Abstract
Background
The methanogenic Archaea Methanosphaera stadtmanae has been detected in the human gut microbiota by both culture and culture-independent methods. Its growth reaches an exponential phase after 5 to 7-day culture in medium 322 (10% vol). Our recent successful isolation of Methanomassiliicoccus luminyensis, a tungstate-selenite-requiring Archaea sharing similar metabolism characteristics with M. stadtmanae prompted us to study the effects of tungsten and selenium on M. stadtmanae growth.
Findings
Addition of 0.2 mg/L sodium tungstate to medium 322 yielded, 48 hours after inoculation, a growth rate equivalent to that obtained after 6 days with control culture as measured by methane monitoring and optical density measurement. Addition of 50 μg/mL sodium selenate had no effect on M. stadtmanae growth. Quantitative real-time PCRs targeting the M. stadtmanae 16S rRNA confirmed these data.
Conclusions
These data provide new information regarding the poorly known nutritional requirements of the human gut colonizing organismsM. stadtmanae. Adding sodium tungstate to basal medium may facilitate phenotypic characterization of this organism and additionally aid the isolation of new Archaeafrom complex host microbiota.
Keywords: Methanogenic Archaea, Methanosphaera stadtmanae, Methanomassiliicoccus luminyensis, Tungsten, Selenium
Findings
Methanosphaera stadtmanae is a spherical-shaped, non-motile archaeon initially isolated from human feces [1]. M. stadtmanae was the first human Archaea to be genome sequenced and analysis of the genome confirmed that M. stadtmanae belonged to Methanobacteriales[2]. PCR-based analyses further indicated that M. stadtmanae-specific sequences could be detected in stool specimen in up to 30% of individuals [3]. However, M. stadtmanae is a fastidious organism, with only one M. stadtmanae isolate reported and accordingly only one M. stadtmanae strain available in public collections. M. stadtmanae oxidizes hydrogen to reduce methanol into methane [1,2]. This metabolic trait has been already reported for M. stadtmanae[4], and more recently for members of the genus Methanobacterium (e.g. M. veterum and M. lacus; [5,6]) within the order Methanobacteriales. We recently isolated Methanomassiliicoccus luminyensis, the first cultured representative of new order of methanoarchaea [7]. This archaeon exhibits a metabolic trait similar to that of M. stadtmanae by using hydrogen as electron donor and methanol as electron acceptor [7]. Unexpectedly, we observed that addition of tungstate-selenite to culture medium had been a key factor for successful isolation of M. luminyensis and that this archaeon indeed required tungstate-selenite as an essential element for growth. We therefore tested the hypothesis that the addition of sodium tungstate or sodium selenate or both to basal culture medium would also enhance the growth of M. stadtmanae.
M. stadtmanae DSMZ 3091 T (ATCC 43021T) purchased from the German Collection of Microorganisms and Cell Cultures (DSMZ, Braunschweig, Germany) was grown on medium 322 (http://www.dsmz.de) incubated at 37°C in Hungate tubes (Dutscher, Issy-les-Moulineaux, France) under 2-bar pressure of a H2/CO2 (80–20) atmosphere. The inoculated medium (10% vol) was incubated at 37°C with shaking. On the exponential phase of this first culture, a second inoculation was performed by 10% vol. in the same basal medium modified or not by the addition of Na2O4W (0.2 mg/L) and/or Na2O4Se (50 μg/L) (Sigma, Saint-Quentin Fallavier, France). Non-inoculated media were used as negative controls and each experiment was repeated ten times.
Growth was assessed by optical microscope observation, parallel methane production measurement and measurement of the optical density of the medium. Methane production measurement used a GC-8A gas chromatograph (Shimadzu, Champs-sur-Marne, France) equipped with a thermal conductivity detector and a Chromosorb WAW 80/100 mesh SP100 column (Alltech, Carquefou, France). N2 at a pressure of 100 kPa was used as the carrier gas. The detector and the injector temperatures were 200°C, and the column temperature was 150°C. H2 consumption and CH4 production were measured every 6 hours for 24 hours and then every 12 hours for 6 days. The optical density at 580 nm was measured by inserting Hungate tubes into the spectrophotometer (Varian Cary50; Agilent Technologies, Massy, France). Experiment was done in triplicate and average optical density value for the three replicates was calculated.
M. stadtmanae DNA extraction, quantification and sequencing were performed as previously described based on specific quantitative real-time PCR targeting 16S rRNA gene [3].
Negative controls (with and without tungstate and selenium) remained negative with no growth occurring after one-week incubation indicating that results herein reported did not merely result from carry-over of organisms. The exponential phase of M. stadtmanae growth cultured in medium 322 was reached at 6-day incubation. At this point microscopic observation disclosed organisms with morphology compatible with M. stadtmanae and no contaminant. Also, qPCR detected an equivalent of 3.22E + 12 ± 1.53E + 11 copies of 16S rRNA gene/mL (Table 1). Sequencing of 16S rRNA gene PCR products from all specimens yielded a sequence similarity of 99-100% with the reference M. stadtmanae DSM 3091 sequence.
Table 1.
M. stadtmanae 16S rDNA gene copy number after 48-hour culture with Na2O4W + Na2O4Se or only Na2O4W and a 6-day culture with no Na2O4W + Na2O4Se or only with Na2O4Se (Mean and standard deviation were calculated for 10 independent culture tests for each condition)
| |
48-hour culture |
6-day culture |
|||
|---|---|---|---|---|---|
| |
without |
with |
with |
with |
without |
| Na2O4W + Na2O4Se | Na2O4W + Na2O4Se | Na2O4W | Na2O4Se | Na2O4W + Na2O4Se | |
| Means |
2.13E + 10 |
4.42E + 12 |
3.93E + 12 |
4.02E + 12 |
3.22E + 12 |
| Standard deviation | 5.56E + 09 | 1.84E + 11 | 3.67E + 11 | 2.23E + 11 | 1.53E + 11 |
The addition of sodium selenate alone has no effect on the growth curve of M. stadtmanae. However, the addition of sodium tungstate alone or in combination with sodium selenate shortened the lag period to 2 days post-inoculation with an equivalent 16S rRNA and rpoB genes copy number and with equivalent rates of methane production (Figure 1). In the absence of tungstate, M. stadtmanae exhibited a 30-hour log phase. Adding tungsate to the culture medium reduced the delay of this log-phase so that it took 47 hours instead of 72 hours to achieve a 0.35 optical density of the culture (Figure 2). These results correlated with the fact that M. stadtmanae genome encodes a formylmethanofuran dehydrogenase comprising of five sub-units (Genes IDs: 3855499-3855500-3855501-3855502-3855503), an enzyme found in methanogenic Archaea. In strict anaerobic micro-organisms, this enzyme catalyzes the reversible dehydrogenation of formylmethanofuran into CO2 and methanofuran. The formylmethanofuran dehydrogenases are either molybdenum- or tungsten-iron-sulfur proteins. The tungsten is likely bound to the same skeleton as the molybdenum in the so-called molybdopterin dinucleotide cofactor [8-10].
Figure 1.
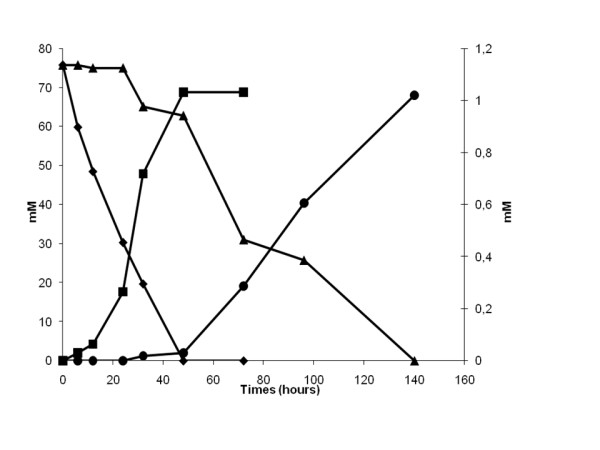
Visualizations of H2used (in mM; left Y axis) and CH4(in mM; right Y axis) produced by M. stadtmanae with and without addition of sodium tungstate solution (Na2O4W) (over 140 hours (X axis). ♦ H2 used with sodium tungstate (Na2O4W), ■ CH4 production with sodium tungstate (Na2O4W), ▴ H2 used without sodium tungstate (Na2O4W), and ● CH4 production without sodium tungstate (Na2O4W).
Figure 2.
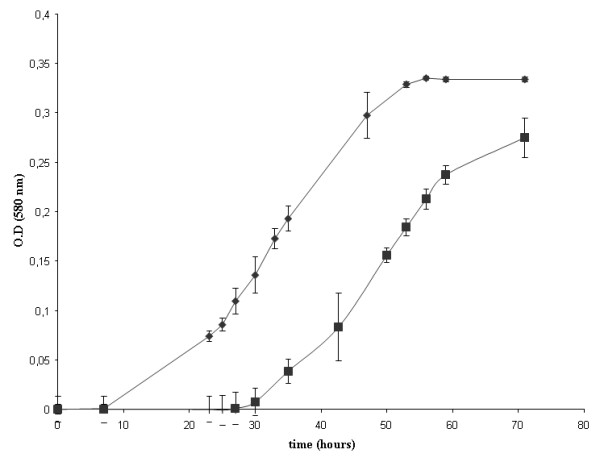
The effect of addition of selenite/tungstate solution on growth of M. stadtmanae. ♦ Growth of M. stadtmanae with tungstate (Na2O4W). ■ Growth of M. stadtmanae without tungstate (Na2O4W).
Previous reports described the requirement of tungsten for growing numerous methanogens including Methanothermobacter wolfei which has an obligate requirement for tungsten to maintain autotrophic growth, Methanococcus vannielii requiring tungsten as a cofactor for the enzyme formate dehydrogenase [11], Methanogenium tatii[12] and Methanocorpusculum parvum[13] also requiring tungsten for growth (Table 2). Selenium has also been reported as stimulatory and may be required for many methanogens, especially members of the genus Methanococcus as Methanococcus vannielii[11], Methanococcus jannaschii[14], Methanococcus maripaludis[15], Methanococcus voltae[16] and Methanococcus thermolithotrophicus[17] (Table 1). Requirement for selenium could have enzymatic basis, since it was reported that M. vannielii possesses a selenium-dependant formate dehydrogenase [18]. Selenium was also reported as a component of both a hydrogenase [19] and tRNA [20].
Table 2.
Requirement of tungsten or/and selenium for growth of methanogens as reported in bibliography
| Species | Tungsten | Selenium | References |
|---|---|---|---|
|
Methanothermobacter wolfei |
YES |
NA |
[21] |
|
Methanococcus vannielii |
YES |
YES |
[11,18] |
|
Methanogenium tatii |
YES |
NA |
[12] |
|
Methanocorpusculum parvum |
YES |
NA |
[13] |
|
Methanococcus jannaschii |
NA |
YES |
[14] |
|
Methanococcus maripaludis |
NA |
YES |
[15] |
|
Methanococcus voltae |
NA |
YES |
[16] |
| Methanococcus thermolithotrophicus | NA | YES | [17] |
In the absence of tungstate, M. stadtmanae exhibited a growth delay of 5–7 days which is long for testing in vitro susceptibility to antibiotics [22]. As we now observed that tungsten enhances the growth of two taxonomically unrelated methanogens, M. stadtmanae and M. luminyensis, we suggest that tungsten-containing media could be incorporated into the panel of media used for the isolation and culture of new methanogens from clinical and environmental specimens, and for testing their in-vitro susceptibility to antibiotics.
Methanogenic Archaea recently emerged as normal components of the human gastrointestinal and oral microbial ecosystems, where they could play important roles in health and diseases [23]. However, the isolation of such organisms requires long incubation times and strict anoxic atmosphere and is hampered by the incomplete knowledge of their nutritional requirements [23]. In fact, the result obtained in the present study may prompt further phenotypic characterization including extended antibiotic susceptibility testing [22] and even allowing isolation of new Archaea in order to assess understanding their contribution in the physiology of complex human microbiomes and their potential role in the course of infections.
Competing interests
The authors declare that they have no competing interests.
Author’s contributions
BD, SK, MLF designed and performed analyses, BO, MD interpreted data and wrote the draft. All authors read and approved the final manuscript.
Contributor Information
Bédis Dridi, Email: dridib10@gmail.com.
Saber Khelaifia, Email: khelaifia.saber@gmail.com.
Marie-Laure Fardeau, Email: arie-laure.fardeau@univmed.fr.
Bernard Ollivier, Email: bernard.ollivier@univmed.fr.
Michel Drancourt, Email: michel.drancourt@univmed.fr.
References
- Miller TL, Wolin MJ. Methanosphaera stadtmaniaegen. nov., sp. nov.: a species that forms methane by reducing methanol with hydrogen. Arch Microbiol. 1985;141:116–122. doi: 10.1007/BF00423270. [DOI] [PubMed] [Google Scholar]
- Fricke WF, Seedorf H, Henne A, Kruer M, Liesegang H, Hedderich R. et al. The genome sequence ofMethanosphaera stadtmanaereveals why this human intestinal archaeon is restricted to methanol and H2 for methane formation and ATP synthesis. J Bacteriol. 2006;188:642–658. doi: 10.1128/JB.188.2.642-658.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dridi B, Henry M, El Khéchine A, Raoult D, Drancourt M. High prevalence ofMethanobrevibacter smithiiandMethanosphaera stadtmanaedetected in the human gut using an improved DNA detection protocol. PLoS One. 2009;4:e7063. doi: 10.1371/journal.pone.0007063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kendall M, Boone D. In: The Prokaryotes. Dworkin M, Falkow S, Rosenberg E, Schleifer KH, Stackebrandt E, editor. Spriger, New York; 2006. The OrderMethanosarcinales; pp. 244–256. [Google Scholar]
- Krivushin KV, Shcherbakova VA, Petrovskaya LE, Rivkina EM. Methanobacterium veterumsp. nov., from ancient Siberian permafrost. Int J Syst Evol Microbiol. 2010;60:455–459. doi: 10.1099/ijs.0.011205-0. [DOI] [PubMed] [Google Scholar]
- Borrel G, Joblin K, Guedon A, Colombet J, Tardy V, Lehours AC, Fonty G. Methanobacterium lacussp. nov., a novel hydrogenotrophic methanogen from the deep cold sediment of a meromictic lake. Int J Syst Evol Microbiol. 2011. Sep 2. [Epub ahead of print] PubMed PMID: 21890730. [DOI] [PubMed]
- Dridi B, Fardeau ML, Ollivier B, Raoult D, Drancourt M. Methanomassiliicoccus luminyensis,gen. nov., sp. nov., a novel methanogenicArchaeaisolated from human feces. Int J Syst Evol Microbiol. 2012;62:1902–1907. doi: 10.1099/ijs.0.033712-0. in press. [DOI] [PubMed] [Google Scholar]
- Karrasch M, Borner G, Enssle M, Thauer RK. The molybdoenzyme formylmethanofuran dehydrogenase fromMethanosarcina barkericontains a pterin cofactor. Eur J Biochem. 1990;194:367–372. doi: 10.1111/j.1432-1033.1990.tb15627.x. [DOI] [PubMed] [Google Scholar]
- Karrasch M, Borner G, Enssle M, Thauer RK. Formylmethanofuran dehydrogenase from methanogenic bacteria, a molybdoenzyme. FEBS Lett. 1989;253:226–230. doi: 10.1016/0014-5793(89)80964-0. [DOI] [PubMed] [Google Scholar]
- Borner G, Karrasch M, Thauer RK. Molybdopterin adenine dinucleotide and molybdopterin hypoxanthine dinucleotide in formylmethanofuran dehydrogenase fromMethanobacterium thermoautotrophicum(Marburg) FEBS Lett. 1991;290:31–34. doi: 10.1016/0014-5793(91)81218-W. [DOI] [PubMed] [Google Scholar]
- Jones JB, Stadtman TC. Methanococcus vannielii: culture and effects of selenium and tungsten on growth. J Bacteriol. 1977;130:1404–1406. doi: 10.1128/jb.130.3.1404-1406.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zabel HP, König H, Winter J. Isolation and characterization of a new coccoid methanogen,Methanogenium tatiispec. nov. from a solfataric field on Mount Tatio. Arch Microbiol. 1984;137:308–315. doi: 10.1007/BF00410727. [DOI] [Google Scholar]
- Zellner G, Alten C, Stackebrandt E, Conway De Macario E, Winter J. Isolation and characterization ofMethanocorpusculum parvumgen. nov., spec. nov., a new tungsten requiring, coccoid methanogen. Arch Microbiol. 1987;147:13–20. doi: 10.1007/BF00492898. [DOI] [Google Scholar]
- Jones WJ, Leigh JA, Mayer F, Woese CR, Wolfe RS. Methanococcus jannaschiisp. nov., an extremely thermophilic methanogen from a submarine hydrothermal vent. Arch Microbiol. 1983;136:254–261. doi: 10.1007/BF00425213. [DOI] [Google Scholar]
- Jones WJ, Paynter MJB, Gupta R. Characterization ofMethanococcus maripaludissp. nov., a new methanogen isolated from salt marsh sediment. Arch Microbiol. 1983;135:91–97. doi: 10.1007/BF00408015. [DOI] [Google Scholar]
- Berghöfer Y, Agha-Amiri K, Klein A. Selenium is involved in the negative regulation of the expression of selenium-free [NiFe] hydrogenases inMethanococcus voltae. Mol Gen Genet. 1994;242:369–373. doi: 10.1007/BF00281785. [DOI] [PubMed] [Google Scholar]
- Belay N, Sparling R, Daniels L. Relationship of formate to growth and methanogenesis byMethanococcus thermolithotrophicus. Appl Environ Microbiol. 1986;52:1080–1085. doi: 10.1128/aem.52.5.1080-1085.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones JB, Stadtman TC. Selenium-dependent and selenium-independent formate dehydrogenases ofMethanococcus vannielii. Separation of the two forms and characterization of the purified selenium-independent form. J Biol Chem. 1981;256:656–663. [PubMed] [Google Scholar]
- Yamazaki S. A selenium-containing hydrogenase fromMethanococcus vannielii, Identification of the selenium moiety as a selenocysteine residue. J Biol Chem. 1982;257:7926–7929. [PubMed] [Google Scholar]
- Ching WM, Wittwer AJ, Tsai L, Stadtman TC. Distribution of two selenonucleosides among the selenium-containing tRNAs fromMethanococcus vannielii. Proc Natl Acad Sci U S A. 1984;81:57–60. doi: 10.1073/pnas.81.1.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- König H, Semmler R, Lerp C, Winter J. Evidence for the occurrence of autolytic enzymes inMethanobacterium wolfei. Arch Microbiol. 1985;141:177–180. doi: 10.1007/BF00423281. [DOI] [Google Scholar]
- Dridi B, Fardeau ML, Ollivier B, Raoult D, Drancourt M. The antimicrobial resistance pattern of cultured human methanogens reflects the unique phylogenetic position of archaea. J Antimicrob Chemother. 2011;66:2038–2044. doi: 10.1093/jac/dkr251. [DOI] [PubMed] [Google Scholar]
- Dridi B, Raoult D, Drancourt M. Archaea as emerging organisms in complex human microbiomes. Anaerobe. 2011;17:56–63. doi: 10.1016/j.anaerobe.2011.03.001. [DOI] [PubMed] [Google Scholar]


