Abstract
Multiple members of the ADAR (adenosine deaminases acting on RNA) gene family are involved in A-to-I RNA editing. It has been speculated that they may form a large multicomponent protein complex. Possible candidates for such complexes are large nuclear ribonucleoprotein (lnRNP) particles. The lnRNP particles consist mainly of four spliceosomal subunits that assemble together with the pre-mRNA to form a large particle and thus are viewed as the naturally assembled pre-mRNA processing machinery. Here we investigated the presence of ADARs in lnRNP particles by Western blot analysis using anti-ADAR antibodies and by indirect immunoprecipitation. Both ADAR1 and ADAR2 were found associated with the spliceosomal components Sm and SR proteins within the lnRNP particles. The two ADARs, associated with lnRNP particles, were enzymatically active in site-selective A-to-I RNA editing. We demonstrate the association of ADAR RNA editing enzymes with physiological supramolecular complexes, the lnRNP particles.
Keywords: adenosine deaminases acting on RNA, double-stranded RNA adenosine deaminase, nucleic acid–protein interactions, large nuclear ribonucleoprotein
ADAR (adenosine deaminases acting on RNA) deaminates multiple adenosines to inosines, specifically in double-stranded RNA (dsRNA) (1, 2). In humans as well as rodents, three separate ADAR genes (ADAR1–ADAR3) have been identified, revealing an ADAR gene family (1, 3–10). Both ADAR1 and ADAR2 appear to be expressed ubiquitously (3–6, 8), whereas ADAR3 is expressed at low levels only in several regions of the brain (7, 10). The members of ADAR gene family are involved in A-to-I RNA editing of transcripts of certain genes such as glutamate receptor (GluR) subunits (11, 12) and serotonin receptor 2C subtype (5-HT2CR) (13). Editing of the “Q/R” site of α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid GluR-B subunit dramatically decreases Ca2+ permeability of the channel (11), whereas significant changes in the G-protein coupling efficiency of 5-HT2CR by A-to-I RNA editing also have been reported (13–15). In view of the ubiquitous expression of ADAR1 and ADAR2, the A-to-I RNA editing by ADAR is likely a widespread phenomenon (16, 17). RNA editing studies in vitro, using purified recombinant ADAR proteins, have indicated a significantly different site selectivity displayed by different ADAR gene family members. For instance, ADAR2a, a splicing isoform of ADAR2, edits GluR-B RNA very efficiently at the Q/R site, whereas ADAR1 barely edits this site but edits an intronic +60 site preferentially (3, 5, 7, 18).
Although RNA editing has been shown to affect only selected gene transcripts, the posttranscriptional processing of most pre-mRNAs requires several common steps such as 5′-end capping, 3′-end processing, and splicing. These processes have been proposed to be carried out within large nuclear ribonucleoprotein (lnRNP) particles that sediment at 200S in sucrose gradients. The composition of the lnRNP particles (19–22) is very similar to that of the in vitro-assembled spliceosome (reviewed in refs. 23 and 24). Specifically, all of the U small nuclear ribonucleoproteins (snRNPs) required for pre-mRNA splicing, as well as several non-snRNP protein splicing factors, including U2AF and the SR protein family (23–25) are present within the lnRNP particles (19, 21, 22).
Three-dimensional image reconstruction of isolated lnRNP particles by automated electron tomography revealed a model that is composed of four major subunits of similar dimensions, which are connected to each other (26, 27). An additional domain is sometimes observed toward the center of the particle (26, 28). Each repeating subunit has a mass of 4.8 ± 0.5 mDa, close to the estimated mass of the fully in vitro-assembled 60S spliceosome, whereas the tetrameric lnRNP particle has a mass of 21.1 ± 1.6 mDa (28). The mass determination studies also revealed a discrepancy of 1.9 mDa between the lnRNP particle's mass (21.1 mDa) and the cumulative mass of its four subunits (4 × 4.8 mDa), which was consistent with the frequent presence of an additional domain (28). This additional domain may be attributed to additional RNA processing activities such as cap binding and 3′-end processing, as components involved in these processes have been detected also in lnRNP particles (O.R., J.S., and R.S., unpublished observations).
The A-to-I editing of certain substrates such as GluR-B and 5-HT2CR RNAs must occur before or simultaneously with splicing, because the dsRNA structure essential for the editing mechanism forms between the exonic editing site and the downstream intron sequences (12, 13, 15, 29, 30). Thus, it is anticipated that ADARs involved in the A-to-I RNA editing mechanism may be included in the lnRNP complexes that constitute the natural pre-mRNA processing machinery. Here we report that enzymatically active ADAR1 and ADAR2 proteins are indeed associated with spliceosomal Sm and SR proteins within the 200S lnRNP particles as physiologically significant supramolecular complexes.
Materials and Methods
Oligonucleotides.
The following oligonucleotides used for PCR were synthesized at the University of Pennsylvania Cancer Center Nucleic Acid Facility and, if necessary, purified in 20% acrylamide-7 M urea gels. All ADAR oligonucleotides correspond to the human sequence (4, 5). The restriction recognition site contained at the 5′ end is shown in bold: DR1PEP1, 5′-CGTGGATCCGGCCCCTCAAAAGCAGGG-3′; DR1PEP2, 5′-AGTCACGATGCGGCCGCTGGGGACCTTGAGAGGA-3′; DR1PEP3, 5′-CGTGGATCCGGCACTGTGGATGGGCCA-3′; DR1PEP4, 5′-AGTCACGATGCGGCCGCATACTATACTGGGCAG-3′; DR2PEP1, 5′-CGTGGATCCAAGCACGCGTTGTACTGT-3′; and DR2PEP2, 5′-AGTCACGATGCGGCCGCCCGGGTCAGGGCGTGA-3′.
Preparation of ADAR-Specific Antibodies.
Different regions of human ADAR1 (amino acids 440–826, dsRNA binding domain and amino acids 1,144–1,226, C terminal) and ADAR2a (amino acids 629–701, C-terminal) were constructed in pGEX-4T-3 vector (Amersham Pharmacia) as glutathione S-transferase (GST) fusion protein (31). The inserts of all constructs were prepared by PCR amplification of human ADAR1 (4) and ADAR2a cDNA plasmids (5) using a set of oligonucleotide primers designed to create NotI and XbaI restriction sites. DR1PEP1 and DR1PEP2 primers were used for PCR amplification of the ADAR1 dsRNA binding domain, whereas DR1PEP3 and DR1PEP4 primers were used for amplification of the ADAR1 C-terminal region. DR2PEP1 and DR2PEP2 were used for PCR amplification of the ADAR2a C-terminal region. The GST-ADAR fusion proteins were expressed in Escherichia coli BL21 and purified by glutathione Sepharose 4B column chromatography (31). mAbs were generated against the purified GST-ADAR fusion proteins. Selected mAbs 15.8.6 (ADAR1 dsRNA binding domain specific), 4G3B9 (ADAR1 C-terminal specific), and HBI-HYB7 (ADAR2a C-terminal specific) were screened for specific binding to ADAR1 and ADAR2a and confirmed not to cross-react with different members of ADAR or GST proteins (D.-S.C.C., S. Kang, T. Sanford, and K.N., unpublished results). Affinity-purified polyclonal antibody P00381 specific to the N-terminus region (amino acids 6–66) of ADAR2a was obtained from Bionostics (Ontario, Canada).
In Vitro RNA Editing Assay.
Editing of a synthetic GluR-B RNA B11 was assayed in vitro by using lnRNP fractions and control recombinant ADAR1 and ADAR2a proteins as described (5, 18). The standard editing reactions containing 20 fmol of B11 RNA, cell-equivalent samples of total lnRNP proteins (10 μg from 200S fractions or 20 μg from 70S fractions), or 10 ng of purified recombinant ADAR proteins were incubated at 30°C for 4 h.
ADAR Enzyme Activity Assay.
The dsRNA-specific A-to-I deamination enzyme activities of lnRNP fractions were assayed in vitro by using 10 fmol of [α-32P]ATP-labeled c-myc synthetic dsRNA as a substrate RNA as described (32).
Preparation of lnRNP Particles.
LnRNP particles were prepared from HeLa cells as described (19, 21). Briefly, nuclear supernatants enriched in lnRNP particles were prepared from clean cell nuclei and fractionated on 15–45% sucrose gradients in 100 mM NaCl, 10 mM Tris⋅HCl (pH 8.0), 2 mM magnesium chloride, and 2 mM vanadyl ribonucleoside. Centrifugations were carried out at 4°C in an SW41 rotor run at 41 krpm for 90 min (or an equivalent ω2t). When a second fractionation was required, 2–3 gradient fractions corresponding to the 200S region of the gradient were combined, dialyzed, concentrated, and refractionated on a second sucrose gradient (19). Peaks of 200S tobacco mosaic virus and 70S bacterial ribosomes, sedimented in parallel gradients, were used as sedimentation references.
Western and Slot Blot Analyses.
For slot blot analysis, aliquots from sucrose gradient fractions were spotted directly onto a nitrocellulose membrane. For Western blot analysis, aliquots were applied directly to the wells of a 10% or 8% polyacrylamide/SDS gel as described (33). Sm proteins were probed with anti-Sm polyclonal antibodies (34) diluted 1:100 and revealed with protein A horseradish peroxidase conjugate diluted 1:3,000 or with anti-Sm mAb Y12 (35). SR proteins were probed with mAb 104 (36) according to Zahler et al. (37). ADAR1 was probed with mAb 15.8.6 or 4G3B9 (see above) diluted 1:2,000 and visualized with horseradish peroxidase conjugated to affinity-pure goat anti-mouse IgG F(ab)′ fragment diluted 1:3,000. ADAR2a was probed with affinity-purified polyclonal Ab P00381 (Bionostics) or mAb HBI-HYB7 diluted 1:1,000 and probed with horseradish peroxidase-conjugated affinity-pure rabbit anti-IgG (H + L) diluted 1:5,000.
Immunoprecipitations.
Indirect immunoprecipitations were performed as described (22) by binding gradient fractions to anti-Sm mAb Y12 coupled to protein G Sepharose beads (Sigma). As controls, (i) gradient fractions were incubated with beads that had not been treated with mAb Y12, and (ii) beads carrying mAb Y12 were analyzed without prior incubation with gradient fractions. The immunoprecipitated proteins were released by treatment with SDS-containing gel sample buffer. Western blotting revealed ADAR proteins as described. The relative quantities of ADAR proteins in the pellet and supernatant fractions were determined by densitometric scanning (model SL-TRFF, Biomed Instruments, Fullerton, CA).
Results
Identification of Different ADARs in lnRNP Particles by Immunochemistry.
We previously have shown that when HeLa cell nuclear supernatants were fractionated in sucrose gradients and probed for a specific pre-mRNA (e.g., β-actin), a relatively broad but distinct peak of the specific RNA was observed at the 200S region of the gradient (38). All of the nucleoplasmic phosphorylated SR proteins also sedimented as one peak at 200S and were associated exclusively with 200S lnRNP particles (22). Other components of the lnRNP particles, such as the U snRNPs, U2AF, and PTB (19, 22) gave a broader distribution across the sucrose gradients and were particularly abundant in smaller complexes that sedimented closer to the top of the gradient. For the latter splicing factors, however, a specific association with 200S lnRNP particles was unambiguously demonstrated by refractionation of the combined 200S peak fractions on a second sucrose gradient, which gave rise to a single peak at the 200S region of the gradient (19, 22). Such experiments indicated the existence of at least two populations of U snRNPs, U2AF, and PTB. One sediments at 200S in association with lnRNP particles. The other, most probably, represents smaller particles or free species that are present in excess of what is required for lnRNP particles assembly or resulted from dissociation of lnRNP particles during preparation.
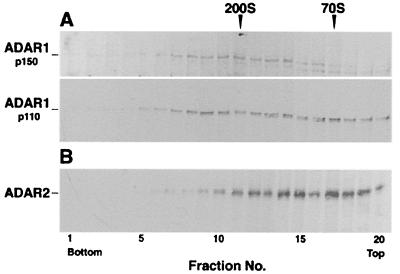
As a first step in analyzing the association of ADARs with lnRNP particles, we examined, by Western blotting, the distribution of different ADAR gene family members in fractionated lnRNP particles. HeLa cell nuclear supernatants enriched in lnRNP particles were fractionated on sucrose gradients, and the distribution of ADAR proteins was probed with Abs recognizing two different regions of ADAR1 (4, 8, 9) or ADAR2 (3, 5, 6). The full-length 150-kDa form of ADAR1 was found in a broad peak around the 200S region of the gradient (Fig. 1A Upper), where specific nuclear pre-mRNAs have been shown to sediment (19, 38–40). The more abundant shorter (110 kDa) form of ADAR1 had a broader distribution, with significant level sedimenting at the 200S and 70S regions of the gradient (Fig. 1A Lower). The 110-kDa ADAR1, another enzymatically active form of the protein, is most likely the translation product initiated from the downstream methionine codon (9). The 90-kDa form of ADAR2 protein also showed a broad distribution, with significant levels sedimenting as large complexes at the 200S and 70S regions of the sucrose gradient (Fig. 1B). The bands detected at the top fractions of the gradient are likely due to ADAR proteins disassembled from lnRNP particles during cell disruption and gradient sedimentation, or due to free nuclear ADAR proteins (also see Discussion). This distribution is reminiscent of that of U snRNPs, U2AF, and PTB as detailed above. Thus, three fractions corresponding to the 200S region of the gradient (Fig. 1, fractions 10, 11, and 12) were combined and refractionated in a second sucrose gradient. Probing the gradient fractions with the respective antibodies revealed a single peak of ADAR1 and ADAR2 proteins that comigrated with the 200S lnRNP particles represented by the Sm and SR proteins (Fig. 2A). These slot blot results were further confirmed by Western blot analyses of ADAR1 and Sm proteins (Fig. 2B). The distinct peak at 200S and the low abundance of ADAR proteins in the top and bottom fractions of the second gradient suggest their specific association with 200S particles.
Figure 1.
The distribution of ADAR proteins in a sucrose gradient. Nuclear supernatants of HeLa cells, enriched in lnRNP particles, were fractionated in a 15–45% sucrose gradient and collected (bottom to top) in 20 fractions. Aliquots from each fraction were analyzed by Western blotting. (A) Probing with anti-ADAR1 mAb 15.8.6. (Upper) The distribution of the 150-kDa form of ADAR1 protein (20-sec exposure). (Lower) The distribution of the 110-kDa form of ADAR1 protein (5-sec exposure). (B) Probing with anti-ADAR2 Abs P00381. The sedimentation position of the 200S TMV and 70S bacterial ribosomes size markers are indicated on the top. Molecular masses of the ADAR1 species are indicated on the left.
Figure 2.
ADAR proteins cosediment with spliceosomal Sm and SR proteins at the 200S region in a sucrose gradient. HeLa cell nuclear supernatant, enriched in lnRNP particles, was fractionated in a 15–45% sucrose gradient. The 200S peak fractions were combined and refractionated in a second sucrose gradient. Aliquots from each fraction were analyzed by slot blotting (A) or Western blotting (B). The distributions of SR proteins, Sm antigens, and ADAR1 and ADAR2 proteins were revealed by their cognate antibodies. Shown are the results of two independent experiments. Similar results were obtained in five different experiments. The sedimentation position of the 200S TMV and 70S bacterial ribosomes size markers are indicated.
ADAR Proteins Are Integral Components of lnRNP Particles.
The association of the ADAR proteins with 200S lnRNP particles was confirmed by indirect immunoprecipitation experiments. The efficacy of this approach already has been demonstrated by immunoprecipitating lnRNP particles using antibodies directed against the Sm antigens or the SR proteins (22, 34). Here, we immunoprecipitated sucrose gradient-fractionated 200S lnRNP particles with Y12 anti-Sm mAbs (35) and probed both the immunoprecipitate (Fig. 3, lane 2) and supernatant (Fig. 3, lane 1) with anti-ADAR1 mAbs. In parallel, fractions from the 70S and top regions of the gradient were analyzed in the same manner (Fig. 3, lanes 5 and 7). The absence of adventitious adhesion of complexes to protein G Sepharose was confirmed by analyses of beads that were incubated with gradient fractions but were not treated with the antibody, which showed no signal of ADAR1 (Fig. 3, lane 3 for the 200S fraction; identical results were obtained for the 70S and top fractions). In a second control, beads were incubated only with the antibody, but not with the antigen, to ensure that no bands resulting from the antibody comigrated with ADAR1 proteins (Fig. 3, lane 8). The relative amount of ADAR1 that was indirectly immunoprecipitated from each of the 200S, 70S, and top fractions of the gradient provides confirmation of the association of the ADAR1 proteins with RNP particles. We found that 40% and 15% of the ADAR1 content of the 200S and 70S fractions, respectively, were precipitated by the anti-Sm mAbs (Fig. 3, lanes 2 and 5), whereas only a small amount (<5%) of the protein was precipitated from the top fraction (Fig. 3, lane 7), where free ADAR1 presumably sediments. Similar results were obtained by indirect immunoprecipitation of lnRNP particles by anti-SR mAbs and probing with anti-ADAR1 or anti-ADAR2 mAbs (data not shown). These results demonstrate specific association of ADAR1 within lnRNP particles where the pre-mRNA, spliceosomal U snRNPs, SR proteins, and other splicing factors are assembled together.
Figure 3.
ADAR proteins are associated with spliceosomal proteins in 200S lnRNP particles. HeLa cell nuclear supernatant, enriched in lnRNP particles, was fractionated in a 15–45% sucrose gradient. Aliquots from the 200S, 70S, and top fractions were immunoprecipitated by anti-Sm Y12 mAbs bound to protein G Sepharose beads. The precipitated proteins (lanes 2, 5, and 7) as well as the proteins left in the supernatant (lanes 1, 4, and 6) were analyzed by Western blotting with anti-ADAR1 mAb 15.8.6. Controls: Lane 3, analysis of beads incubated with the 200S fraction without Y12 mAb. Lane 8, analysis of beads incubated with Y12 mAb alone (no antigen added). Similar results were obtained in five different experiments.
Reconstitution of ADAR Proteins into lnRNP Particles.
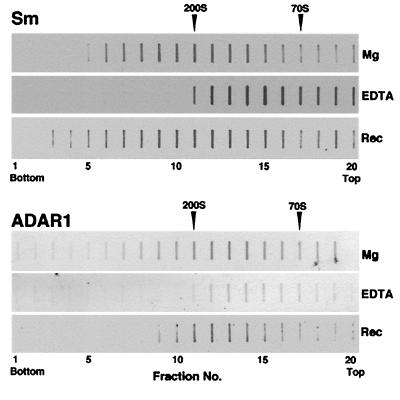
We previously have shown that the stability of the lnRNP complex depends on Mg2+ concentration. The complex dissociates into 70S particles when Mg2+ is removed by chelating agents. Under these conditions the family of essential splicing factors, the SR proteins in their phosphorylated form, stay associated with the pre-mRNA and sediment at 70S. Some of the U snRNPs, such as U1, almost completely dissociate and migrate at the top of the gradient, whereas other U snRNPs, such as U2 and U5, only partially dissociate and partially stay associated with the pre-mRNA in 70S particles (19). The dissociated complex can be reconstituted by readdition of Mg2+ into a complex that is indistinguishable from the original one. The reconstituted complex thus regains the original biochemical composition, and morphology as seen in the electron microscope (19). We thus examined whether ADAR1 can be dissociated from and then reconstituted back into 200S lnRNP particles. Gradient fractions containing 200S lnRNP particles (analogous to fractions 10–12 of Fig. 1) were combined and concentrated, and one-third was dissociated by addition of EDTA and reconstituted by readdition of Mg2+. Another third was just dissociated by addition of EDTA. The remaining untreated third and the two treated thirds were fractionated in parallel sucrose gradients and analyzed for the distribution of spliceosomal Sm proteins (Fig. 4, Sm) and ADAR1 (Fig. 4, ADAR1) by slot blotting. Addition of EDTA caused the migration of ADAR1 proteins as 70S complexes, as was previously shown for the pre-mRNA, the SR proteins and part of the U snRNPs (19). ADAR1 proteins, as well as the Sm antigens, were reconstituted into lnRNP particles after addition of Mg2+ to the dissociated particles, indicating once again that the association of ADARs with lnRNP particles is specific and is physiologically significant.
Figure 4.
Reconstitution of ADAR proteins into 200S lnRNP particles. EDTA-dissociated spliceosomal Sm proteins (Sm) and ADAR1 protein (ADAR1) were reconstituted into 200S lnRNP particles after readdition of Mg2+. Nuclear supernatant of HeLa cells was sedimented in a standard Mg2+-containing sucrose gradient and the 200S peak fractions were combined. One-third was refractionated in the presence of Mg2+ (Top); another third was incubated with EDTA and refractionated in the presence of EDTA (Middle); and the remaining third was incubated with EDTA, then with Mg2+ and fractionated in the presence of Mg2+ (Bottom). Sm and ADAR1 proteins were analyzed, in aliquots taken from the same gradients, by slot immunoblotting. The sedimentation positions of the 200S TMV and 70S bacterial ribosomes size markers are indicated.
Detection of the ADAR Enzymatic Activity in lnRNP Particles.
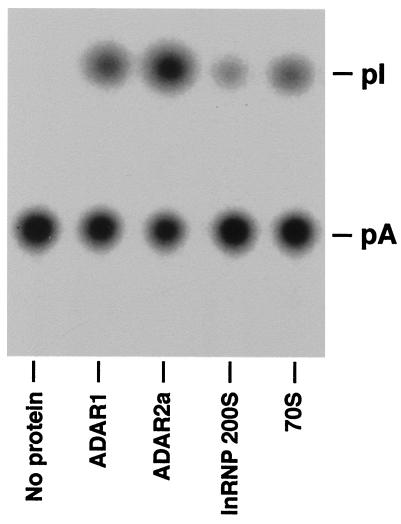
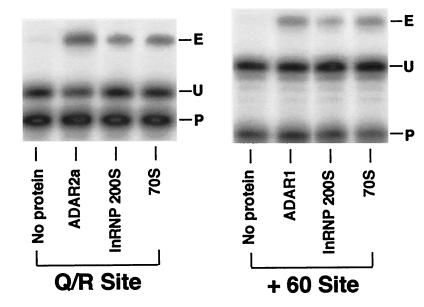
The A-to-I conversion activity of ADARs present in lnRNP particles was examined by using a previously described base modification assay with a synthetic long c-myc dsRNA substrate (32). As positive controls, we analyzed purified recombinant ADAR1 and ADAR2a proteins simultaneously. The results clearly indicated that both 200S and 70S fractions of lnRNP preparation contain enzymatically active ADARs (Fig. 5). The A-to-I base modification assay is useful to identify the activity of ADAR proteins, but it cannot distinguish the activity of different ADAR gene family members. The different ADARs display very distinctive site selectivity for editing of GluR-B or 5-HT2CR RNA at certain sites when they are tested in an in vitro editing assay. Therefore, we tested both 200S and 70S lnRNP fractions by using as substrate GluR-B B11 RNA harboring the Q/R site and the intronic hot spot +60 site (18). ADAR2a and ADAR2b, enzymatically active splicing isoforms of ADAR2, selectively edit the Q/R site, whereas ADAR1 preferentially selects the +60 site (3, 5, 6, 18). Fig. 6 shows that both 200S and 70S fractions contained the enzymatic activities that edit Q/R and +60 sites, confirming the presence of both ADAR1 and ADAR2 activities in these lnRNP particles as indicated by our Western blotting and immunoprecipitation experiments. We conclude that the RNA editing active ADAR1 and ADAR2 are complexed with lnRNP particles.
Figure 5.
Detection of the A-to-I base conversion activity in 200S lnRNP particles and 70S fractions made from HeLa cells. The A-to-I conversion activity of 200S lnRNP particles (10 μg) and 70S fractions (20 μg) was determined by a base modification assay using 10 fmol of [32P]ATP-labeled c-myc dsRNA as a substrate (32). For comparison, purified recombinant ADAR1 and ADAR2a proteins (10 ng each) were tested in the same assay.
Figure 6.
RNA editing site selectivity of lnRNP fractions. Editing site selectivity of 200S lnRNP particles (10 μg) and 70S fractions (20 μg) was examined in vitro at the Q/R and intronic +60 sites of GluR-B RNA. For comparison, purified recombinant ADAR1 and ADAR2a proteins (10 ng) were tested simultaneously. The RNAs were examined by primer extension analysis using 32P-labeled oligonucleotide primers specific for each editing site. The extension primer used (P) and extended bands specific for edited (E) and unedited (U) RNA are indicated.
Discussion
Complex Formation of ADAR Enzymes with lnRNP Particles.
Analysis of specific as well as general polyadenylated populations of nuclear pre-mRNAs by sucrose gradient fractionation revealed a single peak at the 200S region of the gradient. These nuclear pre-mRNAs are packaged in 200S lnRNP particles in a supraspliceosome configuration (20, 26, 28, 38). All of the nucleoplasmic phosphorylated SR proteins, essential for spliceosome assembly and splicing, are found associated exclusively with 200S lnRNP particles (22). These previous studies have established the 200S lnRNP particles as the naturally assembled pre-mRNA splicing machinery. Formation of the dsRNA structure essential for A-to-I editing of GluR and 5-HT2CR RNAs requires base pairing of exon and intron sequences and therefore is expected to occur before or concomitant with splicing. The association of ADAR1 with the RNP matrix on transcriptionally active Xenopus lampbrush chromosomes and its cotranscriptional function has been suggested previously (41). In this study, we have demonstrated directly that enzymatically active ADAR proteins involved in the A-to-I RNA editing mechanism are associated with 200S lnRNP particles.
In addition to their association with the 200S particles, ADAR proteins and activity appear to associate with particles sedimenting in the 70S fractions of the sucrose gradient. The biological significance of the association of ADAR proteins with the 70S fractions remains to be clarified due to our limited understanding of the relationship of 70S and 200S particles. The 70S fractions most likely represent a population of dissociated particles similar to the complexes obtained upon treatment of 200S lnRNP particles with EDTA. These particles have been shown to contain intact pre-mRNA associated with only part of the components of the 200S lnRNP particles (19). Alternatively, they may represent degradation products of the 200S lnRNP particles in which the pre-mRNA is partially degraded. Finally, the 70S fractions could represent intermediate complexes in the assembly of the 200S lnRNP particles. Further studies are required to elucidate whether the 70S fractions and their association with ADARs represent a physiologically significant intermediate in the assembly pathway of the lnRNP particles. The ADAR proteins detected at the top of the gradient may indicate that they are usually present in excess of what is required for pre-mRNA processing complexes. This is similar to what was found for several spliceosomal components (e.g., U snRNPs, and the splicing protein factors Sm, U2AF, and PTB), which are present in the nucleus in excess of what is required for spliceosome assembly and splicing (ref. 22 and references cited therein). Alternatively, the ADAR proteins recovered at the top of the gradient may represent the proteins dissociated from lnRNP particles during sample preparation.
Presence of Both Editing and Splicing Machineries in lnRNP Particles.
ADARs containing both substrate binding and catalytic domains appear to be self-sufficient for executing the site selective A-to-I RNA editing in vitro (5, 6, 18). However, the presence of regulatory factors as well as inhibitors in vivo also has been suggested (18, 42). It is possible that some of these putative regulators and inhibitors of the A-to-I RNA editing may be known components of lnRNP particles. The A-to-I RNA editing mechanism requires a critical dsRNA structure formed around editing site(s) (2). ADAR identifies and binds to the dsRNA structure through its dsRNA binding domain consisting of two or three dsRNA binding motifs (4, 43). Interestingly, the dsRNA structure proven to be essential for editing of certain substrate RNAs such as GluR and 5-HT2CR RNAs (12, 13, 29, 30), is formed between exon sequences around the editing site(s) and downstream intron sequences. It contains the critical dinucleotide GU, which is part of the 5′ splice site consensus sequence AG/GURAGU predominantly found in most mammalian pre-mRNAs (24, 44, 45). Thus, the 5′ splice site sequence serves as a component of the dsRNA structure required for editing of these RNAs as well as an essential element of the splicing machinery. Binding of U1 snRNA to the 5′ splice site is required for spliceosome assembly, and the same consensus sequence is subsequently base-paired with U6 snRNA during the activation of the spliceosome (46, 47). The formation of the dsRNA structure essential for A-to-I RNA editing and spliceosome assembly may, therefore, interfere with each other. If both splicing and RNA editing machineries are kept together, the dsRNA structure critical for A-to-I RNA editing is likely to be constantly involved in two functional interactions. One is binding of U1 snRNA to the 5′ splice site, including the GU nucleotides, followed by initiation of spliceosome assembly; and the other is binding of ADAR followed by initiation of A-to-I RNA editing.
It has been recently reported that resolution of the dsRNA structure involved in A-to-I RNA editing may be regulated by a specific ATP-dependent dsRNA helicase (48). The mutation in the helicase and thus delayed resolution of the dsRNA structure result in the occurrence of a “splicing catastrophe” of the Drosophila para Na+ channel transcripts at the region surrounding the editing sites (48). In addition, cryptic and alternative splicing of 5-HT2CR RNA have been detected at the region surrounding the editing sites (15). These studies indicate an active interaction of the dsRNA structure, as well as the dsRNA helicase activity, with the A-to-I RNA editing and splicing machinery. Therefore, our direct demonstration that both ADAR1 and ADAR2 are present within 200S lnRNP complexes is significant as it confirms spatial proximity of two different posttranscriptional processes (spliceosome assembly and RNA editing) within the naturally assembled pre-mRNA processing machinery.
Acknowledgments
We thank J. A. Steitz and I. Mattaj for anti-Sm Y12 mAbs, T. Sanford, S. Kang, and A. Pecho for excellent technical assistance, and J. M. Murray for critical reading of this manuscript. We also thank the Wistar editorial services department for preparing the manuscript. This work was supported by grants from the National Institutes of Health (GM40536, CA72765, and AG10124) and the Doris Duke Charitable Foundation (to K.N.), the Israel-U.S. Binational Science Foundation (to R.S., J. S. and K. N.), and the Joseph and Ceil Mazer Center for Structural Biology at the Weizmann Institute of Science (to J.S.). D.-S.C.C. was supported by a training grant from the National Cancer Institute (CA09171).
Abbreviations
- ADAR
adenosine deaminases acting on RNA
- lnRNP
large nuclear ribonucleoprotein
- dsRNA
double-stranded RNA
- GluR
glutamate receptor
- 5-HT2CR
serotonin receptor 2C subtype
- snRNP
small nuclear ribonucleoprotein
References
- 1.Bass B L, Nishikura K, Keller W, Seeburg P H, Emeson R B, O'Connell M A, Sameul C E, Herbert A. RNA. 1997;3:947–949. [PMC free article] [PubMed] [Google Scholar]
- 2.Smith H C, Gott J M, Hanson M R. RNA. 1997;3:1105–1123. [PMC free article] [PubMed] [Google Scholar]
- 3.Gerber A, O'Connell M A, Keller W. RNA. 1997;3:453–463. [PMC free article] [PubMed] [Google Scholar]
- 4.Kim U, Wang Y, Sanford T, Zeng Y, Nishikura K. Proc Natl Acad Sci USA. 1994;91:11457–11461. doi: 10.1073/pnas.91.24.11457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lai F, Chen C-X, Carter K C, Nishikura K. Mol Cell Biol. 1997;17:2413–2424. doi: 10.1128/mcb.17.5.2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Melcher T, Maas S, Herb A, Sprengel R, Seeburg P H, Higuchi M. Nature (London) 1996;379:460–464. doi: 10.1038/379460a0. [DOI] [PubMed] [Google Scholar]
- 7.Melcher T, Maas S, Herb A, Sprengel R, Higuchi M, Seeburg P H. J Biol Chem. 1996;271:31795–31798. doi: 10.1074/jbc.271.50.31795. [DOI] [PubMed] [Google Scholar]
- 8.O'Connell M A, Krause S, Higuchi M, Hsuan J J, Totty N, Jenny A, Keller W. Mol Cell Biol. 1995;15:1389–1397. doi: 10.1128/mcb.15.3.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Patterson J B, Samuel C E. Mol Cell Biol. 1995;15:5376–5388. doi: 10.1128/mcb.15.10.5376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen C-X, Cho D-S C, Wang Q, Lai F, Carter K C, Nishikura K. RNA. 2000;6:755–767. doi: 10.1017/s1355838200000170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sommer B, Köhler M, Sprengel R, Seeburg P H. Cell. 1991;67:11–19. doi: 10.1016/0092-8674(91)90568-j. [DOI] [PubMed] [Google Scholar]
- 12.Lomeli H, Mosbacher H, Melcher T, Höger T, Geiger J R P, Kuner T, Monyer H, Higuchi M, Bach A, Seeburg P H. Science. 1994;226:1709–1713. doi: 10.1126/science.7992055. [DOI] [PubMed] [Google Scholar]
- 13.Burns C M, Chu H, Rueter S M, Hutchinson L K, Canton H, Sanders-Bush E, Emeson R B. Nature (London) 1997;387:303–308. doi: 10.1038/387303a0. [DOI] [PubMed] [Google Scholar]
- 14.Niswender C M, Copeland S C, Herrick-Davis K, Emeson R B, Sanders-Bush E. J Biol Chem. 1999;274:9472–9478. doi: 10.1074/jbc.274.14.9472. [DOI] [PubMed] [Google Scholar]
- 15.Wang Q, O'Brien P J, Chen C-X, Cho D-S C, Murray J M, Nishikura K. J Neurochem. 2000;74:1290–1300. doi: 10.1046/j.1471-4159.2000.741290.x. [DOI] [PubMed] [Google Scholar]
- 16.Paul M S, Bass B L. EMBO J. 1998;17:1120–1127. doi: 10.1093/emboj/17.4.1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Q, Khillan J, Gadue P, Nishikura K. Science. 2000;290:1765–1768. doi: 10.1126/science.290.5497.1765. [DOI] [PubMed] [Google Scholar]
- 18.Dabiri G A, Lai F, Drakas R A, Nishikura K. EMBO J. 1996;15:34–45. [PMC free article] [PubMed] [Google Scholar]
- 19.Miriami E, Angenitzki M, Sperling R, Sperling J. J Mol Biol. 1995;246:254–263. doi: 10.1006/jmbi.1994.0081. [DOI] [PubMed] [Google Scholar]
- 20.Sperling R, Sperling J. In: The Eukaryotic Nucleus, Molecular Biochemistry and Macromolecular Assemblies. Strauss P R, Wilson S H, editors. Vol. 2. Caldwell, NJ: Telford; 1990. pp. 453–476. [Google Scholar]
- 21.Sperling R, Sperling J. In: RNP Particles, Splicing, and Autoimmune Diseases. Schenkel J, editor. Berlin: Springer; 1998. pp. 29–47. [Google Scholar]
- 22.Yitzhaki S, Miriami E, Sperling J, Sperling R. Proc Natl Acad Sci USA. 1996;93:8830–8835. doi: 10.1073/pnas.93.17.8830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krämer A. Annu Rev Biochem. 1996;65:367–409. doi: 10.1146/annurev.bi.65.070196.002055. [DOI] [PubMed] [Google Scholar]
- 24.Moore J M, Query C C, Sharp P A. In: The RNA World. Gesteland R F, Atkins J F, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1993. pp. 303–358. [Google Scholar]
- 25.Will C L, Lührmann R. Curr Opin Cell Biol. 1997;9:320–328. doi: 10.1016/s0955-0674(97)80003-8. [DOI] [PubMed] [Google Scholar]
- 26.Sperling R, Koster A J, Melamed-Bessudo C, Rubinstein A, Angenitzki M, Berkovitch-Yellin Z, Sperling J. J Mol Biol. 1997;267:570–583. doi: 10.1006/jmbi.1997.0898. [DOI] [PubMed] [Google Scholar]
- 27.Medalia O, Koster A J, Tocilj A, Angenitzki M, Sperling J, Berkovitch Y Z, Sperling R. J Struct Biol. 1997;120:228–236. doi: 10.1006/jsbi.1997.3926. [DOI] [PubMed] [Google Scholar]
- 28.Müller S, Wolpensinger B, Angenitzki M, Engel A, Sperling J, Sperling R. J Mol Biol. 1998;283:383–394. doi: 10.1006/jmbi.1998.2078. [DOI] [PubMed] [Google Scholar]
- 29.Herb A, Higuchi M, Sprengel R, Seeburg P H. Proc Natl Acad Sci USA. 1996;93:1875–1880. doi: 10.1073/pnas.93.5.1875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Higuchi M, Single F N, Köhler M, Sommer B, Sprengel R, Seeburg P H. Cell. 1993;75:1361–1370. doi: 10.1016/0092-8674(93)90622-w. [DOI] [PubMed] [Google Scholar]
- 31.Guan K L, Dixon J E. Anal Biochem. 1991;192:262–267. doi: 10.1016/0003-2697(91)90534-z. [DOI] [PubMed] [Google Scholar]
- 32.Kim U, Garner T L, Sanford T, Speicher D, Murray J M, Nishikura K. J Biol Chem. 1994;269:13480–13489. [PubMed] [Google Scholar]
- 33.Yitzhaki S, Sperling J. BioTechniques. 1998;24:762–766. doi: 10.2144/98245bm15. [DOI] [PubMed] [Google Scholar]
- 34.Sperling R, Spann P, Offen D, Sperling J. Proc Natl Acad Sci USA. 1986;83:6721–6725. doi: 10.1073/pnas.83.18.6721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lerner E A, Lerner M R, Janeway L A, Steitz J A. Proc Natl Acad Sci USA. 1981;78:2737–2741. doi: 10.1073/pnas.78.5.2737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Roth M B, Murphy C, Gall J G. J Cell Biol. 1990;111:2217–2223. doi: 10.1083/jcb.111.6.2217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zahler A M, Lane W S, Stolk J A, Roth M B. Genes Dev. 1992;6:837–847. doi: 10.1101/gad.6.5.837. [DOI] [PubMed] [Google Scholar]
- 38.Spann P, Feinerman M, Sperling J, Sperling R. Proc Natl Acad Sci USA. 1989;86:466–470. doi: 10.1073/pnas.86.2.466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sperling R, Sperling J, Levine A D, Spann P, Stark G R, Kornberg R D. Mol Cell Biol. 1985;5:569–575. doi: 10.1128/mcb.5.3.569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Miriami E, Sperling J, Sperling R. Nucleic Acids Res. 1994;22:3084–3091. doi: 10.1093/nar/22.15.3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Eckmann C R, Jantsch M F. J Cell Biol. 1999;144:603–615. doi: 10.1083/jcb.144.4.603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lai F, Chen C-X, Lee V M, Nishikura K. J Neurochem. 1997;69:43–52. doi: 10.1046/j.1471-4159.1997.69010043.x. [DOI] [PubMed] [Google Scholar]
- 43.Lai F, Drakas R, Nishikura K. J Biol Chem. 1995;270:17098–17105. doi: 10.1074/jbc.270.29.17098. [DOI] [PubMed] [Google Scholar]
- 44.Nilsen T W. In: RNA Structure and Function. Simons R W, Grunberg-Manago M, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1998. pp. 279–307. [Google Scholar]
- 45.Burge C B, Tuschl T H, Sharp P A. In: The RNA World. 2nd Ed. Gesteland R F, Cech T R, Atkins J F, editors. Plainview, NY: Cold Spring Harbor Lab. Press; 1999. pp. 525–560. [Google Scholar]
- 46.Madhani H D, Guthrie C. Annu Rev Genet. 1994;28:1–26. doi: 10.1146/annurev.ge.28.120194.000245. [DOI] [PubMed] [Google Scholar]
- 47.Murray H L, Jarrell K A. Cell. 1999;96:599–602. doi: 10.1016/s0092-8674(00)80568-1. [DOI] [PubMed] [Google Scholar]
- 48.Reenan R A, Hanrahan C J, Barry G. Neuron. 2000;25:139–149. doi: 10.1016/s0896-6273(00)80878-8. [DOI] [PubMed] [Google Scholar]