Abstract
The F1F0 ATP synthase is the smallest motor enzyme known. Previous studies had established that the central stalk, made of the γ and ɛ subunits in the F1 part and c subunit ring in the F0 part, rotates relative to a stator composed of α3β3δab2 during ATP hydrolysis and synthesis. How this rotation is regulated has been less clear. Here, we show that the ɛ subunit plays a key role by acting as a switch of this motor. Two different arrangements of the ɛ subunit have been visualized recently. The first has been observed in beef heart mitochondrial F1-ATPase where the C-terminal portion is arranged as a two-α-helix hairpin structure that extends away from the α3β3 region, and toward the position of the c subunit ring in the intact F1F0. The second arrangement was observed in a structure determination of a complex of the γ and ɛ subunits of the Escherichia coli F1-ATPase. In this, the two C-terminal helices are apart and extend along the γ to interact with the α and β subunits in the intact complex. We have been able to trap these two arrangements by cross-linking after introducing appropriate Cys residues in E. coli F1F0, confirming that both conformations of the ɛ subunit exist in the enzyme complex. With the C-terminal domain of ɛ toward the F0, ATP hydrolysis is activated, but the enzyme is fully coupled in both ATP hydrolysis and synthesis. With the C-terminal domain toward the F1 part, ATP hydrolysis is inhibited and yet the enzyme is fully functional in ATP synthesis; i.e., it works in one direction only. These results help explain the inhibitory action of the ɛ subunit in the F1F0 complex and argue for a ratchet function of this subunit.
An F1F0 ATP synthase, found in bacterial plasma membranes, mitochondrial inner membranes, and chloroplast thylakoid membranes, produces ATP from ADP and Pi by using the transmembrane proton motive force generated by oxidative phosphorylation or photosynthesis (1, 2). It is composed of two major parts: a cytoplasmic F1 part (α3β3γδɛ) that includes the three catalytic sites for ATP synthesis/hydrolysis and a membrane-embedded F0 part (ab2c10–14) that constitutes a proton channel. These two parts are structurally connected by two stalks, a central stalk of the γ and ɛ subunits that links to the c subunit ring and an outer stalk of δb2, linking α3β3 to the a subunit (3). The alternating arrangement of fully interdigitated α and β subunits around a central γ subunit was first established by electron microscopy studies (4). This picture was confirmed and extended in a landmark crystal structure of the bovine heart F1-ATPase that revealed the coiled-coil structure of the γ subunit (5).
There are three catalytic nucleotide binding sites, one located in each β subunit where it interfaces with the α subunit. These are around 100 Å away from the proton channel, formed by the a subunit and the c subunit ring of the F0 part. Recent evidence indicates that catalytic sites and the proton channel are coupled in a mechanism that includes rotation of the γɛ subunits and c subunit ring together (the rotor) relative to the α3β3δab2 subunits (stator). This rotation of γ and ɛ has been observed by single molecule studies (6–8). For one full (360°) rotation, the three catalytic sites progress through 2 or 3 different conformations, as described by the alternating site mechanism (2). Rotation of the c subunit ring in unison with the γ and ɛ subunits has been tested by using the actin filament method (9–11) and demonstrated conclusively in recent biochemical studies (12, 13).
The structure of the ɛ subunit from F1F0 ATP synthase from Escherichia coli (EF1F0) was determined several years ago (14, 15). It is a two-domain protein. The N-terminal part (residues 1–86) forms a 10-stranded β sandwich structure. The C-terminal domain (residues 91–138) is an α-helix–loop–α-helix structure. Two recent crystal structures have added new information about the arrangement of both the γ and ɛ subunits in the F1 part (16, 17). The structure of the N,N′-dicyclohexylcarbodiimide (DCCD)-inhibited F1-ATPase, the water soluble part of F1F0, from mitochondria (MF1) reveals a δ subunit (equivalent to the ɛ in bacterial F1) that is very similar to that of the isolated bacterial ɛ (16). The two C-terminal α helices face away from the F1 part toward the F0, an arrangement consistent with the δ subunit modeled into the recently determined low resolution x-ray data of a partial F1F0 from yeast containing the F1 subunits and a ring of 10 c subunits (18). A very different arrangement of the ɛ subunit is seen in the newly reported structure of a 1:1 complex of γ and ɛ subunits from the E. coli enzyme (17). This structure shows the two α helices of the C-terminal part of ɛ as separated, and extending up the γ subunit to where this subunit interacts with the α3β3 part, a distance of around 50 Å from the interface of the c-ring in the F1c10 structure.
These recently accumulated structural data raise several interesting questions. For example: Can both arrangements of the ɛ subunit exist in the intact F1F0 and, if so, what role might such large conformational changes of the ɛ subunit have in the functioning of the enzyme complex? Here, we describe cross-linking studies that address these questions.
Materials and Methods
Strains, Plasmids, and Preparation of E. coli Inner Membrane.
E. coli strains used were XL1-Blue (Stratagene) for site directed mutagenesis and for cloning, and the unc− RA1 (19) to express the ATP synthase. Plasmids used were pRA13 (20), pRA100 (21), and pRA197 (12). The two new EF1F0 mutants, pST210 [containing the c-dimer and Cys in positions 117 of ɛ and 42 of the second copy of c in the c-dimer (cc′Q42C/ɛA117C)] and pST111 [containing Cys in positions 99 of γ and 118 of ɛ (γL99C/ɛS118C)], were generated by employing Quick Change Site-directed mutagenesis (Stratagene). E. coli inner membranes were isolated from wild-type and two mutants as described (22).
Formation of the ɛ–cc′ and γ–ɛ Cross-Linked Products.
Inner membranes at a concentration of 0.8 mg/ml in buffer containing 50 mM Mops-NaOH, 5 mM MgCl2, and 10% glycerol (pH 7.0) were treated with 100 μM CuCl2 for 15 min at 23°C. For comparison with non-cross-linked enzyme, 1 mM DTT was added instead of CuCl2. Then, 7.5 mM EDTA was added to terminate the oxidation reaction. Cross-linked products were analyzed by gel electrophoresis (15% polyacrylamide) containing 0.1% SDS in the absence of reducing agent, followed by immunoblotting for identification with monoclonal antibodies against γ, ɛ, and c subunits. The cross-link yield was determined from the decrease of the ɛ subunit band on the Western blotting membrane.
Other Methods.
ATP hydrolysis was measured at 37°C in the presence of an ATP regenerating system. The assay mixture contained 25 mM Hepes-KOH, 25 mM KCl, 5 mM MgCl2, 5 mM KCN, 0.25 mM NADH, 2 mM phosphoenolpyruvate, 2 mM ATP, 30 units/ml pyruvate kinase, and 30 units/ml lactate dehydrogenase (pH 7.5). ATP-dependent proton translocation was determined by measuring the quenching of the 9-amino-6-chloro-2-methoxyacridine (ACMA) in inner membranes. Eighty micrograms of the inner membranes were diluted 10-fold in 900 μl of buffer [50 mM Hepes-KOH/5 mM MgCl2/100 mM KCl/3.6 μM valinomycin/1 μM ACMA (pH 7.5)]. Change of fluorescence at 480 nm (excitation at 410 nm) was monitored in response to 0.5 mM NADH, 10 mM KCN, and 2 mM ATP, followed by the addition of 3.6 μM nigericin. ATP synthesis was determined as follows: 16 μg of inner membranes in 25 mM Tris⋅HCl, 5 mM MgCl2, 10% glycerol, 5 mM ADP, 5 mM K2HPO4, and 2 mM NADH (pH 7.5) were incubated at 37°C for 0, 60, 120, and 180 s, followed by addition of 0.1 M trichloroacetic acid on ice to stop the reaction. The amount of ATP was determined with the luciferin/luciferase system by measuring the emitted light using a chemiluminometer. The value of units/mg corresponds to hydrolyzed or synthesized μmol ATP per min per mg protein. Protein concentrations were determined by using the BCA (bicinchoninic acid) protein assay (Pierce).
Results
Two Very Different Arrangements of the ɛ Subunit Can Be Trapped in the EF1Fo Complex.
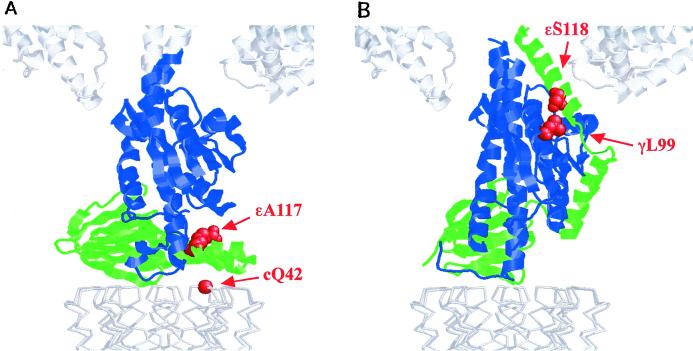
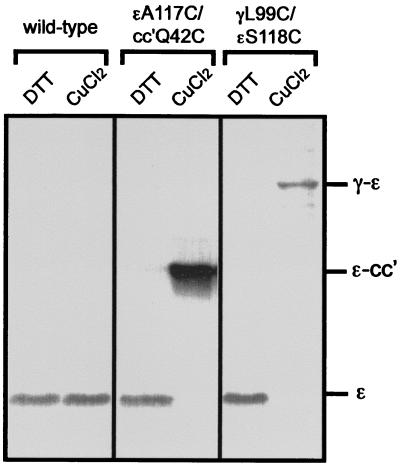
The two arrangements of the ɛ subunit relative to γ found in MF1 (16) and in the E. coli γɛ subunit complex (17), respectively, are shown in Fig. 1. Ala-117 of ɛ and Gln-42 of the c subunit are in close proximity in the structure reported by Gibbons et al. (ref. 16; Fig. 1A), and these residues were chosen for substitution by Cys. However, previous studies had shown that Cys residues at position 42 of the c subunit readily generate c–c subunit cross-linked products. To avoid this and instead favor ɛ–c cross-linking, ɛCys-117 was introduced into the mutant cc′Q42C, which has two c subunits covalently joined through an 11-aa linker and the Cys present only in the second c subunit (12). As shown in Fig. 2, oxidation with CuCl2 generated a high yield of cross-linking between the c and ɛ subunits in the mutant (ɛA117C/cc′Q42C), as confirmed by immunoblotting with the appropriate subunit antibodies. At 100 μM CuCl2, the cross-linking yield was 85–90%, based on the quantitation of the disappearance of the ɛ subunit band in immunoblots stained with anti-ɛ antibody.
Figure 1.
Comparison of the arrangement of the ɛ subunit (mitochondrial δ subunit) between (A) bovine mitochondria and (B) E. coli F1-ATPase. The bottom part of the subunit γ is shown in blue, the subunit ɛ (mitochondrial δ) in green, and c subunit ring in light gray. The positions of Cys residues mutated in this study are shown with space-filling spheres in red. The bottom parts of an α and a β subunit of α3β3 are shown in white. The residues are numbered from the E. coli sequence. The two models were created based on the coordinates of the bovine heart MF1-ATPase (1E79), and E. coli γɛ subunits (1FS0) and the unrefined cα model (1Q01) in the Protein Data Bank.
Figure 2.
Formation of the ɛ–cc′ and γ–ɛ cross-links via disulfide bonds in the EF1F0 mutants. Inner membranes from the wild-type and the two EF1F0 mutants were exposed to 100 μM CuCl2 to induce the cross-linking. As a control, 1 mM DTT was added instead of CuCl2. The samples were loaded on the SDS/PAGE (15%). The dissociation buffer for SDS/PAGE contained 40 mM N-ethylmaleimide but no reducing agent. The cross-linked products were identified with anti-γ, ɛ, and c subunit immunoblotting, respectively (the data from the anti-γ, and c subunit antibodies are not shown.).
In the second arrangement of ɛ, there is close approach of positions 99 of γ and 118 of ɛ as revealed by the structure determination of the γ–ɛ complex of EF1 (F1-ATPase, a cytoplasmic part of F1F0, from E. coli), and the presence of this conformation in EF1F0 was assessed with the mutant γL99C/ɛS118C (Fig. 1B). As shown in Fig. 2, a high yield (90%) of cross-linking between γCys99 and ɛCys118 was obtained under the same conditions (100 μM CuCl2) that gave cross-linking between the Cys at position 117 of ɛ and the c subunit. Thus, both arrangements occur in EF1F0 in E. coli membranes, and the ɛ subunit must be able to switch between the two conformations. The functional significance of this switching was examined in activity studies of the different cross-linked states.
Functional Effects of Trapping the ɛ Subunit in the Two Different Conformations.
EF1F0 from the mutant ɛA117C/cc′Q42C showed an ATPase activity of 32% of that of the wild-type membrane (1.96 units/mg compared with 6.10 units/mg; Fig. 3A). This lowered activity is due to the presence of the c subunit dimer and not related to the introduction of the Cys residues, as reported previously (12). Cross-linking between the ɛ and cc′ subunit in yields of 85–90% stimulated ATPase activity to around 150% of non-cross-linked enzyme. The mutant γL99C/ɛS118C had 92% of the activity of the wild-type (5.70 units/mg vs. 6.10 units/mg). High yield cross-linking of γ to ɛ in this mutant (85–90% yield) reduced the activity to 20% of the original (Fig. 3A). Thus, in one mutant, γA117C/cc′Q42C, there is activation whereas in the other, L99C/ɛS118C, there is strong inhibition of ATP hydrolysis as a result of fixing the different conformations of the ɛ subunit. For both, there was a reversal of the observed effects of cross-linking on addition of 50 mM DTT. This was complete with the mutant γA117C/cc′Q42C. With the mutant γL99C/ɛS118C, there was reactivation to 80% of the activity of non-cross-linked enzyme.
Figure 3.
Effect of cross-linking on the ATP hydrolysis and ATP-driven proton translocation. (A) Effect of the cross-linking on ATPase activity and inhibitor sensitivity. The inner membranes from wild-type and mutants were treated with CuCl2 or DTT as described in Fig. 2. The ATPase activity was measured in the presence of an ATP regenerating system. Samples were also incubated for 60 min at 23°C with 40 μM of DCCD, the specific inhibitor, before ATPase activity was measured. The inhibition was calculated as percentage of the activity without DCCD treatment. Inner membrane reacted with DTT, filled; CuCl2, diagonal stripes. (B) Effect of the cross-linking on ATP-driven proton translocation. The proton pumping ability of the inner membranes from wild-type and mutants was determined by monitoring the decrease of the fluorescence intensity of ACMA. Before the assay, the inner membranes were treated with DTT or CuCl2 as described in Fig. 2. At the time indicated by arrow heads, 0.5 mM NADH, 10 mM KCN, 2 mM ATP, and 3.6 μM nigericin were added respectively. Inner membrane treated with DTT (solid line), with CuCl2 (dotted line). Vertical bar, 30% of relative fluorescence; horizontal bar, 100 s.
DCCD, a well-characterized F1F0 inhibitor, reacts covalently with cD61 (in E. coli sequence), which is responsible for the proton translocation, to irreversibly block both ATP hydrolysis and synthesis (23). Both mutants showed full sensitivity to DCCD, and this inhibition was not altered by either ɛ–cc′ or γ–ɛ cross-linking, indicating that coupling between F1 and F0 was not disrupted by the covalent linking of subunits in either arrangement. As shown in Fig. 3B, ATP-driven proton translocation, examined by using the ACMA quenching assay, was not significantly altered by cross-linking of ɛ to cc′ in the mutant ɛA117C/cc′Q42C. The mutant γL99C/ɛS118C showed a modest pumping activity after the γ–ɛ cross-linking, which can be explained by the much reduced ATPase activity leading to lower steady-state levels of protons inside the membrane vesicles.
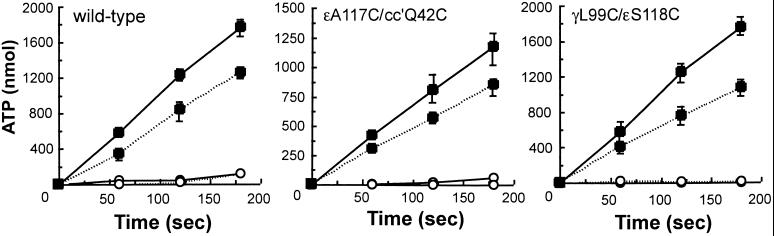
Particularly significant is the effect of cross-linking on ATP synthesis (Fig. 4). Membranes from the mutant ɛA117C/cc′Q42C had 67% of the ATP synthesis activity of the wild type before the cross-linking (0.40 units/mg vs. 0.60 units/mg; again because of the introduction of the c dimer), and the mutant γL99C/ɛS118C showed the same activity as that of the wild type (0.60 units/mg vs. 0.60 units/mg). After high yield (85–90%) cross-linking, both mutants retained 62–72% of the original activity, and these losses do not derive from the cross-linking between the introduced cysteine residues, because the same extent of inhibition was also observed in the wild-type EF1F0 treated with CuCl2. DCCD completely stopped the activity of both cross-linked and non-cross-linked enzyme. In summary, EF1F0 cross-linked to fix the conformation of ɛ reported by Gibbons et al. (16) is a functional ATPase and has normal ATP synthesis. Enzyme cross-linked to favor the conformation determined by Rodgers and Wilce (17) is a very poor ATP hydrolase but can still synthesize ATP normally.
Figure 4.
Effect of cross-linking on ATP synthesis. The inner membranes from wild-type and mutants were exposed to 2 mM NADH at 37°C to generate a proton gradient. The data show the amount of ATP produced by 1 mg of inner membrane protein. Solid line, DTT; dashed line, CuCl2-treated membranes as described in Fig. 2. Before the assay, the samples were reacted with (open circle) or without (filled square) 40 μM DCCD for 60 min at 23°C.
Discussion
The ɛ Subunit Can Exist in Two (or More) Very Different Conformations in F1F0.
Structure determinations of parts of the F1F0 ATP synthase are appearing with increasing regularity. These studies include x-ray structures of the α3β3γ part of the complex from beef heart, rat liver, and E. coli (5, 24, 25), and NMR structures of the isolated ɛ subunit and the c subunit from E. coli (14, 26). In addition, there are substantial data on subunit interactions based on cross-linking studies (3, 27). This accumulated information has been used to develop first generation models of the entire complex. Recently, two structures from Leslie, Walker and their colleagues have greatly improved our understanding of the arrangement of subunits in the intact F1F0. First, Gibbons et al. have provided a high resolution structure of beef heart MF1 as discussed (16). Second, Stock et al. have provided a low resolution structure of a portion of yeast MF1F0 (F1F0 ATP synthase from mitochondria) containing α3β3γδɛ and a ring of c10 subunits (18). In both structures, the δ subunit, i.e., the ɛ subunit in bacteria, is arranged with the C-terminal helix–turn–helix domain pointing away from the F1 part so that it makes close contact with the c subunit ring of the F0 part (Fig. 1A). As pointed out by Gibbons et al., this arrangement is unexpected, based on a considerable body of cross-linking data in EF1F0 that places the ɛ subunit, and specifically residue 108, in contact with α and β subunits (20, 28, 29). The results presented here show that the structure and arrangement of the ɛ subunit seen in MF1 and the partial MF1F0 represent one conformation of this subunit in EF1F0. However, this is not the only arrangement of the ɛ. We were able to generate the same yields of cross-linking of the γ to the ɛ subunit by introducing Cys residues at 99 in γ and 118 in ɛ, establishing that the conformation of the ɛ described recently by Rodgers and Wilce (17) also occurs in EF1F0 (Fig. 1B). That both the “parallel” (relative to the plane of the membrane; Fig. 1A) and “perpendicular” (Fig. 1B) arrangements of the C-terminal part of the ɛ subunit can be stabilized by cross-linking in the intact EF1F0 indicates that the two conformations are in some equilibrium. Our studies of enzyme trapped in the two arrangements of the ɛ show that there is a functional significance to this equilibrium (Fig. 2).
The ɛ Subunit as an Inhibitor of ATPase Activity.
The role of the ɛ subunit in bacterial and chloroplast F1F0 has been much debated. In isolated F1, the evidence that this subunit acts as an inhibitor of ATPase activity is unequivocal (30–32). Moreover, it is clear that most or all of the inhibitory effect is caused by the C-terminal helix–turn–helix domain of this subunit (33). Recent studies have demonstrated a role of the ɛ subunit in inhibition of the ATPase activity of EF1F0, although the effect is not as dramatic as in F1 alone (34). For example, several mutants of EF1F0 have been described in which changes in the C-terminal part of ɛ activate the enzyme. Among these are mutants that affect the association of the two C-terminal α helices, including ones that fix the two helices to the β sandwich part of the ɛ by disulfide bond formation (34). An important feature is that this activation occurs in F1F0 without uncoupling ATPase activity from proton pumping or without effect on ATP synthesis. Cross-linking of the C-terminal helix of the ɛ subunit to the c subunit ring, i.e., in the parallel position here (Fig. 1A), gives a similar effect. There is activation of ATP hydrolysis without effect on ATP synthesis (Figs. 3 and 4).
In contrast, cross-linking of the ɛ subunit to γ in the perpendicular position, as in the structure of Rodgers and Wilce (17), causes a significant inhibition of ATP hydrolysis and the ATP-driven proton pump activity (Fig. 3). The inhibition obtained here is close to the levels of cross-linking. The position of the C-terminal helix in the perpendicular position brings it close to a β subunit, although there must be some displacement relative to that shown in Fig. 1B for steric reasons. The close approach of the ɛ C-terminal α-helix to β subunits explains the cross-linking between ɛCys-108 and both αCys-411 and βCys-381 reported earlier (20, 28, 29). Such close interaction between ɛ and α or β can be expected to restrict or even prevent conformational changes in the β subunits that are central to the rotation of the central stalk (35, 36). Related to this, Hara et al. have recently shown that the electrostatic interaction between the C-terminal helix of the ɛ subunit and the DELSEED motif in the β subunit is essential for the inhibitory effect of the ɛ subunit (37). Under normal functioning, entry and exit into the perpendicular position must occur transiently, but sufficiently often to slow ATP hydrolysis to the levels observed for the enzyme both in E. coli membranes and when isolated. These rates are considerably lower than observed when the perpendicular conformation is prevented (as in the ɛA117C/cc′Q42C described here, or when the two C-terminal α helices are fixed by cross-linking to the N-terminal domain (34)).
A Ratchet Model of ɛ Subunit Functioning in Bacterial F1F0.
The most remarkable and a highly significant feature of the γL99C/ɛS118C mutant is that, after cross-linking to fix the perpendicular position of the C-terminal α-helices, ATP hydrolysis is severely reduced, but ATP synthesis is unaffected (Figs. 3 and 4). These findings are clear evidence that the position of ɛ can determine the efficiency of the forward versus reverse direction of the reaction H2O + ATP = ADP + Pi + H+. A rotational mechanism for the function of F1F0 ATPases provides a rational explanation of how this can occur. Thus, the interaction(s) of the C-terminal α-helix of ɛ with each α–β subunit pair (in turn) could be such that these are readily broken by rotation in one direction (clockwise, when viewed from the F0 part toward the F1), thereby allowing ATP synthesis. However, in the reverse (counterclockwise direction), concerted release of the ɛ subunit with a rotation step, if electrostatically and/or sterically blocked, inhibits ATP hydrolysis (37). Such a mechanism would mean that the ɛ subunit works as a ratchet. Nucleotide-dependent switching of the location of the C-terminal part of ɛ relative to α and β subunits has been seen (29), which may involve transfer of ɛ from β to α to ready the enzyme for the next 120° jump as an ATP is synthesized. Whether it is this β → α movement, or transitioning of α–β subunits between open and closed conformations, or both, that is affected by ɛ remains to be determined.
The ability to selectively switch off ATP hydrolysis, but retain ATP synthesis function, may be relevant in E. coli only under extreme conditions where ATP levels are very low. In bacteria, the predominant role of the F1F0 complex is as an ATPase to provide a proton gradient to drive ion transport activities. Where the ability to regulate the F1F0 complex becomes very important is in mitochondria. The F1F0 complex in mitochondria acts predominantly as a synthase and is tightly regulated, so that ATP is not wasted (for reviews see refs. 38 and 39). As described above, the δ subunit of MF1F0 is the structural equivalent of the bacterial ɛ subunit, and has a similar overall structure. However, in mitochondria, the inhibitory function ascribed to the C-terminal domain of ɛ in bacteria has been taken over by a specialized inhibitor protein (IF1; ref. 40). In MF1F0, the structural data indicate that the δ (ɛ in bacteria) is trapped in the parallel position by a small polypeptide (16) not found in bacterial F1F0 (unfortunately named the ɛ subunit).
Structural studies on the IF1 indicate that it has a predominantly α-helical motif (41, 42), and this subunit could act to regulate the forward versus reverse direction of functioning of the enzyme, as we are postulating here, for the more primitive EF1F0. There is unambiguous evidence that IF1 inhibits ATP hydrolysis, but whether it inhibits ATP synthesis is less clear (43–45). Studies that suggest that IF1 also inhibits ATP synthesis are inconclusive, in part, because they were conducted under conditions in which other factors could have altered functioning of MF1F0 (43, 44). Effects of IF1 on ATP synthesis now need to be reassessed. Switching of MF1F0 between an ATP synthase and an ATP hydrolase mode by a ratchet-like device, as we propose, allows for independent regulation of the reversible reaction steps within this (single) enzyme complex. The importance of such a control device for energy metabolism has been reviewed recently by Vinogradov (46). This control can be effected by cellular modulation of the binding of the IF1, for example by pH or ΔΨ, for which there is already considerable evidence (47).
In summary, we present evidence of two very different conformations of ɛ subunit in EF1F0 by which the ɛ subunit can function as a ratchet to differentially regulate ATP hydrolysis and ATP synthesis. We suggest that this ratchet function is primitive in the bacterial enzyme and more elaborated in the mitochondrial enzyme by separation of the structural and inhibitory roles of the ɛ in two polypeptides, the δ and IF1, respectively.
Acknowledgments
We thank Dr. Robert H. Fillingame for providing the c subunit antibody. This work was supported by the Human Frontier Science Program Organization (RG15/1998-M) and National Institutes of Health Grant HL24526 (to R.A.C.). S.P.T. and A.J.W.R. were supported by a Research Fellowship of the Japan Society for the Promotion of Science for Young Scientists and a Fellowship from the Healy Medical Research Foundation respectively.
Abbreviations
- EF1F0 and MF1F0
F1F0 ATP synthase from Escherichia coli and mitochondria, respectively
- EF1 and MF1 F1-ATPase
a cytoplasmic part of F1F0, from Escherichia coli and the equivalent, water-soluble part of the enzyme from mitochondria, respectively
- IF1
mitochondrial F1F0 inhibitor protein
- DCCD
N,N′-dicyclohexylcarbodiimide
- ACMA
9-amino-6-chloro-2-methoxyacridine
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
References
- 1.Senior A E. Annu Rev Biophys Biophys Chem. 1990;19:7–41. doi: 10.1146/annurev.bb.19.060190.000255. [DOI] [PubMed] [Google Scholar]
- 2.Boyer P D. Annu Rev Biochem. 1997;66:717–749. doi: 10.1146/annurev.biochem.66.1.717. [DOI] [PubMed] [Google Scholar]
- 3.Capaldi R A, Schulenberg B, Murray J, Aggeler R. J Exp Biol. 2000;203:29–33. doi: 10.1242/jeb.203.1.29. [DOI] [PubMed] [Google Scholar]
- 4.Gogol E P, Aggeler R, Sagermann M, Capaldi R A. Biochemistry. 1989;28:4717–4724. doi: 10.1021/bi00437a031. [DOI] [PubMed] [Google Scholar]
- 5.Abrahams J P, Leslie A G W, Lutter R, Walker J E. Nature (London) 1994;370:621–628. doi: 10.1038/370621a0. [DOI] [PubMed] [Google Scholar]
- 6.Noji H, Yasuda R, Yoshida M, Kinosita K., Jr Nature (London) 1997;386:299–302. doi: 10.1038/386299a0. [DOI] [PubMed] [Google Scholar]
- 7.Kato-Yamada Y, Noji H, Yasuda R, Kinosita K, Jr, Yoshida M. J Biol Chem. 1998;273:19375–19377. doi: 10.1074/jbc.273.31.19375. [DOI] [PubMed] [Google Scholar]
- 8.Adachi K, Yasuda R, Noji H, Itoh H, Harada Y, Yoshida M, Kinosita K., Jr Proc Natl Acad Sci USA. 2000;97:7243–7247. doi: 10.1073/pnas.120174297. . (First Published June 6, 2000; 10.1073/pnas.120174297) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sambongi Y, Iko Y, Tanabe M, Omote H, Iwamoto-Kihara A, Ueda I, Yanagida T, Wada Y, Futai M. Science. 1999;286:1722–1724. doi: 10.1126/science.286.5445.1722. [DOI] [PubMed] [Google Scholar]
- 10.Tsunoda S P, Aggeler R, Noji H, Kinosita K, Yoshida M, Capaldi R A. FEBS Lett. 2000;470:244–248. doi: 10.1016/s0014-5793(00)01336-3. [DOI] [PubMed] [Google Scholar]
- 11.Pänke O, Gumbiowski K, Junge W, Engelbrecht S. FEBS Lett. 2000;472:34–38. doi: 10.1016/s0014-5793(00)01436-8. [DOI] [PubMed] [Google Scholar]
- 12.Schulenberg B, Aggeler R, Murray J, Capaldi R A. J Biol Chem. 1999;274:34233–34237. doi: 10.1074/jbc.274.48.34233. [DOI] [PubMed] [Google Scholar]
- 13.Tsunoda S P, Aggeler R, Yoshida M, Capaldi R A. Proc Natl Acad Sci USA. 2001;98:898–902. doi: 10.1073/pnas.031564198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wilkens S, Dahlquist F W, Mclntosh L P, Donaldson L W, Capaldi R A. Nat Struct Biol. 1995;2:961–967. doi: 10.1038/nsb1195-961. [DOI] [PubMed] [Google Scholar]
- 15.Uhlin U, Cox G B, Guss J M. Structure. 1997;5:1219–1230. doi: 10.1016/s0969-2126(97)00272-4. [DOI] [PubMed] [Google Scholar]
- 16.Gibbons C, Montgomery M G, Leslie A G W, Walker J E. Nat Struct Biol. 2000;7:1055–1061. doi: 10.1038/80981. [DOI] [PubMed] [Google Scholar]
- 17.Rodgers A J W, Wilce M C J. Nat Struct Biol. 2000;7:1051–1054. doi: 10.1038/80975. [DOI] [PubMed] [Google Scholar]
- 18.Stock D, Leslie A G, Walker J E. Science. 1999;286:1700–1705. doi: 10.1126/science.286.5445.1700. [DOI] [PubMed] [Google Scholar]
- 19.Aggeler R, Ogilvie I, Capaldi R A. J Biol Chem. 1997;272:19621–19624. doi: 10.1074/jbc.272.31.19621. [DOI] [PubMed] [Google Scholar]
- 20.Aggeler R, Haughton M A, Capaldi R A. J Biol Chem. 1995;270:9185–9191. doi: 10.1074/jbc.270.16.9185. [DOI] [PubMed] [Google Scholar]
- 21.Aggeler R, Chicas-Cruz K, Cai S X, Keana J F W, Capaldi R A. Biochemistry. 1992;31:2956–2961. doi: 10.1021/bi00126a016. [DOI] [PubMed] [Google Scholar]
- 22.Foster D L, Fillingame R H. J Biol Chem. 1979;254:8230–8236. [PubMed] [Google Scholar]
- 23.Hermolin J, Fillingame R H. J Biol Chem. 1989;264:3896–3903. [PubMed] [Google Scholar]
- 24.Bianchet M A, Hullihen J, Pedersen P L, Amzel L M. Proc Natl Acad Sci USA. 1998;95:11065–11070. doi: 10.1073/pnas.95.19.11065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hausrath A C, Grüber G, Matthews B W, Capaldi R A. Proc Natl Acad Sci USA. 1999;96:13697–13702. doi: 10.1073/pnas.96.24.13697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Girvin M E, Rastogi V K, Abildgaard E, Markley J L, Fillingame R H. Biochemistry. 1998;37:8817–8824. doi: 10.1021/bi980511m. [DOI] [PubMed] [Google Scholar]
- 27.Fillingame R H, Jiang W, Dmitriev O Y. J Exp Biol. 2000;203:9–17. doi: 10.1242/jeb.203.1.9. [DOI] [PubMed] [Google Scholar]
- 28.Dallmann H G, Flynn T G, Dunn S D. J Biol Chem. 1992;267:18953–18960. [PubMed] [Google Scholar]
- 29.Aggeler R, Capaldi R A. J Biol Chem. 1996;271:13888–13891. doi: 10.1074/jbc.271.23.13888. [DOI] [PubMed] [Google Scholar]
- 30.Richter M L, Snyder B, McCarty R E, Hammes G G. Biochemistry. 1985;24:5755–5763. doi: 10.1021/bi00342a011. [DOI] [PubMed] [Google Scholar]
- 31.Mendel-Hartvig J, Capaldi R A. Biochemistry. 1991;30:10987–10991. doi: 10.1021/bi00109a025. [DOI] [PubMed] [Google Scholar]
- 32.Kato Y, Matsui T, Tanaka N, Muneyuki E, Hisabori T, Yoshida M. J Biol Chem. 1997;272:24906–24912. doi: 10.1074/jbc.272.40.24906. [DOI] [PubMed] [Google Scholar]
- 33.Kato-Yamada Y, Bald D, Koike M, Motohashi K, Hisabori T, Yoshida M. J Biol Chem. 1999;274:33991–33994. doi: 10.1074/jbc.274.48.33991. [DOI] [PubMed] [Google Scholar]
- 34.Schulenberg B, Capaldi R A. J Biol Chem. 1999;274:28351–28355. doi: 10.1074/jbc.274.40.28351. [DOI] [PubMed] [Google Scholar]
- 35.Tsunoda S P, Muneyuki E, Amano T, Yoshida M, Noji H. J Biol Chem. 1999;274:5701–5706. doi: 10.1074/jbc.274.9.5701. [DOI] [PubMed] [Google Scholar]
- 36.Ren H, Dou C, Stelzer M S, Allison W S. J Biol Chem. 1999;274:31366–31372. doi: 10.1074/jbc.274.44.31366. [DOI] [PubMed] [Google Scholar]
- 37.Hara, K. Y., Kato-Yamada, Y., Kikuchi, Y., Hisabori, T. & Yoshida, M. (2001) J. Biol. Chem.276, in press. [DOI] [PubMed]
- 38.Harris D A, Das A M. Biochem J. 1991;280:561–573. doi: 10.1042/bj2800561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Walker J E. Curr Opin Struct Biol. 1994;4:912–918. doi: 10.1016/0959-440x(94)90274-7. [DOI] [PubMed] [Google Scholar]
- 40.Schwerzmann K, Pedersen P L. Arch Biochem Biophys. 1986;250:1–18. doi: 10.1016/0003-9861(86)90695-8. [DOI] [PubMed] [Google Scholar]
- 41.Van Heeke G, Deforce L, Schnizer R A, Shaw R, Couton J M, Shaw G, Song P S, Schuster S M. Biochemistry. 1993;32:10140–10149. doi: 10.1021/bi00089a033. [DOI] [PubMed] [Google Scholar]
- 42.Van Raaij M J, Orriss G L, Montgomery M G, Runswick M J, Fearnley I M, Skehel J M, Walker J E. Biochemistry. 1996;35:15618–15625. doi: 10.1021/bi960628f. [DOI] [PubMed] [Google Scholar]
- 43.Harris D A, Von Tscharner V, Radda G K. Biochim Biophys Acta. 1979;548:72–84. doi: 10.1016/0005-2728(79)90188-9. [DOI] [PubMed] [Google Scholar]
- 44.Lippe G, Sorgato M C, Harris D A. Biochim Biophys Acta. 1988;933:1–11. doi: 10.1016/0005-2728(88)90050-3. [DOI] [PubMed] [Google Scholar]
- 45.Schwerzman K, Pedersen P L. Biochemistry. 1981;20:6305–6311. doi: 10.1021/bi00525a004. [DOI] [PubMed] [Google Scholar]
- 46.Vinogradov A D. Biochemistry (Moscow) 1999;64:1443–1456. [Google Scholar]
- 47.Cabezon E, Butler P J G, Runswick M J, Walker J E. J Biol Chem. 2000;275:25460–25464. doi: 10.1074/jbc.M003859200. [DOI] [PubMed] [Google Scholar]