Abstract
Inhibitor of growth 4 (ING4) is a member of the ING family and acts as a tumor suppressor protein. To investigate the impact of ING4 on breast cancer proliferation, the present study examined the antitumor effects caused by upregulation in the expression of ING4 and its possible mechanism of effect in MCF-7 cells. A plasmid-based expression system encoding the ING4 gene was used to construct a stable cell line and overexpress ING4 in MCF-7 cells. Real-time PCR and western blot analysis were used to detect the mRNA and protein expression levels of ING4, respectively. Cell growth was examined by methylthiazolyltetrazolium (MTT) assay. Cell cycle distribution and cell apoptosis were measured by flow cytometry. The expression of p21, p53 and bax genes were tested by real-time PCR and western blot analysis. The stably transfected cell lines pcDNA3.1(+)/ING4 (with the ING4 gene) and pcDNA3.1(+) (an empty vector) were established. The expression levels of ING4 mRNA and protein in the stable cell line expressing pcDNA3.1(+)/ING4 were significantly higher than those of the two control cell lines. The cell proliferation of stably transfected cells was inhibited, and the inhibitory rate was 62.58±2.93%. Based on the changes in cell cycle distribution in stably transfected cells compared with two control cell lines, a number of cells were blocked in the G0/G1 phase 67.82±3.78% (P<0.05). In addition, the apoptotic rate was significantly higher, at 31.51±3.02% (P<0.05). Real-time PCR revealed that p21 and bax mRNA expression were increased (P<0.01), but the expression of p53 did not change significantly (P>0.05) in the stably transfected cell lines. Western blot analysis results of p21, bax and p53 were in accordance with real-time PCR results. ING4 upregulation may inhibit breast cancer cell proliferation and accelerate the process of apoptosis. It is suggested that ING4 plays a significant role in the suppression of breast cancer progression.
Keywords: inhibitor of growth 4, breast cancer, MCF-7
Introduction
Breast cancer is the leading cause of cancer mortality in women worldwide (1). Despite advances in diagnosis and chemotherapy, numerous women with breast cancer continue to succumb to this malignancy. The underlying mechanisms that regulate the progression of breast cancer are still poorly understood. A variety of factors and genes are involved in breast cancer pathogenesis. The gene ING4 encodes a protein belonging to the inhibitor of growth (ING) family, which are considered to be candidate tumor suppressor genes, playing a pivotal role in apoptosis, cell cycle regulation, DNA damage repair (2–5) and control of cellular aging (6). Similar to other ING family members, ING4 encodes a 249-amino acid protein containing a highly conserved plant home domain (PHD) finger motif at the C-terminal region, a conserved central region containing the nuclear localization signal (NLS) and a variable N-terminal region (7). Various studies have suggested that ING4 exerts its biological functions via its specific functional domains, which in association with other regulatory molecular partners, such as p53, p300, HBO1, NF-κB p65 and H3K4me (7–9), activate cellular regulation networks. ING4 is also among the few known regulatory proteins that are able to directly interact with chromatin as well as being involved in transcription. In the present study, to explore the role of ING4 in human breast cancer, we used vector-mediated ING4 to upregulate its expression in the human breast cancer cell line MCF-7. We investigated the effect of ING4 overexpression on the proliferation, cell cycle and apoptosis of MCF-7 cells.
Materials and methods
Cells, reagents and vectors
Human breast cancer cell line MCF-7 was obtained from the American Type Culture Collection (ATCC, Manassas, VA, USA). PcDNA3.1(+)/ING4 has been described in previous studies (10). Cell culture medium Dulbecco’s modified Eagle’s medium (DMEM) and fetal calf serum (FCS) were bought from Gibco (Life Technologies, Grand Land, NY, USA). TRIzol reagents and transfection reagent Lipofectamine™ 2000 were provided by Invitrogen Life Technologies (Carlsbad, CA, USA). A microplate reader was purchased from Bio-Rad. Flow cytometry analysis was undertaken with a BD FACS system. PrimeScript™ reverse transcription reagent kit was purchased from Takara (Dalian, China). SYBR-Green I kit was obtained from Roche Diagnostics (Indianapolis, IN, USA). The primary antibody ING4 was obtained from Santa Cruz (Santa Cruz, CA, USA), p21 and p53 were purchased from Signalway Antibody Co. (Preland, TX, USA), bax was purchased from Boster (Wuhan, China) and β-actin was purchased from Bioworld (Dublin, OH, USA). Enhanced chemiluminescence detection reagents were purchased from Applygen Technologies Incorporation (Beijing, China).
Cell culture and transfection
MCF-7 cells were cultured in DMEM supplemented with 10% FCS, 2 mM L-glutamine, 100 U/ml penicillin and 100 μg/ml streptomycin. All cells were subcultured in six-well plates (with cells at 60–70% confluence) at 37°C in 5% CO2 and fed every 2–3 days with complete medium. Transfection was performed using Lipofectamine 2000 according to the manufacturer’s instructions (the ratio of the plasmids to the transfection reagent was 4 μg:10 μl). Three types of cells were utilized, including the specific plasmid-transfected cells (pcDNA3.1(+)/ING4), and the empty plasmid-transfected cells and the untransfected cells were taken as controls. Stable cells were selected with G418 (400 μg/μl) for three weeks. The stably transfected cells were subsequently verified by real-time PCR and western blot analysis.
MTT assay
The three cell types were seeded in 96-well plates at 2.5×103 cells/well in DMEM containing 10% FCS and incubated for five days at 37°C with 5% CO2. During this period, cells were selected from three wells from each group every day at random for methylthiazolyltetrazolium (MTT, 100 μg/well) assay at 37°C for 4 h. The formazan crystals in the cells were solubilized with 150 μl/well DMSO for 15 min. Absorbance of the plates was read using a microplate reader at 570 nm to determine the inhibitory rates of cell growth.
Flow cytometric analysis of the cell cycle
Cells were seeded into 6-well plates at a density of 2×105 cells and cultured for 24 h at 37°C. All harvested cells were washed with ice-cold phosphate-buffered saline (PBS), then fixed with cold 70% (vol/vol) ethanol overnight at 4°C. Cells were resuspended in PBS buffer containing a final concentration of 20 μg/ml RNase A and 20 μg/ml propidium iodide for 20 min. The cell cycle profiles were determined using flow cytometry (FACScan, Beckton Dickinson, Franklin Lakes, NJ, USA) and analyzed using CellQuest software. All of the samples were assayed in triplicate.
Flow cytometric analysis of apoptosis
An annexin V-fluorescein isothiocyanate kit (BD Pharmingen, USA) was used to detect apoptosis. The cultured cells were harvested and washed with ice-cold PBS buffer twice. Binding buffer containing 5 μl FITC-labeled annexin V and 10 μl of 20 μg/ml propidium iodide was added to an aliquot of the cell suspension. Following incubation for 15 min in the dark at room temperature, stained cells were tested by flow cytometry with ModiFit3.0 software. Cells were gated for analysis by a combination of forward scatter channels (FSC) and side scatter channels (SSC). All samples were assayed in triplicate.
Real-time polymerase chain reaction
To further explore the effect of ING4 on cell proliferation, p21, p53 and bax mRNA were analyzed by real-time PCR. Total RNA was extracted from the three types of cells using TRIzol reagent (Invitrogen) according to the manufacturer’s instructions. RNA was quantified by spectrophotometry and analyzed by 1% agarose/formaldehyde denaturing gel electrophoresis to confirm RNA integrity. An equal amount of RNA (500 ng) was used as a template and reverse transcribed into cDNAs using the PrimeScript RT kit, then amplified by using SYBR-Green mix kit. The melting curve data were collected to verify PCR specificity. Each gene was analyzed in triplicate to diminish operation error. The relative gene expression levels were calculated according to housekeeping gene β-actin. Relative gene expression was calculated using the 2-ΔΔCt method. Primers used in the experiments were as follows: β-actin: F, 5′-AAAGACCTGTACGCCAACAC-3′ and R, 5′-GTCATA CTCCTGCTTGCTGAT-3′; p21: F, 5′-GATTAGCAG CGGAACAAGGAGT-3′ and R, 5′-GGAGAAACGGGAA CCAGGACA-3′; p53: F, 5′-CAGATTGCAAGTTCACCT GCCACTA-3′ and R, 5′-GATGAAGCCTGTGTAAGAACC GTCCT-3′; bax: F, 5′-TTTCTGACGGCAACTTCAACTG-3′ and R, 5′-GGAGTCTCACCCACCACCCT-3′; ING4: F, 5′-GAGGCTGATCTCAAGGAGAA-3′ and R, 5′-TCC ACAGGCATATCCAACAC-3′.
Western blot analysis
Collected cells were lysed in RIPA buffer [10 mmol/l Tris-HCl (pH 7.4), 1% deoxycholate, 1% NP40, 150 mmol/l NaCl, 0.1% SDS, 0.2 mmol/l phenylmethy sulfonyl fluoride, 1 μg/ml aprotinin and 1 μg/ml leupeptin] for 30 min on ice, then the lysates were centrifuged at 15,000 × g for 15 min to remove debris. Total protein concentrations in the supernatants were quantified using the Lowry method. Equal amounts (20 μg) of protein were denatured, then separated by 15% SDS-polyacrylamide gel electrophoresis, and transferred to PVDF membrane. Membranes were blocked in 3% bovine serum albumin (10 mM Tris-HCl, 150 mM NaCl, 0.1% Tween-20) for 2 h, then incubated with primary antibodies against ING4 (1:1,000 dilution, Santa Cruz), p21 (1:500 dilution), p53 (1:500 dilution), bax (1:200 dilution) and β-actin (1:5,000 dilution) in 3% bovine serum albumin at 4°C overnight. Subsequently, the blots were incubated with HRP-conjugated secondary antibodies following 3–5 washes with TBST. The protein bands were visualized by enhanced chemiluminescence detection reagents and developed with Kodak film. Normalization was made against β-actin expression.
Statistical analysis
Statistical analysis was carried out using Scheffe’s F test and Student’s t-test. The differences were considered statistically significant at P<0.05 and P<0.01.
Results
ING4 overexpression in MCF-7 cells by transfection
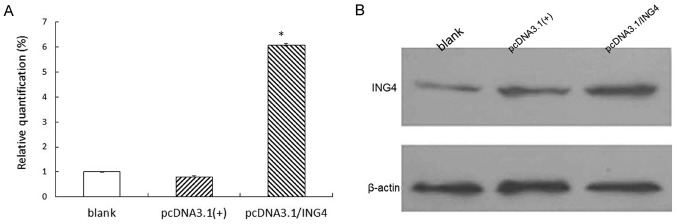
Real-time PCR and western blot analysis were used to evaluate ING4 expression. It was observed that the expression of ING4 in the stable cell group with exogenous ING4 was increased significantly in comparison to the negative group and blank group (P<0.05), while there were no significant differences between the negative group and the blank group (P>0.05; Fig. 1).
Figure 1.
ING4 expression in MCF-7 cells. (A) mRNA expression of ING4 detected by real-time PCR. Total RNA from the three types of cells were reverse transcribed and amplified by the primers from ING4 and β-actin. ING4 mRNA expression in the stable MCF-7 cells was increased significantly in comparison with the negative group and the blank group (P<0.01). Data values and error bars show the means and SD, respectively. (B) Protein expression of ING4 was detected by western blot analysis. Total cell lysates were separated by SDS-polyacrylamide gel electrophoresis and immunoblotted with an antibody against ING4. ING4 expression levels were normalized against β-actin expression. ING4, inhibitor of growth 4.
ING4 inhibites MCF-7 cell growth
The cell growth curve demonstrated that compared with the negative group and the blank group, proliferation in the stably transfected pcDNA3.1(+)/ING4 group was inhibited significantly in a time-dependent manner, and the highest inhibitory rate was 2.58±2.93% (P<0.05) on day 4. However, there was no obvious difference in cell proliferation in the control groups (P>0.05; Fig. 2).
Figure 2.
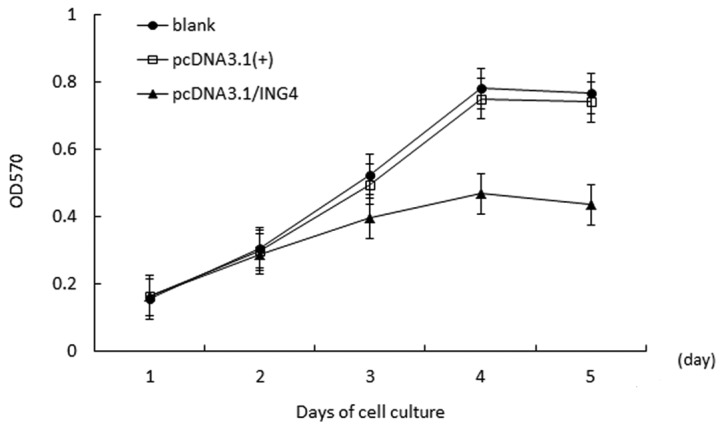
Proliferation curves were determined by MTT assay. Cell growth curves and the results of inhibitory rates of cell growth were applied to absorbance at 570 nm on a microreader (Bio-Rad, USA). The stably transfected cell proliferation was inhibited significantly in a time-dependent manner, and the highest inhibitory rate was 62.58±2.93% (P<0.05) on day 4. All data represent the mean of three independent experiments. ING4, inhibitor of growth 4.
Overexpression of ING4 induces MCF-7 cell cycle arrest
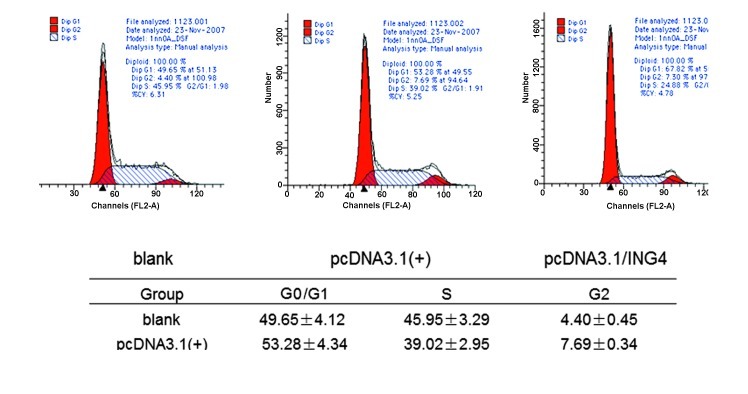
To investigate the underlying mechanism by which ING4 suppresses cell growth, cell cycle distribution analysis by flow cytometry was performed for stably transfected cells and control cells. Compared with control group cells, the stably transfected ING4 cells were blocked in the G0/G1 phase by 67.82±3.78% (P<0.05) and reduced in the S phase by 24.88±3.09% (P<0.05; Fig. 3). These data suggest that ING4 inhibited the cell proliferation by inducing G0/G1 arrest and S phase inhibition in the stably transfected cells.
Figure 3.
The results of cell cycle distribution analysis by flow cytometry. The graphs show cell cycle profiles in the stably transfected cells and control cells. The percentages of cell populations of different cell cycle phases are shown in the upper-right corner. Many stably transfected cells were blocked in the G0/G1 phase (67.82±3.78%; P<0.05), and there was a reduction in the S phase (24.88±3.09%; P<0.05). ING4, inhibitor of growth 4.
Overexpression of ING4 induces MCF-7 cell apoptosis
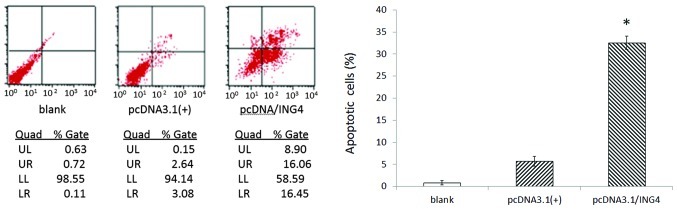
Cell apoptosis detected by flow cytometry revealed that compared with control group cells, the apoptotic rate of stably transfected ING4 cells notably increased to 31.51±3.02% (P<0.05), whereas there were no obvious differences in cell apoptosis in control group cells (Fig. 4).
Figure 4.
The results of flow cytometry in the stably transfected group and controls. Cells were double-stained with FITC and PI, then screened by flow cytometry. The presented data shows that the apoptotic rate of the pcDNA3.1(+)/ING4 group was higher than that of the control group (P<0.05), whereas no apparent differences were observed between the two control groups. The means and SD are shown. ING4, inhibitor of growth 4.
ING4 induces changes in the expression of certain genes involved in the cell cycle and apoptosis
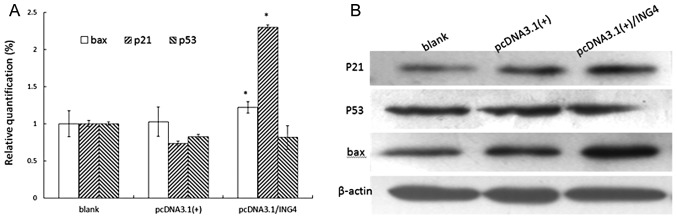
To further elucidate the basic molecular mechanism by which ING4 might influence cell fate, the cell cycle and apoptosis-related genes were investigated. In this study, we investigated the overexpression of ING4 at mRNA level, and protein expression of p21, p53 and bax by real-time PCR and western blot analysis. As shown in Fig. 5, p21 and bax mRNA and protein levels were relatively upregulated in MCF-7 cells under the impact of overexpression of exogenous ING4. However, p53 mRNA and protein levels did not change markedly (Fig. 5).
Figure 5.
ING4-induced expression changes in some of the genes involved in the cell cycle and apoptosis. (A) Real-time PCR analysis showed that compared with control group cells, the transcriptional expressions of p21 and bax were markedly upregulated in the stably transfected ING4 cells (P<0.05), while the transcriptional expression of p53 was not significantly different to control groups. All data represent the mean of three independent experiments. (B) The expression of p21, bax and p53 proteins in MCF-7 cells. The protein lysates were analyzed by western blot analysis with anti-p21, anti-bax and anti-p53 antibodies. The mRNA and protein expression levels were similar in all groups of cells. P21, bax and p53 levels were normalized against β-actin expression.
Discussion
ING4 family proteins are known for their tumor suppression activity. The type II tumor suppressor protein ING4 specifically displays multiple anti-cancer effects by inhibiting cell growth and promoting apoptosis, angiogenesis and cell cycle regulation. Previous studies have shown that ING4 is widely expressed in normal cell lines, while it is downregulated in glioblastoma (11) and melanoma cells (12), as well as in head and neck squamous cell carcinoma (13). ING4 overexpression negatively regulates cell growth, with significant growth arrest at the G2/M stage of the cell cycle, and also enhances apoptosis triggered by serum starvation in a p53-dependent manner (14). Studies exploring ING4 have revealed the frequent deletion or mutational inactivation of ING4 in multiple cancers including prostate, ovarian and breast carcinomas (15).
To investigate the effects of ING4 on cell proliferation in breast cancer cells, we established the stable breast cancer cell (MCF-7) with constitutive overexpression of ING4. Furthermore, we detected the cellular behaviors of the stably transfected cells with exogenous ING4. The results of our study demonstrate that the overexpression of exogenous ING4 in MCF-7 cells inhibited cell proliferation by inducing G0/G1 arrest, inhibiting the S phase and enhancing apoptosis. The exogenous ING4 upregulated endogenous p21 and bax, whereas there was no significant increase of p53 expression in MCF-7 cells.
To explain our experimental results, we referred to previous studies and assumed that ING4 binds directly to p53, which modulates its transcriptional activity and also regulates the expression of p21 and bax (16). Although the expression level of p53 did not change significantly, p53 may be involved via the p53-dependent apoptosis pathway, which is not affected by the amount of p53 present. It has been reported that crosstalk between signaling pathways may be regulated by differing levels and modifications of p53, including acetylation and/or phosphorylation at Lys-382, Ser-15 and Ser-392 residues (7). Therefore, it is unsurprising that in the present study the expression levels of p53 remained constant, whereas those of p21 and bax increased following treatment. The induced overexpression of p21 led to an increase in the number of cells in the G1 phase, as shown by flow cytometry analysis. The p21 protein may interact with proliferating cell nuclear antigen (PCNA) to regulate S phase DNA replication and repair. An increase in the levels of p21 inhibits the activity of cyclin E/CDK2 and cyclin D/CDK4/6, resulting in S phase inhibition (17). It also has been reported that overexpression of p21 results in G2 phase arrest (18), but this was not observed in our study. In addition, p21 is reported to be cleaved by caspase, which leads to activation of CDK2, and may be functional in the execution of apoptosis following caspase activation. Bax has been shown to interact with SH3GLB1 (19), VDAC1 (20), Bcl-2-related protein A1 (21) and others. In this study, we inferred that overexpression of bax might act as a regulator in its classical form. Bax is thought to induce the opening of the mitochondrial voltage-dependent anion channel, resulting in the release of cytochrome C and other pro-apoptotic factors from the mitochondria, which subsequently leads to the activation of caspase via the intrinsic apoptotic pathway.
Together, overexpression of exogenous ING4 significantly induced in vitro cell repression, G1 phase arrest, S phase inhibition and apoptosis in human breast cancer MCF-7 cells. This inhibited tumor cell growth elicited by pcDNA3.1(+)/ING4 was closely associated with upregulation of the cell cycle and apoptosis-related molecules p21 and bax. ING4 plus relevant chemotherapy drugs have been widely reported in in vitro and in vivo studies in cancer cell lines or animal model experiments (5,22). Our study further suggests that ING4 functions as a tumor-suppressor molecule in the human breast cancer cell line MCF-7 and may provide new approaches to using ING4 in breast cancer treatment.
Acknowledgements
This study was supported by a grant from the National Natural Science Foundation of China (No. 81172142).
References
- 1.Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D. Global cancer statistics. CA Cancer J Clin. 2011;61:69–90. doi: 10.3322/caac.20107. [DOI] [PubMed] [Google Scholar]
- 2.Wang Y, Li G. ING3 promotes UV-induced apoptosis via Fas/caspase-8 pathway in melanoma cells. J Biol Chem. 2006;281:11887–11893. doi: 10.1074/jbc.M511309200. [DOI] [PubMed] [Google Scholar]
- 3.Moreno A, Palacios A, Orgaz JL, Jimenez B, Blanco FJ, Palmero I. Functional impact of cancer-associated mutations in the tumor suppressor protein ING4. Carcinogenesis. 2010;31:1932–1938. doi: 10.1093/carcin/bgq171. [DOI] [PubMed] [Google Scholar]
- 4.Li J, Li G. Cell cycle regulator ING4 is a suppressor of melanoma angiogenesis that is regulated by the metastasis suppressor BRMS1. Cancer Res. 2010;70:10445–10453. doi: 10.1158/0008-5472.CAN-10-3040. [DOI] [PubMed] [Google Scholar]
- 5.Xie Y, Sheng W, Miao J, Xiang J, Yang J. Enhanced antitumor activity by combining an adenovirus harboring ING4 with cisplatin for hepatocarcinoma cells. Cancer Gene Ther. 2011;18:176–188. doi: 10.1038/cgt.2010.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Garkavtsev I, Riabowol K. Extension of the replicative life span of human diploid fibroblasts by inhibition of the p33ING1 candidate tumor suppressor. Mol Cell Biol. 1997;17:2014–2019. doi: 10.1128/mcb.17.4.2014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shiseki M, Nagashima M, Pedeux RM, Kitahama-Shiseki M, Miura K, Okamura S, Onogi H, Higashimoto Y, Appella E, Yokota J, Harris CC. p29ING4 and p28ING5 bind to p53 and p300, and enhance p53 activity. Cancer Res. 2003;63:2373–2378. [PubMed] [Google Scholar]
- 8.Garkavtsev I, Kozin SV, Chernova O, Xu L, Winkler F, Brown E, Barnett GH, Jain RK. The candidate tumour suppressor protein ING4 regulates brain tumour growth and angiogenesis. Nature. 2004;428:328–332. doi: 10.1038/nature02329. [DOI] [PubMed] [Google Scholar]
- 9.Shi X, Hong T, Walter KL, Ewalt M, Michishita E, Hung T, Carney D, Peña P, Lan F, Kaadige MR, et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Cai L, Li X, Zheng S, Wang Y, Wang Y, Li H, Yang J, Sun J. Inhibitor of growth 4 is involved in melanomagenesis and induces growth suppression and apoptosis in melanoma cell line M14. Melanoma Res. 2009;19:1–7. doi: 10.1097/CMR.0b013e32831bc42f. [DOI] [PubMed] [Google Scholar]
- 11.Hassler M, Seidl S, Fazeny-Doerner B, Preusser M, Hainfellner J, Rössler K, Prayer D, Marosi C. Diversity of cytogenetic and pathohistologic profiles in glioblastoma. Cancer Genet Cytogenet. 2006;166:46–55. doi: 10.1016/j.cancergencyto.2005.08.021. [DOI] [PubMed] [Google Scholar]
- 12.Li J, Martinka M, Li G. Role of ING4 in human melanoma cell migration, invasion and patient survival. Carcinogenesis. 2008;29:1373–1379. doi: 10.1093/carcin/bgn086. [DOI] [PubMed] [Google Scholar]
- 13.Gunduz M, Nagatsuka H, Demircan K, Gunduz E, Cengiz B, Ouchida M, Tsujigiwa H, Yamachika E, Fukushima K, Beder L, et al. Frequent deletion and down-regulation of ING4, a candidate tumor suppressor gene at 12p13, in head and neck squamous cell carcinomas. Gene. 2005;356:109–117. doi: 10.1016/j.gene.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 14.Zhang X, Xu LS, Wang ZQ, Wang KS, Li N, Cheng ZH, Huang SZ, Wei DZ, Han ZG. ING4 induces G2/M cell cycle arrest and enhances the chemosensitivity to DNA-damage agents in HepG2 cells. FEBS Lett. 2004;570:7–12. doi: 10.1016/j.febslet.2004.06.010. [DOI] [PubMed] [Google Scholar]
- 15.Tapia C, Zlobec I, Schneider S, Kilic E, Güth U, Bubendorf L, Kim S. Deletion of the inhibitor of growth 4 (ING4) tumor suppressor gene is prevalent in human epidermal growth factor 2 (HER2)-positive breast cancer. Hum Pathol. 2011;42:983–990. doi: 10.1016/j.humpath.2010.10.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Soliman MA, Riabowol K. After a decade of study-ING, a PHD for a versatile family of proteins. Trends Biochem Sci. 2007;32:509–519. doi: 10.1016/j.tibs.2007.08.006. [DOI] [PubMed] [Google Scholar]
- 17.Radhakrishnan SK, Feliciano CS, Najmabadi F, Haegebarth A, Kandel ES, Tyner AL, Gartel AL. Constitutive expression of E2F-1 leads to p21-dependent cell cycle arrest in S phase of the cell cycle. Oncogene. 2004;23:4173–4176. doi: 10.1038/sj.onc.1207571. [DOI] [PubMed] [Google Scholar]
- 18.Niculescu AB, III, Chen X, Smeets M, Hengst L, Prives C, Reed SI. Effects of p21 (Cip1/Waf1) at both the G1/S and the G2/M cell cycle transitions: pRb is a critical determinant in blocking DNA replication and in preventing endoreduplication. Mol Cell Biol. 1998;18:629–643. doi: 10.1128/mcb.18.1.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pierrat B, Simonen M, Cueto M, Mestan J, Ferrigno P, Heim J. SH3GLB, a new endophilin-related protein family featuring an SH3 domain. Genomics. 2001;71:222–234. doi: 10.1006/geno.2000.6378. [DOI] [PubMed] [Google Scholar]
- 20.Weng C, Li Y, Xu D, Shi Y, Tang H. Specific cleavage of Mcl-1 by caspase-3 in tumor necrosis factor-related apoptosis-inducing ligand (TRAIL)-induced apoptosis in Jurkat leukemia T cells. J Biol Chem. 2005;280:10491–10500. doi: 10.1074/jbc.M412819200. [DOI] [PubMed] [Google Scholar]
- 21.Zhang H, Cowan-Jacob SW, Simonen M, Greenhalf W, Heim J, Meyhack B. Structural basis of BFL-1 for its interaction with BAX and its anti-apoptotic action in mammalian and yeast cells. J Biol Chem. 2000;275:11092–11099. doi: 10.1074/jbc.275.15.11092. [DOI] [PubMed] [Google Scholar]
- 22.Tzouvelekis A, Aidinis V, Harokopos V, Karameris A, Zacharis G, Mikroulis D, Konstantinou F, Steiropoulos P, Sotiriou I, Froudarakis M, et al. Down-regulation of the inhibitor of growth family member 4 (ING4) in different forms of pulmonary fibrosis. Respir Res. 2009;10:14. doi: 10.1186/1465-9921-10-14. [DOI] [PMC free article] [PubMed] [Google Scholar]