Abstract
The virulence mechanisms that allow pathogens to colonize the intestine remain unclear. Here we show that germ-free animals are unable to eradicate Citrobacter rodentium, a model for human infections with attaching/effacing bacteria. Early in infection, virulence genes were expressed and required for pathogen growth in conventionally raised but not germ-free mice. Virulence gene expression was downregulated during the late phase of infection, which led to relocation of the pathogen to the intestinal lumen where it was out-competed by commensals. The ability of commensals to out-compete C. rodentium was determined, at least in part, by the capacity of the pathogen and commensals to grow on structurally similar carbohydrates. Thus, pathogen colonization is controlled by bacterial virulence and through competition with metabolically related commensals.
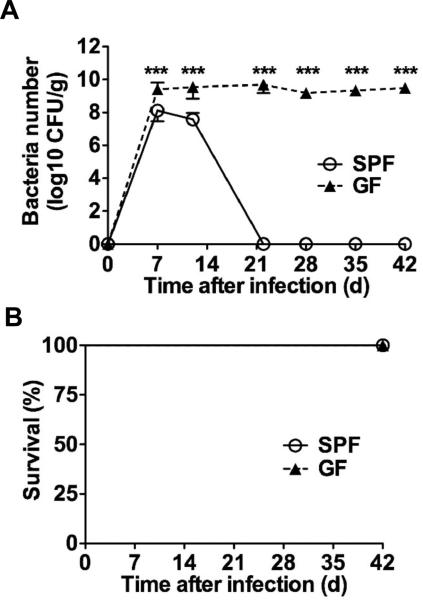
Enterohemorragic Escherichia coli (EHEC) and enteropathogenic E. coli (EPEC) are important causes of diarrhea and mortality worldwide (1, 2). These Gram-negative bacteria attach to and colonize the intestinal tract by inducing attaching and effacing (A/E) lesions on the intestinal epithelium (1, 2). The genomes of A/E pathogens harbor the locus of enterocyte effacement (LEE) that is critical for these bacteria to colonize their hosts and cause pathology (3, 4). Infection with Citrobacter rodentium, a natural pathogen of mice that is used to model human infections with EPEC and EHEC (5, 6), is associated with a significant but reversible decrease in the number of total commensals in the colon (7, 8). To assess the role of the microbiota in this enteric infection, germ-free (GF) and specific pathogen-free (SPF) C57BL/6 mice were orally inoculated with C. rodentium. The pathogen colonized the intestines of SPF mice reaching maximal concentrations in the feces on day 7-10 post infection followed by a decline by day 12 and becoming undetectable by day 22 (Fig. 1A). In contrast, GF mice harbored 10-fold more C. rodentium on day 7-10 post infection, and unlike SPF mice, they were unable to clear C. rodentium even by day 42 when the experiments were terminated (Fig. 1A). Remarkably, all GF infected mice remained alive despite high and persistent pathogen burdens (Fig. 1B). Notably, the recruitment of neutrophils, inflammatory macrophages and CD3+ T cells in response to infection was similar in SPF and GF mice (Fig. S1). Consistently, histological analysis revealed comparable pathology scores on day 12 post-infection which declined on day 22 in both GF and SPF mice (Fig. S1). Furthermore, expression of anti-microbial peptides including RegIIIγ, β-defensin-1, β-defensin-3, and β-defensin-4 was comparable in the colons of infected SPF and GF mice (Fig. S2).
Fig. 1. The microbiota is required for eradication of C. rodentium.
A,B, SPF and GF mice (n= 7) were infected orally with 1×109 cfu of C. rodentium and pathogen load in feces (A) and mouse survival (B) were determined over the indicated time. Data points are given as mean ± SD. Results are representative of at least 3 independent experiments. ***; p<0.001 by Mann-Whitney U test.
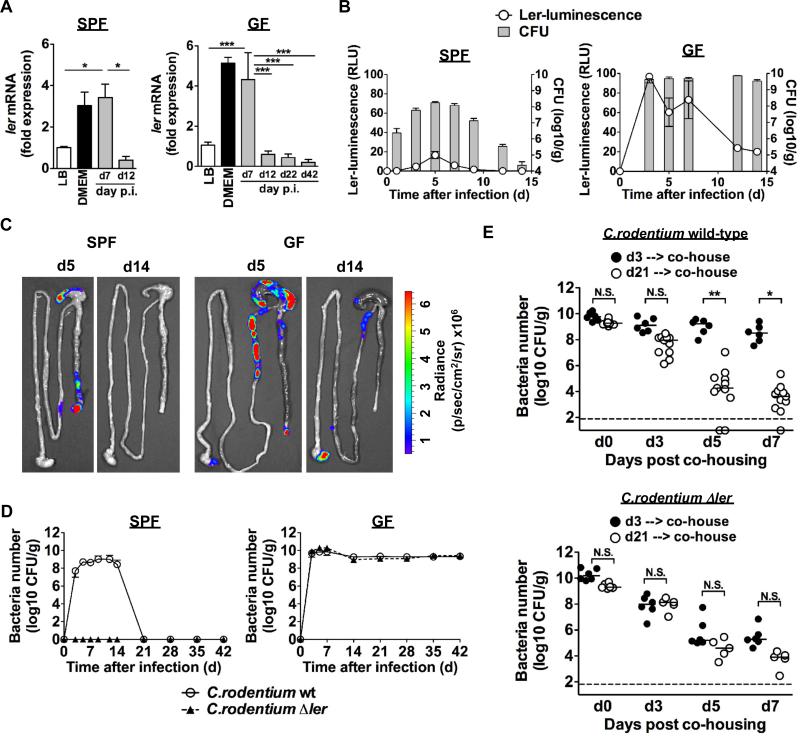
The expression of most LEE genes in C. rodentium is controlled by Ler, a member of the H-NS protein family (3, 4, 9, 10). Ler was expressed in the feces on day 7 post-infection in both GF and SPF mice (Fig. 2A). Importantly, expression of ler and tir, a Ler-regulated gene were both down-regulated on day 12 after infection in both GF and SPF mice and were not expressed at day 42 in GF mice despite robust pathogen colonization (Fig. 2A and Fig. S3). To monitor the expression of ler in the intestine, we engineered a bioluminescent reporter C. rodentium strain in which the ler promoter was fused to the luxCDABE operon of Photorhabdus luminescens (Fig. S4) (11). Ler expression was detected in the feces on day 5, but down-regulated by day 7-12 post-infection (Fig. 2B). Furthermore, ler expression was visualized on day 5 post-infection in the ileum, cecum and distal colon, but was down-regulated on day 14 in both SPF and GF mice (Fig. 2C). The ler luminescent signal was ~ 10-fold higher in GF mice than in SPF mice consistent with higher pathogen load in GF mice. If the C. rodentium were harvested from infected GF mice at day 21 and re-inoculated into SPF mice, the pathogen robustly grew in the intestines and elicited colonic inflammation (Fig. S5).
Fig. 2. Expression and role of ler during C. rodentium infection in SPF and GF mice.
A, ler mRNA levels were determined by qPCR in fecal pellets of SPF and GF mice infected with C. rodentium at the indicated days post-infection. Expression was normalized to that of the kanamycin resistance gene carried by the C. rodentium strain. Control experiments were performed by determining ler mRNA levels in C. rodentium grown under inducing (DMEM) and repressing (LB) in vitro culture conditions (4, 10). Data represent mRNA expression relative to that in C. rodentium cultured in LB medium. Results are given as mean ± SD of individual mice (n=3). Results are representative of at least 2 experiments. *; p<0.05, ***; p<0.001 by Dunnett's multiple comparison test. B, Expression of ler in fecal pellets of SPF and GF mice infected with the reporter ler-lux C. rodentium strain at the indicated day post-infection. Results show luminescence (relative light units) and cfu of ler-lux C. rodentium in the same samples. Data expressed as mean ± SD of individual mice (n=4). Results are representative of at least 2 experiments. C, Bioluminescent imaging of ler expression in the intestines of SPF and GF mice infected with the ler-lux C. rodentium strain. Imaging was performed on day 5 and 14 post-infection and the signal was quantified based on the color scale shown below. Results are representative of 3 individual mice. D, SPF and GF mice (n= 5) were infected orally with 109cfu of WT and Δler mutant C. rodentium, and pathogen load in feces was determined over the indicated time. Data points are given as mean ± SD. Results are representative of at least 2 experiments. E, GF mice were infected with WT and Δler mutant C. rodentium. At day 3 or day 21 post infection, mice were co-housed with SPF mice (1:1). Pathogen load was determined in feces on indicated days after co-housing. Dots represent individual mice. Results are representative of at least 3 experiments. N.S., not significant, *; p<0.05, **; p<0.01 by Dunn's test.
Since expression of virulence LEE genes is essential for pathogen colonization in the intestines of SPF mice (4), we asked whether LEE virulence was required for colonization in GF mice. To assess this, we orally infected GF and SPF mice with wild-type (WT) C. rodentium and isogenic strains deficient in Ler (Δler) or in EscN (ΔescN) a Ler-regulated ATPase required for injection of virulence factors via the type-III secretion system (T3SS) into host cells (4). As expected, WT, but not the Δler or ΔescN strains, grew in the intestine of SPF mice (Fig. 2D). Notably, the Δler and ΔescN mutants grew robustly in GF mice, reaching numbers similar to the WT pathogen (Fig. 2D and Fig. S6). Thus, Ler-dependent LEE virulence is not required for intestinal C. rodentium growth in the absence of the microbiota. To determine whether Ler and the T3SS regulate the ability of the microbiota to out-compete the pathogen, we orally infected GF mice with WT, the Δler or the ΔescN mutant C. rodentium and the infected GF mice were colonized with commensals by cohousing them at either day 3 or day 21 post-infection with SPF mice. Notably, the burden of WT C. rodentium in GF mice harboring the pathogen for 21 days declined significantly after 3 days of co-housing and was further reduced by 5-6 logs by day 7, similarly to the decline observed in mice infected with the Δler or the ΔescN mutant for 3 and 21 days (Fig. 2E and Fig. S6). In contrast, the microbiota could not out-compete the WT bacterium on day 3 post-infection upon co-housing with SPF mice (Fig. 2E), consistent with the observation that ler expression is active in WT C. rodentium at day 3 post-infection (Fig. S7).
C. rodentium induces marked inflammation in the distal colon which requires LEE virulence factors including the T3SS (4). If the T3SS promotes outgrowth of C. rodentium by enabling the pathogen to colonize a niche at or near the epithelium, it would be expected that impaired colonization of the ΔescN mutant could not be rescued when inflammation is provided by other means. Oral administration of dextran sulfate sodium (DSS), a chemical that directly damages the colonic epithelium (12) enhanced the colonization of the WT bacterium by 3 logs but not that of the ΔescN mutant (Fig. S8). In addition, co-infection of the ΔescN mutant with the WT C. rodentium that induces inflammation did not rescue colonization by the T3SS mutant strain (Fig. S8). These results indicate that inflammation promotes colonization of WT C. rodentium, but not in the absence of a functional T3SS. The adaptive immune system including IgG production and Th17 responses against the pathogen are important for eradication of C. rodentium (13-16). Importantly, the production of IgG, IgM, and IgA against C. rodentium was similar in GF and SPF mice (Fig. S9). Furthermore, the generation of colonic IL-17 and interferon-γ-producing T cells in response to C. rodentium was comparable in SPF and GF mice (Fig. S10).
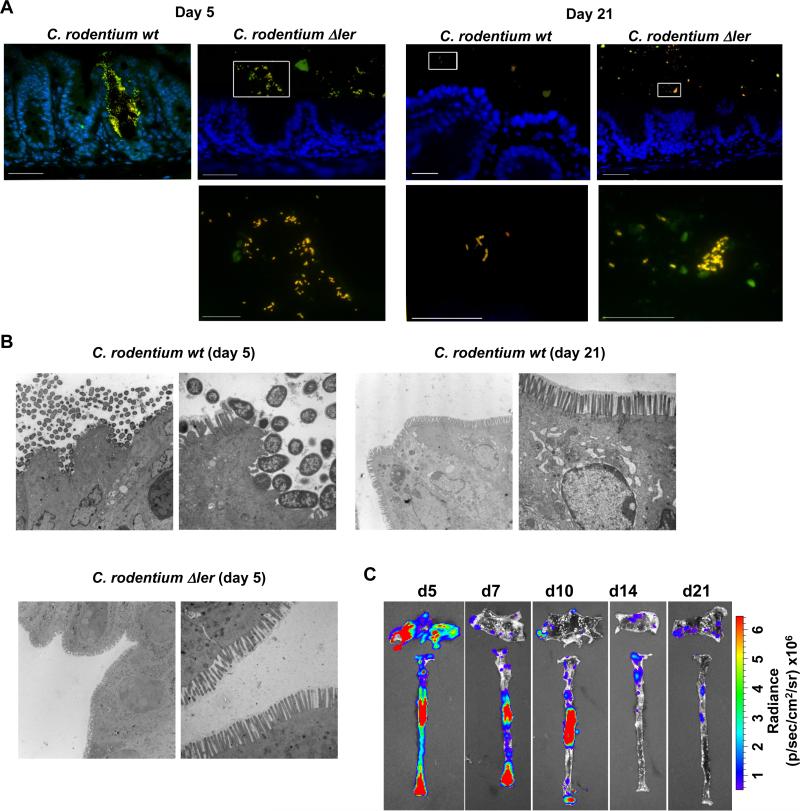
C. rodentium colonizes and infects the surface of the intestinal epithelium, a site largely devoid of commensals (17). At day 5 post-infection, Tir, a marker of C. rodentium attachment to the epithelium, was seen on the intestinal surface of GF mice infected with WT C. rodentium, but not with the Δler strain (Fig. S11). In contrast, Tir labeling was not detected on day 21 post-infection on the intestinal surface of mice infected with either WT or mutant C. rodentium (Fig. S11). Consistently, the WT pathogen was associated with the epithelium whereas the Δler mutant localized to the intestinal lumen on day 5 post-infection as determined by dual fluorescence in-situ hybridization (Fig. 3A). On day 21 post-infection, however, neither WT nor Δler C. rodentium were detected on the epithelial surface, but instead they localized to the intestinal lumen (Fig. 3A). Similarly, transmission electron microscopy revealed C. rodentium at or near the intestinal epithelium on day 5, but not on day 21 post-infection (Fig. 3B). As expected, Δler C. rodentium did not localize at or near the epithelium on day 5 post-infection (Fig. 3B). Consistent with these results, ler-lux-expressing WT C. rodentium found attached to the cecal and colonic epithelium were abundant in the early phase of infection, but dramatically decreased later during infection, even though pathogen burdens in the feces were comparable (Fig. 3C and fig. S12). These results indicate that the localization of the pathogen differs in the early and late phase of infection and this is controlled by LEE virulence gene expression.
Fig. 3. Localization of C. rodentium to intestinal niches is mediated by LEE-encoded virulence factors.
A, Dual FISH staining using DNA probes that label virtually all true bacteria (EUB338, red) and the γ-Proteobacteria class to which C. rodentium belongs (GAM42a, green). Pathogenic bacteria (i.e. EUB338+/GAM42a+ cells) are yellow. Lower panels show high magnification images corresponding to boxed areas (upper panels). Note the yellow color on all the bacteria stained in the cecum infected with the Δler mutant; indicating all bacteria in the lumen are C. rodentium. Scale bar: 50μm (upper panels), 20μm (lower panels), Results are representative of 2 experiments. B, Transmission Electron Micrographs of cecum from infected GF mice at day 5 and day 21 post infection with WT C. rodentium and at day 5 with the Δler mutant. Original magnification: 3,400× (left panel) and 13,500× (right panel). Scale bar: 2 μm (left panel) and 500 nm (right panel). Results are representative of 2 experiments. C, GF mice were infected orally with WT C. rodentium carrying the ler-lux fusion. Cecum and colonic tissues were collected at the indicated day and then washed with PBS to remove non-adherent bacteria. Bioluminescent imaging of ler expression of C. rodentium attached to the cecum (top) and colon (bottom). Results are representative of 2 experiments using 4 different mice.
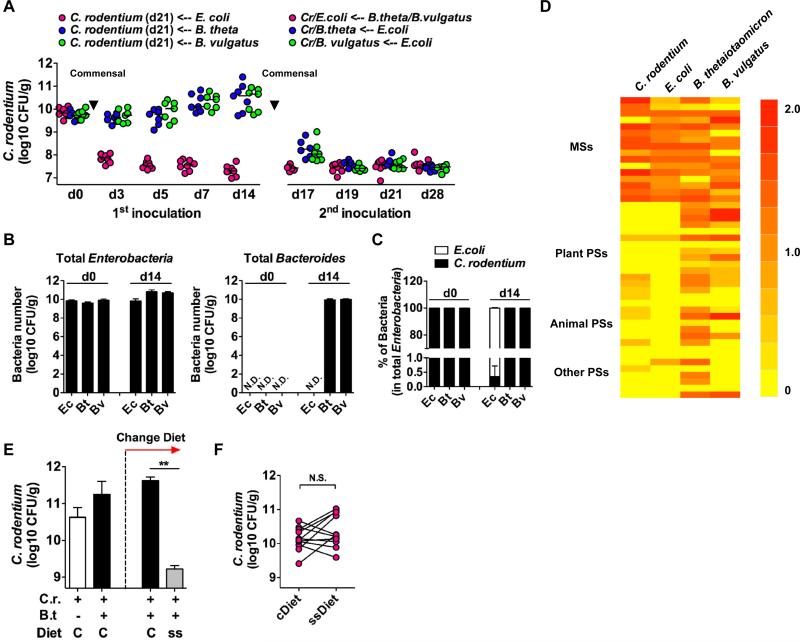
The intestine harbors a large number of bacterial species (18). To determine whether commensal bacteria exhibit different abilities to out-compete C. rodentium, we orally infected GF mice with C. rodentium and on day 21 post-infection the mice were colonized with either E. coli or one of two different anaerobic Bacteroides species, all isolated from the intestines of SPF mice (19). The burden of C. rodentium in GF mice declined about 200-fold by day 3 and more than 500-fold by day 14 upon colonization with E. coli, but not at all, with B. thetaiotaomicron or B. vulgatus (Fig. 4A). Secondary administration of E. coli to GF mice already colonized with B. thetaiotaomicron or B. vulgatus also reduced the number of C. rodentium in the feces about 500-fold (Fig. 4A). In contrast, colonization of mice harboring E. coli with B. thetaiotaomicron or B. vulgatus was not effective in further reducing the burden of C. rodentium in their feces (Fig. 4A). Assessment of total Enterobacteria (E. coli and C. rodentium) and Bacteroides on day 14 post colonization with individual commensal bacteria showed comparable numbers of bacteria in the mouse feces (Fig. 4B). Before inoculation of commensal bacteria (d0), all Enterobacteria detected in the feces were C. rodentium (Fig. 4C). In contrast, > 99% of the total Enterobacteria in the E. coli inoculated group were E. coli after 14 days of colonization (Fig. 4C). We next analyzed the growth abilities of each of these species on a custom carbohydrate growth array that contains most of the common mono- and polysaccharides present in plant and animal tissue (Fig. 4D and Table S1). Both C. rodentium and E. coli exhibited optimal growth on monosaccharides (Fig. 4D). In contrast, both B. thetaiotaomicron and B. vulgatus exhibited broad abilities to catabolize both mono- and polysaccharides (Fig. 4D). Because γ-Proteobacteria such as E. coli specifically accumulates after C. rodentium infection (7), these results suggest that this microbial change may benefit the host by increasing the number of commensals that can out-compete the pathogen. To further assess if competition for glycans between commensals and C. rodentium is important for pathogen eradication, GF mice on a conventional maintenance diet containing both mono- and polysaccharides were orally infected with C. rodentium and on day 21 post-infection the infected GF mice were colonized with B. thetaiotaomicron for 7 days. On day 7 post-colonization with B. thetaiotaomicron, the mice were divided into two groups that were fed the conventional diet or a simple sugar diet containing monosaccharides but not polysaccharides. The burden of C. rodentium in the GF mice on the simple sugar diet rapidly declined ~ 200-fold by day 3, whereas no decline was observed when the mice were fed the conventional diet (Fig. 4E). The results were not explained by the simple sugar diet alone because in the absence of colonization with B. thetaiotaomicron, the number of C. rodentium in feces was comparable between animals fed with the conventional and simple sugar diets (Fig. 4F). These results suggest that under the simple sugar diet, B. thetaiotaomicron cannot use polysaccharides and is forced to compete with C. rodentium for available monosaccharides or another common nutrient that both C. rodentium and B. thetaiotaomicron rely on in the absence of dietary polysaccharides. These findings clarify the tenuous nature of A/E pathogens, in terms of their survival/colonization within hosts, and therefore nutrient or probiotic shifting of the microbiota to promote commensals that directly compete with pathogens for food sources may be a useful therapeutic approach.
Fig. 4. Similar catabolic preferences for saccharides may determine the competing ability of commensal bacteria with the enteric pathogen.
A, GF mice were infected with WT C. rodentium (Cr). At day 21 post infection, E. coli (Ec) or B. thetaiotaomicron (Bt) or B. vulgatus (Bv) were inoculated. As a second inoculation, mixture of B. thetaiotaomicron and B. vulgatus was inoculated into the E. coli harboring group, and E. coli was inoculated into the Bacteroides harboring group, respectively. Pathogen load was determined in feces on the indicated days after inoculation of commensal bacteria. Dots represent individual mice from 2 independent experiments. B, Total Enterobacteria (C. rodentium and E. coli) and Bacteroides culture in feces at day 0 and day 14 (first inoculation in panel A). Data are given as mean ± SD (n=4). Results are representative of at least 2 experiments. C, Number of C. rodentium (kanamycin resistant) in the total Enterobacteria (kanamycin sensitive) are indicated as a percentage. Data are given as mean ± SD (n=4). Results are representative of at least 2 experiments. D, Carbohydrate catabolic profiles of C. rodentium and commensal bacteria strains. Robust growth under supplementation of monosaccharides (MSs) or polysaccharides (PSs) indicated as red, and no growth indicated as yellow. Raw data is provided in Table S1. E, GF mice were infected with WT C. rodentium (Cr). At day 21 post infection, B. thetaiotaomicron (Bt) was inoculated. On day 7 post-colonization with Bt, the mice were divided into two groups that were fed a conventional maintenance diet (C) or a simple sugar diet (ss). Pathogen load was determined in feces on day 3 after diet switching. Results are given as mean ± SD of individual mice (initially n=10, and divided into 2 groups n=5 each). Results are representative of 2 experiments. F, C. rodentium mono-associated GF mice (day 21) were fed with a simple sugar diet (ssDiet) for 7 days. Pathogen load was compared before and after switching to ssDiet from conventional diet (cDiet). Dots represent individual mice and representative of 3 independent experiments **; p<0.01. N.S., denotes not significant by Mann-Whitney U test.
Supplementary Material
Acknowledgements
The authors thank the University of Michigan Germ-free Animal Core, Microscopy and Image Analysis Laboratory, and the Center for Molecular Imaging for support, S. Koonse for animal husbandry, A. Huerta-Saquero for constructing the pler-lux plasmid, N. Pudlo for anaerobic bacteria culture, T. Stappenbeck for mouse commensal strains, J. Rousseau for technical assistance, and M.H. Shaw, G. Chen for expert review of the manuscript. B.A.V. is the Canada Research Chair (Tier 2) in Pediatric Gastroenterology and the CHILD Foundation Chair in Pediatric IBD Research. This work was supported by grants from the NIH grants DK61707 and DK091191 (G.N.), CONACyT (J. L. P.), CIHR (B.A.V.) and the Uehara Memorial Foundation and Crohn's and Colitis Foundation of America Fellowship Awards (N.K). N. K and G. N. hold U.S. Provisional Patent Application no. 61/616,707 regarding inhibition of LEE virulence as a potential therapeutic for infection with A/E pathogens. The data reported in this paper are tabulated in the main paper and in the supplementary materials.
References
- 1.Kaper JB, Nataro JP, Mobley HL. Pathogenic Escherichia coli. Nat Rev Microbiol. 2004;2:123–140. doi: 10.1038/nrmicro818. [DOI] [PubMed] [Google Scholar]
- 2.Mundy R, MacDonald TT, Dougan G, Frankel G, Wiles S. Citrobacter rodentium of mice and man. Cell Microbiol. 2005;7:1697–1706. doi: 10.1111/j.1462-5822.2005.00625.x. [DOI] [PubMed] [Google Scholar]
- 3.Deng W, Li Y, Vallance BA, Finlay BB. Locus of enterocyte effacement from Citrobacter rodentium: sequence analysis and evidence for horizontal transfer among attaching and effacing pathogens. Infect Immun. 2001;69:6323–6335. doi: 10.1128/IAI.69.10.6323-6335.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Deng W, et al. Dissecting virulence: systematic and functional analyses of a pathogenicity island. Proc Natl Acad Sci U S A. 2004;101:3597–3602. doi: 10.1073/pnas.0400326101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Luperchio SA, et al. Citrobacter rodentium, the causative agent of transmissible murine colonic hyperplasia, exhibits clonality: synonymy of C. rodentium and mouse-pathogenic Escherichia coli. J Clin Microbiol. 2000;38:4343–4350. doi: 10.1128/jcm.38.12.4343-4350.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Borenshtein D, McBee ME, Schauer DB. Utility of the Citrobacter rodentium infection model in laboratory mice. Curr Opin Gastroenterol. 2008;24:32–37. doi: 10.1097/MOG.0b013e3282f2b0fb. [DOI] [PubMed] [Google Scholar]
- 7.Lupp C, et al. Host-mediated inflammation disrupts the intestinal microbiota and promotes the overgrowth of Enterobacteriaceae. Cell Host Microbe. 2007;2:204. doi: 10.1016/j.chom.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 8.Hoffmann C, et al. Community-wide response of the gut microbiota to enteropathogenic Citrobacter rodentium infection revealed by deep sequencing. Infect Immun. 2009;77:4668–4678. doi: 10.1128/IAI.00493-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mellies JL, Elliott SJ, Sperandio V, Donnenberg MS, Kaper JB. The Per regulon of enteropathogenic Escherichia coli : identification of a regulatory cascade and a novel transcriptional activator, the locus of enterocyte effacement (LEE)-encoded regulator (Ler) Mol Microbiol. 1999;33:296–306. doi: 10.1046/j.1365-2958.1999.01473.x. [DOI] [PubMed] [Google Scholar]
- 10.Barba J, et al. A positive regulatory loop controls expression of the locus of enterocyte effacement-encoded regulators Ler and GrlA. J Bacteriol. 2005;187:7918–7930. doi: 10.1128/JB.187.23.7918-7930.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bjarnason J, Southward CM, Surette MG. Genomic profiling of iron-responsive genes in Salmonella enterica serovar typhimurium by high-throughput screening of a random promoter library. J Bacteriol. 2003;185:4973–4982. doi: 10.1128/JB.185.16.4973-4982.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cooper HS, Murthy SN, Shah RS, Sedergran DJ. Clinicopathologic study of dextran sulfate sodium experimental murine colitis. Lab Invest. 1993;69:238–249. [PubMed] [Google Scholar]
- 13.Simmons CP, et al. Central role for B lymphocytes and CD4+ T cells in immunity to infection by the attaching and effacing pathogen Citrobacter rodentium. Infect Immun. 2003;71:5077–5086. doi: 10.1128/IAI.71.9.5077-5086.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Maaser C, et al. Clearance of Citrobacter rodentium requires B cells but not secretory immunoglobulin A (IgA) or IgM antibodies. Infect Immun. 2004;72:3315–3324. doi: 10.1128/IAI.72.6.3315-3324.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bry L, Brenner MB. Critical role of T cell-dependent serum antibody, but not the gut-associated lymphoid tissue, for surviving acute mucosal infection with Citrobacter rodentium, an attaching and effacing pathogen. J Immunol. 2004;172:433–441. doi: 10.4049/jimmunol.172.1.433. [DOI] [PubMed] [Google Scholar]
- 16.Ivanov II, et al. Induction of intestinal Th17 cells by segmented filamentous bacteria. Cell. 2009;139:485–498. doi: 10.1016/j.cell.2009.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bergstrom KS, et al. Muc2 protects against lethal infectious colitis by disassociating pathogenic and commensal bacteria from the colonic mucosa. PLoS Pathog. 2010;6:e1000902. doi: 10.1371/journal.ppat.1000902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qin J, et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature. 2010;464:59–65. doi: 10.1038/nature08821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bloom SM, et al. Commensal Bacteroides species induce colitis in host-genotype-specific fashion in a mouse model of inflammatory bowel disease. Cell Host Microbe. 2011;9:390–403. doi: 10.1016/j.chom.2011.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.