Abstract
The composition of the intestinal microbial community is a distinctive individual trait that may divergently influence host biology. Because dendritic cells (DC) regulate the quality of the host response to microbiota, we evaluated DC in mice bearing distinct enteric microbial communities divergent for colitis susceptibility. Surprisingly, a selective, systemic reduction of plasmacytoid dendritic cells (pDC) was observed in isogenic mice with different microbiota: restricted flora (RF) vs specific pathogen free (SPF). This reduction was not observed in germfree mice, suggesting that the pDC deficiency was not simply due to a lack of intestinal microbial products. The microbial action was linked to cytotoxic CD8+ T cells, since pDC in RF mice were preserved in the CD8−/− and perforin−/− genotypes, partially restored by anti-CD8β Ab, and augmented in SPF mice bearing the TAP−/− genotype. Direct evidence for pDC cytolysis was obtained by rapid and selective pDC depletion in SPF mice transferred with RF CD8+ T cells. These data indicate that commensal microbiota, via CTL activation, functionally shape systemic immune regulation that may modify risk of inflammatory disease.
Among APCs, dendritic cells (DC)3 are particularly important in bridging innate and adaptive immunity due to their deployment at barrier sites of Ag encounter, their efficient trafficking to central lymphoid sites of T cell recruitment, and the exquisite tissue and environmental regulation of their Ag-presenting traits (1, 2). Human and murine DC are categorized by their morphology, function, and expression of cell surface markers and cytokines (3). In mice, there are three major subtypes of DC in spleen and lymph nodes: myeloid DC (mDC; CD11chigh CD11b+B220−; also termed conventional DC), CD8+ DC (CD11chighCD11b−B220−CD8α+), and plasmacytoid DC (pDC; CD11clowCD11b−B220+), which often can be further distinguished by two specific markers, 120G8 (4) and mPDCA-1 (5). pDC are distinguished by their large amounts of type I IFN in response to virus, termed natural IFN-producing cells (6, 7). Although only moderately active for Ag uptake and presentation, they promote Th1 and Th2 cell polarization depending on the stimuli (7), and there is growing interest that pDC contribute to induction of the tolerogenic phenotype. For example, human pDC induce CD4+CD25+ T regulatory (Treg) cells (8) and murine pDC stimulated with inhibitory ligands such as CTLA-4-Ig or OX2-Ig induce the IDO, which has a strong inhibitory activity on T cell proliferation (9, 10).
Activation and maturation of DC can be triggered by various microbial stimuli, notably through TLR sensing (11–13). Curiously, when specific pathogen-free (SPF) and germfree (GF) mice are compared, the abundance, maturation state, and Ag presentation proficiency of DC are similar (14). Thus, the effect and mechanism of enteric microbial influence on systemic differentiation and activation of DC in vivo remains poorly understood.
The enteric microbiota form an abundant and highly divergent community comprised of as high as 1010–1011/g contents and as many as 400 distinct species in a human individual (15, 16). Conventional and molecular phylotyping indicate that enteric microbiota is acquired during infancy (predominantly of maternal origin), and its composition is a unique but stable trait of each individual throughout life (17). Most of these microbial taxa are as yet uncultivable and only minimally defined functionally. Accordingly, an important question is whether this abundant, biologically active community shapes functional traits of the host such as immune function and disease susceptibility.
A unique opportunity to model these issues emerged through a mouse colony bearing complex enteric microbiota distinct from typical SPF mice. A segregated mouse colony, established by H. Suit and R. Sedlacek at M.D. Anderson in the 1980s, was constructed by transfer of a small set of nonpathogenic anaerobic bacteria into antibiotic-treated mice and originally termed reduced flora (RF) mice. Derivative colonies using various mouse strains were produced at a number of academic centers by Caesarian section of mouse pups with adoptive RF mothers. Using conventional culture methods, RF mice are readily distinguished from SPF mice by the lack of diverse enteric bacterial aerobes or Gram-negative anaerobes and by a diversity of anaerobic Gram-positive species (see Materials and Methods). A RF colony was established at University of California, Los Angeles (UCLA) more than a decade ago (18). Molecular phylotyping of enteric microorganisms, comparing these mice and their isogenic SPF counterparts, revealed that RF mice, like SPF mice, harbored a large variety of bacteria and fungal phylotypes, but were distinguished by broad-based compositional differences (19). For example, the most abundant phylotypes in RF and SPF belonged to the Firmicutes and Acinetobacter, respectively. Relative differences in Firmicutes vs Bacteroidetes and other taxa are also a common distinguishing trait of enteric microbiota among humans (20, 21). In this report, we used quantitative PCR (qPCR) assays to provide additional useful indices for the distinct enteric microbiota of SPF and RF mouse colonies.
An initial analysis of RF mice revealed surprising systemic changes in T lymphocytes, including reduced naive CD4+ and CD8+ T cells, expanded memory CD8+ T cells, and increased levels of activated, Th1-polarized cells (22). These unusual traits were reversed when RF mice were rederived with SPF microbiota, indicating that the phenotype was not attributable to the genetic drift of the RF mouse colony, but through the action of commensal microbiota. Moreover, RF mice were resistant to colitis under genetic or adoptive transfer conditions that permit disease activity in SPF mice (23). The mechanisms linking these divergent enteric microbiota to systemic T lymphocytes are unknown.
In this study, we compared the effect of SPF vs RF microbiota on the systemic status of DC populations. We found that in RF mice, pDC were selectively deficient in spleen and mesenteric lymph nodes (MLN), accompanied by an increased prevalence of mDC and T cells with a proinflammatory phenotype. The pDC deficiency in RF mice was reversed by depletion of CD8+ T cells and in mice lacking perforin function. Moreover, pDC in SPF were increased in SPF mice deficient in perforin or CD8+ T cells. These findings indicate that microbiota shape the systemic DC population in a process involving recruitment of cytolytic CD8+ T cells.
Materials and Methods
Mice
SPF C57BL/6 wild-type (WT), TAP-1−/− (24), and perforin−/− mice (25) were purchased from The Jackson Laboratory. RF C57BL/6 WT (18, 19), CD8α−/− and CD4−/− mice were obtained from the RF colony of the UCLA Department of Radiation Oncology. This colony was originally established at UCLA in 1992 using adoptive mothers imported from the RF mouse colony of Dr. H. Suit at M.D. Anderson. Perforin−/− breeders on the C57BL/6 background were purchased from The Jackson Laboratory and rederived for breeding by Caesarian section and adoptive maternal rearing in the UCLA Radiation Oncology RF colony.
RF mice were validated quarterly for RF status by aerobic and anaerobic microbial cultures. Aerobic bacterial cultures were prepared immediately following resection of the cecum from each mouse using a sterile, disposable, plastic, calibrated, 1.0-ml inoculating loop (Evergreen Scientific). A uniform random sample of cecum material was transferred to standard, enriched, selective, and differential agar medium used for the isolation of aerobic and facultative anaerobic microorganisms (e.g., 5% sheep blood, colistin naladixic acid, and MacConkey agar plates). The plates were incubated for 48 h at 35°C in 5–10% CO2 and isolates were characterized using standard protocols. In parallel, anaerobic bacterial cultures also were performed in the same fashion, except that they were immediately transferred to standard, prereduced, enriched, selective, and differential agar medium used for the isolation of anaerobic microorganisms (PRAS pack; Anaerobe Systems). The plates were immediately transferred to an AnaeroPack jar and oxygen was removed using the Pack-Anaero system (Mitsubishi Gas Chemical America). The plates were incubated for 5 days at 35°C and isolates were characterized using standard protocols.
An example of such analysis is shown in Table I, determined by conventional selective culture conditions and biochemically characterization using the RapID ANA II system (Innovative Diagnostic Systems). RF mice displayed only two aerobic organisms (probable Lactobacillus and coagulase-negative Staphylococcus), which were also found in SPF mice. In contrast, SPF mice were distinguished by a variety of additional aerobic and facultative anaerobic isolates. RF mice displayed two culturable anaerobic species: one Bacteroides fragilis group (also detected in SPF mice) and one unique anaerobic isolate (a Prevotella-like species featuring unusual Gram-negative rod morphology and vancomycin sensitivity). In contrast, diverse culturable anaerobes were found in SPF mice. Specifically, all four SPF mice harbored Enterococcus spp. and Bacillus spp. unique to their group and three SPF mice had an additional Enterococcus-like isolate and two SPF mice had an additional Enterobacter cloacae.
Table I.
Aerobic, facultative anaerobic, and anaerobic cecal cultures from RF and SPF micea
| Aerobic Culture |
|||||||||
|---|---|---|---|---|---|---|---|---|---|
| Isolate Description | Identification | RF-1 | RF-2 | RF-3 | RF-4 | SPF-1 | SPF-2 | SPF-3 | SPF-4 |
| Gram-positive pleomorphic rod | Lactobacillus spp. | Many | Many | Many | Modb | Mod | Mod | Many | Few |
| Gram-positive, coagulase-negative cocci in clusters | Staphylococci coagulase negative | few | Mod | Few | Mod | Few | Few | Many | Few |
| Gram-positive, γ-hemolytic, catalase-negative cocci in pairs and chains | Enterococcus spp. | Few | Rare | Many | Few | ||||
| Gram positive, á-hemolytic, catalase negative cocci in pairs and chains | Enterococcus-like organism | Rare | Rare | Rare | |||||
| Gram-positive rod (possible two morphotypes wrinkled and mucoid) | Bacillus spp. | Rare | Rare | Rare (mucoid) | Rare (mucoid) | ||||
| Gram-negative, lactose-positive rod (possible two morphotypes) | Enterobacter cloacae | Few | Few | ||||||
| Large Gram-negative rod | B. fragilis group | Mod | Mod | Many | Many | Mod | Mod | Many | Mod |
| Unusual Gram- negative rod but SSR indicating Gram-positive organism | Possible Prevotella spp. | Many | Many | Many | Many | ||||
| Anaerobic Gram -positive rod (SRS, flat regular) | Gram-positive rod 1 | Many | Many | ||||||
| Anaerobic Gram-positive rod (SRS, flecked colony) | Gram-positive rod 2 | Mod | |||||||
| Anerobic Gram -positive rod (SRS, small) | Gram-positive rod 3 | Many | |||||||
| Anaerobic Gram- negative rod (RSS resistant to vancomycin) | Fusobacteriumspp. | Few | |||||||
Each column (RF-1, RF-2, etc.; SPF-1, SPF-2, etc.) presents results from cultures of individual RF and SPF mice, respectively.
Mod, Moderate; S, susceptible and R, resistant to vancomycin (5 μg), colistin (10 μg), and kanamycin (1,000 μg), respectively.
With respect to pathogen-free status, mice from our major commercial source (The Jackson Laboratory) routinely monitored by statistically valid sampling procedures combined with established serology, cecum culture, and molecular diagnostic infectious disease PCR for the absence of a panel of viral, fungal, parasite, and opportunistic organisms and bacterial pathogenic taxa, including Helicobacter spp. (http://jaxmice.jax.org/health/agents_list.html). At the UCLA Center for Health Sciences vivarium, health monitoring for mouse pathogens or the agents with a significant potential for interference with research in SPF or RF mice of different age groups from each animal room were performed by Charles River Diagnostics. All of the animals used for the studies were confirmed to be pathogen free.
Germfree (GF) mice were obtained from the National Institutes of Health Gnotobiotic Resource (College of Veterinary Medicine, North Carolina State University). Sterility of germfree mice was documented on a monthly basis by fecal Gram stain and aerobic and anaerobic cultures of the feces and bedding. For selected mice, sterility of cecal contents was documented Gram stain and cultures at the time of necropsy.
All of the mice used in this study were C57BL/6, female, and 4–9 wk old and were handled according to the guidelines and protocols approved by the UCLA animal research committee.
Antibodies
The following fluorescence-conjugated anti-mouse mAbs were purchased from BD Pharmingen: CD3ε (145-2c11), CD4 (RM4-4), CD8α (53–6.7), CD25 (7D4), CD80 (16-10A1), CD86 (GL1), I-A/I-E (2G9), CD19 (1D3), B220 (RA3-6B2), CD11c (HL3), CD11b (M1/70), H-2Db (KH95), CD16/CD32 (2.4G2), Foxp3 (FJK-16s), IFN-γ (XMG1.2), IL-10 (JES5-16E3), and IL-12p40 (C15.6). Purified anti-NK1.1 (PK136) and anti-CD8β (341) (BD Pharmingen) were used for in vivo depletion. Anti-mPDCA-1 was purchased from Miltenyi Biotec.
Sample collection for molecular microbial analyses
Intestinal lumen contents and intestinal epithelium-associated tissue samples were collected for analysis of enteric bacteria in SPF and RF mice. Briefly, age- and gender-matched SPF and RF mice were euthanized by isoflurane inhalation and the intestines were excised. The intestines were then separated into small and large intestines at the junction between terminal ileum and cecum. Lumen content samples from small and large intestines were collected by squeezing the intestinal tubes with a forceps from one end to another. Approximately 0.5 ml of intestinal tissue along with condensed lumen content was collected into 2-ml screw-cap tubes containing 1 ml of CLS-Y DNA lysis buffer with beads from a FastDNA kit (Qbiogene). After being vortexed vigorously for a few seconds, lumen content samples were immediately frozen on dry ice until DNA isolation.
For the detection of epithelium-associated microorganisms, the small and large intestinal tissues were further processed by releasing epithelium-associated cells with DTT treatment. Briefly, the small and large intestines were cut open longitudinally and washed in DMEM three times. The washed intestinal tissues were then cut into 0.5- to 1-cm segments and placed into 50-ml corning tubes containing 25 ml of DMEM with 1 mM DTT (Sigma-Aldrich). The intestine tissues were treated with DTT by shaking at 220 rpm for 40 min at 37°C. After DTT treatment, the tissues were filtered through a 100-μm cell strainer into another 50-ml conical tube. The DTT- released fractions were then spun down at 3000 rpm for 10 min at 4°C with a Beckman Coulter J6M centrifuge. The epithelium-associated cells were then resuspended in 10 ml of DMEM and 1–1.5 ml of cell suspension was transferred into Eppendorf tubes and spun down at 6000 rpm for 5 min. The pellets were then resuspended with ~0.5 ml of DNA lysis buffer and transferred into to 2-ml screw-cap tubes containing CLS-Y DNA lysis buffer and beads. The rest of the cell suspensions were processed for intraepithelial lymphocyte isolation for immunological analysis.
DNA extraction and qPCR assay for enteric bacteria
DNA was extracted from the intestinal samples described above with the FastDNA Spin Kit using CLS-Y as the lysis buffer and bead-beating the samples in a FastPrep Instrument for 40 s at a setting of 6 (Qbiogene). DNA was further purified and size-fractionated by electrophoresis in 0.6% agarose gels. DNA larger than 3 kb was excised without exposure to UV or ethidium bromide and recovered using the QIAquick Gel Extraction Kit (Qiagen) per the manufacturer's instructions, except that the gel pieces were not exposed to heat and the DNA was eluted in EB buffer diluted 1/10 with water.
Two index bacterial phylotypes, Firmicutes 458 and Lactobacillus 456 were assessed using nucleotide sequences of their rRNA-ITS regions. The Firmicutes 458 and Lactobacillus 456 sequences were deposited in Gen-Bank (National Center for Biotechnology Information) under accession numbers EU375461 and EU375462, respectively. Two bacterial phylotypes were quantified using real-time PCR assays: performed in a Bio-Rad iCycler MyiQ Real-Time Detection System (Bio-Rad). The phylotypes (Firmicutes 458 and Lactobacillus 456) were identified in prior rRNA gene analyses (our unpublished data). Sequence-selective primers were designed using the PRISE software. The amplification reactions were performed in iCycler iQ PCR Plates with Optical Flat 8-Cap Strips (Bio-Rad). Twenty-five-microliter reaction mixtures contained the following reagents: 50 mM Tris (pH 8.3), 500 μg/ml BSA, 2.5 mM MgCl2, 250 μM of each dNTP, 400 nM of each primer, 0.33 μl of intestinal DNA (described above), 2 μl of 10× SYBR Green I (Invitrogen), and 1.25 U TaqDNA polymerase. The Lactobacillus 456 primers were LactoITSF2 (AGGCGAAAGATGATGGAG) and LactoITSR3 (ATGTACTAGCTCTTGAGGT), which target a 117-bp fragment of the rRNA-ITS region from the Lactobacillus 456 phylotype. The Firmicutes 458 primers were TuricITSF12 (CCTAGAATTGATCTTTGAAAAC) and TuricITSR10 (CCGGCTCTTTGCTTACT), which target a 151-bp fragment of the rRNA-ITS region from the Firmicutes 458 phylotype. The thermal cycling conditions for the Lactobacillus assay were 94°C for 5 min; 36 cycles of 94°C for 20 s, 65.3°C for 30 s, and 72°C for 30 s, followed by 72°C for 2 min. The thermal cycling conditions for the Firmicutes assay were 94°C for 5 min, 36 cycles of 94°C for 20 s, 60.7°C for 30 s, and 72°C for 30 s, followed by 72°C for 2 min. At each cycle, accumulation of PCR product was measured by monitoring the increase in fluorescence of the dsDNA-binding SYBR Green dye. rRNA gene levels in the intestinal DNA were quantified from a standard curve comprised of a dilution series of cloned rRNA genes for each of the phylotypes. To increase the likelihood that the real-time signals were produced by amplification of the target sequences, PCR fragments from intestinal DNA were cloned into pGEM-T (Promega), and the nucleotide sequences of two clones for each assay were determined; in all cases, these experiments confirmed that the target sequences were being amplified (data not shown). Differences in the amounts of Firmicutes 458 and Lactobacillus 456 rRNA genes in RF and SPF intestinal samples were assessed using two-tailed Student's t tests.
Preparation of DC fractions
Low-density cell fractions were prepared as previously reported, with some modifications (12). Briefly, spleen or lymph nodes (mesenteric, axillary, or popliteal) were minced in Mg2+- and Ca2+-free HBSS and digested with 1 mg/ml collagenase (Sigma-Aldrich) and 0.2 mg/ml DNase I for 45 min at 37°C. EDTA was adjusted to 30 mM and incubated for 10 min at room temperature. For bone marrow, cell suspension were obtained by flushing femurs with PBS supplemented with 0.5% BSA (Sigma-Aldrich) and resuspended in ACK lysing buffer (BioWhittaker) at room temperature for 1 min to lyse RBC. The cells were then resuspended in HBSS. Tissue lysate or bone marrow cell suspension was filtered through a 100-μm nylon cell strainer and layered onto 15% Nycodenz (Accurate Chemical & Scientific) in RPMI 1640 containing 10% FCS (HyClone). Cells were separated by centrifugation at 450 × g for 20 min without brake. Low-density fractions at interface were collected and washed for analysis.
FACS analysis
Cells for FACS analysis were preincubated with anti-CD16/CD32 to block nonspecific Fc receptor binding and then stained with fluorescent dyeconjugated Abs (FITC, PE, PerCP, allophycocyanin). After staining, the cells were washed twice with FACS buffer (0.5% BSA in PBS buffer) and suspended in 2% paraformaldehyde for FACS analysis. The data were collected by FACSCalibur and analyzed by CellQuest software (BD Biosciences).
Intracellular cytokine staining
For staining cytokines produced by DC, cells in the low-density fraction as mentioned above were harvested and stimulated with 10 ng/ml PMA (Sigma-Aldrich) and 1 μg/ml ionomycin (Sigma-Aldrich) overnight. Brefeldin A (10 μg/ml; Sigma-Aldrich) was added during the last 2 h of culture. Cells were resuspended in FACS buffer and stained with anti-CD11c. Cells were then fixed with Cytofix/Cytoperm (BD Biosciences) and stained with anti-IL-12p40. For T cell cytokine staining, total splenic cell suspension was stimulated with PMA/ionomycin for 6 h followed with brefeldin A treatment during the last 2 h. Cells were surface stained with anti-CD4 or anti-CD8α and then fixed and permeabilized for intracellular staining with anti-IFN-γ or anti-IL-10.
Ab treatment
Purified anti-NK1.1 (PK136) and anti-CD8β (341) Abs containing no preservative were administered i.v. into RF mice at 100 μg/mouse. Mice in control groups were injected with isotype control Abs. Injection was repeated twice by the interval of 10 days. The mice were sacrificed 1 wk after the final injection.
Lumenal bacterial lysate preparation
Intestinal lumen contents of RF mice were collected and resuspended in 1 ml of PBS. In brief, 0.25 ml of buffer (PBS containing 2 mM MgCl2 and 1 mg/ml DNase) and 1 ml of glass beads were added to the lumenal suspension. Suspension was vortexed for 3 min to disrupt bacteria. The glass beads and unlysed cells were removed by centrifugation at 5000 × g for 10 min. Lumenal lysates were sterilized by using a 0.2-μm syringe filtration (Nalgene). Protein concentrations were determined using a Quick Start Bradford protein assay kit (Bio-Rad).
Bone marrow-derived DC induction
Bone marrow cell suspensions prepared from SPF mice were cultured at 2 × 106 cells/ml for 7 days in RPMI 1640 medium and supplements (1 mM sodium pyruvate, 2.5 mM HEPES, penicillin-streptomycin, 50 μM 2-ME; all from Invitrogen), 10% FCS, and 100 ng/ml Flt-3L (R&D Systems). Lumenal lysates prepared from SPF or RF mice lumen contents were added at protein concentrations of 4 μg/ml, and cultures were continued for subsequent 24 h. Yielded mixture of pDC and mDC were stained with CD11c-allophycocyanin, CD11b-PerCP, B220-FITC, and CD86-PE.
Adoptive transfer of CD8+ T cells
Splenic single cells were prepared and RBC were lysed by ACK lysing buffer (BioWhittaker). Cells were resuspended in PBS buffer containing 0.5% BSA and 2 mM EDTA. CD8α+ T cells were isolated using a CD8α+ T cell isolation kit (Miltenyi Biotec) according to the manufacturer's instructions. Purity of CD8α+ T cells was >95%. Purified CD8α+ T cells from RF mice were i.v. injected to SPF mice at 4 × 106 cells/mouse. Lysates prepared from intestinal lumen contents of RF mice were i.p. injected at 50 μg/mouse. Control SPF mice were injected with the same volume of saline. Mice were sacrificed 4 days after CD8+ T cell transfer, and splenic DC were prepared and stained for CD11c and mPDCA-1.
Statistics
Statistical analysis was conducted by using Prism software (GraphPad). Each sample group was tested and confirmed for normality (Gaussian distribution) before further analysis was conducted. Each data set was next subjected to a two-sided Student t test for two groups or ANOVA for three or more groups, with a 95% confidence interval. A value of p ≤ 0.05 was determined to be significant.
Results
Quantitative molecular microbial indices for RF mice
The microbial composition of RF mice has been routinely monitored from SPF mice by assessment with qualitative bacterial culture methods (see Materials and Methods). This culture assessment is only an index of the RF microbiota, because the actual makeup of the RF microbiota is complex (19), and with current technologies the comprehensive molecular assessment of enteric microbiota for colony monitoring is impractical. However, quantitative molecular microbial indices would be a useful addition for monitoring RF mouse colonies for the integrity of their distinct microbial state. Accordingly, we established qPCR assays for representative 16S bacterial phylotypes that might serve as quantitative indices distinguishing RF and SPF mice.
An example of such indicator molecular phylotypes is presented in Table I. Among cloned enteric bacterial 16S sequences isolated in our previous study (19), we selected two bacterial 16S sequences (here termed the Firmicutes 458 and Lactobacillus 456 phylotypes) that were recurrently cloned from SPF and RF mice, respectively. The nucleotide sequences of the rRNA-ITS regions from the Firmicutes 458 and Lactobacillus 456 phylotypes were deposited in GenBank (National Center for Biotechnology Information) under accession numbers EU375461 and EU375462, respectively. qPCR measurements for each of them were performed in RF and SPF intestinal samples (Table II). In samples of luminal contents, the Lactobacillus 456 phylotype was selectively elevated in RF mice. In epithelial-associated samples, the Firmicutes 458 phylotype was selectively reduced in RF mice. These data demonstrate that these two phylotypes can be used as quantitative indices for in the distinct intestinal microbiota between RF and SPF mice (19).
Table II.
Abundance of two bacterial phylotypes in RF and SPF intestinal samples measured by sequence-selective qPCR
| Sample Type | Phylotype | Mouse Type | n | Meana | SE | p b |
|---|---|---|---|---|---|---|
| Lumen | Lactobacillus 456 | RF | 20 | 45,851.7 | 1,645.2 | 0.026 |
| SPF | 12 | 5,695.4 | 275.6 | |||
| Epithelium associated | Lactobacillus 456 | RF | 14 | 793.0 | 196.8 | 0.097 |
| SPF | 6 | 394.1 | 116.3 | |||
| Lumen | Firmicutes 458 | RF | 20 | 2.5 | 0.2 | 0.083 |
| SPF | 12 | 199.3 | 10.32 | |||
| Epithelium associated | Firmicutes 458 | RF | 14 | 1.7 | 1.2 | 0.031 |
| SPF | 6 | 9.5 | 2.6 |
Means are copy number of rRNA genes per 1/1000 of the intestinal sample.
Mean comparisons were performed using Student's t test.
Frequency and differentiation of DC subset in microbiota-divergent mice
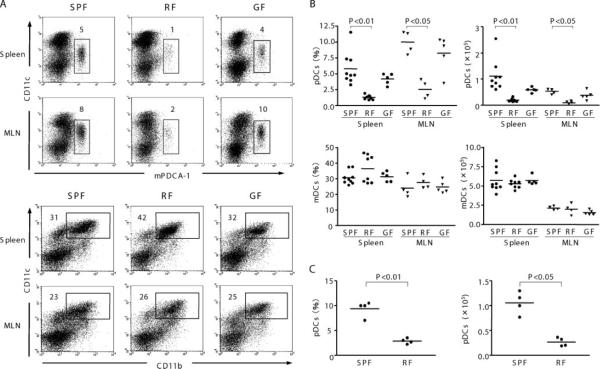
To assess whether distinct microbiota affect DC subsets, the frequency and absolute number of CD11chighCD11b+ (mDC) and CD11clowmPDCA-1+ (pDC) were determined in SPF, RF, and GF mice (Fig. 1, A and B). As previously reported (4), pDC were a minor population in C57BL/6 SPF mice. When compared with SPF mice, pDC were quite decreased in RF mice both in frequency (22 and 32% of SPF levels in spleen and MLN, respectively) and absolute number (17 and 24%, respectively). In contrast, no significant changes were observed for the frequency or absolute numbers of mDC in RF and GF mice compared with SPF mice. In addition, CD8+ DC (CD11chighCD11b−CD8α+) were also present in comparable levels in SPF and RF mice (14.3 ± 4.5% and 15.2 ± 0.6% of total DC in SPF and RF mice, respectively).
FIGURE 1.
Comparison of pDC and mDC in spleen and MLN from mice with different resident microbiota. Low-density fractions of spleen or MLN from SPF, RF, and GF mice were isolated and stained as described in Materials and Methods. A, pDC were stained with allophycocyanin-conjugated anti-CD11c and FITC-conjugated anti-mPDCA-1. mDC was stained with PerCP-conjugated anti-CD11b and allophycocyanin-conjugated CD11c. Numbers in dot plots represent percentages of total low-density cells from four to nine mice per group. B, Tabulation of pDC and mDC frequency (percentage of low-density cells) and absolute number in spleen. Values of p are calculated using ANOVA, comparing each group with RF mice. C, Tabulation of pDC frequency and absolute number in peripheral (axillary and popliteal) lymph nodes. Nonsignificant values (p ≥ 0.05) are not listed.
These data show that among DC subsets, pDC are selectively reduced in RF mice. We considered the possibility that this pDC deficit was due to the requirement for certain commensal microbial species that was absent in the RF microbial community. However, when we evaluated GF mice, which lack any detectable resident bacteria, the frequency and absolute numbers of pDC and mDC were similar to SPF mice (Fig. 1, A and B). These findings indicate that commensal microbiota were not required for the normal formation of pDC (or mDC) in peripheral tissues, as previously reported (14).
We also considered that the pDC deficiency in RF mice might be restricted to sites draining the intestinal tract. To assess this, pDC frequency in popliteal and axillary lymph node was also examined. As shown in Fig. 1C, pDC frequency and absolute number in peripheral lymph nodes of RF mice were lower than that of SPF mice. This observation further confirms that the pDC deficiency in RF mice was systemic.
Because enteric microbial products can be delivered to the bone marrow (26, 27), we wondered whether RF microbiota might inhibit pDC formation in the bone marrow. mPDCA-1 was undetectable in any cell population of the bone marrow; therefore, B220 was used in conjunction with other markers for the delineation of bone marrow pDC progenitors. In SPF bone marrow, cells with a pDC phenotype (CD11c+B220+CD11b−) were quite abundant at levels greater than in SPF spleen and MLN (Fig. 2, A and B). However, the same pDC frequency and absolute number were also observed in RF and GF mice. mDC (CD11c+B220−CD11b+) also were detectable in SPF bone marrow (at lower levels than pDC), and their frequency and absolute number were unaffected by the RF or GF state. A small population of bone marrow CD11b+B220+ double-positive DC was observed at similar levels in SPF, RF, and GF mice. This is notable, because cells with this phenotype have been attributed to progenitor DC populations (26). To further investigate whether RF microbiota might affect DC differentiation or maturation, the frequency of DC precursor (CD11c+MHCII−) and its maturation state were determined in SPF and RF mice (Fig. 2, C and D). However, no significant change was observed in RF mice. These data suggest that bone marrow delivery of Ag from RF microbiota is unlikely to affect DC formation in the bone marrow.
FIGURE 2.
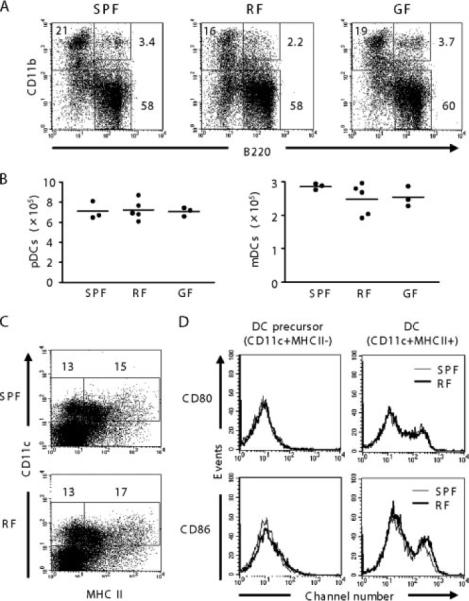
Effect of RF microbiota on bone marrow DC. A, Comparison of DC populations in bone marrow. DC were stained with allophycocyanin-conjugated anti-CD11c, FITC-conjugated anti-B220, and PerCP-conjugated anti-CD11b. B220 vs CD11b plot is shown after gating on CD11c+ cells. Numbers represent percentages of total CD11c+ cells from three to five mice per group. B, Tabulation of pDC and mDC absolute numbers in bone marrow. C, DC precursor (CD11c+MHCII−) and DC (CD11c+MHCII+) in bone marrow were compared between SPF and RF mice. Numbers represent percentages of total low-density cells. D, Maturation state of DC precursor and DC. Cells were gated for either CD11c+MHCII− or CD11c+MHCII+ and evaluated for CD80 and CD86 expression. Thin line, SPF; bold line, RF mice.
DC maturation and T cell polarization in RF mice
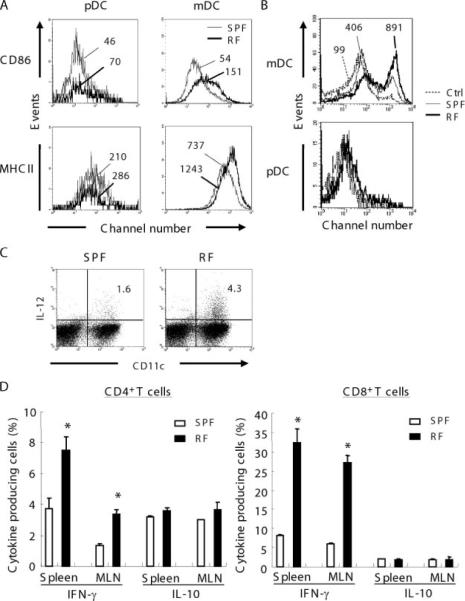
To assess the maturational state of the DC populations in RF mice, CD86 and MHCII expression levels in DC populations of the spleen and MLN were examined. CD86 and MHCII expression in pDC was equivalent in both SPF and RF mice (Fig. 3A). However, CD86 and MHCII expression was selectively increased in mDC from RF mice. These data suggest that the mDC population was shifted to a more mature state in RF mice.
FIGURE 3.
mDC maturation/activation phenotype and IFN-γ-producing T cells in RF mice. A, CD86 and MHCII expression on pDC or mDC were compared between SPF and RF mice. Thin line represents SPF and bold line represents RF mice. Numbers indicate MFI. B, Effect of lumenal lysate on bone marrow-derived DC. Bone marrow cells were cultured for 1 wk in the presence of Flt-3L, and lumenal lysate prepared from SPF or RF mice lumen contents were added and cultured for 24 h. CD86 data were gated on pDC (CD11c+B220+CD11b−) and mDC (CD11c+B220−CD11b+). Dotted line represents vehicle control and thin line and bold line represent SPF lumenal lysate and RF lumenal lysate, respectively. Numbers indicate MFI. C, IL-12 production from CD11chigh DC. Low-density cells from spleen of SPF or RF mice were cultured, stimulated with PMA/ionomycin, and stained with anti-IL-12 and anti-CD11c. Numbers are percentages of total low-density cells. Data are representative of at least three mice in two separate experiments. D, IFN-γ or IL-10 productions from T cells were compared between SPF and RF mice. Spleen cells prepared from SPF or RF mice were stimulated with PMA/ionomycin and stained with anti-IFN-γ or anti-IL-10. CD3ε+CD4+ cells were gated as CD4+ T cells and CD3ε+CD8+ were gated as CD8+ T cells. Values are the percentage of cytokine-producing cells among total CD4+ or CD8+ T cells. *, p < 0.01 for Student's t test comparing each group to SPF mice. Other comparisons are nonsignificant values (p ≥ 0.05).
To clarify whether mDC was a direct action of RF microbial Ags, cecal lumen lysates from SPF or RF mice were used to stimulate Flt-3L-induced bone marrow-derived DC. For mDC, both RF and SPF lumenal lysates increased CD86 expression, but induction was much higher with RF lysates (Fig. 3B). For pDC, neither RF nor SPF lumenal lysates substantially induced CD86. To assess the native mDC population, intracellular IL-12 was determined in the spleen low-density fraction after PMA/ionomycin. As shown in Fig. 3C, IL-12-expressing CD11chigh DC were 2.7-fold increased in RF vs SPF mice. It should be noted that this native DC preparation includes non-DC populations, which may directly respond to PMA/ionomycin and secondarily induce DC IL-12 expression. However, these data provide three lines of evidence that RF microbiota, in part through direct action on newly differentiated mDC, stimulate mDC maturation and IL-12 production.
CD8+ T cells primed with pDC produce substantial amount of IL-10 (28), suggesting that such IL-10 production might be reduced in RF mice, due to their deficiency in pDC. If so, reduced IL-10 production in RF mice might also enhance lymphocyte IFN-γ production, activating CTL or NK cell cytotoxicity. Therefore, IFN-γ and IL-10 production from T cell in SPF and RF mice were compared. As shown in Fig. 3D, IFN-γ production by CD4+ T cells in RF mice was 2.5-fold increased both in spleen and MLN compared with SPF mice. However, IL-10+CD4+ T cells in RF mice in splenic or MLN were unchanged from SPF control mice. IFN-γ+CD8+ T cells in RF mice were 4.4-fold increased in spleen and 4.5-fold in MLN compared with SPF mice. Again, IL-10+ CD8+ T cells were unchanged. These data suggest IFN-γ-producing CD4+ and CD8+ T cells are activated in RF mice, but do not implicate deficient IL-10 production in this process.
A requirement for CD8+ T cells in the pDC deficiency of RF mice
As IFN-γ production from CD4+ and CD8+ T cell were both elevated in RF mice, we tested whether CD4+ or CD8+ T cells were required for pDC deficiency. To do so, CD4 or CD8α null mice were rederived bearing RF microbiota and assessed for DC percentage and absolute number. As shown in Fig. 4A, RF CD8α−/− mice were significantly increased both in pDC percentage and absolute numbers compared with RF WT mice. In contrast, the pDC percentage in RF CD4−/− mice was not increased compared with WT RF mice. Curiously, pDC absolute numbers were significantly increased in CD4−/− vs WT RF mice. This discordance was due to a substantial expansion in absolute numbers of all residual splenic cells (B cells, CD8+ T cells, and DC) in RF CD4−/− mice. The reason for this complex population change in RF CD4−/− mice is uncertain. However, these data suggest that cells expressing the CD8α molecule are involved in pDC deficiency in RF mice.
FIGURE 4.
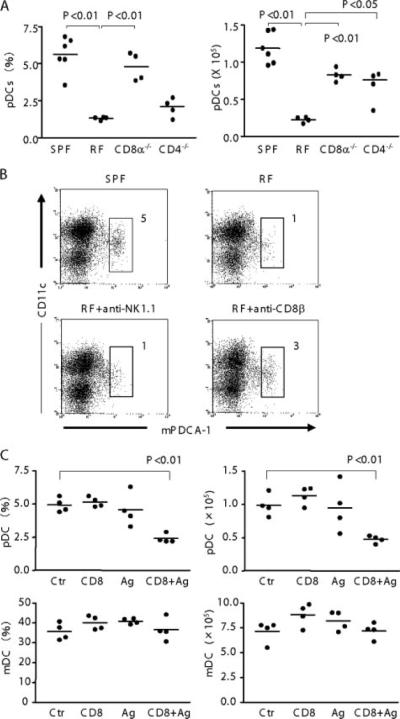
Role of CD8+ T cells in the pDC deficiency of RF mice. A, Low-density cells were prepared from spleen of SPF WT, RF WT, RF CD8α−/−, and RF CD4−/− mice. Cells were stained for pDC as in Fig. 1. pDC frequency (percentage of low-density cells) and absolute numbers in spleen are tabulated; p values were obtained using Student's t test for group comparisons. B, RF mice were treated with anti-NK1.1 and anti-CD8β to deplete NK cells or CD8αβT cells; irrelevant isotype treatment was used as a negative control. Mice received Ab injection twice and were sacrificed 1 wk later after the final injection. Low-density splenocytes were isolated and stained for flow analysis; numbers indicate percentage of total low-density cells. Data are representative of three mice per group. C, SPF mice were injected i.v. with CD8+ T cells from RF mice and injected i.p. with lumenal lysate prepared from RF mice (Ag) or saline (Ctr). Mice were sacrificed 4days later and the frequency (percentage of low-density cells) and absolute numbers of splenic pDC and mDC were determined. Values of p were obtained using Student's t test comparing each group with control mice. Nonsignificant p values (p ≥ 0.05) are not listed.
CD8α can be expressed not only on classic CD8αβ T cells, but also on other T cell and DC populations (29, 30). To assess which population is responsible for pDC deficiency, RF mice were treated with anti-CD8β or anti-NK1.1 Abs were treated against RF mice. Mice were treated with anti-CD8β, anti-NK1.1, or with iso-type-matched control Abs twice at 1-wk intervals. One week after the last challenge, mice were sacrificed and assayed for pDC in the spleen. CD8αβT cell or NK cell were confirmed for their depletion (data not shown). As shown in Fig. 4B, anti-CD8β Ab partially restored pDC but not anti-NK1.1 restored RF pDC levels. Similarly, isotype-matched Abs also did not restore RF pDC (data not shown). These findings indicate that pDC depletion in RF mice is dependent on CD8αβT cells.
CD8+ T cell cytotoxicity is required for the pDC deficiency in RF mice
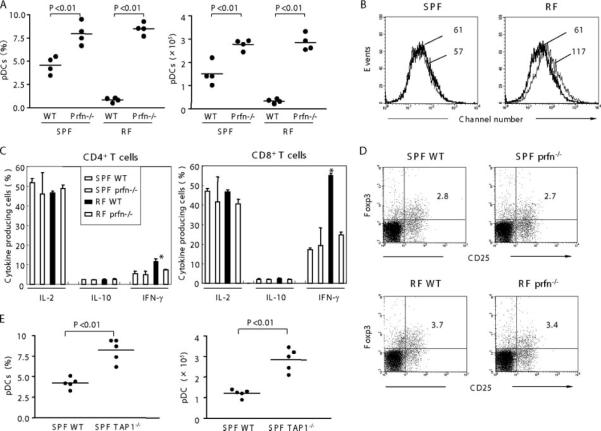
In terms of CD8+ T cell function, the pDC deficiency in RF mice may be due to CD8+ T cell cytolytic activity or other cytokines or cell-cell interactions which may impair pDC differentiation, homing, or survival. To test the former possibility, we rederived perforin−/− mice (25) bearing RF microbiota and examined them for pDC levels. As shown in Fig. 5A, splenic pDC levels in RF perforin−/− mice were significantly increased compared with RF WT mice, reaching levels exceeding that of SPF WT mice. The number of CD8+ T cells in perforin−/− mice bearing SPF or RF microbiota were comparable to their WT mice (data not shown). These data demonstrate that pDC depletion by CD8+ T cells involves a perforin-dependent CTL process.
FIGURE 5.
Effect of perforin null mutation on pDC in RF mice. A, Low-density cells from perforin−/− mice (Prfn−/−) and WT mice bearing either SPF or RF microbiota were prepared from spleen and stained for pDC. Splenic pDC frequency (percentage of low-density cells) and absolute numbers are tabulated. B, mDC was gated and compared for CD86 expression in WT mice and perforin−/− mice bearing either SPF or RF microbiota. Thin line, WT; bold line, perforin−/−. Numbers are MFI, representative of data from four mice. C, Cytokine production from WT and perforin−/− mice bearing either SPF or RF microbiota. D, Frequency of Foxp3+ Treg cells were determined in Prfn−/− and WT. Splenic cells were stained for CD3, CD4, CD25, and intracellular Foxp3. Data shown are gated on CD3+CD4+. E, Low-density cells from SPF TAP-1−/− mice and SPF WT mice were prepared from spleen and stained for pDC. Splenic pDC frequency (percentage of low-density cells) and absolute numbers are shown. Values of p were obtained using Student's t test (A and E) or ANOVA (C), comparing each group with WT SPF mice. *, In Fig. 5C indicates p < 0.05.
Mechanistically, the simplest explanation for these findings is that RF CD8+ T cells directly kill pDC. To test this idea, an in vivo cytotoxicity experiment was performed. Splenic CD8+ T cells from RF mice were sorted and injected into SPF mice, and the levels of splenic pDC were measured after 4 days. In some cases, RF lumenal microbial Ag (or saline control) was also administered. As shown in Fig. 4C, SPF pDC were rapidly reduced in mice administered RF CD8+ T cells and microbial Ag (50 and 33% of control frequency and absolute number, respectively). This reduction was dependent on both CD8+ T cells and microbial lysate, because neither alone affected pDC levels (Fig. 4C). At the same time, mDC was not significantly affected by either RF Ag nor RF CD8+ T cells. These findings provide evidence for cytotoxicity of RF CD8αβ T cells selectively against peripheral pDC. The requirement for lumenal microbial lysate also suggests that antigenic challenge is required for efficient T cell cytotoxicity.
RF perforin−/− mice were also notable for reductions of mDC CD86 expression and IFN-γ+ T cells to normal (SPF-like) levels (Fig. 5, B and C). This suggests that the elevated abundance of mature phenotype mDC and IFN-γ- producing T cells in RF mice might reflect a regulatory role of pDC on these populations. In this context, it is notable that pDC are reported to prime CD8+ T cells for IL-10 production and can promote CD4+CD25+ Treg cell formation (8, 28). To assess these possibilities, IL-10 production from T cells and Foxp3+ Treg cells were compared between perforin−/− mice and WT (Fig. 5, C and D); however, no significant change was observed. This suggests that pDC does not regulate IFN-γ expression via expansion of Treg or Tr1 populations.
Evidence for Ag-specific elimination of pDC by CD8+ T cells in SPF mice
We sought to determine whether the cytolytic elimination of pDC involved an Ag-specific CD8+ T cell process. The presentation of peptide Ag in association with MHC class I molecules is deficient in TAP-1−/− mice, with concomitant depletion of CD8+ T cells (24). We therefore evaluated DC levels in TAP-1−/− mice bearing SPF microbiota. Compared with SPF mice, pDC in TAP-1−/− mice bearing SPF microbiota were increased 2-fold in percentage and 2.5-fold in number (Fig. 5E). Similarly, pDC levels were significantly increased in SPF perforin−/− mice compared with WT SPF mice (Fig. 5A). These data suggest that CD8+ T cell-dependent pDC elimination is not restricted to RF mice and that pDC turnover by CD8+ T cells may also be a housekeeping event in SPF mice.
Discussion
In the natural population, individuals are divergent in their composition of resident commensal microbiota. This study demonstrates that commensal microbiota shape systemic levels of pDC. Systemic depletion of pDC was selectively observed in mice bearing RF microbiota. This depletion was associated with maturation and elevated constitutive IL-12 production of mDC and increased levels of IFN-γ-expressing CD4+ T cell and CD8+ T cells. Unexpectedly, several lines of evidence indicated pDC depletion was mediated by cytolytic CD8+ T cells. These data suggest a surprising interplay of resident commensal microbiota and cytolytic CD8+ T cells in systemic immunoregulation.
There are few precedents for selective DC deficiency for the pDC subset. For example, mice bearing a null mutation for the transcription factor Ikaros (IkL/L) lack peripheral pDC but not other DC subsets (31). With this in mind and the detectable delivery of microbial products to the bone marrow environment (26), it is conceivable that Ikaros expression by pDC progenitors might be dependent on such microbial products. However, pDC were not deficient in GF mice, indicating that normal pDC formation does not require the stimulus of resident microbiota. It is possible that products of RF microbiota might inhibit pDC formation. However, abundant CD11c+B220+CD11b− cells were present in bone marrow from RF mice. Thus, it appears that the pDC deficiency in RF mice is not due to impairment of pDC differentiation, but instead due to their elimination in peripheral tissues.
This study's central finding is the surprising role of cytotoxic CD8+ T cells mediating the effect of commensal microbiota on the pDC population. First, the pDC deficiency was averted when RF microbiota were introduced into mice with CD8α−/− or perforin−/− genotypes, whereas the pDC percentage remained deficient when they were introduced into CD4−/− mice. Second, pDC were rapidly restored in RF mice treated with anti-CD8β but not anti-NK1.1, implicating CD8αβ T cells, but not NK cells, as the functional basis for these genetic phenotypes (29, 30). Third, pDC were rapidly and selectively reduced in SPF mice transferred with RF CD8β+ T cells and lumenal Ag, providing evidence for their direct cytotoxicity against pDC in vivo. Fourth, pDC were increased in SPF mice bearing the TAP-1−/− genotype, suggesting that pDC depletion by CD8+ T cells involves cognate peptide/MHC class I recognition and occurs at detectable levels in SPF mice.
It is uncertain whether the interplay of RF microbiota and cytolytic CD8+ T cells involves direct antigenic targeting of pDC or is mediated through an intermediate cytolytic target affecting pDC formation, homing, or tissue persistence. These four lines of evidence, in particular, the rapid and selective pDC depletion in SPF mice by RF CD8+ T cells, are most simply explained by direct cytotoxic action of the pDC population. As a precedent, recent work has demonstrated that DC loaded with specific Ag can be eliminated by CTL in a perforin-dependent manner (32–34). One might imagine that such a process may account for the systemic delivery of RF microbial products to DC populations, resulting in their targeting by cognate CD8+ T cells. We note that this would not simply explain why the depletion selectively targets pDC vs mDC populations, unless pDC selectively express receptors suitable for uptake of the relevant RF Ags (35, 36).
Alternatively, the pDC population may involve novel cross-reactive targeting. For example, a conserved bacterial peptide presented on class Ib molecule can elicit cognate CD8+ T cells targeting host cells presenting endogenous cross-reactive Qa-1-restricted peptide (37) and invariant NK T cells target cross-reactive microbial and endogenous ligands associated with CD1 (38–40). Because APCs vary widely in expression of MHC class 1b molecules according to cell subset and anatomic location (41), it is conceivable that such antigenic specificity may be an unappreciated feature of the pDC subset of DC.
The mDC population was numerically unaffected in RF mice, but shifted to a more mature phenotype, with an associated elevation of IFN-γ+CD4+ and CD8+ T cells. The pDC population can attenuate DC maturation and effector T cell expansion through production of IDO, IFN-α, and recruitment or induction of invariant NKT or Foxp3+ Treg cells (35, 42–44). It is thus plausible that reduction of the pDC population in RF mice might contribute to the observed elevation in the basal abundance of mature mDC and activated T cell populations.
Surprisingly, functional studies of RF mice reveal their resistance to immune colitis induced by CD4+CD45RBhigh or Gαi2−/− T cells (23) and their increased colonization but reduced colonic inflammation in Campylobacter jejuni infection (45). CD4+ Treg cells play an important role in attenuating immune colitis (46, 47), notably by blocking differentiation or activation of Th17- and IL-23-producing cells (48, 49). Although pDC Ag presentation favors formation of CD4+ Treg cells (44, 50–52), pDC also can promote intestinal inflammation, in part through local IFN-α production (53). Accordingly, the immune functional consequence of pDC deficiency or other associated host immune changes in RF mice might promote intestinal homeostasis. Alternatively, CD8+ T cells may directly elicit immunoregulation (32–34, 54), a process attributed to depletion of Ag-presenting DC. In the present study, DC bearing enteric commensal Ags may be depleted by CD8+ T cells in RF mice, thus removing the antigenic drive for Th17 cell formation and activation.
In summary, this study reveals that the alternate commensal microbial communities, representative of the human divergence of enteric microbiota, can shape the systemic DC population through a novel mechanism involving cytolytic CD8+ T cells. Because such mice are also divergent in their susceptibility to immune or infectious colitis, this observation suggests an unexpected acquired trait that shapes host immunoregulation and immune disease susceptibility.
Acknowledgments
We thank El Khansa Kaicer for skillful technical support.
Footnotes
This work was supported by National Institutes of Health DK46763 (to J.B. and M.K.), DK69434 (to J.B. and B.W.), GM07185 (to M.M.), AI52031 (to S.B.), and CA016042 (Jonsson Comprehensive Cancer Center Flow Cytometry); the Crohn's and Colitis Foundation of America (to B.W.); and the Broad Medical Research Foundation (to J.B.). The germfree animal facility at the Center for Gastrointestinal Biology and Disease, North Carolina State University, was supported by Grant P30 DK349870 from National Institutes of Health.
Abbreviations used in this paper: DC, dendritic cell; mDC, myeloid DC; GF, germ-free; MLN, mesenteric lymph node; pDC, plasmacytoid DC; RF, restricted flora; SPF, specific pathogen free; MHCII, MHC class II.
Disclosures The authors have no financial conflict of interest.
References
- 1.Banchereau J, Steinman RM. Dendritic cells and the control of immunity. Nature. 1998;392:245–252. doi: 10.1038/32588. [DOI] [PubMed] [Google Scholar]
- 2.Villadangos JA, Schnorrer P. Intrinsic and cooperative antigen-presenting functions of dendritic-cell subsets in vivo. Nat. Rev. Immunol. 2007;7:543–555.2. doi: 10.1038/nri2103. [DOI] [PubMed] [Google Scholar]
- 3.Shortman K, Liu YJ. Mouse and human dendritic cell subtypes. Nat. Rev. Immunol. 2002;2:151–161. doi: 10.1038/nri746. [DOI] [PubMed] [Google Scholar]
- 4.Asselin-Paturel C, Brizard G, Pin JJ, Briere F, Trinchieri G. Mouse strain differences in plasmacytoid dendritic cell frequency and function revealed by a novel monoclonal antibody. J. Immunol. 2003;171:6466–6477. doi: 10.4049/jimmunol.171.12.6466. [DOI] [PubMed] [Google Scholar]
- 5.Fallarino F, Orabona C, Vacca C, Bianchi R, Gizzi S, Asselin-Paturel C, Fioretti MC, Trinchieri G, Grohmann U, Puccetti P. Ligand and cytokine dependence of the immunosuppressive pathway of tryptophan catabolism in plasmacytoid dendritic cells. Int. Immunol. 2005;17:1429–1438. doi: 10.1093/intimm/dxh321. [DOI] [PubMed] [Google Scholar]
- 6.Liu YJ. Dendritic cell subsets and lineages, and their functions in innate and adaptive immunity. Cell. 2001;106:259–262. doi: 10.1016/s0092-8674(01)00456-1. [DOI] [PubMed] [Google Scholar]
- 7.Colonna M, Trinchieri G, Liu YJ. Plasmacytoid dendritic cells in immunity. Nat. Immunol. 2004;5:1219–1226. doi: 10.1038/ni1141. [DOI] [PubMed] [Google Scholar]
- 8.Moseman EA, Liang X, Dawson AJ, Panoskaltsis-Mortari A, Krieg AM, Liu YJ, Blazar BR, Chen W. Human plasmacytoid dendritic cells activated by CpG oligodeoxynucleotides induce the generation of CD4+CD25+ regulatory T cells. J. Immunol. 2004;173:4433–4442. doi: 10.4049/jimmunol.173.7.4433. [DOI] [PubMed] [Google Scholar]
- 9.Mellor AL, Baban B, Chandler P, Marshall B, Jhaver K, Hansen A, Koni PA, Iwashima M, Munn DH. Induced indoleamine 2,3 dioxygenase expression in dendritic cell subsets suppresses T cell clonal expansion. J. Immunol. 2003;171:1652–1655. doi: 10.4049/jimmunol.171.4.1652. [DOI] [PubMed] [Google Scholar]
- 10.Fallarino F, Asselin-Paturel C, Vacca C, Bianchi R, Gizzi S, Fioretti MC, Trinchieri G, Grohmann U, Pucceti P. Murine plasmacytoid dendritic cells initiate the immunosuppressive pathway of tryptophan catabolism in response to CD200 receptor engagement. J. Immunol. 2004;173:3748–3754. doi: 10.4049/jimmunol.173.6.3748. [DOI] [PubMed] [Google Scholar]
- 11.Agrawal S, Agrawal A, Doughty B, Gerwitz A, Blenis J, Van Dyke T, Pulendran B. Cutting edge: different Toll-like receptor agonists instruct dendritic cells to induce distinct Th response via differential modulation of extracellular signal-regulated kinase-mitogen-activated protein kinase and c-Fos. J. Immunol. 2003;171:4984–4989. doi: 10.4049/jimmunol.171.10.4984. [DOI] [PubMed] [Google Scholar]
- 12.Sparwasser T, Koch ES, Vabulas RM, Heeg K, Lipford GB, Ellwart JW, Wagner H. Bacterial DNA and immunostimulatory CpG oligonucleotides trigger maturation and activation of murine dendritic cells. Eur. J. Immunol. 1998;28:2045–2054. doi: 10.1002/(SICI)1521-4141(199806)28:06<2045::AID-IMMU2045>3.0.CO;2-8. [DOI] [PubMed] [Google Scholar]
- 13.Edwards AD, Manickasingham SP, Sporri R, Diebold SS, Schulz O, Sher A, Kaisho T, Akira S, Reis e Sousa C. Microbial recognition via Toll-like receptor-dependent response of murine dendritic cells subsets to CD40 triggering. J. Immunol. 2002;169:3652–3660. doi: 10.4049/jimmunol.169.7.3652. [DOI] [PubMed] [Google Scholar]
- 14.Walton KLW, He J, Kelsall BL, Sartor RB, Fisher NC. Dendritic cells in germ-free and specific pathogen-free mice have similar phenotypes and in vitro antigen presenting function. Immunol. Lett. 2006;102:16–24. doi: 10.1016/j.imlet.2005.07.001. [DOI] [PubMed] [Google Scholar]
- 15.Savage DC. Microbial ecology of the gastrointestinal tract. Annu. Rev. Microbiol. 1977;31:107–133. doi: 10.1146/annurev.mi.31.100177.000543. [DOI] [PubMed] [Google Scholar]
- 16.Conway P-L. Microbial ecology of the human large intestine. In: Gibson GR, Macfarlane GT, editors. Human Colonic Bacteria: Role in Nutrition, Physiology and Pathology. CRC; Boca Raton, FL: 1995. pp. 1–24. [Google Scholar]
- 17.Mackie RI, Sghir A, Gaskins HR. Developmental microbial ecology of the neonatal gastrointestinal tract. Am. J. Clin. Nutr. 1999;69:1035S–1045S. doi: 10.1093/ajcn/69.5.1035s. [DOI] [PubMed] [Google Scholar]
- 18.Camerini V, Sydora BC, Aranda R, Nguyen C, MacLean C, McBride WH, Kronenberg M. Generation of intestinal mucosal lymphocytes in SCID mice reconstituted with mature, thymus-derived T cells. J. Immunol. 1998;160:2608–2618. [PubMed] [Google Scholar]
- 19.Scupham AJ, Presley LL, Wei B, Bent E, Griffith N, McPherson M, Zhu F, Oluwadara O, Rao N, Braun J, Borneman J. Abundant and diverse fungal microbiota in the murine intestine. Appl. Environ. Microbiol. 2006;72:793–801. doi: 10.1128/AEM.72.1.793-801.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eckburg PB, Bik EM, Bernstein CN, Purdom E, Dethlefsen L, Sargent M, Gill SR, Nelson KE, Relman DA. Diversity of the human intestinal microbial flora. Science. 2005;308:1635–1638. doi: 10.1126/science.1110591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ley RE, Turnbaugh PJ, Klein S, Gordon JI. Microbial ecology: human gut microbes associated with obesity. Nature. 2006;444:1022–1023. doi: 10.1038/4441022a. [DOI] [PubMed] [Google Scholar]
- 22.Huang T, Wei B, Velazquez P, Borneman J, Braun J. Commensal microbiota alter the abundance and TCR responsiveness of splenic naive CD4+ T lymphocytes. Clin. Immunol. 2005;117:221–230. doi: 10.1016/j.clim.2005.09.012. [DOI] [PubMed] [Google Scholar]
- 23.Aranda R, Sydora BC, McAllister PL, Binder SW, Yang HY, Targan SR, Kronenberg M. Analysis of intestinal lymphocytes in mouse colitis mediated by transfer of CD4+, CD45RBhigh T cells to SCID recipients. J. Immunol. 1997;158:3464–3473. [PubMed] [Google Scholar]
- 24.Aldrich CJ, Ljunggren HG, Van Kaer L, Ashton-Rickardt PG, Tonegawa S, Forman J. Positive selection of self- and alloreactive CD8+ T cells in Tap-1 mutant mice. Proc. Natl. Acad. Sci. USA. 1994;91:6525–6528. doi: 10.1073/pnas.91.14.6525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.van den Broek ME, Kagi D, Ossendorp F, Toes R, Vamvakas S, Lutz WK, Melief CJ, Zinkernagel RM, Hengartner H. Decreased tumor surveillance in perforin-deficient mice. J. Exp. Med. 1996;184:1781–1790. doi: 10.1084/jem.184.5.1781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nagai Y, Garrett KP, Ohta S, Bahrun U, Kouro T, Akira S, Takatsu K, Kincade PW. Toll-like receptors on hematopoietic progenitor cells stimulate innate immune system replenishment. Immunity. 2006;24:801–812. doi: 10.1016/j.immuni.2006.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tacke F, Ginhoux F, Jakubzick C, Rooijen NV, Merad M, Randolph GJ. Immature monocytes acquire antigens from other cells in the bone marrow and present them to T cells after maturing in the periphery. J. Exp. Med. 2006;203:583–597. doi: 10.1084/jem.20052119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Gilliet M, Liu YJ. Generation of human CD8 T regulatory cells by CD40 ligand-activated plasmacytoid dendritic cells. J. Exp. Med. 2002;195:695–704. doi: 10.1084/jem.20011603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.O'Keeffe M, Hochrein H, Vremec D, Caminschi I, Miller JL, Anders EM, Wu L, Lahoud MH, Henri S, Scott B, et al. Mouse plasmacytoid cells: long-lived cells, heterogeneous in surface phenotype and function, that differentiate into CD8+ dendritic cells only after microbial stimulus. J. Exp. Med. 2002;198:1307–1319. doi: 10.1084/jem.20021031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Emoto M, Zerrahn J, Miyamoto M, Perarnau B, Kaufmann SHE. Phenotypic characterization of CD8+ NKT cells. Eur. J. Immunol. 2000;30:2300–2311. doi: 10.1002/1521-4141(2000)30:8<2300::AID-IMMU2300>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- 31.Allman D, Dalod M, Asselin-Paturel C, Delale T, Robbins SH, Trinchieri G, Biron CA, Kastner P, Chan S. Ikaros is required for plasmacytoid dendritic cell differentiation. Blood. 2006;108:4025–4034. doi: 10.1182/blood-2006-03-007757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Yang J, Huck SP, McHugh RS, Hermans IF, Ronchese F. Perforin-dependent elimination of dendritic cells regulates the expansion of antigen-specific CD8+ T cells in vivo. Proc. Natl. Acad. Sci. USA. 2006;103:147–152. doi: 10.1073/pnas.0509054103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guarda G, Hons M, Soriano SF, Huang AY, Polley R, Martin-Fontecha A, Stein JV, Germain RN, Lanzavecchia A, Sallusto F. L-selectin-negative CCR7- effector and memory CD8+ T cells enter reactive lymph nodes and kill dendritic cells. Nat. Immunol. 2007;8:743–752. doi: 10.1038/ni1469. [DOI] [PubMed] [Google Scholar]
- 34.Ludewig B, Bonilla WV, Dumrese T, Odermatt B, Zinkernagel RM, Hengartner H. Perforin-independent regulation of dendritic cell homeostasis by CD8+ T cells in vivo: implications for adaptive immunotherapy. Eur. J. Immunol. 2001;31:1772–1779. doi: 10.1002/1521-4141(200106)31:6<1772::aid-immu1772>3.0.co;2-8. [DOI] [PubMed] [Google Scholar]
- 35.Montoya CJ, Jie HB, Al-Harthi L, Mulder C, Patino PJ, Rugeles MT, Krieg AM, Landay AL, Wilson SB. Activation of plasmacytoid dendritic cells with TLR9 agonists initiates invariant NKT cell-mediated cross-talk with myeloid dendritic cells. J. Immunol. 2006;177:1028–1039. doi: 10.4049/jimmunol.177.2.1028. [DOI] [PubMed] [Google Scholar]
- 36.Cao W, Rosen DB, Ito T, Bover L, Bao M, Watanabe G, Yao Z, Zhang L, Lanier LL, Liu YJ. Plasmacytoid dendritic cell-specific receptor ILT7-Fc1RI inhibits Toll-like receptor-induced interferon production. J. Exp. Med. 2006;203:1399–1405. doi: 10.1084/jem.20052454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lo WF, Woods AS, DeCloux A, Cotter RJ, Metcalf ES, Soloski MJ. Molecular mimicry mediated by MHC class Ib molecules after infection with Gram-negative pathogens. Nat. Med. 2000;6:215–218. doi: 10.1038/72329. [DOI] [PubMed] [Google Scholar]
- 38.Borg NA, Wun KS, Kjer-Nielsen L, Wilce MC, Pellicci DG, Koh R, Besra GS, Bharadwaj M, Godfrey DI, McCluskey J, Rossjohn J. CD1d-lipid-antigen recognition by the semi-invariant NKT T-cell receptor. Nature. 2007;448:44–49. doi: 10.1038/nature05907. [DOI] [PubMed] [Google Scholar]
- 39.Wu D, Xing GW, Poles MA, Horowitz A, Kinjo Y, Sullivan B, Bodmer-Narkevitch V, Plettenburg O, Kronenberg M, Tsuji M, et al. Bacterial glycolipids and analogs as antigens for CD1d-restricted NKT cells. Proc. Natl. Acad. Sci. USA. 2005;102:1351–1356. doi: 10.1073/pnas.0408696102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mattner J, Debord KL, Ismail N, Goff RD, Cantu C, III, Zhou D, Saint-Mezard P, Wang V, Gao Y, Yin N, et al. Exogenous and endogenous glycolipid antigens activate NKT cells during microbial infections. Nature. 2005;434:525–529. doi: 10.1038/nature03408. [DOI] [PubMed] [Google Scholar]
- 41.Hansen TH, Huang S, Arnold PL, Fremont DH. Patterns of nonclassical MHC antigen presentation. Nat. Immunol. 2007;8:563–568. doi: 10.1038/ni1475. [DOI] [PubMed] [Google Scholar]
- 42.Ochando JC, Homma C, Yang Y, Hidalgo A, Garin A, Tacke F, Angeli V, Li Y, Boros P, Ding Y, et al. Alloantigen-presenting plasmacytoid dendritic cells mediate tolerance to vascularized grafts. Nat. Med. 2006;7:652–662. doi: 10.1038/ni1333. [DOI] [PubMed] [Google Scholar]
- 43.Palucka AK, Blanck JP, Bennett L, Pascual V, Banchereau J. Cross-regulation of TNF and IFN-α in autoimmune diseases. Proc. Natl. Acad. Sci. USA. 2005;102:3372–3377. doi: 10.1073/pnas.0408506102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bilsborough J, George TC, Norment A, Viney JL. Mucosal CD8α+ DC, with a plasmacytoid phenotype, induce differentiation and support function of T cells with regulatory properties. Immunology. 2003;108:481–492. doi: 10.1046/j.1365-2567.2003.01606.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Chang C, Miller JF. Campylobacter jejuni colonization of mice with limited enteric flora. Infect. Immun. 2006;74:5261–5271. doi: 10.1128/IAI.01094-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ziegler SF. FOXP3: of mice and men. Annu. Rev. Immunol. 2006;24:209–226. doi: 10.1146/annurev.immunol.24.021605.090547. [DOI] [PubMed] [Google Scholar]
- 47.Izcue A, Coombes JL, Powrie F. Regulatory T cells suppress systemic and mucosal immune activation to control intestinal inflammation. Immunol. Rev. 2006;212:256–271. doi: 10.1111/j.0105-2896.2006.00423.x. [DOI] [PubMed] [Google Scholar]
- 48.Weaver CT, Harrington LE, Mangan PR, Gavrieli M, Murphy KM. Th17: an effector CD4 T cell lineage with regulatory T cell ties. Immunity. 2006;24:677–688. doi: 10.1016/j.immuni.2006.06.002. [DOI] [PubMed] [Google Scholar]
- 49.Uhlig HH, McKenzie BS, Hue S, Thompson C, Joyce-Shaikh B, Stepankova R, Robinson N, Buonocore S, Tlaskalova-Hogenova H, Cua DJ, Powrie F. Differential activity of IL-12 and IL-23 in mucosal and systemic innate immune pathology. Immunity. 2006;25:309–318. doi: 10.1016/j.immuni.2006.05.017. [DOI] [PubMed] [Google Scholar]
- 50.Vlad G, Cortesini R, Suciu-Foca N. License to heal: bidirectional interaction of antigen-specific regulatory T cells and tolerogenic APC. J. Immunol. 2005;174:5907–5914. doi: 10.4049/jimmunol.174.10.5907. [DOI] [PubMed] [Google Scholar]
- 51.Wingender G, Garbi N, Schumak B, Jungerkes F, Endl E, von Bubnoff D, Steitz J, Striegler J, Moldenhauer G, Tuting T, et al. Systemic application of CpG-rich DNA suppresses adaptive T cell immunity via induction of IDO. Eur. J. Immunol. 2006;36:12–20. doi: 10.1002/eji.200535602. [DOI] [PubMed] [Google Scholar]
- 52.Katakura K, Lee J, Rachmilewitz D, Li G, Eckmann L, Raz E. Toll-like receptor 9-induced type I IFN protects mice from experimental colitis. J. Clin. Invest. 2005;115:695–702. doi: 10.1172/JCI22996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Obermeier F, Dunger N, Strauch UG, Hofmann C, Bleich A, Grunwald N, Hedrich HJ, Aschenbrenner E, Schlegelberger B, Rogler G, et al. CpG motifs of bacterial DNA essentially contribute to the perpetuation of chronic intestinal inflammation. Gastroenterology. 2005;129:913–927. doi: 10.1053/j.gastro.2005.06.061. [DOI] [PubMed] [Google Scholar]
- 54.Wei B, Velazquez P, Turovskaya O, Spricher K, Aranda R, Kronenberg M, Birnbaumer L, Braun J. Mesenteric B cells centrally inhibit CD4+ T cell colitis through interaction with regulatory T cell subsets. Proc. Natl. Acad. Sci. USA. 2005;102:2010–2015. doi: 10.1073/pnas.0409449102. [DOI] [PMC free article] [PubMed] [Google Scholar]