Abstract
We describe the application of a novel HIV confirmatory testing algorithm to determine the primary efficacy endpoint in a large Phase III microbicide trial. 9385 women were enrolled between 2005 and 2009. Of these women, 537 (6%) had at least one positive HIV rapid test after enrolment. This triggered the use of the algorithm which made use of archived serum and Buffy Coat samples. The overall sample set was >95% complete. 419 (78%) of the rapid test positive samples were confirmed as primary endpoints using a combination of assays for the detection of HIV-specific antibodies (EIA's and Western Blot), and for components of the virus itself (PCR for the detection of nucleic acids and EIA for p24 antigen). 63 (12%) cases were confirmed as being HIV-positive at screening or enrolment and 55 (10%) were confirmed as HIV negative. The testing algorithm confirmed the endpoint at the same visit as that of the first positive rapid test in 90% of cases and at the time of the preceding visit in 10% of cases. Of the 63 cases which were subsequently confirmed to be HIV-1 positive at or before enrolment, 54 specimens contained no detectable HIV antibodies at screening or enrolment. However, 43 were positive using an EIA which detects both HIV antigen and antibody and also had a positive p24 antigen or HIV PCR test, which was highly suggestive of acute infection. There were 6 unusual cases which had undetectable HIV-1 DNA or RNA. In 4 of the 6 cases the presence of HIV-1-specific antibodies was confirmed by Western Blot. One of these cases with an indeterminate Western Blot was a previous vaccine trial participant. The algorithm served the objectives of the study well and can be recommended for use in determining HIV as an endpoint in clinical trials.
Trial Registration
ISRCTN.org ISRCTN 64716212
Introduction
Accurate and reliable detection of HIV infection is of obvious intrinsic benefit but is also crucial to ensure the validity and quality of clinical trials in which HIV infection is an endpoint. The Centre for Disease Control (CDC) criteria define HIV infection status on the basis of repeated reactive Enzyme Immunoassays (EIA) and a positive confirmatory Western Blot (WB) or Immunofluoresence Assay (IFA) for the detection of specific HIV antibodies [1]. This type of algorithm is commonly employed in countries where the prevalence of HIV is low (<1%).
CDC and World Health Organization (WHO) guidelines advocate the use of rapid tests in a clinic setting for the purposes of cost-effective patient management [1]–[5]. It is also recommended that newly diagnosed cases have a second sample collected and tested in order to exclude clerical or technical errors. P24 antigen (P24 Ag) testing and HIV viral load (VL) testing do not constitute primary diagnostic tests, but are useful supplementary tests in helping to resolve inconclusive serological results.
For settings in which resources are limited, the WHO advocates the use of serial rapid tests for the detection of HIV infection [3]. This applies to many parts of Africa, where the necessary infrastructure and skills required for laboratory based assays such as Western blotting and PCR are limited, and it is only feasible to use less technically demanding assays. Areas with the highest prevalence of HIV are frequently poor in resources and, provided assay performance has been verified, reactive results obtained using 2 different rapid tests is routinely used to indicate evidence of HIV infection.
“Parallel testing”, in which 2 rapid tests are carried out concurrently, has also been advocated [3]. This strategy for HIV screening is popular in the clinical trial setting as the method contains intrinsic quality control and the extent of result concordance can be used to evaluate accuracy. This approach can be used to trigger further evaluation of discordant results, as these may indicate very early seroconversion.
Many laboratories in Africa offer HIV testing using EIA. These methods can supplement the Rapid tests carried out in clinical settings both for confirmatory testing purposes or to resolve the results of inconclusive rapid tests. Third generation HIV EIAs (3rd gen HIV EIA) detect HIV-specific IgM antibodies which typically develop 3–4 weeks after infection [6], [7]. More recently developed fourth generation HIV EIAs (4th gen HIV EIA), detecting both free p24 Ag and Ag complexed with specific antibodies (Ag/Ab), have enabled detection of infection up to 2 weeks earlier [8], [9]. Ly et al. showed that this relatively superior performance can be explained by an ability to detect very low concentrations of P24 Ag [9]. Ag/Ab assays are now recommended for use in first-line screening in many areas of the world, although only one has been approved by the United States Food and Drug Administration (FDA) to date [10], [11] . There is concern about the performance of some EIAs [12]–[15] and rapid tests [16]–[18] currently in use in certain parts of Africa. False positive results have been reported, and these have been shown to be particularly associated with early generation assays and cross reactions with other infectious agents and immune responses to them.
In a recent study in adolescents in Tanzania, Everett and colleagues explored the basis for false positive results which they obtained using the 4th generation Murex Ag/Ab EIA [15]. After accounting for clinical, sociological and immunological variables, conditional logistic regression showed that false positivity was strongly associated with levels of specific IgG antibodies against Schistosoma spp and also with Rheumatoid factor (RF) suggesting cross reactivity. Trypanosomiasis and Leishmaniasis have also been similarly associated with false positive serology [19], [20]. It is therefore important to evaluate assays locally, to use assays with acceptable sensitivity and specificity and to use more specific assays in a confirmatory algorithm.
It is now considered highly desirable to test for acute HIV infection; both for reasons of public health and individual patient management. The acutely infected population is believed to be a major driver of new infections, as the viral load is particularly high during early stages of infection before the viral set point is reached [21]. Currently, the inclusion of PCR assays for the detection of HIV RNA in plasma or HIV DNA in cells (Nucleic acid amplification -NAAT) is advised as there is a period of about 3 to 4 weeks after infection when HIV-1 specific antibodies are undetectable and HIV rapid or EIA tests are uninformative [22]–[25].
The detection of viral nucleic acid is more sensitive and cost-effective than the detection of p24 Ag [26], [27]. Although molecular assays are not considered as “gold standard” diagnostic assays for HIV diagnosis in adults by the CDC, they are playing an increasingly important role in the field of HIV diagnosis, especially for the detection of acute infection and/or the resolution of inconclusive antibody results.
MDP 301 was a phase 3, multi-centre, randomized, double-blind, placebo-controlled trial to evaluate the efficacy and safety of 0.5% and 2% PRO 2000/5 microbicide gels. The study commenced in October 2005 and was completed end of September 2009. This trial is registered at http://isrctn.org, number ISRCTN 64716212 (grant number G0100137).
The trial was conducted at 6 research institutions in Africa: 3 in South Africa (Johannesburg, Durban and Mtubatuba) and one each in Zambia (Mazabuka), Tanzania (Mwanza) and Uganda (Masaka). A cohort of 9385 HIV negative women were enrolled and followed up for 52 weeks at all sites, except in Uganda, where women were followed-up for 2 years. The primary objective of the trial was to assess the safety and efficacy of both 2% and 0.5% PRO2000 gel in reducing vaginally acquired HIV infection. The results of the study have been published [28]. In February 2008, the 2% arm was discontinued for reasons of no benefit, when data was analyzed by the independent data monitoring committee. In this paper the HIV endpoint data is presented and includes results from the 2% arm. An HIV endpoint was defined as having occurred during the trial when a participant was confirmed to be HIV uninfected at enrolment by having a negative HIV EIA and PCR test and subsequently determined as HIV positive according to the algorithm at any follow up visit on the basis of the testing of 2 different samples.
The Medical Research Council (MRC) Clinical Trials Unit in London contracted “Contract Laboratory Services” (CLS), linked to the School of Pathology of the University of the Witwatersrand in Johannesburg South Africa, to function as the central laboratory. The key functions of CLS were: HIV endpoint confirmation, overall quality assurance and Good Clinical and Laboratory Practise (GCLP) implementation at local laboratories.
Methods
Ethics Statement
The main paper for this study has already been published [28]: “The protocol was approved by local and national ethics committees, in all participating countries and in the UK. Authorisation was obtained from the national regulatory authority in all participating countries and the US Food and Drug Administration. Participants indicated their consent by signature or witnessed thumbprint.”
Participants and design
MDP301 was a phase 3, randomised, double-blind, and parallel-group trial. Full details of trial design, sample size, research sites, study populations, study conduct including the randomization and masking and data underpinning the sample size calculations have been reported elsewhere [29]. Participants were enrolled by 6 research institutions in Africa (three in South Africa and one each in Tanzania, Uganda and Zambia). Details of eligibility are described elsewhere [28] but the importance of this analysis required participants to be HIV negative at enrolment.
For the purposes of this study, all data obtained from the algorithmic testing was captured in an Excel format. The statistical approach was based on working out percentages for the testing outcomes. The denominators were total number of enrolled participants per site. The data were held in a single centralized database, were analyzed descriptively and presented as proportions.
HIV rapid testing and local sample storage
HIV screening was carried out using parallel rapid testing at all but two sites (Table 1).
Table 1. Summary of rapid tests used by MDP Centres.
| Assay | Approval | Centre |
| Determine HIV 1/2 (Abbott Laboratories, Illinois, USA) | WHO | All sites |
| Uni-Gold HIV test (Trinity Biotech, Wicklow, Ireland) | WHO | JHB, Mtubatuba |
| OraQuick Advance HIV 1/2 (OraSure Technologies, Bethlehem USA) | FDA | Durban |
| Capillus HIV-1/HIV-2 (Trinity Biotech, Wicklow, Ireland) | WHO | Tanzania |
| Genie II HIV-1/HIV-2 (BioRad) | WHO | Zambia |
Abbreviations: JHB – Johannesburg.
Tanzania was in the progress of validating rapid testing [30], and was still relying on laboratory based EIA testing for screening purposes when the study commenced. In Uganda confirmation of a single positive rapid test was done by HIV EIA and HIV WB testing due to the poor specificity of HIV rapid tests reported in that geographical location [31]. The criteria for the selection of suitable HIV rapid tests were that all sites had to use WHO [32] or FDA [33] approved HIV rapid tests that were validated in each site. Before the study commenced an HIV testing validation exercise was performed. All sites were subjected to testing of a 30 member blinded panel (15 positive and 15 negative samples) using their HIV rapid and EIA kits. This panel was prepared by the NICD (National Institute for Communicable Diseases, South Africa) and was characterized using a panel of HIV EIAs, rapid tests, HIV-1 WB for the detection of HIV 1/2 antibodies, and an EIA for p24 Ag testing. The study sites had to achieve a score of > = 29/30 to qualify for competence for on-site HIV rapid and EIA testing.
HIV rapid testing was conducted at screening, weeks 12, 24, 40 and 52 (and weeks 66, 76, 88, 100 and 104 in Uganda). Serum samples were obtained up to 6 weeks before enrolment, at enrolment and then at weeks 4,12,24,40 and 52 (and week 104 in Uganda) and stored at −20°C at the local laboratory repository. Buffy Coat (BC) was collected and stored at −70°C at enrolment, weeks 24, 40, 52 (for all sites except Uganda) and at week 104 (Uganda only), (Table 2).
Table 2. Schedule of visits and corresponding blood draws.
| Week | −6 | 0 | 4 | 12 | 24 | 40 | 52 | 662 | 762 | 882 | 1002 | 1042 |
| Rapids | X | X | X | X | X | X | X | X | X | X | ||
| Serum storage | X | X | X | X | X | X | X1 | X | X | |||
| BC storage | X | X | X | X | X | X |
All sites except Uganda.
Uganda only weeks 66 through to 104.
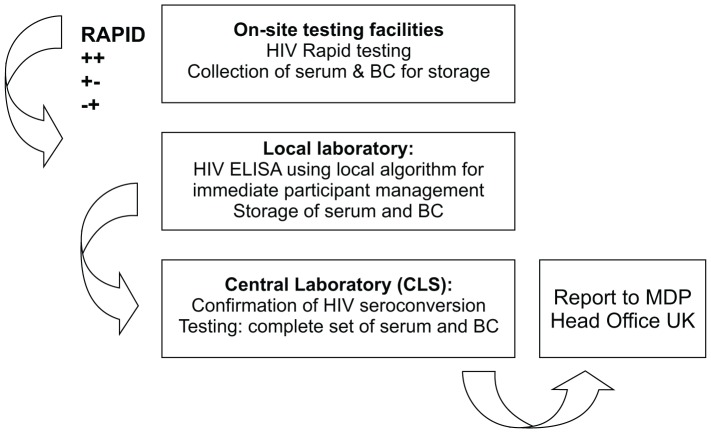
Women who tested positive at screening had this status confirmed using the local country-specific algorithm and were classified as “not eligible” to join MDP301. Women with a positive or discordant rapid test after enrolment triggered the MDP 301 algorithm (Figure 1).
Figure 1. Overview of the HIV testing procedures.
This figure describes the diagnostic tasks for the clinic, local laboratory and central laboratory.
Confirmation of HIV status
The algorithm was triggered by a positive rapid test result at time after enrolment. The complete set samples (serum and BC) collected during any scheduled or unscheduled visit from screening up to and including the visit which triggered the algorithm were shipped on dry ice from the local laboratory repository to CLS (Figure 2). If HIV infection was confirmed using this set of samples, a second specimen was requested. This second (serum) sample was used to verify the seroconversion and was collected at the visit subsequent to the one which had triggered the algorithm.
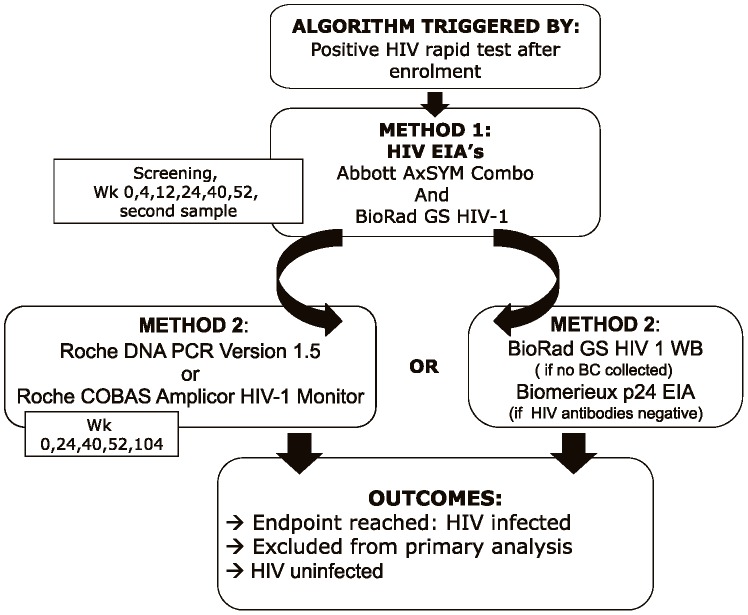
Figure 2. Confirmatory HIV testing algorithm used for MDP301.
This figure details the diagnostic methods used at the central laboratory for all potential seroconverters and the possible outcomes.
MDP301 used a novel algorithm for the detection of HIV infection.The algorithm had to be able to confirm HIV infection based on two separate samples. At each endpoint, 2 different methods of diagnosis had to be available: 2 different EIAs and a confirmatory positive HIV qualitative DNA PCR, p24 EIA or HIV-1 WB. The WB was utilized as a second method of confirmation for those visits at which no BC was collected for DNA PCR (at week 4 and 12 for all sites; week 52, 76, for Uganda only). Where the BC was negative or failed to give a result (due to poor sample quality) a VL assessment was performed on the residual serum sample. The algorithm had to also allow for the detection of acute HIV infection to reliably diagnose those cases at or before enrolment.
At CLS, serum samples were tested for HIV-1 antibodies with Abbott AxSYM HIV Ag/Ab Combo (Wiesbaden, Germany), Bio-Rad HIV-1 Genetic Systems rLAV EIA (Redmond, USA), and Bio-Rad Genetic Systems HIV-1 Western Blot (Redmond, USA) assays. We used Biomerieux Vironostika HIV-1 antigen EIA (Boxtel, Netherlands) as a confirmatory assay for p24 testing. BC samples were tested for the presence of HIV-1 DNA using the Roche qualitative DNA PCR Version 1.5 assay (Roche Diagnostic Systems Inc, Branchburg, NJ, US). The Roche COBAS Amplicor HIV-1 Monitor (Roche Diagnostic Systems Inc, Branchburg, NJ, US) was used for the detection of HIV-1 RNA if the BC specimen was not satisfactory. A second serum sample was drawn for serological confirmation of the result at the next visit if HIV infection was indicated after the first round of testing.
The amplification of DNA over RNA was preferred because DNA is intrinsically more stable and easier to prepare and ship. In the South African setting this assay has a sensitivity and specificity of 99% and 99.8% respectively for HIV diagnosis in infants [34].
HIV seroconversion was defined as having occurred during the trial when a participant tested negative by rapid test, HIV EIA and PCR at enrolment and subsequently tested positive according to the algorithm at any follow up visit. An acute HIV infection was defined as a positive PCR or 4th generation HIV EIA (Abbott AxSYM Combo) with a positive p24 result in the absence of HIV antibodies using the rapid test or 3rd generation HIV EIA (Bio-Rad EIA). All results were peer reviewed by a panel of diagnostic experts, constituting the “HIV endpoint committee”. This included an independent expert, who was not involved in the study.
Results
From October 2005 through to September 2009, samples from 537 participants were positive by rapid test at a follow up visit after enrolment. Over this period, 2946 serum samples and 1332 BC samples were shipped to CLS. No major problems were encountered with the shipping processes that could have impacted on sample viability. The samples were more than 95% complete: all 537 sets of samples were received in good condition. 17 of 419 expected second samples were not collected. This was typically because the participant was lost to follow-up. 6% (n = 74) of the BC samples were either inadequate (insufficient BC or a clotted sample) or because an incorrect sample type was collected. At one site for example, 40 plasma samples were collected instead of cells in error.
Of the 537 participants, who had positive rapid tests after enrolment, 66 were discordant and 471 were positive on both rapid tests. The stored sera and BC samples from all were tested according to the HIV confirmatory algorithm: 419 (78%) were confirmed as endpoints, 55 (10%) were confirmed to be HIV uninfected and 63 (12%) were confirmed as HIV infected at screening or enrolment (Table 3). A considerable number of participants at Africa Centre were found to be HIV infected prior to enrolment (n = 18, 24%) and this could not be explained by the existence of a longer enrolment window.
Table 3. Breakdown by site and outcome of the 537 samples which generated at least one positive rapid test result and were processed according to the MDP testing algorithm.
| Site | Durban | JHB | Mtubatuba | Masaka | Mazabuka | Mwanza | Total |
| Number (n) | 194 | 129 | 75 | 62 | 50 | 27 | 537 |
| 1Excluded | 19 (10%) | 13 (10%) | 18 (24%) | 10 (16%) | 1 (2%) | 2 (7%) | 63 |
| 2Endpoint reached | 144 (74%) | 113 (88%) | 46 (61%) | 52 (84%) | 47 (94%) | 17 (63%) | 419 |
| 3Endpoint not reached | 31 (16%) | 3 (2%) | 11 (15%) | 0 (0%) | 2 (4%) | 8 (30%) | 55 |
HIV infected at screening or enrolment.
HIV infected after enrolment.
HIV uninfected.
Uninfected cases
Of the 55 uninfected cases, 51 had a discordant rapid test result and in 3 cases both rapid tests were positive and in one case the rapid test was negative. The latter case was picked up as the sample was a random quality control sample that had an EIA test which gave a weakly positive result.
False positive rapid results
55 of 537 cases which triggered the algorithm were not confirmed as endpoints. Mwanza generated the highest percentage of false positive rapid test results (n = 8, 30%) and this may have been due to errors in performing the assays as the site had just introduced the method. Numerically, most false positive rapids came from Durban (n = 31) and Africa centre (n = 11) sites, both sites located in KwaZulu Natal. The rate of false positive rapid tests in Durban and Africa Centre was significantly higher than the false positive rate from the Johannesburg site: 16% and 15% compared to 2%, respectively.
Endpoints
Of the 419 confirmed endpoints, the algorithm detected seroconversion at the same visit as the rapid test in 83% (n = 349) of cases. For 16% (n = 68) of endpoints, the algorithm detected infection at an earlier visit. However, 28 of the 68 endpoints were acute infections and would not be expected to be detected by rapid tests which depend on the detection of HIV-specific antibodies. When these were excluded, the algorithm and rapid testing detected the endpoint at the same visit in 89% (349/391) of cases. In 2 cases a discordant rapid result was confirmed as positive at the following visit.
Acute infections
There were 54 cases of acute HIV-1 infection. This was defined on the basis of being negative for the presence of HIV-1 specific antibodies but positive for the presence of HIV DNA or HIV RNA or p24 antigen. All these cases seroconverted at a later visit when antibodies could be detected using the conventional methods. 28 of the 63 (44%) seroconversions occurring before or at enrolment were acute cases.
43 of the 54 acute cases (80%) generated a positive HIV Combo EIA result that was confirmed by HIV PCR or p24 Ag testing. 11 had a negative HIV Combo result but had a positive PCR or p24 Ag test.
Unusual cases
There were 6 cases that generated unusual and/or unexpected combinations of test results (Table 4).
Table 4. Unusual cases which generated “unexpected” sets of results using the algorithm.
| Case | Positive EIA | WB | DNA PCR (week) | RNA PCR (week) | Endpoint |
| 1 | Week 24 | Pos | Neg ( 0, 24) | <50 c/ml (2nd ) | Yes |
| 2 | Week 52 | Pos | Neg (0,24,40, 2nd ) | <400 c/ml (52) | Yes |
| 3 | Week 52 | Pos | Neg (0,24,40) | <400 c/ml (52) | Yes |
| 4 | Week 12, 52, 2nd | Ind | Neg (0, 2nd ) | <50 c/ml (2nd) | No |
| 5 | Week 24,40,52, 2nd | Ind | Neg (0,24,40,52) | Not done | No* |
| 6 | Week 24 | Not done | Pos (0, 24) | <400 c/ml (0) | Yes |
Abbreviations: Neg: negative; Pos: positive; Ind: indeterminate; c/ml: copies/ml; 2nd: second sample collected after positive EIA.
Participant co-enrolled in an HIV vaccine trial.
Cases 1 to 3 demonstrated clear evidence of seroconversion and this was confirmed by WB for the detection of HIV- specific antibodies. However, PCR testing did not confirm infection. None the less, these 3 cases were defined as endpoints based on confirmation by WB testing.
Cases 4 and 5 generated positive EIA's with indeterminate non-evolving WB banding patterns and negative PCR. Case 5 was discovered to be co-enrolled into an HIV vaccine trial and this probably explained the existence of EIA/WB reactivity. This might also have explained the inconclusive results seen for Case 4 but could not be confirmed. Neither of these cases was classified as endpoints.
Case 6 was a week 24 seroconversion with a positive PCR result at enrolment. The HIV endpoint committee consensus was that the positive enrolment PCR result was due to a sample error; however a delayed seroconversion could not be excluded.
Discussion
Accurate endpoint determination for HIV infection is important in the context of clinical trials. This analysis of the HIV seroconversions which occurred during MDP301 confirms that the testing algorithm was user-friendly, accurate and had the additional benefit of being able to retrospectively detect acute HIV infection. The HPTN035 trial also evaluated the efficacy of PRO2000 and a similar algorithm was employed [35]. HPTN differed in the following ways: confirmation was performed by WB on 2 separate specimens, while the MDP algorithm confirmed HIV infection with two methods: HIV EIA and HIV PCR testing or by Western Blotting. Both studies collected a second sample for confirmation. The CAPRISA algorithm [36] was different again and HIV infection was based on 2 separate positive PCR results. In all 3 studies, the algorithms were able to detect acute infections at enrolment, which is crucial for the accurate diagnosis of HIV endpoints, the primary outcome of the trials.
The data was >95% complete and this reflected good accounting of samples at all centres. Monthly sample reconciliation was performed and this allowed for real-time tracking of sample movement to the repository. In addition, each site was assessed annually and random sample storage checks were performed. These quality control procedures, including the use of a reputable courier, ensured completeness of the sample set.
Although sites were provided with a standard operating procedure for the collection and processing of BC samples; further verification of the methodology, by rehearsing the procedure prior to the start of the study, might have prevented inappropriate samples being collected at one of the study sites.
Our data show that the algorithm was able to make a reliable diagnosis of HIV infection status in 531 of the 537 cases with a positive rapid test at a follow-up visit confirming its accuracy.
This was facilitated by the fact that a complete set of serum and BC samples was tested for each potential endpoint from screening through to seroconversion, using 2 different methods of diagnosis plus testing of a second serum specimen. In addition, all cases were reviewed by the “HIV endpoint committee”.
HIV rapid testing correlated with confirmatory endpoint testing in 90% of cases. 51 of the remaining 55 cases (10%) had a discordant rapid result which was confirmed as negative for HIV by the algorithm. Inaccurate reading of rapid test results or technical operator related problems could not be excluded. Mwanza had just introduced rapid testing and so had relatively little experience of the technique which may have led to “over reading” and the relatively high rate of false positive results at this site. It has been pointed out previously in the literature that false positive rapid and EIA results resulting in suboptimal specificity are real concerns in areas with endemic tropical diseases, such as East African countries [15], [19], [20].
As it is essential to exclude HIV infection at enrolment in order to correctly assess efficacy of any product, the use of the HIV DNA PCR together with use of tests for the detection of HIV antibodies testing using the archived samples collected at enrolment was critical. This was reinforced by the relatively high number of seroconverters identified at screening and enrolment (n = 63, 12%). Testing of such a high proportion of samples at the first follow up visit provides evidence that these women were already seroconverting as they entered the study. Just under half (44%) of these were confirmed as acute infections.
The 4th generation HIV EIA performed well for the diagnosis of all acute infections, detecting 80% of them, confirming the finding of Ly et al [9]. Branson [10] and Skidmore et al [37] also recommend the use of this assay for improved HIV screening outcomes and earlier detection of HIV, especially of acute cases and in settings of delayed seroconversion. These assays are therefore an appropriate tool for HIV screening in clinical trials. It would have been too costly to screen all women by HIV PCR for early HIV infection at enrolment, but testing all women at enrolment with a 4th generation EIA would have enabled early diagnosis and potential intervention as well as exclusion from the trial. It is likely that the future of HIV testing for clinical trials will include a point-of-care HIV Ag/Ab and RNA Rapid assay.
Unusual diagnostic results of HIV infection have been previously described, including delayed seroconversion and negative HIV PCR results [37]–[41].
During MDP301 we have described cases which were confirmed to have seroconverted but which were negative for viral RNA and DNA using the specified assays. It is possible that these individuals were infected by clades not detected by the Roche assay or that these discrepancies were due to technical errors. Jackson et al [42] have previously found the Western Blot to be more reliable for confirming HIV infection.
In the latter study 94% of patients with AIDS had HIV DNA positive pellets using the Roche HIV-1 AMPLICOR test. However, on repeat testing of the same pellet a positive test DNA test was obtained.
As demonstrated in Case 5, it is essential to ask participants if they have or currently are participating in an HIV vaccine trial. Vaccine-induced seropositivity (VISP) has been described by Cooper et al in the HVTN trials [44]. Approximately 42% of HIV uninfected participants who had previously participated in HIV vaccine trials were positive for HIV-specific antibodies. Apart for possible problematic diagnostic and social consequences, these antibodies may persist for many years [43]. The rate of VISP varied significantly with the assay used. Where an HIV WB was performed, 10% of those who had been vaccinated had a positive WB and 66% had an indeterminate WB.
Delayed seroconversion may have accounted for the results seen for case 6, although the endpoint committee felt that a sample error had occurred at enrolment.
We have shown that study samples can be reliably stored and retrieved at local laboratories repositories in African study sites before shipping to the central Laboratory. Lessons learnt included the essential need for close monitoring of the sample preparation and storage at the beginning of the study and ongoing sample storage checks.
In conclusion, this study demonstrated that HIV endpoint testing for MDP301 was linked to an accurate, robust, user friendly algorithm in which all results were independently reviewed. Similar algorithms can be recommended for microbicides studies where HIV infection is the endpoint.
Acknowledgments
The authors would like to acknowledge the MDP collaborators, Mitzy Gafos for supporting the internal review of the paper and also the managers and staff at local laboratories and clinics.
Funding Statement
The Microbicides Development Programme 301, a partnership of African, UK and Spanish academic government/institutions and commercial organisations was funded by the British Government Department for International Development (DIFD) and the UK Medical Research Council. This trial is registered at http://isrctn.org, number ISRCTN 64716212 (grant number G0100137). The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1. CDC (2001) Revised Guidelines for HIV Testing, Counselling and Referral. MMWR 50 (RR19): 1–58. [PubMed] [Google Scholar]
- 2. WHO (1997) Revised recommendations for the selection and use of HIV antibody tests. Wkly Epid Rec 12: 81–88. [PubMed] [Google Scholar]
- 3.WHO (2004) Rapid HIV Tests: Guidelines for use in HIV testing and counselling services in resource-constrained settings. ISBN 92 4 159181 1. [Google Scholar]
- 4. CDC (2006) Revised Recommendations for HIV Testing of Adults, Adolescents, and Pregnant Women in Health Care Settings. MMWR 55 (RR14) 1–26. [PubMed] [Google Scholar]
- 5. Delaney KP, Branson BM, Uniyal A, Phillips S, Candal D, et al. (2011) Evaluation of the Performance Characteristics of 6 Rapid HIV Antibody Tests. Clin Infect Dis 52: 257–263. [DOI] [PubMed] [Google Scholar]
- 6. Busch MP, Satten GA (1997) Time course of Viraemia and Antibody Seroconversion following Human Immunodeficiency Virus Exposure. Am J Med 102 (5B) 117–124. [DOI] [PubMed] [Google Scholar]
- 7. Hecht FM, Busch MP, Rawal B, Webb M, Rosenberg E, et al. (2002) Use of laboratory tests and clinical symptoms for identification of primary HIV infection. AIDS 16: 1119–1129. [DOI] [PubMed] [Google Scholar]
- 8. Ly TD, Laperche S, Couroucé AM (2001) Early Detection of Human Immunodeficiency Virus Infection Using Third- and Fourth-Generation Screening Assays. Eur J Clin Microbiol Infec Dis 20: 104–110. [DOI] [PubMed] [Google Scholar]
- 9. Ly TD, Laperche S, Brennan C, Vallari A, Ebel A, et al. (2004) Evaluation of the sensitivity and specificity of six HIV combined p24 antigen and antibody assays. J Virol Methods 122: 185–194. [DOI] [PubMed] [Google Scholar]
- 10. Branson BM (2010) The Future of HIV Testing. J Acquir Immune Defic Syndr 55: S102–105. [DOI] [PubMed] [Google Scholar]
- 11.Food and Drug Administration (2010) Approval Letter - ARCHITECT HIV Ag/Ab Combo. http://www.fda.gov/BiologicsBloodVaccines/BloodBloodProducts/ApprovedProducts/LicensedProductsBLAs/BloodDonorScreening/InfectiousDisease/ucm216300.htm.
- 12. Bredberg-Rådén U, Kiango J, Mhalu F, Biberfeld G (1988) Evaluation of commercial immunoassays for anti-HIV-1 using East African sera. AIDS 2: 281–285. [DOI] [PubMed] [Google Scholar]
- 13. Behets F, Disasi A, Ryder RW, Bishagara K, Piot P, et al. (1991) Comparison of Five Commercial Enzyme-linked Immunosorbent Assays and Western Immunoblotting for Human Immunodeficiency Virus Antibody Detection in Serum Samples from Central Africa. J Clin Micro 29: 2280–2284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Everett DB, Weiss HA, Changalucha, Anemona A, Chirwa T, et al. (2007) Low specificity of the Murex fourth-genenration HIV enzyme immunoassay in Tanzanian adolescents. Trop Med and Intern Health 12: 1323–1326. [DOI] [PubMed] [Google Scholar]
- 15. Everett DB, Baisley KJ, McNerney R, Hambleton I, Chirwa T, et al. (2010) Association of Schistosomiasis with False Positive HIV Test Results in an African Adolescent Population. J Clin Micro 48: 1570–1577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Claasen M, van Zyl GU, Korsman SNJ, Smit L, Cotton MF, et al. (2006) Pitfalls in rapid HIV antibody testing in HIV-infected children in the Western Cape, South Africa. J Clin Virol 37: 68–71. [DOI] [PubMed] [Google Scholar]
- 17. Madhivanan P, Krupp K (2007) Technological challenges in diagnosis and management of HIV infection in resource limited settings. BMJ 335: 165–166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Bruzzone B, Bisio F, Ventura A, Nigro N, Miguel LM, et al. (2008) HIV serologic screening in a population of pregnant women in the Republic of Congo: suitability of different assays. Trop Med Int Health 13: 900–903. [DOI] [PubMed] [Google Scholar]
- 19. Lejon V, Ngoyi DM, Ilunga M, Beelaert G, Maes I, et al. (2010) Low specificities of HIV Diagnostic Tests Caused by Trypanosomiasis brucei gambiense Sleeping Sickness. J Clin Micro 48: 2836–2839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Salinas A, Gorgolas M, Fernandez-Guerrero M (2007) Refrain from telling bad news: patients with leishmaniasis can have false positive HIV test results. Clin Infect Dis 45: 139–140. [DOI] [PubMed] [Google Scholar]
- 21. McMichael AJ, Burrow P, Tomaras GD, Goonetilleke N, Haynes BF (2010) The immune response during acute HIV-1 infection: clues for vaccine development. Nature Reviews Immunology 10: 11–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Morandi P-A, Schockmel GA, Yerly S, Burgisser P, Erb P (1998) Detection of Human Immunodeficiency Virus Type 1 (HIV-1) RNA in Pools of Sera Negative for Antibodies to HIV-1 and HIV-2. J Clin Microbiol 36: 1534–1537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Quinn TC, Brookmeyer R, Kline R, Shepherd M, Paranjape R, et al. (2000) Feasibility of pooling sera for HIV-1 viral RNA to diagnose acute primary HIV-1 infection and estimate HIV incidence. AIDS 14: 2751–2757. [DOI] [PubMed] [Google Scholar]
- 24. Pilcher CD, McPherson JT, Leone PA, Smurzynski M, Owen-O'Dowd J, et al. (2002) Universal Screening for Acute HIV Infection in a Routine HIV Counselling and Testing Population. JAMA 288: 216–221. [DOI] [PubMed] [Google Scholar]
- 25. Pilcher CD, Eron JJ, Gavin S, Gay C, Cohen MS (2004) Acute HIV revisited: new opportunities for treatment and prevention. J Clinl Invest 113: 937–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Henrard DR, Phillips J, Windsor I, Fortenberry D, Korte L, et al. (1994) Detection of human immunodeficiency virus type 1 p24 antigen and plasma RNA: relevance to indeterminate serologic tests. Transfusion 34: 376–380. [DOI] [PubMed] [Google Scholar]
- 27. Schacker T, Collier AC, Hughes J, Shea T, Corey L (1996) Clinical and Epidemiologic Features of Primary HIV Infection. Ann Intern Med 125: 257–264. [DOI] [PubMed] [Google Scholar]
- 28. McCormack S, Ramjee G, Kamali A, Rees H, Crook AM, et al. (2010) PRO2000 vaginal gel for prevention of HIV-1 infection (Microbicides Development Programme 301): a phase 3, randomised, double-blind, parallel-group trial. Lancet 2010, published online DOI:10.1016/S0140-6736(10)61086-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nunn A, McCormack S, Crook AM, Pool R, Rutterford C, et al. (2009) Microbicides Development Programme; design of a Phase III trial to measure the efficacy of the vaginal microbicide PRO2000/5 for HIV prevention. Trials 10: 99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Everett DB, Kathy B, Changalucha J, Vallely A, Watson-Jones D, et al. (2009) Suitability of Simple Human Immunodeficiency Virus Rapid Tests in Clinical Trials in Community-Based Clinic Settings. J Clin Microbiol 47: 1058–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gray RH, Makumbi F, Serwadda D, Lutalo T, Nalugoda F, et al. (2007) Limitations of rapid HIV-1 tests during screening for trials in Uganda: diagnostic test accuracy study. BMJ| ONLINE FIRST bmj.com 335: 1–4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.WHO, UNAIDS (2002) HIV Simple/Rapid Assays: Operational Characteristics (Phase 1) Report 12, Whole blood specimens. [Google Scholar]
- 33.CDC (2010) FDA Approved Rapid HIV Antibody Screening Tests. http://www.cdc.gov/hiv/topics/testing/rapid/rt-comparison.htm
- 34. Stevens W, Erasmus L, Moloi M, Taleng T, Sarang Somaya (2008) Performance of a Novel Human Immunodeficiency Virus (HIV) Type 1 Total Nucleic Acid-Based Real-Time PCR Assay Using Whole Blood and Dried Blood Spots for Diagnosis of HIV in Infants. J Clin Micro 46: 3941–3945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Abdool Karim SS, Richardson BA, Ramjee G, Hoffman IF, Chirenje ZM, et al. (2011) Safety and effectiveness of vaginal microbicides BufferGel and 0,5% PRO 2000 Gel for the prevention of HIV infection in women. AIDS 25: 957–966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Abdool Karim Q, Abdool Karim SS, Frohlich JA, Grobler AC, Baxter C, et al. (2010) Effectiveness and Safety of Tenofovir Gel, an Antiretroviral Microbicide, for the Prevention of HIV Infection in Women. CAPRISA. Science Express 1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Skidmore S, Devendra S, Weaver J, Short J, Osman H, et al. (2009) A case study of delayed HIV-1 seroconversion highlights the need for Combo assays. Int J STD AIDS 20: 205–206. [DOI] [PubMed] [Google Scholar]
- 38. Jenny-Avital ER, Beatrice ST (2001) Erroneously low or undetectable plasma human immunodeficiency virus type 1 (HIV-1) ribonucleic acid load, determined by polymerase chain reaction, in West African and American patients with non-B subtype HIV-1 infection. Clin Infect Dis 32: 1227–1230. [DOI] [PubMed] [Google Scholar]
- 39. Zaman MM, Recco RA, Haag R (2002) Infection with Non-B Subtype HIV Type 1 Complicates Management of Established Infection in Adult Patients and Diagnosis of Infection in Newborn Infants. Clin Infect Dis 34: 417–418. [DOI] [PubMed] [Google Scholar]
- 40. Trezi R, Niero F, Iemoli E, Capetti A, Coen M, et al. (2007) Late HIV seroconversion after non-occupational postexposure prophylaxis against HIV with concomitant Hepatitis C virus seroconversion. AIDS 21: 262–263. [DOI] [PubMed] [Google Scholar]
- 41. Bøgh M, Machuca R, Gerstoft J, Pedersen C, Obel N, et al. (2001) Subtype-specific problems with qualitative Amplicor HIV-1 DNA PCR test. J Clin Virol 20: 149–153. [DOI] [PubMed] [Google Scholar]
- 42. Brooks Jackson J, Parsons JS, Nichols LS, Knoble N, Kennedy S, et al. (1997) Detection of Human Immunodeficiency Virus Type 1 (HIV-1) Antibody by Western Blotting and HIV-1 DNA by PCR in Patients with AIDS. J Clin Microbiol 35: 1118–1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Silbermann B, Tod M, Desaint C, Pialoux G, Petitprez K, et al. (2008) Long-Term Persistence of Vaccine-Induced HIV Seropositivity among Healthy Volunteers. AIDS Res Hum Retroviruses 24: 1–4. [DOI] [PubMed] [Google Scholar]
- 44. Cooper CJ, Metch B, Dragavon J, Coombs RW, Baden LR (2010) Vaccine-Induced HIV Seropositivity/Reactivity in Noninfected HIV Vaccine Recipients. JAMA 304: 275–283. [DOI] [PMC free article] [PubMed] [Google Scholar]