Abstract
Common genetic variants have been recently associated with fasting glucose and insulin levels in white populations. Whether these associations replicate in pre-diabetes is not known. We extended these findings to the Diabetes Prevention Program, a clinical trial in which participants at high risk for diabetes were randomized to placebo, lifestyle modification or metformin for diabetes prevention. We genotyped previously reported polymorphisms (or their proxies) in/near G6PC2, MTNR1B, GCK, DGKB, GCKR, ADCY5, MADD, CRY2, ADRA2A, FADS1, PROX1, SLC2A2, GLIS3, C2CD4B, IGF1, and IRS1 in 3,548 Diabetes Prevention Program participants. We analyzed variants for association with baseline glycemic traits, incident diabetes and their interaction with response to metformin or lifestyle intervention. We replicated associations with fasting glucose at MTNR1B (P<0.001), G6PC2 (P = 0.002) and GCKR (P = 0.001). We noted impaired β-cell function in carriers of glucose-raising alleles at MTNR1B (P<0.001), and an increase in the insulinogenic index for the glucose-raising allele at G6PC2 (P<0.001). The association of MTNR1B with fasting glucose and impaired β-cell function persisted at 1 year despite adjustment for the baseline trait, indicating a sustained deleterious effect at this locus. We also replicated the association of MADD with fasting proinsulin levels (P<0.001). We detected no significant impact of these variants on diabetes incidence or interaction with preventive interventions. The association of several polymorphisms with quantitative glycemic traits is replicated in a cohort of high-risk persons. These variants do not have a detectable impact on diabetes incidence or response to metformin or lifestyle modification in the Diabetes Prevention Program.
Introduction
Glucose homeostasis is tightly regulated. Control of its variation in non-diabetic individuals is influenced by familial factors, many of which are presumed to be heritable [1], [2]. In searching for genetic determinants of quantitative glycemic traits, candidate gene and genome-wide association studies (GWAS) conducted in populations of European descent have identified associations of fasting glucose with genetic variants in or near the genes that encode glucokinase (GCK; [3]), the glucose-6-phosphatase catalytic subunit (G6PC2; [4], [5]) and the melatonin receptor 1b (MTNR1B; [6], [7]). The Meta-Analysis of Glucose and Insulin-related traits Consortium (MAGIC) recently performed a global meta-analysis of 21 GWAS cohorts followed by replication in 26 studies, totaling >122,000 non-diabetic individuals for fasting glucose and >98,000 non-diabetic individuals for fasting insulin [8]. These efforts confirmed the GCK, G6PC2 and MTNR1B associations, and uncovered associations of fasting glucose with single nucleotide polymorphisms (SNPs) in or near DGKB, GCKR, ADCY5, MADD, CRY2, ADRA2A, FADS1, PROX1, SLC2A2, GLIS3, C2CD4B and the type 2 diabetes genes TCF7L2 and SLC30A8. In addition, SNPs in or near IGF1, GCKR and perhaps IRS1 have been found to influence fasting insulin concentrations, a surrogate for insulin resistance. Of these loci, only GCK, MTNR1B, DGKB, GCKR, ADCY5 and PROX1 (besides TCF7L2 and SLC30A8) were associated with type 2 diabetes at genome-wide significance levels, with several others (but not all) showing a consistent trend but not meeting the same stringent statistical threshold. This work has illustrated that genetic associations with quantitative intermediate traits may lead to the discovery of type 2 diabetes loci, but also that not all genetic loci that influence fasting glucose levels in healthy individuals necessarily contribute to type 2 diabetes pathogenesis.
The MAGIC investigators have also performed more detailed characterization of the mechanisms of glucose regulation influenced by these loci in white individuals [9]. In the Third National Health and Nutrition Examination Survey (NHANES III), a genetic risk score constructed with the glucose-raising alleles was shown to have consistent effects in other ethnic groups representative of the US population [10]. The Gene × Lifestyle interactions And Complex traits Involved in Elevated disease Risk (GLACIER) investigators showed that several of these loci associate with impaired fasting glucose (IFG) cross-sectionally and prospectively, and some have a progressively deleterious effect on fasting glucose [11]. Shortly thereafter, the Whitehall II investigators reported that a genetic risk score constructed with these variants was strongly associated with fasting glucose and remained stable over time [12]. Finally, we have recently shown that different genetic variants influence type 2 diabetes risk at distinct stages of the normoglycemia to IFG to type 2 diabetes progression, with MTNR1B and GCK exerting their effects preferentially in the normoglycemia to IFG transition [13].
To understand why some loci raise fasting glucose but do not increase type 2 diabetes risk, it is critical to establish whether their glucose-raising effects remain evident in the setting of impaired glucose tolerance (IGT), as glycemic context may modulate the strength of the genetic effect [13]. Furthermore, the impact of these loci on the prospective development of diabetes has not yet been reported. Finally, establishing whether and how distinct preventive interventions modulate these effects may facilitate the clinical translation of these findings and illuminate the specific genes and mechanisms by which these loci affect glycemic homeostasis. We concentrated on SNPs associated with fasting glucose, rather than those associated with 2-hour glucose [14], because 1) the two 2-hour glucose SNPs that are not already captured by fasting glucose-associated variants (GIPR and VPS13C) have no detectable impact on type 2 diabetes [15], 2) the ascertainment of DPP participants by the strict IGT definition is likely to bias the distribution of 2-hour glucose alleles, 3) longitudinal changes in 2-hour glucose among carriers of the 2-hour glucose-raising alleles have already been reported in a better suited population cohort [16], and 4) evidence obtained by the MAGIC investigators argues against an interaction of known 2-hour glucose loci with physical activity or body mass index (BMI) (Robert Scott, personal communication). We therefore genotyped the fasting glucose-associated SNPs in the multi-ethnic cohort of the Diabetes Prevention Program (DPP), and analyzed their relationships with glycemic measures at baseline and one year, the development of diabetes, and their potential interaction with preventive interventions on diabetes incidence.
Methods
The Diabetes Prevention Program
The DPP study design and baseline characteristics of the participants have been described previously [17], [18]. Briefly, the DPP was designed to test whether intensive lifestyle modification or pharmacologic interventions with metformin or troglitazone prevent or delay the onset of diabetes in individuals at high risk. The trial, conducted from 1996 to 2001 in 27 US-based medical centers, included 3,234 participants randomized to intensive lifestyle modification (goal >7% weight loss and >150 min/week of physical activity), metformin (850 mg twice daily), or placebo; the fourth arm, comprising 585 additional participants randomized to troglitazone, was terminated early because of concerns with hepatotoxicity. For enrollment, participants had to have a fasting glucose between 95–125 mg/dL and IGT (2h-glucose between 140–199 mg/dL after a 75-gram oral glucose tolerance test [OGTT]). Of the total 3,819 DPP participants, 3,548 had DNA and consented to genetic investigation: 56.4% were of European descent, 20.2% African American, 16.8% Hispanic, 4.3% Asian and 2.4% American Indian by self-report. Their mean age was 51 years and mean BMI was 34.0 kg/m2. The primary endpoint (diabetes incidence, ascertained biannually and confirmed on a second occasion) was reached in nearly 38% of participants randomized to the placebo arm after a mean of 3.2 years of follow-up; there was a 58% reduction of diabetes incidence in the lifestyle intervention group and a 31% reduction in the metformin group compared to placebo [19]. For the purposes of this study, participants randomized to troglitazone were excluded, leaving a total of 2,890 individuals with valid genotypes for analysis. Institutional Review Board approval was obtained by each participating site, and all participants included in this report provided written informed consent for the main study and for subsequent genetic investigations.
Quantitative Glycemic Traits
We calculated the insulin sensitivity index (ISI) as 22.5/[(fasting insulin × fasting glucose)/18.01]; the ISI is the reciprocal of insulin resistance calculated by homeostasis model assessment (HOMA-IR) [20]. We estimated insulin secretion by the insulinogenic index using the formula [(insulin at 30 min)-(insulin at 0 min)]/[(glucose at 30 min)-(glucose at 0 min)]. The oral disposition index (DIo) was calculated as 1/fasting insulin × insulinogenic index [21]. We studied genetic associations with these measures at baseline and at 1 year: we chose one year because changes in weight were most pronounced at that time point, and it contained the highest number of participants with available measures.
SNP Selection and Genotyping
We genotyped the index SNPs associated with fasting glycemic traits reported by the MAGIC investigators [8]. Where assay design failed we selected proxies based on linkage disequilibrium in the HapMap CEU population: rs573225 for rs560887 in G6PC2, r2 = 0.961; rs917793 for rs4607517 in GCK, r2 = 1.0; and rs855228 for rs35767 in IGF1, r2 = 0.915. DNA was extracted from peripheral blood leukocytes and quantitated as previously described [22]. Genotyping was carried out by allele-specific primer extension of multiplex amplified products and detection using matrix-assisted laser desorption ionization time-of-flight mass spectrometry on a Sequenom iPLEX platform [23]. Genotyping success rate was ≥98.5%. Because results for the two previously known type 2 diabetes genes TCF7L2 and SLC30A8 have been reported elsewhere [22], [24], [25], they are not presented here.
Statistical Analyses
We used Cox proportional hazards regression models with genotype, intervention and their interactions as the independent variables predicting time to diabetes over mean 3.2 years follow-up. We adjusted for gender, age at enrollment, ethnicity, treatment arm, and baseline BMI. For the quantitative glycemic traits, we employed generalized mixed models to test additive effects of genotype on baseline log-transformed quantitative traits, and on the same traits after one year of intervention adjusted for the baseline value, age, sex, self-reported ethnicity, BMI and treatment arm. We note that these SNPs have been associated with glycemic traits at genome-wide levels of significance, and therefore their prior probability of true effects is many orders of magnitude higher than the genome average. As our analyses represent further characterization of each of these established loci, we selected a P value threshold of 0.05. Finally, we also tested for any evidence of epistatic interactions between the MTNR1B SNP rs10830963 and the G6PC2 SNP rs573225, both of which have significant effects on fasting glucose in the DPP, by including appropriate interaction terms at baseline and one year.
Results
Baseline Associations
The SNPs genotyped, their chromosomal location, the nearest gene and their allele frequencies in the five DPP ethnic groups are shown in Table 1. Allele frequencies were comparable to those previously reported by MAGIC in Europeans [8] and NHANES III in non-Hispanic whites, African Americans and US Hispanics [10].
Table 1. SNPs genotyped and their allele frequencies by ethnic group.
| Allele frequencies (%) | |||||||||
| SNP | Chromosome | Position(NCBI 36) | Nearest gene | Alleles(effect/other) | White(n = 1,617) | African-American(n = 592) | Hispanic(n = 475) | Asian(n = 125) | American Indian (n = 81) |
| Fasting glucose | |||||||||
| rs340874 | 1 | 184833918 | PROX1 * | C/T | 55.9 | 19.8 | 41.2 | 42.3 | 35.4 |
| rs573225 | 2 | 161653734 | G6PC2 | A/G | 71.7 | 91.8 | 85.4 | 90.3 | 92.0 |
| rs11708067 | 3 | 120438894 | ADCY5 * | A/G | 79.4 | 85.9 | 77.3 | 91.2 | 70.0 |
| rs11920090 | 3 | 168087406 | SLC2A2 | T/A | 87.1 | 67.3 | 86.6 | 91.1 | 94.4 |
| rs2191349 | 7 | 14947780 | DGKB * | T/G | 55.7 | 57.9 | 48.1 | 64.0 | 24.1 |
| rs917793 | 7 | 44131132 | GCK * | T/A | 19.5 | 23.7 | 32.3 | 21.6 | 48.1 |
| rs7034200 | 9 | 4244098 | GLIS3 | A/C | 48.9 | 64.2 | 57.3 | 46.0 | 65.6 |
| rs10885122 | 10 | 106670840 | ADRA2A | G/T | 88.0 | 35.6 | 84.3 | 86.8 | 88.9 |
| rs11605924 | 11 | 45579933 | CRY2 | A/C | 49.3 | 87.0 | 47.7 | 68.0 | 51.9 |
| rs7944584 | 11 | 47035421 | MADD | A/T | 71.7 | 95.0 | 83.7 | 90.4 | 98.1 |
| rs174550 | 11 | 57899714 | FADS1 | T/C | 68.0 | 91.4 | 43.5 | 55.2 | 11.1 |
| rs10830963 | 11 | 88799685 | MTNR1B * | G/C | 28.8 | 9.1 | 22.7 | 41.2 | 24.1 |
| rs11071657 | 15 | 39256547 | C2CD4B | A/G | 64.4 | 86.9 | 53.7 | 70.0 | 37.0 |
| Fasting insulin | |||||||||
| rs4675095 | 2 | 219495543 | IRS1 | A/T | 93.3 | 98.5 | 84.8 | 85.6 | 69.1 |
| rs855228 | 12 | 99957291 | IGF1 | T/C | 84.3 | 40.9 | 76.1 | 65.4 | 79.0 |
| Fasting glucose and insulin | |||||||||
| rs780094 | 2 | 27483120 | GCKR * | C/T | 59.6 | 81.7 | 62.2 | 66.8 | 88.9 |
Loci previously associated with type 2 diabetes at genome-wide levels of statistical significance. The allele previously associated with higher levels of the trait (effect allele) is shown first; allele frequencies correspond to the effect allele. Gene names: PROX1, prospero homeobox 1; G6PC2, glucose-6-phosphatase, catalytic, 2; ADCY5, adenylate cyclase 5; SLC2A2, solute carrier family 2, member 2; DGKB, diacylglycerol kinase, beta 90 kDa; GCK, glucokinase; GLIS3, GLIS family zinc finger 3; ADRA2A, adrenergic, alpha-2A-, receptor; CRY2, cryptochrome 2; MADD, MAP-kinase activating death domain; FADS1, fatty acid desaturase 1; MTNR1B, melatonin receptor 1B; C2CD4B, C2 calcium-dependent domain containing 4B; IRS1, insulin receptor substrate 1; IGF1, insulin-like growth factor 1; GCKR, glucokinase regulator.
We tested associations of these SNPs with baseline fasting glucose, fasting insulin, fasting proinsulin adjusted for fasting insulin, the insulinogenic index, the ISI and the DIo in this multiethnic cohort of individuals with IGT. We replicated associations with fasting glucose at G6PC2 (P = 0.002), MTNR1B (P<0.001) and GCKR (P = 0.001). We also replicated associations of the glucose-raising allele with reduced insulinogenic index at MTNR1B and increased insulinogenic and disposition indices at G6PC2. We again noted a strong association of MADD with fasting proinsulin levels, adjusted for concomitant insulin (P<0.001). All nominally significant (P<0.05) associations and corresponding trait distributions are shown in Table 2.
Table 2. Nominal genotypic associations with quantitative traits at baseline.
| SNP | Nearest gene | Alleles (effect/other) | Trait | LS Means (95% CI) | Additive P value | Pairwise P values |
| rs573225 | G6PC2 | A/G | FG (mg/dL) | AA 106.7 (106.2–107.2) | 0.002 | AA vs AG 0.002 |
| AG 105.6 (104.9–106.3) | AA vs GG 0.31 | |||||
| GG 105.8 (104.5–107.1) | AG vs GG 0.73 | |||||
| Fins (µU/mL) | AA 24.44 (23.65–25.26) | 0.006 | AA vs AG 0.31 | |||
| AG 24.97 (23.87–26.11) | AA vs GG 0.005 | |||||
| GG 27.71 (25.56–30.05) | AG vs GG 0.02 | |||||
| Ins Index | AA 1.25 (1.20–1.31) | 0.002 | AA vs AG 0.16 | |||
| AG 1.20 (1.13–1.28) | AA vs GG 0.003 | |||||
| GG 1.04 (0.92–1.17) | AG vs GG 0.03 | |||||
| ISI | AA 0.155 (0.15–0.161) | 0.03 | AA vs AG 0.62 | |||
| AG 0.154 (0.147–0.161) | AA vs GG 0.01 | |||||
| GG 0.138 (0.127–0.15) | AG vs GG 0.03 | |||||
| DIo | AA 0.049 (0.047–0.051) | <0.001 | AA vs AG 0.03 | |||
| AG 0.046 (0.043–0.049) | AA vs GG <0.001 | |||||
| GG 0.037 (0.033–0.042) | AG vs GG <0.001 | |||||
| rs11708067 | ADCY5 | A/G | Fins (µU/mL) | AA 24.05 (23.24–24.88) | 0.001 | AA vs AG 0.001 |
| AG 25.85 (24.79–26.95) | AA vs GG 0.53 | |||||
| GG 25.29 (23.12–27.67) | AG vs GG 0.64 | |||||
| ISI | AA 0.158 (0.153–0.164) | 0.004 | AA vs AG 0.003 | |||
| AG 0.148 (0.141–0.154) | AA vs GG 0.72 | |||||
| GG 0.151 (0.138–0.166) | AG vs GG 0.72 | |||||
| rs11920090 | SLC2A2 | T/A | DIo | AA 0.042 (0.037–0.049) | 0.006 | AA vs AT 0.27 |
| AT 0.046 (0.043–0.049) | AA vs TT 0.04 | |||||
| TT 0.049 (0.047–0.051) | AT vs TT 0.03 | |||||
| rs7944584 | MADD | A/T | Proins (pmol/L) | AA 16.4 (15.9–16.92) | <0.001 | AA vs AT <0.001 |
| AT 14.98 (14.34–15.65) | AA vs TT <0.001 | |||||
| TT 13.53 (12.46–14.68) | AT vs TT 0.01 | |||||
| rs174550 | FADS1 | T/C | Fins (µU/mL) | TT 23.78 (22.83–24.78) | 0.008 | TT vs CT 0.06 |
| CT 24.97 (23.96–26.03) | TT vs CC 0.06 | |||||
| CC 25.52 (24.25–26.86) | CT vs CC 0.47 | |||||
| ISI | CC 0.149 (0.141–0.157) | 0.01 | TT vs CT 0.09 | |||
| CT 0.153 (0.146–0.159) | TT vs CC 0.09 | |||||
| TT 0.160 (0.153–0.167) | CT vs CC 0.46 | |||||
| rs10830963 | MTNR1B | G/C | FG (mg/dL) | GG 108.7 (107.6–109.9) | <0.001 | GG vs CG 0.02 |
| CG 107.3 (106.7–108.0) | GG vs CC <0.001 | |||||
| CC 105.6 (105.1–106.2) | CG vs CC <0.001 | |||||
| Proins (pmol/L) | GG 15.88 (14.80–17.03) | 0.009 | GG vs CG 0.66 | |||
| CG 15.43 (14.85–16.04) | GG vs CC 0.66 | |||||
| CC 16.44 (15.90–17.00) | CG vs CC 0.003 | |||||
| Ins Index | GG 1.17 (1.05–1.29) | 0.01 | GG vs CG 0.74 | |||
| CG 1.19 (1.12–1.25) | GG vs CC 0.21 | |||||
| CC 1.27 (1.21–1.33) | CG vs CC 0.05 | |||||
| rs855228 | IGF1 | T/C | FG (mg/dL) | TT 106.1 (105.5–106.7) | 0.01 | TT vs CT 0.37 |
| CT 106.4 (105.8–107.0) | TT vs CC 0.02 | |||||
| CC 107.7 (106.6–108.7) | CT vs CC 0.43 | |||||
| rs780094 | GCKR | C/T | FG (mg/dL) | CC 106.8 (106.2–107.3) | 0.001 | CC vs CT 0.12 |
| CT 106.3 (105.6–106.9) | CC vs TT 0.003 | |||||
| TT 105.2 (104.3–106.1) | CT vs TT 0.04 |
FG, fasting glucose; Fins, fasting insulin; Ins Index, insulinogenic index; ISI, insulin sensitivity index; DIo, oral disposition index; Proins, fasting proinsulin adjusted for fasting insulin. To convert glucose mg/dL to mmol/L, divide by 18.01. To convert insulin µU/ml to pmol/L to, multiply by 6.0.
Associations at One Year
We tested whether the metformin or lifestyle preventive interventions interacted with each SNP to modulate quantitative glycemic traits at one year. We adjusted one-year traits for the corresponding baseline trait, to indicate change in each variable during active treatment. Where no nominally significant interaction with treatment was found, SNP main effects on the one-year trait were tested in the whole cohort with an adjustment for treatment arm; if an interaction was detected at P<0.05, analyses were stratified by treatment arm (Table 3). Nominally significant interactions were found for DGKB and fasting insulin, GLIS3 and both fasting insulin and ISI, and both MADD and C2CD4B and fasting glucose. Least-square means for each genotype group and the corresponding pairwise comparisons are shown in Table 4.
Table 3. Associations with quantitative traits at one year.
| FG | Fins | Proins | Ins Index | ISI | DIo | |||||||||
| SNP | Nearest gene | Alleles(effect/other) | P int | P assoc | P int | P assoc | P int | P assoc | P int | P assoc | P int | P assoc | P int | P assoc |
| rs340874 | PROX1 | C/T | 0.81 | 0.47 | 0.90 | 0.15 | 0.99 | 0.08 | 0.42 | 0.99 | 0.89 | 0.14 | 0.63 | 0.22 |
| rs573225 | G6PC2 | A/G | 0.96 | 0.17 | 0.08 | 0.88 | 0.34 | 0.91 | 0.77 | 0.20 | 0.11 | 0.66 | 0.81 | 0.08 |
| rs11708067 | ADCY5 | A/G | 0.80 | 0.27 | 0.22 | 0.98 | 0.31 | 0.85 | 0.46 | 0.32 | 0.20 | 0.80 | 0.86 | 0.52 |
| rs11920090 | SLC2A2 | T/A | 0.88 | 0.49 | 0.24 | 0.53 | 0.79 | 0.38 | 0.59 | 0.98 | 0.25 | 0.49 | 0.69 | 0.89 |
| rs2191349 | DGKB | T/G | 0.79 | 0.41 | 0.04 | – | 0.09 | 0.80 | 0.50 | 0.55 | 0.07 | 0.84 | 0.84 | 0.99 |
| rs917793 | GCK | T/A | 0.07 | 0.12 | 0.39 | 0.12 | 0.08 | 0.31 | 0.86 | 0.23 | 0.24 | 0.08 | 0.42 | 0.07 |
| rs7034200 | GLIS3 | A/C | 0.72 | 0.98 | 0.02 | – | 0.25 | 0.56 | 0.94 | 0.94 | 0.03 | – | 0.13 | 0.64 |
| rs10885122 | ADRA2A | G/T | 0.82 | 0.14 | 0.20 | 0.60 | 0.16 | 0.46 | 0.29 | 0.18 | 0.25 | 0.44 | 0.31 | 0.03 |
| rs11605924 | CRY2 | A/C | 0.20 | 0.56 | 0.13 | 0.46 | 0.76 | 0.80 | 0.40 | 0.28 | 0.13 | 0.40 | 0.21 | 0.58 |
| rs7944584 | MADD | A/T | 0.04 | – | 0.73 | 0.80 | 0.63 | 0.30 | 0.11 | 0.10 | 0.63 | 0.83 | 0.36 | 0.29 |
| rs174550 | FADS1 | T/C | 0.58 | 0.20 | 0.87 | 0.09 | 0.73 | 0.23 | 0.76 | 0.97 | 0.86 | 0.07 | 0.64 | 0.65 |
| rs10830963 | MTNR1B | G/C | 0.68 | 0.003 | 0.17 | 0.37 | 0.27 | 0.41 | 0.69 | 0.002 | 0.16 | 0.90 | 0.96 | 0.08 |
| rs11071657 | C2CD4B | A/G | 0.04 | – | 0.96 | 0.42 | 0.55 | 0.97 | 0.38 | 0.09 | 0.98 | 0.54 | 0.35 | 0.048 |
| rs4675095 | IRS1 | A/T | 0.21 | 0.99 | 0.67 | 0.26 | 0.68 | 0.59 | 0.53 | 0.55 | 0.71 | 0.30 | 0.62 | 0.62 |
| rs855228 | IGF1 | T/C | 0.39 | 0.60 | 0.15 | 0.27 | 0.52 | 0.87 | 0.36 | 0.72 | 0.15 | 0.24 | 0.63 | 0.75 |
| rs780094 | GCKR | C/T | 0.57 | 0.76 | 0.07 | 0.22 | 0.25 | 0.38 | 0.17 | 0.92 | 0.09 | 0.26 | 0.43 | 0.28 |
FG, fasting glucose; Fins, fasting insulin; Ins Index, insulinogenic index; ISI, insulin sensitivity index; DIo, oral disposition index; Proins, fasting proinsulin adjusted for fasting insulin. P int denotes the P value for the genotype × intervention interaction test; P assoc denotes the P value for the main effect association in the full cohort when P int >0.05.
Table 4. Levels of quantitative glycemic traits at one year by genotype and treatment arm at loci with a nominally significant interaction.
| Placebo | Metformin | Lifestyle | ||||||
| SNP gene | Alleles (effect/other) | Trait | LS Means (95% CI) | P values | LS Means (95% CI) | P values | LS Means (95% CI) | P values |
| rs2191349 | T/G | Fins | GG 24.71 (22.96–26.59) | GG/GT: 0.99 | GG 22.62 (21.02–24.34) | GG/GT: 0.21 | GG 18.43 (17.03–19.94) | GG/GT: 0.99 |
| DGKB | (µU/mL) | GT 24.99 (23.59–26.48) | GG/TT: 0.99 | GT 21.30 (20.06–22.62) | GG/TT: 0.04 | GT 19.01 (17.82–20.28) | GG/TT: 0.99 | |
| TT 25.71 (24.00–27.54) | GT/TT: 0.99 | TT 20.41 (19.03–21.88) | GT/TT: 0.21 | TT 19.05 (17.71–20.51) | GT/TT: 0.99 | |||
| rs7034200 | A/C | Fins | AA 24.86 (23.27–26.55) | AA/AC: 0.99 | AA 22.54 (21.06–24.12) | AA/AC: 0.12 | AA 18.01 (16.76–19.36) | AA/AC: 0.28 |
| GLIS3 | (µU/mL) | AC 25.23 (23.83–26.73) | AA/CC: 0.99 | AC 21.14 (19.87–22.50) | AA/CC: 0.05 | AC 19.20 (18.02–20.45) | AA/CC: 0.28 | |
| CC 25.07 (23.18–27.12) | AC/CC: 0.99 | CC 20.45 (19.00–22.01 | AC/CC: 0.37 | CC 19.47 (17.96–21.11) | AC/CC: 0.74 | |||
| ISI | AA 0.153 (0.142–0.164) | AA/AC: 0.99 | AA 0.175 (0.163–0.189) | AA/AC: 0.14 | AA 0.222 (0.205–0.240) | AA/AC: 0.28 | ||
| AC 0.152 (0.143–0.162) | AA/CC: 0.99 | AC 0.187 (0.175–0.200) | AA/CC: 0.06 | AC 0.207 (0.193–0.221) | AA/CC: 0.28 | |||
| CC 0.151 (0.139–0.165) | AC/CC: 0.99 | CC 0.194 (0.179–0.210) | AC/CC: 0.36 | CC 0.204 (0.186–0.222) | AC/CC: 0.74 | |||
| rs7944584 | A/T | FG | AA 106.8 (105.5–108.1) | AA/AT: 0.008 | AA 102.4 (101.3–103.5) | AA/AT: 0.99 | AA 102.1 (101.0–103.2) | AA/AT: 0.99 |
| MADD | (mg/dL) | AT 104.3 (102.6–106.1) | AA/TT: 0.50 | AT 102.7 (101.1–104.2) | AA/TT: 0.99 | AT 101.5 (99.98–103.1) | AA/TT: 0.99 | |
| TT 104.8 (101.4–108.3) | AT/TT: 0.78 | TT 101.3 (98.45–104.3) | AT/TT: 0.99 | TT 101.4 (98.65–104.2) | AT/TT: 0.99 | |||
| rs11071657 | A/G | FG | AA 107.1 (105.6–108.7) | AA/AG: 0.41 | AA 102.3 (101.0–103.6) | AA/AG: 0.99 | AA 101.9 (100.6–103.2) | AA/AG: 0.96 |
| C2CD4B | (mg/dL) | AG 105.9 (104.5–107.4) | AA/GG: 0.41 | AG 102.7 (101.4–104.0) | AA/GG: 0.99 | AG 101.7 (100.4–103.0) | AA/GG: 0.96 | |
| GG 105.3 (103.1–107.6) | AG/GG: 0.63 | GG 102.0 (100.2–103.9) | AG/GG: 0.99 | GG 102.7 (100.8–104.7) | AG/GG: 0.96 | |||
P values for pairwise comparisons between genotypic groups are shown, with groups separated by a “/”. Fins, fasting insulin (µU/mL); ISI, insulin sensitivity index; FG, fasting glucose (mg/dL). To convert glucose mg/dL to mmol/L, divide by 18.01. To convert insulin µU/ml to pmol/L to, multiply by 6.0.
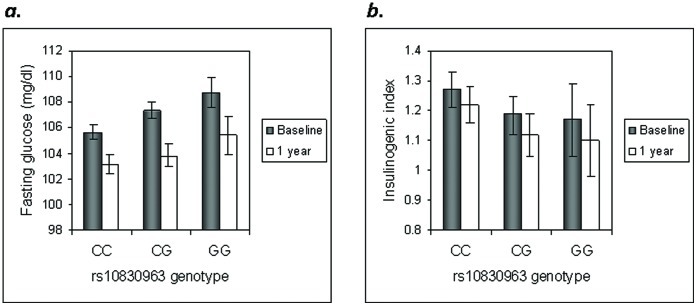
At MTNR1B, the glucose-raising allele continued to have a significant main effect on raising fasting glucose and lowering the insulinogenic index at one year (Figure 1). Because one-year traits are adjusted for the baseline level, this effect is indicative of a worsening deleterious effect of this locus on β-cell function. We further explored the concordant effects of SNPs at MTNR1B and G6PC2 on fasting glucose but discordant effects for insulinogenic index by testing for epistatic interactions between the two on fasting glucose at baseline and one year: the interaction terms were not statistically significant.
Figure 1. Effect of genotype at MTNR1B rs10830963 on glycemic traits at baseline and one year.
Fasting glucose is shown in panel (a) and the insulinogenic index is shown in panel (b). Because no significant SNP × intervention interaction was found, the full cohort was analyzed in aggregate. Fasting glucose is higher (P = 0.003) and the insulinogenic index is lower (P = 0.002) in carriers of the G risk allele after one year, even after adjustment for the corresponding baseline levels. Least-square means (±95% CI) are shown. To convert glucose mg/dL to mmol/L, divide by 18.01.
Diabetes Incidence
We tested whether the metformin or lifestyle preventive interventions interact with each SNP on the risk of developing diabetes during 3.2 years of mean follow-up. As no nominal interactions were found, the effects of each SNP on diabetes incidence were evaluated in the full cohort while adjusting for treatment arm; stratified analyses are also shown (Table 5). The only nominal association with diabetes incidence was found for the glucose-lowering allele at PROX1 (P = 0.02), in a direction opposite to that reported in case-control analyses in MAGIC, where the C allele increased type 2 diabetes risk (odds ratio 1.07 [95% CI 1.05–1.09], P = 7.2×10−10) [8].
Table 5. Diabetes incidence by genotype at each locus, in the overall cohort and stratified by treatment arm.
| SNP | Nearest gene | Alleles | SNP * Tx | Treatment adjusted HR(95% CI) | P-value | PLACEBO HR(95% CI) | P-value | METFORMIN HR(95% CI) | P-value | LIFESTYLE HR(95% CI) | P-value |
| rs340874 | PROX1 * | C (vs T) | N | 0.88 (0.78–0.98) | 0.02 | 0.85 (0.71–1.01) | 0.06 | 0.92 (0.75–1.12) | 0.39 | 0.86 (0.68–1.08) | 0.20 |
| rs573225 | G6PC2 | A (vs G) | N | 1.11 (0.97–1.27) | 0.14 | 0.96 (0.77–1.19) | 0.70 | 1.18 (0.94–1.47) | 0.15 | 1.27 (0.98–1.64) | 0.07 |
| rs11708067 | ADCY5 * | A (vs G) | N | 1.06 (0.92–1.23) | 0.38 | 1.04 (0.84–1.28) | 0.73 | 1.08 (0.84–1.35) | 0.60 | 1.10 (0.82–1.47) | 0.51 |
| rs11920090 | SLC2A2 | T (vs A) | N | 1.02 (0.88–1.19) | 0.75 | 1.08 (0.86–1.33) | 0.56 | 1.00 (0.77–1.30) | 0.99 | 0.99 (0.71–1.37) | 0.93 |
| rs2191349 | DGKB * | T (vs G) | N | 1.06 (0.94–1.18) | 0.34 | 1.05 (0.88–1.27) | 0.56 | 1.10 (0.90–1.33) | 0.34 | 1.01 (0.80–1.27) | 0.96 |
| rs917793 | GCK * | T (vs A) | N | 0.96 (0.84–1.10) | 0.59 | 0.87 (0.70–1.07) | 0.20 | 1.14 (0.90–1.44) | 0.29 | 0.92 (0.69–1.22) | 0.56 |
| rs7034200 | GLIS3 | A (vs C) | N | 1.00 (0.89–1.12) | 1.00 | 0.90 (0.75–1.08) | 0.22 | 1.04 (0.85–1.27) | 0.68 | 1.15 (0.91–1.47) | 0.25 |
| rs10885122 | ADRA2A | G (vs T) | N | 1.03 (0.91–1.16) | 0.63 | 1.01 (0.84–1.22) | 0.89 | 1.03 (0.84–1.28) | 0.76 | 1.08 (0.84–1.39) | 0.56 |
| rs11605924 | CRY2 | A (vs C) | N | 1.01 (0.90–1.12) | 0.91 | 0.93 (0.78–1.10) | 0.40 | 1.06 (0.88–1.28) | 0.56 | 1.09 (0.87–1.37) | 0.47 |
| rs7944584 | MADD | A (vs T) | N | 0.93 (0.80–1.08) | 0.29 | 0.86 (0.69–1.08) | 0.20 | 0.89 (0.69–1.14) | 0.35 | 1.11 (0.84–1.47) | 0.47 |
| rs174550 | FADS1 | T (vs C) | N | 0.94 (0.83–1.05) | 0.26 | 0.95 (0.80–1.14) | 0.57 | 0.98 (0.80–1.19) | 0.81 | 0.86 (0.67–1.09) | 0.21 |
| rs10830963 | MTNR1B * | G (vs C) | N | 1.07 (0.94–1.22) | 0.29 | 1.20 (0.98–1.47) | 0.07 | 1.01 (0.80–1.26) | 0.95 | 0.95 (0.73–1.24) | 0.69 |
| rs11071657 | C2CD4B | A (vs G) | N | 0.93 (0.83–1.05) | 0.26 | 0.93 (0.77–1.12) | 0.43 | 0.92 (0.75–1.14) | 0.42 | 0.96 (0.75–1.22) | 0.72 |
| rs4675095 | IRS1 | A (vs T) | N | 0.96 (0.78–1.18) | 0.68 | 0.93 (0.67–1.30) | 0.69 | 1.16 (0.84–1.59) | 0.37 | 0.71 (0.44–1.15) | 0.17 |
| rs855228 | IGF1 | T (vs C) | N | 1.09 (0.97–1.23) | 0.14 | 1.04 (0.87–1.25) | 0.66 | 1.16 (0.95–1.43) | 0.15 | 1.12 (0.88–1.43) | 0.38 |
| rs780094 | GCKR * | C (vs T) | N | 0.96 (0.85–1.08) | 0.48 | 0.91 (0.75–1.10) | 0.33 | 1.01 (0.82–1.23) | 0.93 | 0.96 (0.75–1.22) | 0.72 |
Loci previously associated with type 2 diabetes. Effect allele denotes the allele associated with higher glucose or insulin levels in MAGIC. There are no significant SNP × treatment interactions. One nominally significant P value for association with diabetes incidence is not consistent with the expected direction of effect.
Discussion
The MAGIC investigators reported a number of loci that influence fasting glucose and fasting insulin levels in non-diabetic populations of European descent; only a few of the loci were also associated with type 2 diabetes at genome-wide levels of significance [8]. The authors speculated that it is not the mere elevation in fasting glucose, but how fasting glucose is raised, that determines overall β-cell dysfunction and future type 2 diabetes risk. However, whether these loci exert their action on fasting glucose in the initial stages of diabetes progression (e.g. from normoglycemia to impaired glucose regulation) or later (e.g. from IGT to type 2 diabetes) is not known. In the GLACIER cohort, eleven loci (including the known type 2 diabetes genes TCF7L2 and SLC30A8) were nominally associated with IFG cross-sectionally, and MTNR1B and G6PC2 were also associated with development of IFG in longitudinal analyses [11]. We have recently shown that among type 2 diabetes-associated loci, risk alleles at MTNR1B, GCK and SLC30A8 confer a stronger rate of progression from normoglycemia to IFG than from IFG to type 2 diabetes [13]. Here we extend these findings by testing these SNPs from the IGT to type 2 diabetes transition, and by assessing their effects on quantitative glycemic traits at baseline and one year in a multiethnic cohort of persons with IGT.
We have demonstrated that the three loci with the strongest reported effect on fasting glucose (MTNR1B, GCKR and G6PC2) have consistent effects in the DPP. All three were known to be associated with fasting glucose prior to the MAGIC GWAS meta-analysis [4], [5], [6], [7], [26], [27], [28]. Power may have been limiting to detect the other reported associations [24].
We have also confirmed that the glucose-raising allele at MTNR1B is associated with a reduced insulinogenic index, as measured during the initial phase of insulin secretion during an OGTT [9], [29]. As shown by Lyssenko and coworkers, the deleterious effects of this allele on β-cell function persist over time; while they noted such worsening over 24 years of follow-up [29], here we see such effects over a much shorter time span (one year). In GLACIER a similar non-significant trend was noted over 10 years of follow-up [11], although a consistent effect was not detected in the Whitehall II study [12]. Because MTNR1B does increase risk of type 2 diabetes [8], this pattern of sustained deterioration suggests that identifying these individuals early in their glycemic progression may be beneficial in prevention efforts.
In contrast, the glucose-raising allele at G6PC2 is associated with superior β-cell function on dynamic testing; this has been shown previously [9], [30], and is consistent with the role of this gene product in regulating hepatic glucokinase and its null effect on type 2 diabetes risk [8]. We found no evidence in support of a non-additive interaction between MTNR1B and G6PC2 on fasting glucose at baseline or one year. The strong effect of the MADD locus on fasting proinsulin levels is also confirmed [9], [31]; because this association is adjusted for concomitant insulin levels, it reflects an increased secretion of insulin precursors out of proportion to the degree of basal insulin resistance. The other nominal associations newly reported here do not withstand correction for the multiple statistical tests performed, and should be considered hypothesis-generating requiring confirmation in independent studies.
In summary, the strongest effects of genetic loci on fasting glucose in non-diabetic individuals of European descent are also evident in a multiethnic cohort with IGT. The deleterious influence of the glucose-raising allele at MTNR1B on β-cell function appears to worsen with time, and this effect is evident in as short a time as one year. Genetic testing may identify a subset of patients with IGT more likely to respond to preventive interventions [32].
Supporting Information
DPP Research Group.
(DOC)
Acknowledgments
The Investigators gratefully acknowledge the commitment and dedication of the participants of the DPP. We thank the MAGIC investigators for access to pre-publication data.
Funding Statement
The National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK) of the National Institutes of Health provided funding to the clinical centers and the Coordinating Center for the design and conduct of the study, and the collection, management, analysis, and interpretation of the data, through Award Number U01DK048489. The Southwestern American Indian Centers were supported directly by the NIDDK and the Indian Health Service. The General Clinical Research Center Program, National Center for Research Resources, and the Department of Veterans Affairs supported data collection at many of the clinical centers. Funding for data collection and participant support was also provided by the Office of Research on Minority Health, the National Institute of Child Health and Human Development, the National Institute on Aging, the Office of Research on Women’s Health, the Centers for Disease Control and Prevention, and the American Diabetes Association. This research was also supported, in part, by the intramural research program of the NIDDK, and by R01 DK072041 to JCF, KAJ and ARS. PWF was supported by the Swedish Research Council, Novo Nordisk, Swedish Diabetes Association, and the Swedish Heart-Lung Foundation. The Investigators gratefully acknowledge the commitment and dedication of the participants of the DPP. The opinions expressed are those of the investigators and do not necessarily reflect the official views of the funding agencies. A complete list of Centers, investigators, and staff can be found in the Appendix. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. Meigs JB, Panhuysen CIM, Myers RH, Wilson PWF, Cupples LA (2002) A genome-wide scan for loci linked to plasma levels of glucose and HbA1c in a community-based sample of Caucasian pedigrees: the Framingham Offspring Study. Diabetes 51: 833–840. [DOI] [PubMed] [Google Scholar]
- 2. Panhuysen CIM, Cupples LA, Wilson PWF, Herbert AG, Myers RH, et al. (2003) A genome scan for loci linked to quantitative insulin traits in persons without diabetes: the Framingham Offspring Study. Diabetologia 46: 579–587. [DOI] [PubMed] [Google Scholar]
- 3. Weedon MN, Clark VJ, Qian Y, Ben-Shlomo Y, Timpson N, et al. (2006) A common haplotype of the glucokinase gene alters fasting glucose and birth weight: association in six studies and population-genetics analyses. Am J Hum Genet 79: 991–1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Bouatia-Naji N, Rocheleau G, Van Lommel L, Lemaire K, Schuit F, et al. (2008) A polymorphism within the G6PC2 gene is associated with fasting plasma glucose levels. Science 320: 1085–1088. [DOI] [PubMed] [Google Scholar]
- 5. Chen W-M, Erdos MR, Jackson AU, Saxena R, Sanna S, et al. (2008) Association studies in Caucasians identify variants in the G6PC2/ABCB11 region regulating fasting glucose levels. J Clin Invest 118: 2620–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Prokopenko I, Langenberg C, Florez JC, Saxena R, Soranzo N, et al. (2009) Variants in MTNR1B influence fasting glucose levels. Nat Genet 41: 77–81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Bouatia-Naji N, Bonnefond A, Cavalcanti-Proenca C, Sparso T, Holmkvist J, et al. (2009) A variant near MTNR1B is associated with increased fasting plasma glucose levels and type 2 diabetes risk. Nat Genet 41: 89–94. [DOI] [PubMed] [Google Scholar]
- 8. Dupuis J, Langenberg C, Prokopenko I, Saxena R, Soranzo N, et al. (2010) New genetic loci implicated in fasting glucose homeostasis and their impact on type 2 diabetes risk. Nat Genet 42: 105–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ingelsson E, Langenberg C, Hivert MF, Prokopenko I, Lyssenko V, et al. (2010) Detailed physiologic characterization reveals diverse mechanisms for novel genetic Loci regulating glucose and insulin metabolism in humans. Diabetes 59: 1266–1275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Yang Q, Liu T, Shrader P, Yesupriya A, Chang MH, et al. (2010) Racial/ethnic differences in association of fasting glucose-associated genomic loci with fasting glucose, HOMA-B, and impaired fasting glucose in the U.S. adult population. Diabetes Care 33: 2370–2377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Renstrom F, Shungin D, Johansson I, Florez JC, Hallmans G, et al. (2011) Genetic predisposition to long-term nondiabetic deteriorations in glucose homeostasis: Ten-year follow-up of the GLACIER study. Diabetes 60: 345–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Jensen AC, Barker A, Kumari M, Brunner EJ, Kivimaki M, et al. (2011) Associations of common genetic variants with age-related changes in fasting and postload glucose: Evidence from 18 years of follow-up of the Whitehall II cohort. Diabetes 60: 1617–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Walford GA, Green T, Neale B, Isakova T, Rotter JI, et al.. (2011) Common genetic variants differentially influence the transition between clinically-defined states of fasting glucose metabolism. Diabetologia (epub ahead of print). [DOI] [PMC free article] [PubMed]
- 14. Saxena R, Hivert MF, Langenberg C, Tanaka T, Pankow JS, et al. (2010) Genetic variation in GIPR influences the glucose and insulin responses to an oral glucose challenge. Nat Genet 42: 142–148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Voight BF, Scott LJ, Steinthorsdottir V, Morris AP, Dina C, et al. (2010) Twelve type 2 diabetes susceptibility loci identified through large-scale association analysis. Nat Genet 42: 579–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Jensen AC, Barker A, Kumari M, Brunner EJ, Kivimaki M, et al. (2011) Associations of common genetic variants with age-related changes in fasting and postload glucose: evidence from 18 years of follow-up of the Whitehall II cohort. Diabetes 60: 1617–1623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. The Diabetes Prevention Program Research Group (1999) The Diabetes Prevention Program. Design and methods for a clinical trial in the prevention of type 2 diabetes. Diabetes Care 22: 623–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. The Diabetes Prevention Program Research Group (2000) The Diabetes Prevention Program: baseline characteristics of the randomized cohort. The Diabetes Prevention Program Research Group. Diabetes Care 23: 1619–1629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. The Diabetes Prevention Program Research Group (2002) Reduction in the incidence of type 2 diabetes with lifestyle intervention or metformin. N Engl J Med 346: 393–403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Matthews DR, Hosker JP, Rudenski AS, Naylor BA, Treacher DF, et al. (1985) Homeostasis model assessment: insulin resistance and beta-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 28: 412–419. [DOI] [PubMed] [Google Scholar]
- 21. Utzschneider KM, Prigeon RL, Faulenbach MV, Tong J, Carr DB, et al. (2009) Oral disposition index predicts the development of future diabetes above and beyond fasting and 2-h glucose levels. Diabetes Care 32: 335–341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Florez JC, Jablonski KA, Bayley N, Pollin TI, de Bakker PIW, et al. (2006) TCF7L2 polymorphisms and progression to diabetes in the Diabetes Prevention Program. N Engl J Med 355: 241–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Tang K, Fu DJ, Julien D, Braun A, Cantor CR, et al. (1999) Chip-based genotyping by mass spectrometry. Proc Natl Acad Sci U S A 96: 10016–10020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Moore AF, Jablonski KA, McAteer JB, Saxena R, Pollin TI, et al. (2008) Extension of type 2 diabetes genome-wide association scan results in the Diabetes Prevention Program. Diabetes 57: 2503–2510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Majithia AR, Jablonski KA, McAteer JB, Mather KJ, Goldberg RB, et al. (2011) Association of the SLC30A8 missense polymorphism R325W with proinsulin levels at baseline and after lifestyle, metformin or troglitazone intervention in the Diabetes Prevention Program. Diabetologia 54: 2570–2574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Diabetes Genetics Initiative of Broad Institute of Harvard and MIT, Lund University and Novartis Institutes for BioMedical Research (2007) Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science 316: 1331–1336. [DOI] [PubMed] [Google Scholar]
- 27. Sparso T, Andersen G, Nielsen T, Burgdorf KS, Gjesing AP, et al. (2008) The GCKR rs780094 polymorphism is associated with elevated fasting serum triacylglycerol, reduced fasting and OGTT-related insulinaemia, and reduced risk of type 2 diabetes. Diabetologia 51: 70–75. [DOI] [PubMed] [Google Scholar]
- 28. Orho-Melander M, Melander O, Guiducci C, Perez-Martinez P, Corella D, et al. (2008) Common missense variant in the glucokinase regulatory protein gene is associated with increased plasma triglyceride and C-reactive protein but lower fasting glucose concentrations. Diabetes 57: 3112–3121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lyssenko V, Nagorny CL, Erdos MR, Wierup N, Jonsson A, et al. (2009) Common variant in MTNR1B associated with increased risk of type 2 diabetes and impaired early insulin secretion. Nat Genet 41: 82–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Rose CS, Grarup N, Krarup NT, Poulsen P, Wegner L, et al. (2009) A variant in the G6PC2/ABCB11 locus is associated with increased fasting plasma glucose, increased basal hepatic glucose production and increased insulin release after oral and intravenous glucose loads. Diabetologia 52: 2122–2129. [DOI] [PubMed] [Google Scholar]
- 31. Strawbridge RJ, Dupuis J, Prokopenko I, Barker A, Ahlqvist E, et al. (2011) Genome-wide association identifies nine common variants associated with fasting proinsulin levels and provides new insights into the pathophysiology of type 2 diabetes. Diabetes 60: 2624–2634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hivert MF, Jablonski KA, Perreault L, Saxena R, McAteer JB, et al. (2011) Updated genetic score based on 34 confirmed type 2 diabetes loci is associated with diabetes incidence and regression to normoglycemia in the Diabetes Prevention Program. Diabetes 60: 1340–1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
DPP Research Group.
(DOC)