of intracellular signals including Rho/ROCK, MAP kinase and calcium, myocardin associates with SRF and the myocardin/SRF complex binds to CArG boxes activating transcription of SMC contractile genes.1, 4 Remarkably, forced expression of myocardin in embryonic stem cells is necessary and sufficient to activate endogenous SMC genes.5 Moreover, vascular SMCs are not observed in the aorta Myocd−/− null embryos.6 Ablation of the Myocd gene in neural crest-derived SMCs populating the great arteries causes a cell autonomous block in expression of genes encoding SMC contractile proteins.7 Consistent with these findings, ablation of the Myocd gene in primary aortic SMCs is associated with a dramatic decrease in SMC contractile protein expression accompanied by increased expression of extracellular matrix.7
Despite much progress over the past decade,1, 2, 4 fundamental questions related to the molecular mechanisms regulating SMC phenotype remain to be addressed. What triggers silencing of genes encoding SMC contractile proteins in response to arterial injury? Is modulation of SMC phenotype solely dependent upon binding of myocardin/SRF complexes to SMC CArG boxes? What role, if any, do transcriptional repressors play in silencing expression of genes encoding SMC contractile proteins? How do environmental signals impact chromatin structure in response to arterial injury? What role, if any, do microRNAs play in this process? Is suppression of SMC genes functionally coupled to the induction of genes associated with cell migration, adhesion and/or proliferation? The gradient of SMC phenotypes observed during embryonic development and in pathological circumstances mandates a responsive, finely-regulated, nuanced, molecular mechanism that coordinates SMC transcription and gene expression.
Previous studies have shown that SMC phenotype is an actively regulated via transduction of growth factors and biomechanical signals to the nucleus resulting in altered gene expression.1, 2, 4 Platelet derived growth factor (PDGF) plays a critical role in SMC differentiation and modulation of SMC phenotype.1, 2 PDGF-BB ligand binds to the PDGF receptor-β activating a Ras/Raf/MEK/ERK kinase cascade leading to phosphorylation of the ETS domain ternary complex factor, ELK-1.8 Phosphorylated ELK-1 (pELK-1) displaces myocardin from SRF leading to repression of SMC contractile genes.8 KLF-4, a member of the Kruppel-like family of transcriptional repressors,9 also plays a critical role in the molecular program regulating SMC phenotype.10 KLF-4 is induced following PDGF-BB exposure and PDGF signaling has been shown attenuate SRF/myocardin binding to SMC CArG boxes.10 In addition, KLF-4 has been implicated in epigenetic re-programming of SMCs.11 Recently it has been shown that microRNAs are also involved in regulating SMC differentiation and modulation of SMC phenotype.12 For instance, miRNA-143/145 targets KLF-4 and miRNA-143/145 has been implicated in the pathogenesis of pulmonary hypertension.12, 13
Several years ago, the Owens laboratory identified a novel G/C repressor element in the SM22α promoter which they demonstrated was involved in suppression of the SM22α gene in response to experimental arterial injury and atherosclerosis in murine models.14, 15 However, the mechanism(s) of G/C repressor-mediated transcriptional silencing was unclear. In this issue of Circulation Research, Salmon and colleagues utilized sophisticated methodologies to define the mechanism of action(s) of this G/C repressor element in vivo.16 As schematically illustrated in the figure, they show that in response to growth factors, oxidized phospholipids and experimental carotid artery injury a nuclear complex containing KLF4, pELK-1 and HDAC2 binds to the G/C repressor element resulting in epigenetic modification and suppression of the SM22α locus Moreover, they show that occupation of the G/C repressor by this tripartite complex is linked to the dissociation of myocardin from SRF and decreases SRF binding to the SM22α promoter. Of note, putative G/C repressor elements were also identified in transcriptional regulatory elements controlling other CArG box-dependent SMC contractile genes including SM-α-actin (ACTA2) and SM-myosin heavy chain (MYH11) genes suggesting, but as yet not proving, that in response to arterial injury the G/C repressor element may act to counter myocardin-mediated SMC contractile gene activation.
The coordinate activation of a transcriptional complex containing a ternary complex factor, ELK-1, a transcriptional repressor, KLF4, and a histone modifying enzyme, HDAC2, represents an efficient mechanism for the inducible suppression of a set of SMC genes in response to arterial injury. Chromatin immunoprecipitation (ChIP) assays coupled with protein proximity ligation assays, which detects in vivo protein-protein interactions at 40 nm resolution, revealed that in response to PDGF-BB or oxidized phospholipid KLF4 binds cooperatively with pELK-1 to the G/C repressor element. This is the first evidence that pELK-1-induced transcriptional repression of SM22α transcription extends beyond the capacity of pELK-1 to displace myocardin from SRF bound to SM22α CArG boxes. Consistent with the report of Wang et al.,8 the authors confirmed that in vivo, in response to PDGF signaling (and oxidized phospholipid), myocardin dissociates from SRF and SRF binding to the SM22α promoter is abrogated. These findings explain how a transcriptional activator, such as pELK-1, can via association with a transcriptional repressor, KLF4, repress transcription. Taken together these data demonstrate that following vascular injury the newly discovered G/C repressor element in concert with previously described SM22α CArG boxes mediate critical, and complementary, functions required for suppression of SM22α gene transcription, a mechanism possibly shared with other SMC contractile genes.
Epigenetic regulation of gene expression via post-translational modification of histones is a powerful mechanism to reversibly modulate, or reprogram, gene expression.17 Owens and colleagues have shown that following arterial injury histone modifications, mediated via HDAC2, 4 and 5, leads to hypo-acetylation and suppression of SMC contractile genes.11 Salmon et al extend these observations demonstrating that the association of HDAC2 with a KLF4-pELK-1 complex bound to the G/C repressor element plays a critical role in epigenetic regulation of the SM22α gene transcription.16 Sequential H3 acetylation ChIP assays show clearly that following vascular injury KLF4, pELK-1 and HDAC2 are present within the same chromatin fragments of the SM22α promoter which is dependent upon expression of each factor and an intact G/C repressor element. Moreover, this higher order complex was also enriched in chromatin fragments spanning the Acta2 and Myh11 promoters following carotid ligation suggesting this may represent a generalized epigenetic mechanism employed in response to arterial injury. Taken together, these data support a model wherein following arterial injury intracellular signals converge upon the conserved G/C repressor element facilitating occupation by a tripartite complex consisting of KLF-4 pELK-1 and HDAC2 (Fig. 1). This, in turn, leads to alterations in chromatin structure and epigenetic silencing the SM22α locus as well as other SMC contractile genes sharing the conserved G/C repressor element.
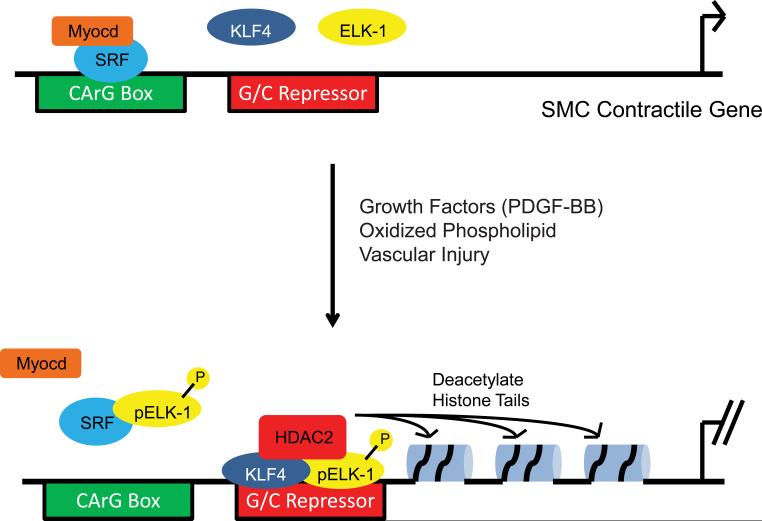
Figure. Molecular model of G/C Repressor-mediated transcriptional silencing of SMC contractile genes.
(Upper panel) In response to intracellular signals including Rho/ROCK, MAP kinase and calcium, the SMC-restricted transcriptional coactivator, myocardin (Myocd) (orange) physically associates with the transription factor, SRF (blue). The Myocd/SRF complex, in turn, binds to CArG boxes (green rectangle) activating transcription of multiple SMC contractile genes. (Lower panel) In response to growth factors, including PDGF-BB, oxidizied phospholipid and vascular injury, the ETS domain transcription factor ELK-1 (yellow) is phosphorylated via a MAP kinase signaling cascade promoting its association with the transcriptional repressor, KLF4 (dark blue), and the chromatin-modifying enzyme, HDAC2 (red). This tripartite complex binds to the G/C Repressor element (red rectangle) in the SM22 promoter suppressing transcription. In addition, HDAC2 deacetylates histone tails altering chromatin structure and suppressing transcription. Phosphorylated ELK-1 (pELK-1) also associates with SRF displacing myocardin and abrogating binding of SRF to SMC CArG boxes. Putative G/C Repressor elements have also been identified in the Acta2 and Myh11 promoters.
These unanticipated findings provide exciting new insights into the molecular mechanisms regulating suppression of the contractile SMC program in the setting of arterial injury. However, further research is needed to determine whether this represents a generalized mechanism involving the full repertoire of SMC contractile genes, or is restricted to a subset of genes encoding SMC contractile proteins. It will be interesting to examine if, and how, microRNAs regulate activity and/or expression of KLF-4, pELK-1 and/or HDAC2 following arterial injury. This still leaves open the question of how the induction of genes encoding extracellular matrix and factors involved cell migration, adhesion and proliferation is regulated and coordinated in development and vascular disease. In this regard it is noteworthy that conditional ablation of the Myocd gene in neural crest-derived SMCs not only suppressed genes encoding SMC contractile proteins, but led to ultrastructural changes indicative of increased protein synthetic function and secretion of extracellular matrix,7 suggesting that at some level the SMC contractile and synthetic gene programs may be functionally coupled. Ultimately, each of these avenues of investigation will be necessary to identify critical nodal points in the molecular circuitry regulating SMC phenotype in order to develop targeted therapies for acquired and heritable forms of vascular disease.
Acknowledgments
Sources of Funding This manuscript was supported in part from NIH R01-HL102968 and R01-HL094520 to M.S.P.
Footnotes
Disclosures None.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Owens GK, Kumar MS, Wamhoff BR. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol Rev. 2004;84:767–801. doi: 10.1152/physrev.00041.2003. [DOI] [PubMed] [Google Scholar]
- 2.Mack CP. Signaling mechanisms that regulate smooth muscle cell differentiation. Arterioscler Thromb Vasc Biol. 2011;31:1495–1505. doi: 10.1161/ATVBAHA.110.221135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Parmacek MS. Myocardin-related transcription factors: Critical coactivators regulating cardiovascular development and adaptation. Circ Res. 2007;100:633–644. doi: 10.1161/01.RES.0000259563.61091.e8. [DOI] [PubMed] [Google Scholar]
- 4.Pipes GC, Creemers EE, Olson EN. The myocardin family of transcriptional coactivators: Versatile regulators of cell growth, migration, and myogenesis. Genes Dev. 2006;20:1545–1556. doi: 10.1101/gad.1428006. [DOI] [PubMed] [Google Scholar]
- 5.Du KL, Ip HS, Li J, Chen M, Dandre F, Yu W, Lu MM, Owens GK, Parmacek MS. Myocardin is a critical serum response factor cofactor in the transcriptional program regulating smooth muscle cell differentiation. Mol Cell Biol. 2003;23:2425–2437. doi: 10.1128/MCB.23.7.2425-2437.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li S, Wang DZ, Wang Z, Richardson JA, Olson EN. The serum response factor coactivator myocardin is required for vascular smooth muscle development. Proc Natl Acad Sci U S A. 2003 doi: 10.1073/pnas.1233635100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Huang J, Cheng L, Li J, Chen M, Zhou D, Lu MM, Proweller A, Epstein JA, Parmacek MS. Myocardin regulates expression of contractile genes in smooth muscle cells and is required for closure of the ductus arteriosus in mice. J Clin Invest. 2008;118:515–525. doi: 10.1172/JCI33304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wang Z, Wang DZ, Hockemeyer D, McAnally J, Nordheim A, Olson EN. Myocardin and ternary complex factors compete for srf to control smooth muscle gene expression. Nature. 2004;428:185–189. doi: 10.1038/nature02382. [DOI] [PubMed] [Google Scholar]
- 9.Haldar SM, Ibrahim OA, Jain MK. Kruppel-like factors (klfs) in muscle biology. J Mol Cell Cardiol. 2007;43:1–10. doi: 10.1016/j.yjmcc.2007.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Liu Y, Sinha S, McDonald OG, Shang Y, Hoofnagle MH, Owens GK. Kruppel-like factor 4 abrogates myocardin-induced activation of smooth muscle gene expression. J Biol Chem. 2005;280:9719–9727. doi: 10.1074/jbc.M412862200. [DOI] [PubMed] [Google Scholar]
- 11.McDonald OG, Owens GK. Programming smooth muscle plasticity with chromatin dynamics. Circ Res. 2007;100:1428–1441. doi: 10.1161/01.RES.0000266448.30370.a0. [DOI] [PubMed] [Google Scholar]
- 12.Albinsson S, Sessa WC. Can micrornas control vascular smooth muscle phenotypic modulation and the response to injury? Physiol Genomics. 2011;43:529–533. doi: 10.1152/physiolgenomics.00146.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Caruso P, Dempsie Y, Stevens H, McDonald RA, Long L, Lu R, White K, Mair K, McClure JD, Southwood M, Upton P, Xin M, van Rooij E, Olson E, Morrell NW, Maclean MR, Baker AH. A role for mir-145 in pulmonary arterial hypertension. Circ Res. 2012 doi: 10.1161/CIRCRESAHA.112.267591. [DOI] [PubMed] [Google Scholar]
- 14.Wamhoff BR, Hoofnagle MH, Burns A, Sinha S, McDonald OG, Owens GK. A g/c element mediates repression of the sm22alpha promoter within phenotypically modulated smooth muscle cells in experimental atherosclerosis. Circ Res. 2004;95:981–988. doi: 10.1161/01.RES.0000147961.09840.fb. [DOI] [PubMed] [Google Scholar]
- 15.Regan CP, Adam PJ, Madsen CS, Owens GK. Molecular mechanisms of decreased smooth muscle differentiation marker expression after vascular injury. J Clin Invest. 2000;106:1139–1147. doi: 10.1172/JCI10522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Salmon M, Gomez D, Greene E, Shankman L, Owens GK. Cooperative binding of klf4, pelk-1 and hdac2 to a g/c repressor element in the sm22alpha promoter mediates transcriptional silencing during smc phenotypic switching in vivo. Circulation Research. 2012 doi: 10.1161/CIRCRESAHA.112.269811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chang CP, Bruneau BG. Epigenetics and cardiovascular development. Annu Rev Physiol. 2012;74:41–68. doi: 10.1146/annurev-physiol-020911-153242. [DOI] [PubMed] [Google Scholar]