Abstract
Humans with Hermansky-Pudlak Syndrome (HPS) or ocular albinism (OA1) display abnormal aspects of organelle biogenesis. The multigenic disorder HPS displays broad defects in biogenesis of lysosome-related organelles including melanosomes, platelet dense granules, and lysosomes. A phenotype of ocular pigmentation in OA1 is a smaller number of macromelanosomes, in contrast to HPS, where in many cases the melanosomes are smaller than normal. In these studies we define the role of the Mregdsu gene, which suppresses the coat color dilution of Myo5a, melanophilin, and Rab27a mutant mice in maintaining melanosome size and distribution. We show that the product of the Mregdsu locus, melanoregulin (MREG), interacts both with members of the HPS BLOC-2 complex and with Oa1 in regulating melanosome size. Loss of MREG function facilitates increase in the size of micromelanosomes in the choroid of the HPS BLOC-2 mutants ruby, ruby2, and cocoa, while a transgenic mouse overexpressing melanoregulin corrects the size of retinal pigment epithelium (RPE) macromelanosomes in Oa1ko/ko mice. Collectively, these results suggest that MREG levels regulate pigment incorporation into melanosomes. Immunohistochemical analysis localizes melanoregulin not to melanosomes, but to small vesicles in the cytoplasm of the RPE, consistent with a role for this protein in regulating membrane interactions during melanosome biogenesis. These results provide the first link between the BLOC pathway and Oa1 in melanosome biogenesis, thus supporting the hypothesis that intracellular G-protein coupled receptors may be involved in the biogenesis of other organelles. Furthermore these studies provide the foundation for therapeutic approaches to correct the pigment defects in the RPE of HPS and OA1.
Introduction
These studies define the role of the Mregdsu gene [1], which suppresses the coat color dilution of Myo5a, melanophilin [2], and Rab27a [3] mutant mice in maintaining melanosome size and distribution. Analysis of cellular abnormalities in patients and mice with HPS reveals that these genes function in biogenesis of lysosome-related organelle complexes (BLOCS) to mediate vesicle budding and transport from the trans-Golgi network to nascent organelles [4], [5]. In OA1, defects are limited to melanosomes, and pathological features are primarily restricted to the eye [6]. Findings suggest that OA1 may function as an intracellular G-protein coupled receptor regulating both melanosome size and maturation [7]. Indeed, Gαi3 has recently been identified as the first downstream component in Oa1 signaling in RPE melanosomes [8]. Individuals with either OA1 or HPS display visual system defects common to albinism, including reduced visual acuity and loss of stereoscopic vision from misrouting of retinal axons at the optic chiasm [7].
Genetic observations of natural mutants, including coat color in mouse, continue to yield a wealth of information about genes involved in many processes, including vesicle transport and organelle formation. Many of these mouse coat color mutants also display abnormalities of other systems, such as the platelet dense granule defects in HPS. Melanoregulin loss of function, by contrast, presents with no observable coat color phenotype, and the dilute suppressor (dsu) mutation was originally identified as an unlinked suppressor of the gray coat color of the dilute (Myo5a), ashen (Rab27a), and leaden (Mlph) mutants [9]–[11]. Myo5a, Mlph, and Rab27a act together as a motor complex to mediate melanosome transport in melanocytes of the skin [12], [13]. The gene encoding the Mregdsu loss of function mutation has been identified as a vertebrate-specific, highly charged 214 amino acid protein, without functional domains or homology to other proteins, but containing a cysteine-rich N-terminal sequence suggestive of a membrane binding domain [1]. Melanoregulin functions in the melanosome transport system in the skin by influencing transfer of melanosomes from melanocytes to keratinocytes, presumably at the level of the plasma membrane or the melanosome membrane [1]. Recent studies suggest that melanoregulin may act as a cargo receptor in melanosome transport [14], although the identity of the cargo remains unknown.
Over a dozen genes are involved in HPS in mouse and at least seven orthologs have been identified in human. Many of these genes have recently been cloned [15]–[24]. With the exception of members of the adaptor protein (AP)-3 and HOPS complexes known to be involved in vesicle transport, most HPS genes encode vertebrate-specific proteins with ubiquitous expression and without homology or obvious functional domains. Electron microscopy studies of skin melanocytes in the various mouse mutants of HPS have yielded clues as to which genes are required for various stages of melanosome biogenesis; mutants of genes that act together have similar phenotypes [25], [26]. Yeast two hybrid and co-immunoprecipitation studies have confirmed the ultrastructural analyses in delineating the HPS proteins that interact in specific BLOC complexes, and showing that HPS3 (cocoa), HSP5 (ruby2J), and HPS6 (ruby-eyed) are components of BLOC-2 [22], [27], [28]. HPS3 (cocoa), a member of the BLOC-2 complex, binds to clathrin-coated vesicles in the cytoplasm of human skin melanocytes [29].
Cytoplasmic transport vesicles appear to be a key component of organelle, specifically melanosome, biogenesis. Melanogenic proteins are transported to melanosomes using a variety of methods, including the AP-3 and AP-1 pathways [30], and arrive at different stages in melanosome development. For example, tyrosinase relocates from the late endosome directly to stage II melanosomes, while Pmel17/gp100/silver is transported from AP-3 vesicles to stage I melanosomes, skipping endosomes altogether [31]. It is possible that HPS genes of unknown function are involved in the transport of other components of nascent melanosomes.
Like the HPS genes, Oa1/OA1 was identified by positional cloning [32]–[34] and displays structural characteristics typical of transmembrane G-protein coupled receptors [35]. Further studies have shown that OA1 indeed functions as an intracellular G-protein coupled receptor in both Saccharomyces cerevisiae [36] and mammalian cells [37], and L-DOPA was suggested as a ligand for OA1 but the signal transduction mechanisms triggered by L-DOPA is still unclear [38]. OA1 is localized at the membrane of melanosomes at all stages of maturation but higher amounts are found in melanosomes at early stages of maturation (stage I and II) and in late endosomes [39]. Because no spontaneous mutant exists for Oa1, likely due to the lack of an observable phenotype, a knockout mouse was created to study its function [40]. This mouse has features of human ocular albinism (OA1), including the macromelanosome in pigment epithelium and skin melanocytes a reduction of melanosome number and a reduced uncrossed retinal projection [40] [41], similar likewise to humans with oculocutaneous albinism and HPS, and similar to other hypopigmented mouse strains such as Oca1Tyr<c>, Hps6ru, Hps2pe, and Hps1ep [42], [43]. Although hypopigmented individuals with RPE melanosomal abnormalities from many genetic causes share the same visual system abnormalities, a common mechanism among these disorders is lacking.
We here provide the first evidence for a molecule that links the HPS and Oa1 pathways in melanosome biogenesis, and provide evidence that modulating the levels of melanoregulin can partially correct the melanosomal defects in the HPS BLOC-2 mutants Hps6ruby, Hps5ruby2J, and Hps3coa, and in the Oa1 knockout mouse.
Results
Melanoregulin affects eye color of HPS mutants in a dose-dependent manner
Melanoregulin loss of function suppresses the eye color defect in ru2J (Hps6ru2J), ruby (Hps5ruby), and cocoa (Hps3cocoa) mice from dark red to black; overexpression of wildtype melanoregulin lightens the eye color of these same HPS mutants to red, without affecting coat color (ruby2 shown in Figure 1A). Melanoregulin loss of function rescues the eye color in ru2J, ruby, and cocoa mice by increasing pigmentation of the choroid (Figure 1B–G). Mice carrying the melanoregulin loss of function (Mregdsu) mutation on a wildtype background, transgenic mice overexpressing the wildtype melanoregulin gene (MregTg), and Oa1 knockout mice have no visible eye or coat color defects alone or in combination; all have black eyes and a black coat on a C57BL/6 background (data not shown).
Figure 1. Melanoregulin levels modulate the eye color of HPS BLOC-2 mutant mice.
A: Three month-old male mice of the genotypes indicated were photographed in ambient light. Note nearly black color of eyes of Hps5ruby2J.Mregdsu mice, darker than the eyes of Hps5ruby2J mice, which are darker than those of the light ruby red Hps5ruby2J.MregTg eyes. B–G: Electron micrographs of sections through the RPE and choroid of Hps6ruby, Hps5ruby2J, and HPS3coa mice, showing the lighter eye color in (A) to be secondary to decreased pigmentation of these layers. Note the effect of a melanoregulin loss of function (Mregdsu) allele in enhancing pigmentation of the choroid in all three BLOC-2 mutants, Hps6ruby, Hps5ruby2J, and Hps3coa. Scale bar = 2 µm.
Loss of melanoregulin increases while overexpression decreases pigment incorporation into melanosomes of the RPE
In wildtype retinal pigment epithelium, melanosomes constitute a major component of the cytoplasm, ranging in size and cross-sectional shape from round to elongated. The width of both long and round melanosomes is consistently ≤0.5 µm (Figure 2E). In melanoregulin loss of function, (labeled dsu), little change is observed in the cross-sectional area, shape, or size distribution of melanosomes (Figure 2D), while the average density of melanosomes is slightly decreased as quantified in Figure 3A. By contrast, overexpression of melanoregulin via a BAC transgenic on a wild type background, (labeled Tg) results in a small decrease in both the average cross-sectional area of RPE melanosomes and density (Figures 2F and 3A, B). Overall, either loss or overexpression of melanoregulin has only minor consequences for melanosome size, shape, and density on an otherwise wildtype background (Figure 2D–F and Figure 3A–C).
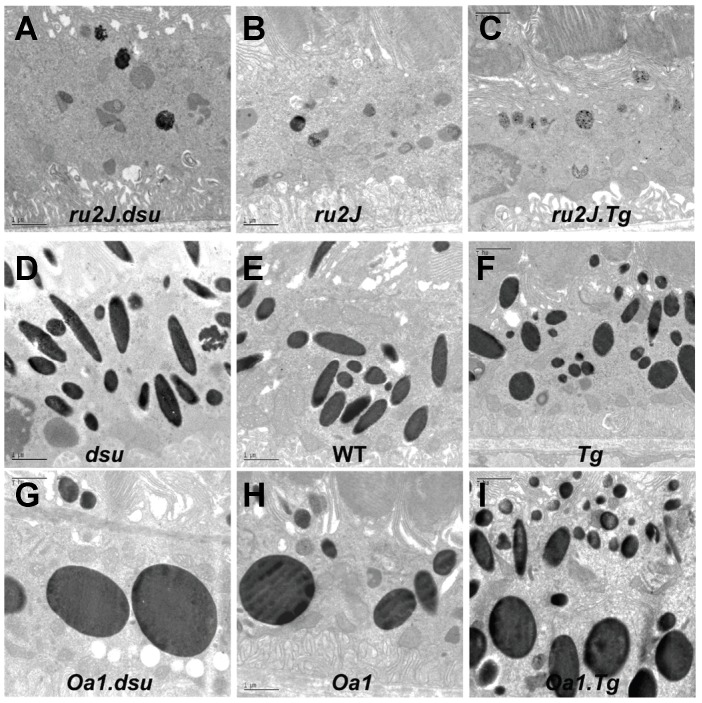
Figure 2. Size, shape, and density of melanosomes in the RPE of Hps and Oa1 loss of function mouse mutants are altered by loss or overexpression of melanoregulin.
A–I: Electron microscopic images of the RPE in mice of the genotypes indicated. Orientation is with photoreceptor outer segments toward the top and Bruch's membrane/choroid toward the bottom of each image. Abbreviations: ru2J = Hps5ru2J; dsu = Mregdsu; WT = wildtype; Tg = MregTg ; Oa1 = Oa1ko. Scale bar = 1 µm.
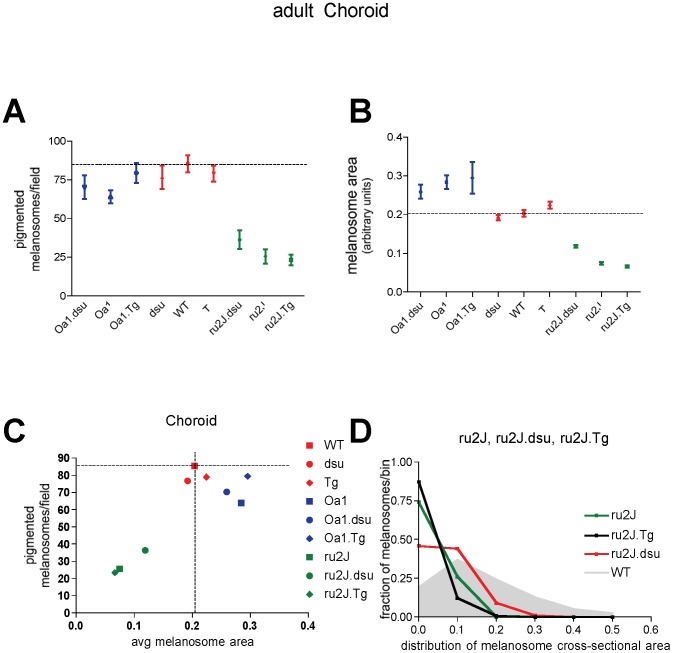
Figure 3. Melanosome density, cross sectional area, and size distribution exhibit a dosage-dependent effect of melanoregulin in the Oa1 null mouse.
Quantitation and analysis of the data shown in Figure 2. Data points are mean values ± SEM. Note the nearly inverse linear relationship between melanoregulin expression and A. Melanosome number and B. Melanosome area on an Oa1 null background. On an Oa1 background, both melanosome number and area show a statistically significant change as a function of melanoregulin expression, with p<0.05 as indicated by asterisks (*). No statistically significant difference was observed in Hps5ru2J mice, p>0.05. Mregdsu mice show a significant decrease in number of melanosomes, p<0.05 with no change in melanosome area, p>0.05. . C. The number of pigmented melanosomes per field is compared to the average melanosomal area in wildtype (C57BL6/J) mice. Abbreviations: ru2J = Hps5ru2J; dsu = Mregdsu; WT = wildtype; Tg = MregTg; Oa1 = Oa1ko.
Lack of HPS5 expression results in a drastic decrease in number, size, and pigmentation of RPE melanosomes in the ru2J mouse (Figures 2B, 3A). By contrast, loss of Oa1 expression results in a dramatic increase in the cross-sectional area of many RPE melanosomes, while others fall within the wildtype size range (Figure 2H). The average size and pigmentation of these melanosomes is greatly increased relative to wildtype, while average number is correspondingly decreased (Figure 3A,B). The normal elongated shape of melanosomes is strikingly lost in the Oa1ko/ko mice. While melanosomes vary in size and longest diameter in approximately the same range as wildtype, very few of the melanosomes are elongated. Those that reach a diameter similar to the longest diameter in wildtype mice are almost invariably round (Figure 2H).
To evaluate how melanoregulin contributes to melanosome biogenesis, we generated mice carrying Hps5ru2J or Oa1ko/ko alleles in combination with either two loss of function or two overexpression alleles of melanoregulin. In the RPE of Hps5ru2J.Mregdsu (ru2J.dsu) double mutant mice, melanosomes are the same size, shape, and density as in ru2J mice; however, some melanosomes appear to be more heavily pigmented (Figures 2A vs. 2B; 3A vs. 3B). In Hps5ru2J.MregTg (ru2J.Tg) mice, we again find little effect on melanosome size or shape; however, the number of pigmented melanosomes is negligible (Figures 2C vs. 2B; 3A,B).
Similarly, in Oa1ko/ko mice, loss of melanoregulin function results in a decrease in melanosome number along with an increase in size of the macromelanosomes compared to Oa1 loss of function alone (Figure 2G vs. 2H; 3A,B). As predicted from the results presented thus far, overexpression of melanoregulin decreases the average cross-sectional area of the largest melanosomes and also results in smaller than normal melanosomes (Figure 2I vs. 2H, 2E; 3A,B). In some cells, as shown in Figure 2I, melanoregulin appears to act in a graded fashion, with a stronger influence on melanosome size in the apical portion of the cell and a weaker influence on basal melanosomes.
Together these results show a consistent association between MREG expression and melanosome biogenesis in the RPE, whereby loss of melanoregulin function facilitates pigment incorporation while overexpression results in reduced melanization. This function of melanoregulin is most pronounced in the presence of another mutation that it either suppresses or enhances, or both, depending on the level of melanoregulin and the function of the other gene. In the case of Hps5ru2J, loss of melanoregulin has little effect, whereas overexpression of melanoregulin exacerbates the lack of melanosome pigmentation (Figure 3C). Conversely, on an Oa1ko/ko background, melanoregulin loss of function greatly worsens the macromelanosome phenotype, while overexpression almost completely rescues the average size and number of RPE melanosomes (Figure 3C).
Because the size distribution of melanosomes in Oa1.Tg RPE appears more uneven than in wildtype RPE, the distribution of melanosome cross-sectional areas was plotted for each of the Oa1-related mutants compared to wildtype (Figure 3D). Based on this view of the data, Oa1ko/ko;MregTg (Oa1.Tg) is shown to have a greater fraction of small melanosomes and also more large melanosomes than wildtype; however, the distribution is closer to wildtype in having a smaller fraction of macromelanosomes than either Oa1ko/ko alone or Oa1ko/ko;Mregdsu (Oa1ko/ko.dsu). The size distribution for melanoregulin loss of function or overexpression alone does not vary significantly from wildtype (not shown; Figure S1).
Melanosome defects in the choroid of HPS5ru2J mice are rescued by loss of melanoregulin function
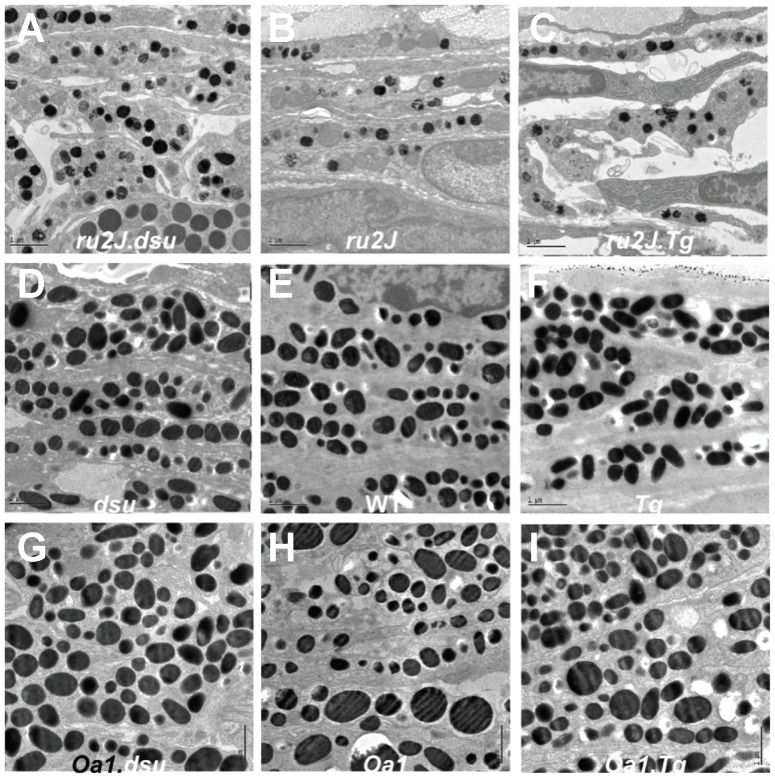
The melanosomes in the choroid of wildtype mice are on average less than half the size (measured as cross-sectional area) of RPE melanosomes and have a smaller size distribution and higher density per cell (Figures 4E and 5A,B). Neither loss nor overexpression of melanoregulin has a substantial effect on size, shape, or density of choroidal melanosomes on an otherwise wildtype background (Figures 4D,F and 5A,B). In the HPS5ru2J mutant, the number and size of choroidal melanosomes are significantly retarded (Figures 4B and 5A,B); melanoregulin overexpression has almost no effect (Figures 4C and 5A,B), while melanoregulin loss of function results in a significant increase in both the size and number of pigmented melanosomes (Figures 4A and 5A,B). In Oa1ko/ko mice, the situation is a mirror image of HPS, where loss of melanoregulin has little effect on size or density of choroidal melanosomes while overexpression results in a slight rescue of melanosome density (Figures 4H,I,J and 5A,B). Overall, the results point to an effect of melanoregulin loss of function facilitating pigment incorporation into maturing melanosomes in the choroid.
Figure 4. Loss of melanoregulin function partially rescues melanosome size and density in the choroid of Hps5ru2J mutant mice.
A–I: Electron microscopic images of the pigment choroid layer in mice of the genotypes indicated. Orientation is with RPE toward the top of each image. Abbreviations: ru2J = Hps5ru2J; dsu = Mregdsu; WT = wildtype; Tg = MregTg ; Oa1 = Oa1ko. Scale bar = 1 µm.
Figure 5. Melanoregulin levels influence melanosome characteristics in the choroid of Hps5ru2J and Oa1 mutant mice.
Quantitation and analysis of the data shown in Figure 4. Data points are mean values ± SEM. A. Melanosome number and B. Melanosome area on an Oa1 null background do not show a statistically significant change as a function of melanoregulin dose; all p values are p>0.05. In contrast, in Hps5ru2J mice, both melanosome number and area increase, p<0.05. Mregdsu mice show a significant increase in melanosome area, p<0.05, with melanoregulin over-expression (MregTg) and a decrease from wildtype in melanoregulin loss of function mice (Mregdsu), p<0.05, with no significant change in number of melanosomes in either case, p>0.05. C. The number of pigmented melanosomes per field is compared to the average melanosomal area in wildtype (C57Bl6/J) mice.
The effects of melanoregulin expression on melanosome biogenesis in the choroid are not as pronounced as in the RPE, in that all three Oa1 mutants are more similar to each other than to melanoregulin loss or overexpression on a wildtype background (Figure 5C). Even the rescue in HPS5ru2J.dsu leaves this mutant with more similarity to HPS5ru2J or Hps5ru2J.Tg than to wildtype in terms of both number and size of melanosomes (Figure 5C). Most prominently, an evaluation of the size distribution of melanosomes in the Hps5ru2J mutants reveals a significant correction in the range of melanosome sizes in Hps5ru2J.dsu compared to either Hpsru2J or Hps5ru2J.Tg (Figure 5D), which is enough to result in significant coloration of the eye (Figure 1A).
Melanoregulin protein localizes to cytoplasmic vesicles in the RPE
Melanoregulin is distributed in the basolateral RPE of wild type mice as shown in Fig. 6A. Within individual RPE cells melanoregulin is localized to small cytoplasmic vesicle-like structures less than 100 µm in diameter (Figure 6B) and [44]. This pattern is similar to that found for transfected HPS3-GFP, recently found to colocalize with cytoplasmic, clathrin-coated vesicles in cultured skin melanocytes [29].
Figure 6. Melanoregulin protein localizes to vesicle-like structures in the RPE cytoplasm.
A: Frozen 7 um sections of 4-month-old mouse retinas were stained with anti-Mreg antibody. Nuclei were visualized with DAPI. Images were captured on a Nikon A1R live cell confocal imaging system. B: ARPE19 cells were fixed and stained with anti-MREG mouse mAb, (Novus Biologics) 1∶250, anti-alpha tubulin rabbit pAb (Thermo) 1∶200, washed and incubated with secondary antibodies: Alexa Fluor 488 donkey anti- mouse IgG (1∶1000), Alexa Fluor 594 donkey anti-rabbit (1∶1000) and Hoechst 33258 (1∶10,000). Images were captured on a Nikon A1R laser scanning confocal microscope with a 100× oil objective and processes using Nikon's Elements software.
Discussion
The reciprocal relationship between the level of melanoregulin expression and melanosome size suggests that this protein may act as a negative regulator of membrane fusion, either directly in the fusion process or indirectly by facilitating the transport of vesicles (Figure 7). This interpretation is consistent with the findings that dsu loss of function alone has no grossly visible phenotype, and that overexpression results in a decrease in melanosome size without loss of pigmentation. While other forms of albinism affect both skin (hair follicles) and retinal pigment, the effects of MREG and OA1 appear to be primarily restricted to the eyes. Changes in melanosomes do occur in hair follicles as well as in eyes of Mregdsu mutants (R. Rachel, unpublished results). However, these changes are less clear because the alterations in hair follicles involve not only melanogenesis but also incorporation of melanin packets into the hair shaft. As such, a specific effect on melanogenesis is not quantifiable.
Figure 7. Model of melanoregulin function in melanosome biogenesis.
These results are consistent with a hypothetical and testable model in which melanoregulin functions as a negative regulator of vesicle fusion with cytoplasmic components required for melanosome biogenesis. The panels in the diagrams are analogous to the panels in Figures 2 and 4, and show both a multivesicular body/endosome and a mature/pigmented melanosome as two stages in the biogenesis of melanosomes. HPS BLOC-2 forms a putative vesicle transport complex, or part of a vesicle system required for moving proteins required for melanosome biogenesis. Melanoregulin acts as a negative regulator of membrane fusion between BLOC-2 and the late endosome/early melanosome. Oa1 acts as a permissive gatekeeper, allowing components of melanosome biogenesis to accumulate up to a certain point, at which time it initiates (or turns off) a signalling cascade that prevents further accumulation of pigment-producing components. These features are compatible with the results presented here, and predict the results of future experiments that can be performed to test the tenets of this model.
Melanoregulin could function in concert with BLOC complexes transporting vesicles to the early melanosome, by preventing these transport vesicles from fusing with developing organelles under certain conditions, e.g., melanosome size or levels of other proteins. Melanoregulin may serve as a linker molecule between the BLOC-2 complex and Oa1, which has been shown to regulate melanosome protein trafficking at early stages of melanosome biogenesis [39]. Localization of Melanoregulin in cytoplasmic vesicles may suggest an interaction with Oa1, which regulates correct targeting of melanosomal protein loaded vesicles to proper stages of melanosomes during their maturation. This hypothesis is supported by a series of findings showing a role for OA1 in melanosome biogenesis via vesicle budding or fusion in the late endosomal compartment [45]–[47]. MREG has been localized to Syn-7 and LAMP-2 rich late endosomal compartments, consistent with a role in this process [44]. In fact, OA1 loss of function causes mislocalization of melanosomal protein Tyrp1 in stage I/II melanosomes causing melanin deposit in not yet mature organelles [39]. This interpretation is consistent with the recent observation that Melanoregulin interacts with RILP (p150glued) [14].
These findings have potential implications for treatment for HPS and OA1 in that inhibition or enhancement of melanoregulin, respectively, may be beneficial in modulating the ocular pigment phenotype of these individuals. What remains to be determined is the extent to which modulating the pigment phenotype in each disorder will correct the visual system defects that accompany it. Interestingly, the melanosomes in both HPS and OA1, irrespective of being undersized or oversized, cause similar defects in the neural retina to complete lack of pigment due to loss of the melanogenic enzyme tyrosinase (for review, see [7]); this suggests that the melanosome is required for signaling to the neural retina and that normal melanosome function is important for this signaling, not just the presence or absence of pigment [48]. This places the melanosome in a more dynamic role: that of active participant in the function of the RPE, rather than being a passive medium for light absorption. A polyacetylene, melanin in intact melanosomes has been shown to exhibit semi-conductor properties, emitting a flash of light when it switches from low to high conductivity [49]. This feature suggests that the conversion of electrical energy to heat or light may provide a novel mechanism for activation or deactivation of biological processes. Generalizing the findings with HPS and melanoregulin linking to OA1 as a size sensor in melanosome biogenesis, we might expect other G-protein coupled receptors to play similar roles in the formation of other intracellular organelles.
Experimental Procedures
Materials
Commercially available antibodies were purchased as follows: anti-MREG mouse mAb, (Novus Biologics) 1∶250; anti-alpha tubulin rabbit pAb (Thermo) 1∶200; AlexaFluor 488 donkey anti- mouse IgG (1∶1000); AlexaFluor 594 donkey anti-rabbit (1∶1000) and Hoechst 33258 (1∶10,000).
Generation of mouse lines
dsu/dsu, dv/dv mice [11] were obtained from Jackson Laboratories and have been maintained at NCI-Frederick for 90+ generations. To obtain mice carrying only the dsu mutation, dsu/dsu, dv/dv mice were backcrossed to C57Bl/6J for 10 generations, followed by intercrossing and selecting for mice wildtype at the dilute locus and carrying dsu/dsu. Some of the data shown here derives from mice homozygous for dsu at the N5 backcross generation while the rest is from the N10 backcross line. Crosses between dsu and ru2J were carried out with B6.dsu mice at the N5 generation, while crosses to Oa1 were done with B6.dsu mice at the N10 generation. Mice were genotyped using the following primers: Primers for dv: Right, common b7: TCCTCTGTGGTCATCACTGG, Left, wildtype: TGGAATCCCAGCAGTGGTA, 179 bp product. dv, Left, viral allele: CCCGTGTATCCAATAAAGCC, 198 bp product. Primers for dsu: Right, common dsu5′brk-f27 CTGGGAGTTCAAGGTTGGTCTG, Left wildtype, dsu5′brk-b24 GCAGGAGAGGCTGGGAAAAAAC, 230 bp product wildtype, Left dsu, dsu3′brk-b CCACAGTCTCAAGTCTTTCCTG, dsu ∼140 bp. Transgenic mice overexpressing wildtype dsu (in the text, Tg) were generated as described [1]. dsu and Tg alleles were separately bred onto lines of mice carrying either ru2J (HPS5), ru (HPS6), or coa (HPS3) mutations, or onto the B6. Oa1ko/ko knockout line. Ru2J/ru2J mice were selected based on coat color, while Oa1ko/ko mice were genotyped according to [40]. Mice were housed under cyclic light conditions: 12-h light/12-h dark and fed ad libitum. All procedures involving animals were in accordance with institutional ACUC-approved protocols and with the Association for Research in Vision and Ophthalmology (ARVO) guidelines for use of animals.
Electron microscopy
Standard procedures for thin-section transmission electron microscopy (TEM) are previously described in detail [50]. A few minor modifications of the procedure were used for mouse eyes. Adult mice were perfused with 0.1 M phosphate buffered saline followed by 2% glutaraldehyde/4% paraformaldehyde fixative in cacodylate buffer (0.1 M, pH. 7.4). The eyes were dissected out. After post-fixation, the cornea and lens were removed, marking the orientation with a slit at the ventral pole. Eyes were rinsed thoroughly with cacodylate buffer and post-fixed in 1% osmium for 1 hr at room temperature, then dehydrated in graded ethanol (e.g., 35%, 50%, 70%, 95%, and 100%) and propylene oxide (100%). Eyes were infiltrated overnight in a 1∶1 mixture of propylene oxide and epoxy resin and embedded in pure resin and cured at 55°C for 48 hrs. The cured block was trimmed and made semi-thin sections (0.5 µm) and stained in Toluidine Blue-O solution to determine proper orientation for thin-section EM analysis. Thin-sections (60 nm) were mounted on naked 200-mesh copper grids, and stained in uranyl acetate and lead citrate. For measuring the number and size of melanosomes, RPE and choroid were imaged on a Hitachi H7000 (Tokyo, Japan) transmission electron microscope at 3000× and images obtained with a Gatan (Pleasanton, CA) digital camera and imaging software. For adults, counts were made on a 40 µm2 area on 10–20 images from each of 2–3 animals of each genotype. Counts on P5, P16, and P28 animals were done on 10 or more images from one animal of each genotype at each age.
MREG localization in retinal sections and human ARPE19 cells
Immunohistochemistry was performed on frozen sections of 4-month-old mouse retinas. The eyecups were fixed in 4% paraformaldehyde in PBS (pH 7.4) overnight at 4°C, cryoprotected in 30% sucrose, and embedded in OCT. The 7 um sections were stained with anti-MREG mAb (Novus Biologicals) at 1∶200, washed 3× and stained with secondary antibody Alexa 488 anti mouse, dilution 1∶500. Controls were incubated with the secondary antibodies only. Nuclei were visualized with DAPI. Images were captured on a Nikon A1R live cell confocal imaging system. Data were analyzed using Nikon Elements AR Software 3.2.
ARPE19 cells grown on glass coverslips for 48 hours were rinse in PBS and then fixed and permeabilized in 4% buffered paraformaldehyde, with 0.2% Triton X100 for 10 minutes at room temperature. After three successive washes in PBS with 0.05%Triton X100 (PBST) the coverslips were blocked in PBST containing 4% Bovine serum albumin for 60 mins at 37°C. After blocking, coverslips were incubated at 37°(-include degree symbol)C for 60 minutes with primary antibodies: anti-MREG mouse mAb, (Novus Biologics) 1∶250, anti-alpha tubulin rabbit pAb (Thermo) 1∶200. After the incubation with primary antibodies, the coverslips were washed 3× in PBST and then incubated with secondary antibodies: Alexa Fluor 488 donkey anti-mouse IgG (1∶1000), Alexa Fluor 594 donkey anti-rabbit (1∶1000) and Hoechst 33258 (1∶10,000) at 37°C for 60 minutes. Three successive PBST washes followed the incubation with secondary antibodies. In preparation for microscopy, the coverslips were mounted in Cytoseal (Electron Microscopy Sciences). Images were captured on a Nikon A1R laser scanning confocal microscope with a 100× oil objective and processes using Nikon's Elements software.
Statistical Analysis
Data are reported as the mean ± SEM with statistical analysis using a one-way ANOVA with appropriate post-hoc test. Results with a p<0.05 were considered significant. Significance of analyses is indicated in the figure legends.
Supporting Information
Melanoregulin loss of function or overexpression have little effect on the size distribution of melanosomes in either the RPE or choroid. Data quantitated as shown in Figures 3D and 5D. The shape of the curves is nearly identical in wildtype, melanoregulin loss of function, and in melanoregulin overexpression. Data shown are an average ± SEM; differences are not statistically significant with p>0.05.
(TIFF)
Acknowledgments
We thank the following individuals for expert technical assistance, with animals: Deborah Swing, Erika Truffer, Stephanie Springer, Holly Morris and Rob Koogle; with genotyping: Deborah Householder and Anna Trivett; with electron microscopy: Michelle Gignac; Alvina Bragin for expert technical assistance. We thank the Vision of Children for facilitating this collaboration and for funding productive meetings, and Drs. Nancy Jenkins and Neal Copeland for supporting the majority of the work in their laboratory at the National Cancer Institute.
Funding Statement
This project was funded by the Intramural Research Program of the National Institutes of Health, National Cancer Institute, Center for Cancer Research, extramural grants from the National Eye Institute, EY-10420 and EY018705 (KBB), federal funds from the National Cancer Institute under contract HHSN26120080001E (KN), and by the Vision of Children Foundation (VM). The funders had no role in study design, data collection and analysis, decision to publish or preparation of the manuscript.
References
- 1. O'Sullivan TN, Wu XS, Rachel RA, Huang JD, Swing DA, et al. (2004) dsu functions in a MYO5A-independent pathway to suppress the coat color of dilute mice. Proc Natl Acad Sci U S A 101: 16831–16836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Matesic LE, Yip R, Reuss AE, Swing DA, O'Sullivan TN, et al. (2001) Mutations in Mlph, encoding a member of the Rab effector family, cause the melanosome transport defects observed in leaden mice. Proc Natl Acad Sci USA 98: 10238–10243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Wilson SM, Yip R, Swing DA, O'Sullivan TN, Zhang Y, et al. (2000) A mutation in Rab27a causes the vesicle transport defects observed in ashen mice. Proc Natl Acad Sci USA 97: 7933–7938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Di Pietro SM, Dell'Angelica EC (2005) The cell biology of Hermansky-Pudlak syndrome: recent advances. Traffic 6: 525–533. [DOI] [PubMed] [Google Scholar]
- 5. Gunay-Aygun M, Huizing M, Gahl W (2004) Molecular defects that affect platelet dense granules. Semin Thromb Hemost 30: 537–547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Schnur RE, Wick PA, Bailey C, Rebbeck T, Weleber RG, et al. (1995) Phenotypic variability in X-linked ocular albinism: relationship to linkage genotypes. Am J Hum Genet 1994 55: 484–496. [PMC free article] [PubMed] [Google Scholar]
- 7. Oetting WS, King RA (1999) Molecular basis of albinism: mutations and polymorphisms of pigmentation genes associated with albinism. Hum Mutat 13: 99–115. [DOI] [PubMed] [Google Scholar]
- 8. Young A, Jiang M, Wang Y, Ahmedli NB, Ramirez J, et al. (2011) Specific interaction of Galphai3 with the Oa1 G-protein coupled receptor controls the size and density of melanosomes in retinal pigment epithelium. PloS one 6: e24376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Moore KJ, Swing DA, Rinchik EM, Mucenski ML, Buchberg AM, et al. (1988) The murine dilute suppressor gene dsu suppresses the coat-color phenotype of three pigment mutations that alter melanocyte morphology, d, ash and ln. Genetics 119: 933–941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Moore KJ, Seperack PK, Strobel MC, Swing DA, Copeland NG, et al. (1988) Dilute suppressor dsu acts semidominantly to suppress the coat color phenotype of a deletion mutation, dl20J, of the murine dilute locus. Proc Natl Acad Sci USA 85: 8131–8135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Sweet HO (1983) Dilute suppressor, a new suppressor gene in the house mouse. J Hered 74: 305–306. [DOI] [PubMed] [Google Scholar]
- 12. Wu X, Wang F, Rao K, Sellers JR, Hammer JA 3rd (2002) Rab27a is an essential component of melanosome receptor for myosin Va. Mol Biol Cell 13: 1735–1749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Wu XS, Rao K, Zhang H, Wang F, Sellers JR, et al. (2002) Identification of an organelle receptor for myosin-Va. Nat Cell Biol 4: 271–278. [DOI] [PubMed] [Google Scholar]
- 14. Ohbayashi N, Maruta Y, Ishida M, Fukuda M (2012) Melanoregulin regulates retrograde melanosome transport through interaction with the RILP-p150Glued complex in melanocytes. Journal of Cell Science 125: 1508–1518. [DOI] [PubMed] [Google Scholar]
- 15. Anikster Y, Huizing M, White J, Shevchenko YO, Fitzpatrick DL, et al. (2001) Mutation of a new gene causes a unique form of Hermansky-Pudlak syndrome in a genetic isolate of central Puerto Rico. Nat Genet 28: 376–380. [DOI] [PubMed] [Google Scholar]
- 16. Gardner JM, Wildenberg SC, Keiper NM, Novak EK, Rusiniak ME, et al. (1997) The mouse pale ear (ep) mutation is the homologue of human Hermansky-Pudlak syndrome. Proc Natl Acad Sci USA 94: 9238–9243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gwynn B, Ciciotte SL, Hunter SJ, Washburn LL, Smith RS, et al. (2000) Defects in the cappuccino (cno) gene on mouse chromosome 5 and human 4p cause Hermansky-Pudlak syndrome by an AP-3-independent mechanism. Blood 96: 4227–4235. [PubMed] [Google Scholar]
- 18. Gwynn B, Martina JA, Bonifacino JS, Sviderskaya EV, Lamoreux ML, et al. (2004) Reduced pigmentation (rp), a mouse model of Hermansky-Pudlak syndrome, encodes a novel component of the BLOC-1 complex. Blood 104: 3181–3189. [DOI] [PubMed] [Google Scholar]
- 19. Suzuki T, Li W, Zhang Q, Karim A, Novak EK, et al. (2002) Hermansky-Pudlak syndrome is caused by mutations in HPS4, the human homolog of the mouse light-ear gene. Nat Genet 30: 321–324. [DOI] [PubMed] [Google Scholar]
- 20. Suzuki T, Li W, Zhang Q, Novak EK, Sviderskaya EV, et al. (2001) The gene mutated in cocoa mice, carrying a defect of organelle biogenesis, is a homologue of the human Hermansky-Pudlak syndrome-3 gene. Genomics 78: 30–37. [DOI] [PubMed] [Google Scholar]
- 21. Suzuki T, Oiso N, Gautam R, Novak EK, Panthier JJ, et al. (2003) The mouse organellar biogenesis mutant buff results from a mutation in Vps33a, a homologue of yeast vps33 and Drosophila carnation. Proc Natl Acad Sci USA 100: 1146–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Zhang Q, Li W, Novak EK, Karim A, Mishra VS, et al. (2002) The gene for the muted (mu) mouse, a model for Hermansky-Pudlak syndrome, defines a novel protein which regulates vesicle trafficking. Hum Mol Genet 11: 697–706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Zhang Q, Zhao B, Li W, Oiso N, Novak EK, et al. (2003) Ru2 and Ru encode mouse orthologs of the genes mutated in human Hermansky-Pudlak syndrome types 5 and 6. Nat Genet 33: 145–153. [DOI] [PubMed] [Google Scholar]
- 24. Zhen L, Jiang S, Feng L, Bright NA, Peden AA, et al. (1999) Abnormal expression and subcellular distribution of subunit proteins of the AP-3 adaptor complex lead to platelet storage pool deficiency in the pearl mouse. Blood 94: 146–155. [PubMed] [Google Scholar]
- 25. Nguyen T, Novak EK, Kermani M, Fluhr J, Peters LL, et al. (2002) Melanosome morphologies in murine models of hermansky-pudlak syndrome reflect blocks in organelle development. J Invest Dermatol 119: 1156–1164. [DOI] [PubMed] [Google Scholar]
- 26. Nguyen T, Wei ML (2004) Characterization of melanosomes in murine Hermansky-Pudlak syndrome: mechanisms of hypopigmentation. J Invest Dermatol 122: 452–460. [DOI] [PubMed] [Google Scholar]
- 27. Di Pietro SM, Falcon-Perez JM, Dell'Angelica EC (2004) Characterization of BLOC-2, a complex containing the Hermansky-Pudlak syndrome proteins HPS3, HPS5 and HPS6. Traffic 5: 276–283. [DOI] [PubMed] [Google Scholar]
- 28. Gautam R, Chintala S, Li W, Zhang Q, Tan J, et al. (2004) The Hermansky-Pudlak syndrome 3 (cocoa) protein is a component of the biogenesis of lysosome-related organelles complex-2 (BLOC-2). J Biol Chem 279: 12935–12942. [DOI] [PubMed] [Google Scholar]
- 29. Helip-Wooley A, Westbroek W, Dorward H, Mommaas M, Boissy RE, et al. (2005) Association of the Hermansky-Pudlak syndrome type-3 protein with clathrin. BMC Cell Biol 6: 33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Theos AC, Tenza D, Martina JA, Hurbain I, Peden AA, et al. (2005) Functions of adaptor protein (AP)-3 and AP-1 in tyrosinase sorting from endosomes to melanosomes. Mol Biol Cell 16: 5356–5372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Hearing VJ (2005) Biogenesis of pigment granules: a sensitive way to regulate melanocyte function. J Dermatol Sci 37: 3–14. [DOI] [PubMed] [Google Scholar]
- 32. Bassi MT, Incerti B, Easty DJ, Sviderskaya EV, Ballabio A (1996) Cloning of the murine homolog of the ocular albinism type 1 (OA1) gene: sequence, genomic structure, and expression analysis in pigment cells. Genome Res 6: 880–885. [DOI] [PubMed] [Google Scholar]
- 33. Newton JM, Orlow SJ, Barsh GS (1996) Isolation and characterization of a mouse homolog of the X-linked ocular albinism (OA1) gene. Genomics 37: 219–225. [DOI] [PubMed] [Google Scholar]
- 34. Schiaffino MV, Baschirotto C, Pellegrini G, Montalti S, Tacchetti C, et al. (1996) The ocular albinism type 1 gene product is a membrane glycoprotein localized to melanosomes. Proc Natl Acad Sci USA 93: 9055–9060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Schiaffino MV, d'Addio M, Alloni A, Baschirotto C, Valetti C, et al. (1999) Ocular albinism: evidence for a defect in an intracellular signal transduction system. Nat Genet 23: 108–112. [DOI] [PubMed] [Google Scholar]
- 36. Staleva L, Orlow SJ (2006) Ocular albinism 1 protein: trafficking and function when expressed in Saccharomyces cerevisiae. Exp Eye Res 82: 311–318. [DOI] [PubMed] [Google Scholar]
- 37. Innamorati G, Piccirillo R, Bagnato P, Palmisano I, Schiaffino MV (2006) The melanosomal/lysosomal protein OA1 has properties of a G protein-coupled receptor. Pigment Cell Res 19: 125–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Lopez VM, Decatur CL, Stamer WD, Lynch RM, McKay BS (2008) L-DOPA is an endogenous ligand for OA1. PLoS Biol 6: e236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Giordano F, Bonetti C, Surace EM, Marigo V, Raposo G (2009) The ocular albinism type 1 (OA1) G-protein-coupled receptor functions with MART-1 at early stages of melanogenesis to control melanosome identity and composition. Human Molecular Genetics 18: 4530–4545. [DOI] [PubMed] [Google Scholar]
- 40. Incerti B, Cortese K, Pizzigoni A, Surace EM, Varani S, et al. (2000) Oa1 knock-out: new insights on the pathogenesis of ocular albinism type 1. Hum Mol Genet 9: 2781–2788. [DOI] [PubMed] [Google Scholar]
- 41. Cortese K, Giordano F, Surace EM, Venturi C, Ballabio A, et al. (2005) The ocular albinism type 1 (OA1) gene controls melanosome maturation and size. Invest Ophthalmol Vis Sci 46: 4358–4364. [DOI] [PubMed] [Google Scholar]
- 42. LaVail JH, Nixon RA, Sidman RL (1978) Genetic control of retinal ganglion cell projections. J Comp Neurol 182: 399–421. [DOI] [PubMed] [Google Scholar]
- 43. Balkema GW, Drager UC (1990) Origins of uncrossed retinofugal projections in normal and hypopigmented mice. Vis Neurosci 4: 595–604. [DOI] [PubMed] [Google Scholar]
- 44. Damek-Poprawa M, Diemer T, Lopes VS, Lillo C, Harper DC, et al. (2009) Melanoregulin (MREG) modulates lysosome function in pigment epithelial cells. J Biol Chem 284: 10877–10889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Samaraweera P, Shen B, Newton JM, Barsh GS, Orlow SJ (2001) The mouse ocular albinism 1 gene product is an endolysosomal protein. Exp Eye Res 72: 319–329. [DOI] [PubMed] [Google Scholar]
- 46. Shen B, Rosenberg B, Orlow SJ (2001) Intracellular distribution and late endosomal effects of the ocular albinism type 1 gene product: consequences of disease-causing mutations and implications for melanosome biogenesis. Traffic 2: 202–211. [DOI] [PubMed] [Google Scholar]
- 47. Shen B, Samaraweera P, Rosenberg B, Orlow SJ (2001) Ocular albinism type 1: more than meets the eye. Pigment Cell Res 14: 243–248. [DOI] [PubMed] [Google Scholar]
- 48. Rachel RA, Dolen G, Hayes NL, Lu A, Erskine L, et al. (2002) Spatiotemporal features of early neuronogenesis differ in wild-type and albino mouse retina. J Neurosci 22: 4249–4263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. McGinness J, Corry P, Proctor P (1974) Amorphous semiconductor switching in melanins. Science 183: 853–855. [DOI] [PubMed] [Google Scholar]
- 50. Tobin GJ, Nagashima K, Gonda MA (1996) Immunologic and Ultrastructural Characterization of HIV Pseudovirions Containing Gag and Env Precursor Proteins Engineered in Insect Cells. Methods 10: 208–218. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Melanoregulin loss of function or overexpression have little effect on the size distribution of melanosomes in either the RPE or choroid. Data quantitated as shown in Figures 3D and 5D. The shape of the curves is nearly identical in wildtype, melanoregulin loss of function, and in melanoregulin overexpression. Data shown are an average ± SEM; differences are not statistically significant with p>0.05.
(TIFF)