Abstract
ExoT is a type III secreted effector protein found in almost all strains of Pseudomonas aeruginosa and is required for full virulence in an animal model of acute pneumonia. It is comprised of an N-terminal domain with GTPase activating protein (GAP) activity towards Rho family GTPases and a C-terminal ADP ribosyltransferase (ADPRT) domain with minimal activity towards a synthetic substrate in vitro. Consistent with its activity as a Rho family GTPase, ExoT has been shown to inhibit P. aeruginosa internalization into epithelial cells and macrophages, disrupt the actin cytoskeleton through a Rho-dependent pathway, and inhibit wound repair in a scrape model of injured epithelium. We have previously shown that mutation of the invariant arginine of the GAP domain to lysine (R149K) results in complete loss of GAP activity in vitro but only partially inhibits ExoT anti-internalization and cell rounding activity. We have constructed in-frame deletions and point mutations within the ADPRT domain in order to test whether this domain might account for the residual activity observed in ExoT GAP mutants. Deletion of a majority of the ADPRT domain (residues 234 to 438) or point mutations of the ADPRT catalytic site (residues 383 to 385) led to distinct changes in host cell morphology and substantially reduced the ability of ExoT to inhibit in vitro epithelial wound healing over a 24-h period. In contrast, only subtle effects on the efficiency of ExoT-induced bacterial internalization were observed in the ADPRT mutant forms. Expression of each domain individually in Saccharomyces cerevisiae was toxic, whereas expression of each of the catalytically inactive mutant domains was not. Collectively, these data demonstrate that the ADPRT domain of ExoT is active in vivo and contributes to the pathogenesis of P. aeruginosa infections.
Pseudomonas aeruginosa is one of the leading causes of nosocomial infections in humans. Acute P. aeruginosa infections, such as burn or surgical infections, corneal ulcer formation at the sites of contact lens-associated trauma, cystitis in catheterized patients, and pneumonia in mechanically ventilated patients, are associated with preexisting epithelial tissue damage and/or host immunocompromise (reviewed in reference 8). P. aeruginosa boasts an impressive array of cell-associated and secreted virulence factors. As is true for many other gram-negative pathogens, the secretion of some of these virulence factors requires the type III secretion system (reviewed in reference 12). This remarkable protein export apparatus allows bacteria to directly translocate a set of toxins, termed effector proteins, into the eukaryotic host cell, where they modulate signal transduction pathways to subvert the host immune response (reviewed in reference 26). While the secretion machinery itself is highly conserved, the effector proteins differ between species and even, in the case of P. aeruginosa, among individual clinical isolates (10, 11, 24). This genetic diversity no doubt accounts for at least some of the difference in pathogenic potential of different microorganisms.
Four type III secreted effector molecules have been identified in P. aeruginosa (8), although almost no strain examined thus far encodes or secretes all four of them (10). ExoU is a potent cytotoxin that causes rapid cell death by necrosis, probably through a phospholipase activity (11, 24, 45). It contributes to virulence in animal models of disease (11, 24), and its presence in the genome or secretion in vitro correlates with poor outcome in human infections (23, 44). ExoY is a host factor-dependent adenylate cyclase that can cause cell rounding in some host cell lines (53). Its exact role in the pathogenesis of disease remains uncertain (49).
ExoS and ExoT are closely related bifunctional proteins (74% identity at the amino acid level) (52). ExoS has been more extensively studied. Its N-terminal domain possesses an arginine finger motif characteristic of GTPase activating proteins (GAPs) (16). ExoS exhibits GAP activity towards Rho, Rac, and Cdc42 in vitro and in vivo and has been shown to be sufficient to disrupt the actin cytoskeleton (22, 33, 39). Its GAP domain is an example of an expanding group of bacterial GAPs that appear to have arisen by convergent evolution (47).
The C-terminal domain of ExoS possesses ADP ribosyltransferase (ADPRT) activity towards Ras, Ral, and various Rab family GTPases and requires a eukaryotic 14-3-3 protein, factor-activating exoenzyme S, for activity (2, 17, 18, 35, 43, 54). This activity interferes with eukaryotic DNA synthesis and endocytosis and causes cytotoxicity and cell death of mammalian cells (2, 38). In vitro, ExoS auto-ADP ribosylates the critical arginine residue (arginine 146) in its GAP domain, leading to downregulation of its GAP activity (42).
While only some strains of P. aeruginosa possess the ExoS gene, all strains examined thus far encode ExoT (10). Consistent with an important role in pathogenesis, ExoT is required for full virulence in a mouse model of acute pneumonia (19). Like ExoS, ExoT has N-terminal GAP activity in vitro and in vivo towards Rho, Rac, and Cdc42 (29, 32). This activity contributes to (i) disruption of the actin cytoskeleton, resulting in cell rounding (but not cytotoxicity), (ii) prevention of bacterial internalization through its inhibition of Rho family GTPases, and (iii) inhibition of wound healing (5, 19, 21). The C terminus of ExoT appears to possess minimal ADPRT activity in vitro (0.2% compared to ExoS) toward a synthetic substrate in vitro. This finding potentially implies that it is nonfunctional in vivo, although the use of a synthetic substrate may have underestimated the catalytic activity of the ADPRT domain. We have found that mutation of the invariant arginine of the ExoT GAP domain to lysine (R149K) resulted in complete loss of GAP activity in vitro but only partial diminution of anti-internalization and cell rounding activity without affecting efficiency of translocation (19). These unexpected findings suggested that the ADPRT domain contributes to the biological activity of ExoT. To test this hypothesis, we have constructed strains of P. aeruginosa that produce ExoT with wild-type or inactive GAP and/or ADPRT domains and tested the phenotypes in biologically relevant assays. In addition, we have introduced the various forms of ExoT into eukaryotic cells and into Saccharomyces cerevisiae. Our results conclusively demonstrate that the ADPRT domain of ExoT is functionally active in vivo, exerts effects on host cell morphology that are distinct from those of the GAP domain, and likely contributes to the pathogenesis of P. aeruginosa infections. ExoT thus joins the growing list of bifunctional type III secreted proteins that are important for bacterial virulence.
MATERIALS AND METHODS
Bacterial strains and media.
Bacterial strains were routinely cultured in Luria-Bertani broth (LB) or Vogel-Bonner minimal medium with antibiotics as needed for cloning purposes. The following antibiotics were used at the indicated concentrations: ampicillin, 100 μg/ml for Escherichia coli; carbenicillin, 200 μg/ml for P. aeruginosa; tetracycline, 100 μg/ml for P. aeruginosa. All antibiotics and chemicals were purchased from Sigma Chemical Company except where noted. A list of bacterial strains and plasmids used in this study is shown in Table 1. All intermediate cloning steps were performed using XL2-Blue ultracompetent E. coli (Stratagene).
TABLE 1.
Bacterial strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| PA103pscJ::Tn5 | Tn5 Gmr inserted into pscJ, defective in type III secretion | 28 |
| PA103ΔUΔT | PA103ΔU with a xylE/aacC1 cassette replacing amino acids 36-348 of exoT, Gmr | 19 |
| PA103ΔU/T(R149K) | PA103ΔU with a point mutation in ExoT (R149K) | 19 |
| PA103ΔU/T(AAA) | PA103ΔU with a point mutations in ExoT (EQE383-385AAA) | This study |
| PA103ΔU/T(R149K)(AAA) | PA103ΔU with point mutations in ExoT (R149K and EQE383-385AAA) | This study |
| PA103ΔU/T(ΔGAP) | PA103ΔU with amino acids 80-228 of ExoT deleted in frame | This study |
| PA103ΔU/T(ΔADPRT) | PA103ΔU with amino acids 234-438 of ExoT deleted in frame | This study |
| PA103ΔUΔT+pUCP20 | PA103ΔUΔT transformed with pUCP20, Gmr Cbr | 19 |
| PA103ΔUΔT+pExoT | PA103ΔUΔT transformed with pUCP20/exoT, Gmr Cbr | 19 |
| PA103ΔUΔT+pExoT(R149K) | PA103ΔUΔT transformed with pUCP20/T(R149K), Gmr Cbr | 19 |
| PA103ΔUΔT+pExoT(AAA) | PA103ΔUΔT transformed with pUCP20/T(AAA), Gmr Cbr | This study |
| PA103ΔUΔT+pExoT(R149K)(AAA) | PA103ΔUΔT transformed with pUCP20/T(R149K)(AAA), Gmr Cbr | This study |
| PA103ΔUΔT+pExoT(ΔGAP) | PA103ΔUΔT transformed with pUCP20/T(Δ80-228), Gmr Cbr | This study |
| PA103ΔUΔT+pExoT(ΔADPRT) | PA103ΔUΔT transformed with pUCP20/T(Δ234-438), Gmr Cbr | This study |
| S17.1 | E. coli strain used for mating constructs into P. aeruginosa thi pro hsdR recA RP4-2 (Tet::Mu) (Km::Tn7) | 46 |
| XL2-Blue | E. coli strain used for cloning; recA1 endA1 gyrA96 thi-1 hsdR17 supE44 relA1 lacF′ proAB lac1qZΔM15 Tn10 (Tetr) Amy Camr | Stratagene |
| pGEM-T | Vector for cloning PCR products, Apr | Promega |
| pGEM-T/T flank | pGEM-T with a 3.6-kb PCR product containing the exoT gene | This study |
| pGEM-T/T flank(AAA) | pGEM-T/Tflank with ExoT amino acids 383-385 changed from EQE to AAA | This study |
| pGEM-T/T flank(R149K)(AAA) | pGEM-T/Tflank with ExoT amino acids R149K and amino acids 383-385 changed from EQE to AAA | This study |
| pGEM-T/T flank(Δ80-228) | pGEM-T/Tflank with ExoT amino acids 80-228 deleted in frame | This study |
| pGEM-T/T flank (Δ234-438) | pGEM-T/Tflank with ExoT amino acids 234-438 deleted in frame | This study |
| pJEN34 | pRIC380 in which the β-lactamase gene is replaced by Tet | Jennifer Sargent |
| pIRESKII-EGFP | pIRESKII-EGFP (Clontech) with bases 1870-1910 removed | Marcus Mohrs and Richard Locksley |
| pIRESKII-EGFP/T | pIRESKII-EGFP with the ExoT open reading frame cloned into the EcoRI site | This study |
| pIRESKII-EGFP/T(R149K) | pIRESKII-EGFP/T with the R149K mutation | This study |
| pIRESKII-EGFP/T(AAA) | pIRESKII-EGFP/T with the EQE383-385AAA mutation | This study |
| pIRESKII-EGFP/T(R149K)(AAA) | pIRESKII-EGFP/T with the R149K and EQE383-385AAA mutations | This study |
| pIRESKII-EGFP/T(2537) | pIRESKII-EGFP with amino acids 1-232 followed by a stop codon | This study |
| pIRESKII-EGFP/T(2537)(R149K) | pIRESKII-EGFP with amino acids 1-232 followed by a stop codon with R149K mutation | This study |
| pIRESKII-EGFP/T(2638) | pIRESKII-EGFP with amino acids 230-457 preceded by a start codon | This study |
| pIRESKII-EGFP/T(2638)(AAA) | pIRESKII-EGFP with amino acids 230-457 preceded by a start codon with the EQE383-385AAA mutation | This study |
| pUCP20 | P. aeruginosa expression vector, Ap (Cbr) | 51 |
| pUCP20/exoT | pUCP20 with a 1,666-bp insert containing exoT | 19 |
| pUCP20/ExoT(R149K) | pUCP20/exoT with the R149K mutation; previously called pBK151 | 19 |
| pUCP20/ExoT(AAA) | pUCP20/exoT with the EQE383-385AAA mutation | This study |
| pUCP20/ExpT(R149K)(AAA) | pUCP20/exoT with the R149K and EQE383-385AAA mutations | This study |
| pUCP20/ExoT(Δ51-123) | pUCP20/exoT with amino acids 51-123 deleted in frame | This study |
| pUCP20/ExoT(R149K)(Δ51-123) | pUCP20/exoT with the R149K mutation and amino acids 51-123 deleted in frame | This study |
| pUCP20/ExoT(Δ80-238) | pUCP20/exoT with amino acids 80-238 deleted in frame | This study |
| pUCP20/ExoT(Δ160-228) | pUCP20/exoT with amino acids 160-228 deleted in frame | This study |
| pUCP20/ExoT(R149K)(Δ160-228) | pUCP20/exoT with the R149K mutation and amino acids 160-228 deleted in frame | This study |
| pUCP20/ExoT(Δ234-438) | pUCP20/exoT with amino acids 234-438 deleted in frame | This study |
| pUCP20/ExoT(R149K)(Δ234-438) | pUCP20/exoT with the R149K mutation and amino acids 234-438 deleted in frame | This study |
Except where otherwise noted, strains expressing wild-type or mutated versions ExoT cloned into a plasmid were grown without shaking in 2 ml of LB medium containing carbenicillin (200 μg/ml) for 18 h at 37°C. Strains expressing ExoT or mutant versions recombined into the ExoT locus were grown in MinS-Ca2+ medium (24) with shaking at 37°C overnight. Bacterial CFU were determined by optical density and confirmed by plating serial dilutions for colony counts.
Construction of plasmid-borne mutant versions of ExoT.
Primer bases that are shown in lowercase type below are not homologous to the chromosomal sequence. All mutation constructs were confirmed by DNA sequencing. Expression plasmids encoding mutated versions of ExoT were used to transform PA103ΔUΔT by electroporation.
Mutations within the ExoT GAP domain (residues 1 to 234) were created as follows. Residues 51 to 123 were deleted from the wild type and the GAP active-site mutant of ExoT by PCR amplification of pUCP20/ExoT and pUCP20/T(R149K), respectively, with primers LKG13 (5′-ttggatCCCTTCGGTGCTTCCTGGCG-3′) and LKG14 (5′-aaggatcCGAGCTGGCGACTTTGACAG-3′). The PCR products were digested with BamHI and self ligated to create pUCP20/T(Δ51-123) and pUCP20/T(R149K)(Δ51-123). ExoT residues 141 to 152, comprising the GAP arginine finger motif, were deleted by PCR amplification of pUCP20/ExoT with primers LKG15 (5′-aaggtaccCGTGTGATCCTTCGCCAG-3′) and LKG16 (5′-aaggtACCGCCCTGGTCGGGATTCGC-3′). The PCR product was digested with KpnI and self-ligated to create pUCP20/T(Δ141-152). Residues 160 to 228 were deleted from the wild-type and the GAP active-site mutant of ExoT by PCR amplification of pUCP20/ExoT and pUCP20/T(R149K), respectively, with primers LKG17 (5′-aaagatctAATCCCGACCAGGGCGGTG-3′) and LKG18 (5′-aaagatctGAGGTAAAGGGCGAGCCTG-3′). The PCR products were digested with BglII and self-ligated to create pUCP20/T(Δ160-228) and pUCP20/T(R149K)(Δ160-228). ExoT residues 80 to 228 were deleted by PCR amplification of pUCP20/ExoT with primers LKG18 and LKG36 (5′-aaagatctGCTCCCCAGCAGTTTGCC-3′). The PCR product was digested with BglII and self-ligated to create pUCP20/T(Δ80-228).
Mutations within the ExoT ADPRT domain (residues 235 to 457) were created as follows. The ExoT ADPRT active site residues EQE at amino acids 383 to 385 were changed to AAA by PCR amplification of pUCP20/ExoT with primers LKG29 (5′-ataagctgcagcaATCCTCTACGACAAGG-3′) and LKG30 (5′-aaatagctgcagCATCGCCCTCGATCG-3′). The PCR product was digested with PstI and self-ligated to create pUCP20/T(AAA). The double active-site mutant of ExoT was created by PCR amplification of pUCP20/T(R149K) with primers LKG29 and LKG30, digestion of the product with PstI, and self-ligation to create pUCP20/T(R149K)(AAA). Residues 234 to 438 were deleted from the wild type and the GAP active-site mutant of ExoT by PCR amplification of pUCP20/ExoT and pUCP20/T(R149K), respectively, with primers LKG19 (5′-gtggatCCCTTTACCTCGCTCTCTACCGC-3′) and LKG21 (5′-ttggaTCcGGCAAGCCCCAGGAACAG-3′). The PCR products were digested with BamHI and self-ligated to create pUCP20/T(Δ234-438) and pUCP20/T(R149K)(Δ234-438).
Construction of chromosomal ExoT mutants.
Using primers LKG31 (5′-aaaaactagtACGAGGATGCGCGGGAAC-3′) and LKG32 (5′-aaaaactagtATCGACCGCGTTGACCAAC-3′) a 3.6-kb product was PCR amplified from PA103ΔU genomic DNA that included the ExoT gene and 1 kb of 5′ and 3′ flanking DNA. The PCR product was digested with SpeI and cloned into the unique SpeI site of pGEM-T (Promega), creating vector pGEM-T/T flank. The ADPRT active-site (AAA) mutant was created by PCR amplification of pGEM-T/T flank using primers LKG29 and LKG30, digestion of the product with PstI (the PstI site in pGEM-T had been removed), and self-ligation to make pGEM-T/T flank (AAA). The R149K mutation was moved into pGEM-T/T flank by swapping in a 936-bp XmaI fragment from pUCP20/T(R149K) that contains the mutation. The Δ80-228 deletion was created in pGEM-T/T flank by PCR amplification with primers LKG18 and LKG36, followed by digestion with BglII and self-ligation to create pGEM-T/T flank (Δ80-228). The Δ234-438 deletion was constructed in pGEM-T/T flank by PCR amplification with primers LKG19 and LKG21, followed by digestion with BamHI and self-ligation to create pGEM-T/T flank (Δ234-438). The inserts of pGEM-T/T flank (AAA), pGEM-T/T flank (R149K)(AAA), pGEM-T/T flank (Δ80-228), and pGEM-T/T flank (Δ234-438) were released by SpeI digestion and subcloned into the unique SpeI site of the sacB-based allelic exchange vector pJEN34 (pJEN34 is pRIC380 [1] in which the β-lactamase gene is replaced with a gene encoding tetracycline resistance). E. coli S17.1 carrying the pJEN34 constructs with the mutant versions of ExoT was mated with PA103ΔUexoT::gent, in which the ExoT gene is partially replaced by a gentamicin resistance cassette (19). In a two-step procedure, exconjugants in which the allelic exchange vector had integrated into the ExoT locus were selected by growth on Vogel-Bonner minimal medium agar plates containing tetracycline. To select for excision of the integrated vector, bacteria were streaked onto LB plates containing 5% sucrose. Colonies that grew in the presence of sucrose (and thus had presumably lost the sacB gene) were then screened for loss of gentamicin resistance (to track introduction of the desired ExoT mutation) and were further verified by Southern blot analysis.
Yeast strains, plasmids, and growth inhibition assays.
To express ExoT or its individual domains constitutively in yeast, appropriate fragments were subcloned into the vector pGBKT7 (Clontech, Palo Alto, Calif.). The plasmids were transformed into S. cerevisiae strain AH109 using a modified lithium acetate-based method according to the manufacturer's directions. Transformants were selected on synthetic medium plates lacking tryptophan. Yeast strains and expression plasmids used in this study are summarized in Table 2.
TABLE 2.
Yeast strains and plasmids used in this study
| Strain or plasmid | Relevant characteristics | Source or reference |
|---|---|---|
| AH109 | MATatrp1-901 leu2-3, 112 ursa3-52 his3-200 gal4Δ gal80Δ LYS2::GAL1UAS-GAL1TATA-HIS3 GAL2UAS-GAL2TATA-ADE2 URA3::MEL1UAS-MEL1TATA-lacZ | 27 |
| L4852 | MATacan1-100 GAL+ ura3-1 leu2-3, 112 his3-11, 15 ade2-1 trp1-1 | |
| pGBKT7 | Vector for constitutive expression in yeast | Clontech |
| pGBKT7-GAP | GAP domain of ExoT cloned into the SalI site of pGBKT7 | This study |
| pGBKT7-GAP(R149K) | GAP domain of ExoT with a R149K mutation subcloned into the SalI site of pGBKT7 | This study |
| pGBKT7-ADPRT | ADPRT domain of ExoT cloned into the EcoRI site of pGBKT7 | This study |
| pGBKT7-ADPRT(AAA) | ADPRT domain of ExoT with an EQE383-385AAA mutation subcloned into the EcoRI site of pGBKT7 | This study |
| pRS424-GAL1 | Vector for galactose-inducible expression | 36 |
| pRS424-GAL1/GAP | GAP domain of ExoT subcloned into the EcoRI site of pRS424-GAL1 | This study |
| pRS424-GAL1/GAP(R149K) | GAP domain of ExoT with a R149K mutation subcloned into the EcoRI site of pRS424-GAL1 | This study |
| pRS424-GAL1/ADPRT | ADPRT domain of ExoT subcloned into the EcoRI site of pRS424-GAL1 | This study |
| pRS424-GAL1/ADPRT(AAA) | ADPRT domain of ExoT with an EQE383-385AAA mutation subcloned into the EcoRI site of pRS424-GAL1 | This study |
The wild-type and mutant versions of the GAP and the ADPRT domains of ExoT were also subcloned as EcoRI fragments into the inducible yeast vector pRS424-GAL1 (36). These plasmids were transformed into S. cerevisiae strain L4852 using a lithium acetate-based method (in accordance with the yeast protocols handbook [Clontech]). Transformants were selected on minimal medium lacking tryptophan and containing 1% glucose. The expression of the proteins was under the control of a GAL1 promoter.
For growth inhibition assays, cultures were grown overnight to saturation in a minimal medium containing glucose as a carbon source (noninducing conditions). The cultures were washed three times in sterile distilled water and resuspended to an optical density at 600 nm (OD600) of 1.0. After 10-fold serial dilutions, 5 μl of each dilution was spotted onto selective medium plates containing 1% glucose (noninducing) or 1% galactose (inducing) as a carbon source. The plates were incubated at 30°C for 3 days before the results were recorded.
Cell wounding assays.
For wounding assays, MDCK cells were grown in 24-well tissue culture dishes containing Dulbecco's modified Eagle medium with glucose at a concentration of 4.5 g/liter (DME-H21) (University of California, San Francisco, Cell Culture Facility) supplemented with glucose (4.5 g/liter), l-glutamine (0.584 g/liter), NaHCO3 (3.7 g/liter), and 10% FBS at a density of ∼2.7 × 105 cells/well. A confluent monolayer was obtained after 24 h of incubation at 37°C in a humidified atmosphere containing 5% CO2.
Wound healing in MDCK cells was measured as described previously (21). Experiments were done in triplicate. Confluent cell monolayers of MDCK epithelial cells were washed twice with DME-H21 to remove serum. A linear wound was then made with a plastic pipette tip. After wounding, the cells were washed three times with DME-H21 to remove the cell debris. The bacterial strains were grown overnight at 37°C in LB medium without shaking. Bacteria were diluted with DME-H21 supplemented with glucose (4.5 g/liter), l-glutamine (0.584 g/liter), NaHCO3 (3.7 g/liter), and 10% fetal bovine serum (FBS). An inoculum of 100 μl of bacteria per well (24-well plate) at an OD600 of 0.05 corresponded to a multiplicity of infection (MOI) of ca. 10. The infected wounded MDCK cells were grown in 1 ml of DME-H21 supplemented with 10% FBS and 2 mM l-glutamate maintained at 37°C in 5% CO2 during the experiment.
The area of the denuded surface was measured immediately after wounding and before infecting the cells, followed by measurements 2, 4, 6, 8, and 20 h after infection. The cells were photographed using an inverted microscope (Axiovert 35; Zeiss) connected to an NEC C2400 charge-coupled device (CCD) camera. The images were subsequently captured by an image-analyzing frame-grabber card (LG-3 Scientific Frame Grabber; Scion) and analyzed using image analysis software (NIH Image 1.55). Wound repair was expressed as a percentage of initial wound area in each well.
HeLa cell rounding assay.
HeLa cells (obtained from the American Type Culture Collection) were grown in DME-H21 and 10% heat-inactivated FBS (Gibco BRL). Cells were plated at 2 × 105 cells/well in 24-well plates and grown overnight at 37°C in the presence of 5% CO2. Cells were washed twice with minimal essential medium-Eagle medium containing Hanks' buffered saline solution (MEM) (Sigma Chemical Co.), 20 mM HEPES buffer (pH 8.0), 3.5% sodium bicarbonate, and 0.6% bovine serum albumin (MEM-etc.). HeLa cells were inoculated with 5 × 106 bacteria and incubated for 3 h at 37°C in room air. Rounding was assessed by microscopic inspection of the wells at magnifications of ×100 and ×200 on a Nikon Eclipse TE200 microscope. Images were captured using a CCD camera and SPOT imaging software (Diagnostic Instruments).
HeLa cell morphological studies using time lapse microscopy.
For video microscopy assays, HeLa cells were grown in six-well tissue culture dishes containing MEM-Eagle medium supplemented with 5% FBS, l-glutamine (0.292 g/liter), glucose (1.0 g/liter), and NaHCO3 (2.2 g/liter) at a density of ∼1.0 × 106 cells/well. After 24 h of incubation at 37°C in a humidified atmosphere containing 5% CO2, confluent monolayers of HeLa cells were obtained, which were then washed twice with the same medium. Bacteria were prepared as described above in the cell wounding assay. The MOI was determined by titrating the dilution; an inoculum of 500 μl of bacteria per well (six-well plate) at an OD600 of 0.05 corresponded to an MOI of ca. 20. The infected HeLa cells were grown in 5 ml of MEM-Eagle medium supplemented with 5% FBS, l-glutamine (0.292 g/liter), glucose (1.0 g/liter), and NaHCO3 (2.2 g/liter) at 37°C in 5% CO2 during the experiment. Video images were captured every 5 min using a Zeiss Axiovert microscope connected to a Cohu video camera and analyzed using the Openlab software (Improvision). Video microscopy was performed at the UCSF Cancer Center. The video movies are available upon request.
Fluorescence microscopy of infected HeLa cells.
Cell were fixed as previously described (19). Coverslips were placed cell side down on a 50-μl drop of phosphate-buffered saline (PBS) with 0.7% (wt/vol) fish scale gelatin, 0.005% (wt/vol) saponin, and RNase A (100 μg/ml) RNase A (PBS-FSG-SAP) with unpurified anti-ExoU polyclonal antisera (diluted 1:500) in a sealed humidified container at 37°C for 2 h. Although the strains used in these experiments do not produce ExoU, the antiserum contains additional antibodies, likely to lipopolysaccharide, which allows visualization of the bacteria. Coverslips were returned to their original wells and washed with PBS-FSG-SAP for 10 min four times, with rocking at 37°C. Coverslips were placed face down on 50-μl drops of PBS-FSG-SAP with 1:500 goat anti-rabbit-fluorescein isothiocyanate (Jackson Laboratories) and 1:200 Texas Red X-phalloidin (Molecular Probes) in a sealed humid container at 37°C for 45 min. Coverslips were returned to their original wells and washed with PBS-FSG-SAP twice for 10 min with rocking at 37°C. The coverslips were then washed with PBS containing 0.1% (wt/vol) Triton X-100 twice for 5 min with rocking at 37°C. Coverslips were rinsed once with PBS and dipped in distilled H2O before mounting on glass slides with the ProLong Antifade Kit (Molecular Probes). Cells were observed and images were captured at a magnification of ×1,000 under oil immersion using a Nikon Eclipse E800 fluorescence microscope. Images were captured using a CCD camera and SPOT imaging software.
HeLa cell invasion assay.
Invasion assays were performed as described previously (19) with the following modifications. HeLa cells were seeded in a 24-well plate at a density of 1.5 × 105 per well and incubated for 24 h at 37°C with 5% CO2. Cells were washed once with MEM-etc., and 2 × 107 to 3 × 107 bacteria were added to the wells. The bacteria were incubated with the cells at 37°C for 2 h in room air. Amikacin was added to the wells at a final concentration of 400 μg/ml, and the culture plate was incubated 37°C for another 2 h in room air. To ensure that invasion assays were not affected by loss of rounded, nonadherent cells, the contents of each well were collected in an Eppendorf tube. To remove the remaining adherent cells, 100 μl of trypsin was added to each well, and the plate was incubated at room temperature for 10 min. The trypsinized cells were added to the nonadherent cells, and the total cell pellet was collected by centrifugation (Eppendorf microcentrifuge) at 14,000 rpm for 1 min. To release the internalized bacteria, the supernatant was removed by aspiration, the pellet was resuspended in 1 ml of Ca2+- and Mg2+-free Hanks' buffered saline solution (UCSF Cell Culture Facility) with 0.25% Triton X-100 (Sigma), and the lysis mixtures were returned to the original culture wells. After 30 min of incubation at room temperature the lysates were plated on LB agar and incubated at 37°C for 18 h to quantify the CFU of internalized bacteria. The invasion assays were normalized to the number of cells in each well, as determined by measuring the total amount of lactate dehydrogenase (using the CytoTox 96 Nonradioactive Cytotoxicity Assay kit [Promega] according to the manufacturer's specification) in a duplicate sample treated with 1% Triton X-100.
Construction of plasmids to express ExoT in mammalian cells.
The complete ExoT open reading frames from constructs pUCP20/ExoT, pUCP20/ExoT(R149K), pUCP20/ExoT(AAA), and pUCP20/ExoT(R149K)(AAA) were amplified by PCR using primers LKG25(5′-aaaagaattcATGCATATTCAATCATCTCAGC-3′) and LKG26 (5′-aaaagaattcTCAGGCCAGGTCGAGGC-3′). To clone the GAP domain alone for expression (amino acids 1 to 232) primer LKG37 (5′-aaaagaattctcaCTTTACCTCGCTCTCTAC-3′) was designed to introduce a stop codon after amino acid 232. The wild-type GAP domain and the active-site mutant version were amplified by PCR with primers LKG25 and LKG37 from templates pUCP20/ExoT and pUCP20/ExoT(R149K), respectively. To clone the ADPRT domain alone for expression (amino acids 230 to 457) primer LKG38 (5′-aaaagaattcatgGTAAAGGGCGAGCCTGTC-3′) was designed to introduce a start codon in front of amino acid 230. The wild-type and active-site mutant version of the ADPRT domain were amplified by PCR with primers LKG26 and LKG38 from templates pUCP20/ExoT and pUCP20/ExoT(AAA), respectively. All PCR products were cloned into pGEM-T, their DNA sequences were confirmed, and they were released by digestion with EcoRI and cloned into the unique EcoRI site of pIRESKII-EGFP in which bases 1870 to 1910 of pIRES-EGFP have been deleted to reduce enhanced green fluorescent protein (enhanced GFP) expression.
HeLa cell transfections.
HeLa cells were plated in a 24-well tissue culture plate (4.5 × 104 cells/well) onto 12-mm-diameter acid-etched glass coverslips and incubated for 24 h at 37°C with 5% CO2. Transfections were performed using Effectene (Qiagen) according to the manufacturer's instructions using 0.2 μg of DNA/well. After 18 h of incubation at 37°C with 5% CO2, the wells were washed twice with ice-cold PBS and fixed by rocking in 0.4% paraformaldehyde in PBS at room temperature for 30 min. After rinsing once with PBS, the samples were quenched in PBS containing 75 mM NH4Cl and 20 mM glycine for 10 min with gentle agitation at room temperature. The fixed cells were stored in PBS for 18 h at 4°C. The cells were permeabilized in PBS-FSG-SAP as described above. The coverslips were then placed with the cell surface facing down on a 50-μl drop of PBS-FSG-SAP with rabbit anti-ExoT polyclonal antisera (diluted 1:500) in a sealed humid container at 37°C for 2 h. Coverslips were returned to their original wells and washed four times with PBS-FSG-SAP for 10 min with gentle rocking at 37°C. The coverslips were then placed face down on 50-μl drops of PBS-FSG-SAP containing goat anti-rabbit-Cascade blue (Molecular Probes) (1:500) and Texas Red X-phalloidin (Molecular Probes) (1:200) in a sealed humid container at 37°C for 45 min. The coverslips were returned to their original wells, washed with PBS-FSG-SAP twice for 10 min with rocking at 37°C, washed with PBS containing 0.1% (wt/vol) Triton X-100 twice for 5 min of rocking at 37°C, rinsed once with PBS, and dipped in distilled H2O before mounting on glass slides with the ProLong Antifade kit (Molecular Probes). Cells were observed using a Nikon Eclipse E800 fluorescence microscope. Images were captured at a magnification of ×1,000 under oil immersion using a CCD camera and SPOT imaging software.
Immunoblot analysis.
Five milliliters of bacteria grown overnight with shaking in LB medium at 37°C was centrifuged at 6,000 × g at 4°C for 20 min. Proteins were recovered from the supernatants by ammonium sulfate precipitation (final concentration of 55%). After incubation on ice for 18 h, precipitated proteins were concentrated by centrifugation at 13,000 × g at 4°C for 20 min. The pellet was boiled in 50 μl of 10 mM NaCl and 50 μl of 2× sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE) sample buffer for 5 min, and 20 μl of each sample was electrophoresed on a sodium dodecyl sulfate-8% polyacrylamide gel. Proteins were electrotransferred to a polyvinylidene difluoride membrane (Millipore) for immunoblot analysis, and ExoT was detected using a rabbit polyclonal ExoT antiserum at a 1:10,000 dilution (25). Goat anti-rabbit immunoglobulin G horseradish peroxidase conjugate (Jackson ImmunoResearch Laboratories) diluted 1:5,000 was used as a secondary antibody. Detection was performed using the chemiluminescent ECL system (Amersham Corp.).
Translocation assays.
HeLa cells were grown in MEM-Eagle medium with Earle's buffered salt solution medium to a density of approximately 1.0 × 106 cells in a 10-cm-diameter dish. Just prior to the addition of the bacteria, the cells were washed twice with PBS and 8.5 ml of MEM-Lite was added to each plate.
The ExoT chromosomal mutants were grown overnight in 2 ml of LB medium at 37°C with agitation. On the day of the experiment, the cultures were diluted 1:20 in fresh LB medium and allowed to grow for one hour at 37°C with agitation. Bacteria (1.5 ml) that had been diluted in MEM Lite to an OD600 of 0.1 was added dropwise to the HeLa cells. The plates were incubated for 90 min at 37°C in ambient air. Cells were then washed twice in PBS and lysed in 500 μl of PBS containing 1% Triton X-100 and one tablet of a protease inhibitor cocktail (Complete Mini; Roche Molecular Biosciences). Translocated proteins were visualized by Western blotting, using a rabbit anti-ExoT polyclonal antibody, as described above.
Western blot detection of transfected ExoT.
HeLa cells were plated into 24-well dishes and transfected as above without coverslips in the wells. At 18 h after transfection, wells were washed twice with PBS. The rounded, nonadherent cells contained in the media and washes were collected by centrifugation at 1,000 rpm for 5 min. The cell pellet and remaining adherent cells were lysed with 50 μl of 1% Triton X-100 in PBS with Complete Mini protease inhibitor cocktail (Roche) at room temperature for 10 min. The lysis mixture was centrifuged at 14,000 rpm for 15 min at room temperature. The supernatant was combined with 50 μl of 2× SDS-PAGE sample buffer. A 40-μl volume of the supernatant in sample buffer was electrophoresed on an SDS-8% polyacrylamide gel. Proteins were transferred to polyvinylidene difluoride membranes and immunoblotted for ExoT as described above. Duplicate membranes were immunoblotted with anti-anti-glyceraldehyde-3-phosphate dehydrogenase antibodies (Chemicon) as a measure of total cellular cytoplasmic proteins loaded.
Mass spectrometry analyses.
Bacteria were grown overnight in 5 ml of MinS-Ca2+. Bacteria were spun down, and the supernatant containing the type III secreted proteins was precipitated using 55% ammonium sulfate. Precipitated proteins were collected after overnight incubation on ice and resuspended in 10 mM NaCl. Sample buffer (2× SDS-PAGE buffer) was added, and the samples were loaded on an SDS-8% polyacrylamide gel. After Coomassie brilliant blue staining and destaining (10% methanol-10% glacial acetic acid), relevant bands were cut out and sent for mass spectrometry analyses. Briefly, the gel pieces were washed with 50 mM ammonium bicarbonate (pH 7.8)-acetonitrile (60:40) for 1 h at room temperature. The solution was removed and the gel pieces were vacuum dried for 25 min in a SpeedVac (Savant Instruments, Holbrook, N.Y.). Gel spots were rehydrated in 12 μl of sequencing-grade modified trypsin digest solution (12 ng/μl in 50 mM ammonium bicarbonate) at 4°C for 1 h. Excess trypsin solution was removed and the gel pieces were suspended in 20 to 30 μl of 50 mM ammonium bicarbonate and incubated overnight at 37°C. Trypsin-cleaved peptides eluted from the gel pieces were applied to target plates in two ways; firstly via a dried-droplet method, or following concentration and desalting using C18 Zip-Tips (Millipore, Bedford, Mass.). For a dried droplet, a 1-μl aliquot of eluted peptide sample was placed on the target plate and an equal volume of cyano-hydroxycinnamic acid matrix solution (10 mg/ml) was placed on top and allowed to air dry. For concentration and desalting, the Zip-Tips were activated and washed with 20 μl of acetonitrile and then acidified with 20 μl of 10% formic acid. Peptide solution was then slowly passed through the column using gentle air pressure. For peptide mass mapping, bound peptides were washed with 10% formic acid and then eluted from the Zip-Tip column onto the target plate with matrix solution.
Matrix-assisted laser desorption ionization-time of flight mass spectrometry peptide mass mapping was performed using a PerSeptive Biosystems (Framingham, Mass.) Voyager DE-STR instrument equipped with a 337-nm nitrogen laser. Mass spectra were obtained in reflectron/delayed extraction and linear modes, averaging 256 laser shots per sample. The ExoT protein sequence was subjected to a theoretical cleavage using the program PeptideMass (www.expasy.ch). Parameters for the database search were as follows: one missed cleavage and cysteine-acrylamide and methionine sulfoxide modifications allowed. The tryptic peptide digests yielded approximately 70% coverage of the protein.
RESULTS
Regions outside the GAP domain contribute to the biological activity of ExoT.
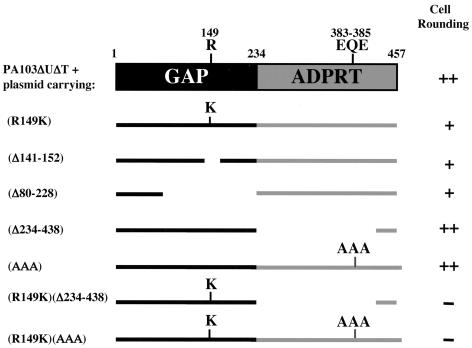
Earlier studies have demonstrated that the ADPRT domain of ExoT has very little activity in vitro towards a synthetic substrate (35). However, previous studies from our laboratory showed that mutation of the critical arginine residue (R149) of the GAP domain of ExoT results in loss of GAP activity in vitro but surprisingly resulted in only partial loss of cell rounding and anti-internalization activity (19). This observation suggested that the ADPRT domain or others regions of the GAP domain could account for this residual biological activity.
To address this question, deletion mutations throughout the GAP domain of ExoT were constructed and tested for biological activity. Because the effect of P. aeruginosa on host cell functions are difficult to assess in a cytotoxic strain, the effect of the various forms of ExoT on the host cell was studied using a strain in which the exoU gene and the exoT genes are deleted (PA103ΔUΔT). Deletion mutants of ExoT were cloned into a plasmid, introduced into PA103ΔUΔT, and tested for type III secretion and translocation, cell rounding and anti-internalization activity upon cocultivation with HeLa cells. Most of the mutants, except for ExoTΔ80-228, were translocated as efficiently as wild-type ExoT (data not shown but see also Fig. 1B). Deletions throughout the GAP domain of ExoT diminished but did not eliminate the cell rounding (summarized in Fig. 1) and anti-internalization activity (data not shown). Despite its decreased translocation, ExoTΔ80-228, which contains the putative membrane localization domain but is otherwise missing the entire GAP domain, had a phenotype indistinguishable from ExoT(R149K) (summarized in Fig. 1). Likewise, bacteria expressing ExoT harboring both the R149K mutation in conjunction with a deletion in other portions of the GAP domain did not exhibit any additional loss of activity (data not shown).
FIG. 1.
Summary of ExoT constructs and their phenotypes. A diagram of the various ExoT deletion and mutated constructs is shown on the left. On the right is a summary of the extent of host cell rounding after 3 h of cocultivation of HeLa cells with PA103ΔUΔT expressing the various ExoT constructs from a plasmid. Symbols: ++, >80% of the cells were rounded by 3 h; +, between 10 and 80% of the cells were rounded by 3 h; −, <10% of the cells were rounded by 3 h.
These findings indicated that the residual cell rounding and anti-internalization activity observed in the ExoT(R149K) mutant does not derive from other portions of the GAP domain and that the ADPRT domain likely contributes to these activities. To further test this hypothesis, we deleted the entire ADPRT domain from wild-type ExoT or GAP active-site mutant [ExoTΔ234-438 and ExoT(R149K)Δ234-438] and expressed it from a plasmid in PA103ΔUΔT. In these assays, which span 2 to 3 h, elimination of the ADPRT domain alone had minimal changes in cell rounding (summarized in Fig. 1) or anti-internalization activity (data not shown). Unexpectedly, when PA103ΔUΔT expressing ExoT with mutations in both the GAP and ADPRT domains [ExoT(R149K)Δ234-438] was cocultivated with HeLa cells, no anti-internalization or cell rounding activity was observed (summarized in Fig. 1). This finding further supports the notion that the ADPRT domain contributes to the biological activity of ExoT.
The ADPRT domain inhibits repair of wounded epithelium.
A key feature of acute P. aeruginosa infections is the presence of preexisting epithelial cell damage (7). Initial steps of wound repair, which largely involve cell migration, can be modeled by measuring the kinetics of wound closure in an epithelium that has been mechanically wounded (20, 31). In previous studies we have demonstrated that PA103 is able to inhibit repair of the scrape-wounded alveolar epithelial A549 cell line through a mechanism that required the GAP activity of ExoT (21). We wished to examine the contribution of the ADPRT domain to the inhibitory effect of ExoT on wound repair.
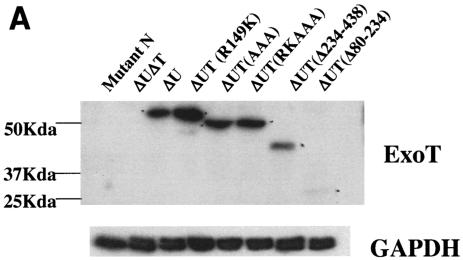
To this end, we constructed ExoT derivatives in which the ADPRT domain was either deleted or in which the catalytic site was altered in the chromosome by allelic exchange and measured their ability to inhibit wound repair in scrape-wounded A549 and MDCK cells. The MDCK model system yielded the most reproducible results, and they are reported here; similar results were observed with wounded A549 monolayers (data not shown). Using site-directed mutagenesis, we disrupted the catalytic site of the ADPRT domain of ExoT by changing the critical glutamic acid residues at amino acids 383 and 385 to alanine (40). To facilitate the site-directed mutagenesis, the glutamine residue at amino acid 384 was also changed to alanine. The resulting ADPRT catalytic site mutation is referred to as “AAA” (Fig. 1). A similar mutant, in which glutamic acid residues 383 and 385 have each been changed to aspartic acid, results in complete loss of ADPRT activity in vivo (48). Likewise, alterations of the corresponding glutamic acid residues in ExoS result in complete loss of ADPRT activity in vitro (40) and inhibit its effects on host cell changes in vivo (13). When expressed from the chromosome, the point mutants were all translocated into HeLa cells as efficiently as wild-type ExoT (Fig. 2A). Full-length ExoT carrying the AAA substitution migrated faster on SDS-polyacrylamide gels for reasons that are not clear and are currently under investigation. However, DNA sequence analysis of the ExoT(AAA) gene mutation and mass spectrometry analysis of the corresponding proteins confirmed the presence of the full-length protein (data not shown).
FIG. 2.
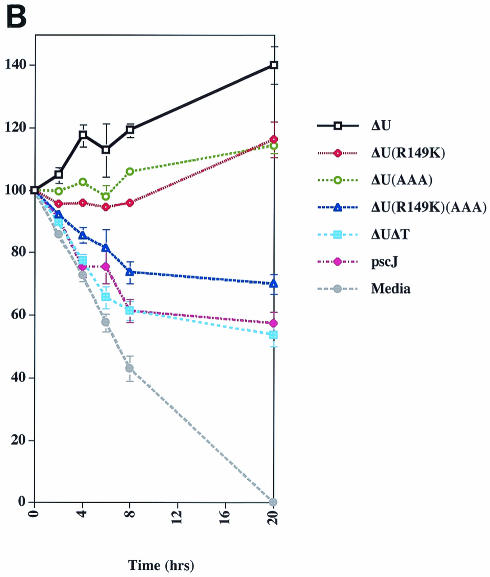
(A) Translocation efficiency of selected mutants integrated into the chromosome. The indicated strains were cocultivated with HeLa cells for 1.5 h, cytoplasmic extracts were prepared, and translocated ExoT was visualized by immunoblot analysis using a polyclonal ExoT antiserum. To control for cell loss and loading efficiency, the gels were also probed with anti-glyceraldehyde-3-phosphate dehydrogenase antiserum. ExoTΔ234-438 was translocated less efficiently than the other ExoT proteins. (B) Both domains of ExoT are required to impair epithelial wound healing. MDCK cells grown as confluent monolayers were mechanically wounded 1 h prior to infection with the indicated isogenic strains of PA103 in which the various ExoT mutants were expressed from the chromosome (MOI of ∼10). The percent wound area remaining (y axis) was measured at 0, 2, 4, 6, 8, and 20 h postwounding as described in Materials and Methods. The combined results from three separate experiments are shown. The area of the wound is expressed as mean percent of the initial wound area ± standard error of the mean (error bars) (for some time points, the error bars are too small to be visible). Wild-type ExoT results in increased wound area whereas the type III secretion mutant partially inhibits wound closure. The R149K and AAA mutants have similar phenotypes and prevent wound healing more than the type III secretion mutant but less than the wild-type ExoT.
Figure 2B shows the kinetics of MDCK cell wound repair after addition of strains of PA103ΔU bacteria expressing wild-type and mutant forms of ExoT encoded on the chromosome 1 h after wounding. Under the conditions of this experiment, an uninfected wound closed within approximately 20 h. Differences in the rate of wound closure could be detected within 4 h after addition of the bacteria, but were most prominent by 24 h. The type III secretion mutant PA103pscJ or the ExoT-null mutant (PA103ΔUΔT) partially inhibited wound closure (57% ± 3% and 54% ± 4%, respectively, of wound area remaining at 20 h; P < 0.01 by Student's two-tailed t test), consistent with previous reports by us (21) and others (6) that P. aeruginosa can inhibit epithelial wound healing in a type III secretion-independent manner. Remarkably, exposure of the wounded epithelium to PA103ΔU expressing wild-type ExoT led to extensive cell rounding and loss, and enlargement of the wound. By 20 h, the wound area had increased to 140% ± 6%, whereas untreated wounds closed fully. PA103ΔU expressing the double GAP and ADPRT active-site mutant [PA103ΔU/ExoT(R149K)(AAA)] allowed partial wound closure (70% ± 3%). In this particular set of experiments we observed a small difference in the amount of wound repair between the null mutant (PA103ΔUΔT) and the double ExoT mutant [PA103ΔU/ExoT(R149K)(AAA)], but this was not routinely observed in the other multiple times that this experiment was carried out and its significance is unknown. Strikingly, PA103ΔU expressing ExoT with an inactive ADPRT domain [PA103ΔU/ExoT(AAA)] or deleted ADPRT domain (PA103ΔU/ExoTΔ234-438 [data not shown]) showed intermediate inhibition of wound repair (final wound area of 114% ± 3%) compared to wild-type ExoT (P < 0.02 by Student's two-tailed t test) and the double mutant (P < 0.0004 by Student's two-tailed t test). Interestingly, the degree of inhibition was similar to an isogenic strain expressing ExoT with a catalytically inactive GAP domain [PA103ΔU/ExoT(R149K)] (final wound area of 116% ± 6%). Together, these experiments provide clear evidence that the ADPRT domain of ExoT plays a role in the inhibition of epithelial wound healing.
The GAP and ADPRT domains have distinct effects on the host cytoskeleton.
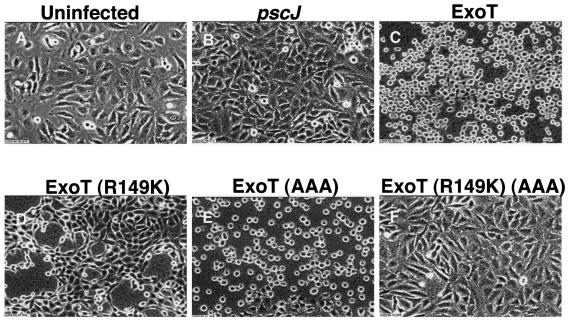
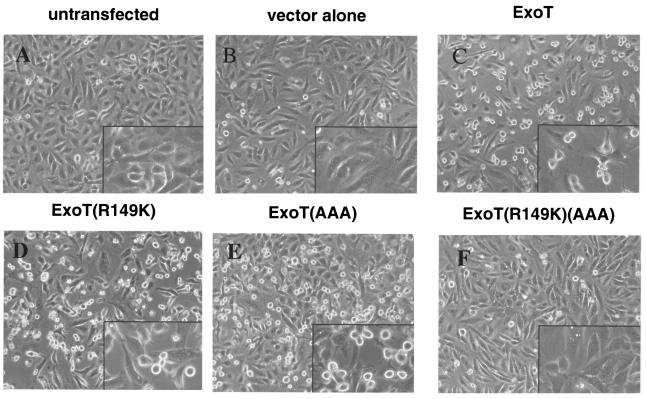
In previous studies of the GAP mutant [ExoT(R149K)], we observed partial loss of cell rounding activity (19). To further investigate the role of the ADPRT domain, the various ExoT chromosomal mutants were incubated with HeLa cells for 6 h and morphological changes were recorded by video microscopy. Figure 3 shows individual pictures taken at the end of the 6 h of cocultivation. Over this time period, the effects of cocultivation with PA103pscJ (Fig. 3B) and PA103ΔU/ExoT(R149K)(AAA) (Fig. 3F) were indistinguishable from uninfected cells (Fig. 3A). The cells were motile and cell division was observed (data not shown). Upon incubation with PA103ΔU/ExoT(AAA) (Fig. 3E), the cells rounded up, though some elongated processes were still observed. In contrast, PA103ΔU/ExoT(R149K) caused the HeLa cells to lose contact with the substrate while maintaining cell-cell contact (Fig. 3D and data not shown). The remaining cells had an elongated appearance and were separated by areas devoid of host cells. The morphology of HeLa cells exposed to PA103ΔU, which produced wild-type ExoT, appeared to be a combination of the effects seen with each separate active domain (Fig. 3C and data not shown). Interestingly, the kinetics of the cell rounding induced by wild-type ExoT was slower than what was observed with ExoT(AAA) (data not shown). These studies clearly indicate that the ADPRT domain affects host cell morphology and appears to affect a pathway distinct of that modulated by the GAP domain.
FIG. 3.
Both domains of ExoT contribute to cell rounding. The indicated bacterial strains were incubated with HeLa cells (MOI of 20) for 6 h. Video images were captured every 5 min using a Zeiss Axiovert microscope connected to a Cohu video camera, analyzed using the Openlab software (Improvision), and compressed using Adobe Premier 6.0. Shown are the picture frames obtained at the end of the 6-h incubation of each strain with HeLa cells. No changes in cell morphology were observed when HeLa cells were cocultivated with PA103pscJ (B) or PA103ΔU/ExoT(R149K)(AAA) (F). Cell rounding was observed with PA103ΔU (C) or PA103ΔU/ExoT(AAA) (E). Cell-cell contacts were maintained but cell-substrate binding was disrupted with PA103ΔU/ExoT(R149K) (D). The video movies are available upon request.
Both domains of ExoT are sufficient to disrupt the actin cytoskeleton.
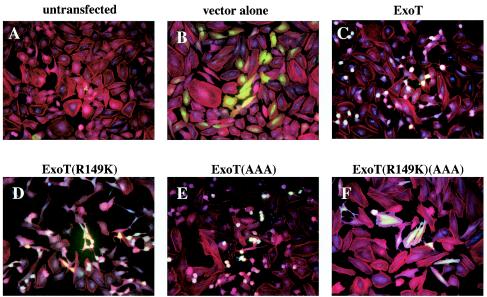
The results presented thus far show that both domains of ExoT are required for full cell rounding activity, but they do not rule out the possibility that other bacterial proteins contribute to this process. To investigate this, the various forms of ExoT were cloned under control of a strong constitutive promoter into a eukaryotic expression vector pIRES-EGFP and transiently transfected into HeLa cells. This construct allows for simultaneous expression and translation of both ExoT and GFP from a bicistronic transcript in the same cell. This approach ensures that all transfected cells, as evidenced by GFP expression, express ExoT. Cells transfected with a plasmid encoding wild-type ExoT or the ADPRT mutant showed extensive rounding (Fig. 4C and E and 5C and E) and loss of stress fibers (Fig. 5C and E). In contrast, cells transfected with a plasmid encoding the ExoT GAP mutant showed partial rounding (Fig. 4D) and partial loss of stress fibers (Fig. 5D). Convincingly, cells transfected with a plasmid encoding the ExoT double GAP and ADPRT active-site mutant (Fig. 4F and 5F) appeared similar to untransfected cells (Fig. 4A and 5A) or cells transfected with the control enhanced GFP vector (Fig. 4B and 5B) in both their flat shape and the presence of stress fibers. Western blot analysis of transfected cells revealed that the lack of effect of the double GAP and ADPRT active-site mutant was not due to a decrease in the expression of the protein (data not shown).
FIG. 4.
Both domains of ExoT are sufficient to mediate cell rounding in transfected cells. HeLa cells plated on coverslips were transfected with the indicated constructs for 18 h. This experiment was conducted in parallel with the immunofluorescence data presented in Fig. 5. The cells were observed by phase-contrast microscopy. Although we cannot distinguish in these micrographs which cells are transfected, it is clear that the extent of cell rounding is consistent with which ExoT construct was used. Cells transfected with plasmids encoding wild-type ExoT (C) or ExoT(AAA) (E) show rounding. (D) Cells transfected with a plasmid encoding ExoT(R149K) show partial rounding. Cells transfected with a plasmid encoding ExoT(R149K)(AAA) (F) have a similar morphology to empty vector-transfected (B) or untransfected (A) cells.
FIG. 5.
Both domains of ExoT contribute to actin cytoskeleton disruption in transfected cells. HeLa cells plated on coverslips were transfected with the indicated constructs for 18 h prior to fixation. All samples were stained with Phalloidin (red) to visualize the actin cytoskeleton. Transfected cells could be identified by GFP (green) and ExoT production was revealed by staining with a rabbit polyclonal antiserum against ExoT followed by staining with goat anti-rabbit-Cascade blue (blue). There is cross-reactivity of the ExoT antibody with the HeLa cell nucleus, but cells expressing ExoT can be clearly identified by their blue cytoplasmic staining. Cells transfected with plasmids encoding wild-type ExoT (C) or ExoT(AAA) (E) show rounding. (D) Cells transfected with a plasmid encoding ExoT(R149K) show partial rounding. Cells transfected with a plasmid encoding ExoT(R149K)(AAA) (F) have a similar morphology to empty vector-transfected (B) or untransfected (A) cells.
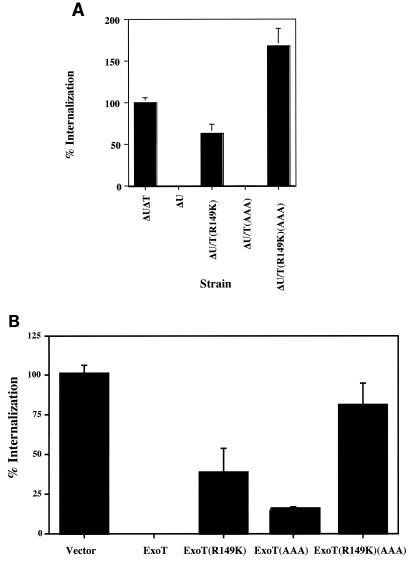
Both domains are required for full anti-internalization activity.
An important biological function of ExoT is its ability to prevent bacterial internalization into eukaryotic cells. Our previous studies had suggested a role for the GAP domain of ExoT in the inhibition of internalization (19). Using both the chromosomal and plasmid-encoded mutants, the role of the ADPRT domain in anti-internalization activity was directly tested. HeLa or MDCK cells were incubated with P. aeruginosa expressing the wild-type or mutant forms of ExoT for 3 h, extracellular bacteria were killed by addition of amikacin, and adherent and nonadherent cells were collected and quantified prior to release of intracellular bacteria by Triton lysis. The number of internalized bacteria in different strains was normalized to the total host cell number. Figure 6A shows that in HeLa cells, bacteria expressing wild-type ExoT (ΔU) were internalized over 300-fold less efficiently than the bacteria that lacked ExoT (ΔUΔT). The GAP active-site mutant [ΔU/ExoT(R149K)] was internalized almost as efficiently as the mutant that lacked ExoT, confirming that the GAP activity is crucial in preventing PA103 internalization. In contrast, the ADPRT active-site mutant [ΔU/ExoT(AAA)] was internalized poorly, suggesting that the ADPRT active site mutant retained close to wild-type anti-internalization activity. We note that when overexpressed from a plasmid, the ExoT(AAA) mutant had diminished anti-internalization activity compared to wild-type ExoT overexpressed from a plasmid (Fig. 6B). Strikingly, whether expressed from a plasmid or from the chromosome, the GAP/ADPRT double mutant (ΔU/ExoT(R149K)(AAA) had complete loss of anti-internalization activity (Fig. 6). Similar results were observed with MDCK cells (data not shown). There was no difference in growth of the various mutants or adherence of the different strains of bacteria to the HeLa or MDCK cells and all mutants were equally sensitive to the antibiotic amikacin (data not shown).
FIG. 6.
Both domains of ExoT contribute to its anti-internalization activity. HeLa cells were infected at an MOI of 25 for 3 h with the indicated strains expressing various forms of ExoT from the chromosome (A) or from a plasmid (B). Bacterial invasion was quantified using a standard aminoglycoside exclusion assay as described in Materials and Methods. The results shown are the average ± standard error of the mean (error bars) for six wells. The percent invasion is normalized to that of PA103ΔUΔT (plus vector for panel B) and to total lactate dehydrogenase units per well to compensate for cell loss due to ExoT-mediated cell rounding and loss of attachment.
Both domains of ExoT are toxic when expressed in yeast.
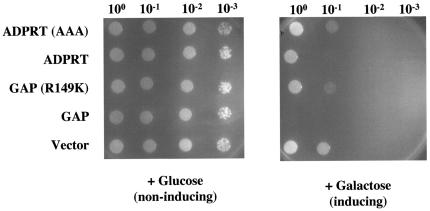
Many proteins and signaling pathways found in higher eukaryotes are conserved in S. cerevisiae, and the ease of genetic manipulation in this organism makes it an attractive model system. Several type III secreted effectors have been expressed in yeast and are localized correctly (34). For example YopE, a bacterial GAP produced by Yersinia species, disrupts the actin cytoskeleton when expressed in S. cerevisiae, resulting in toxicity (50). To further address whether the ADPRT domain of ExoT is active, we cloned the wild-type and active-site mutant domains under control of constitutive and inducible promoters and introduced them into yeast. For comparison, both the wild-type and active-site mutant GAP domains were also introduced into yeast. When expressed under control of a constitutive promoter, transformation of yeast with a plasmid expressing the wild-type GAP domain yielded very few colonies (transformation efficiency of 102 per μg of DNA). In contrast, yeast transformed with a plasmid expressing the mutant GAP domain (R149K) yielded 105 colonies per μg of DNA. As determined by Western blot analysis, the few colonies of yeast transformed with the wild-type GAP domain that grew no longer expressed ExoT (data not shown). Transformation of yeast with a plasmid expressing either the wild-type or the mutant ADPRT domain yielded abundant colonies (105/μg DNA). The colonies transformed with the mutant ADPRT domain were much smaller, suggesting that the ADPRT domain was expressed and functional, though the levels of the wild-type ADPRT were below detection by immunoblot analysis (data not shown). Together, these results suggest that expression of either the GAP or the ADPRT domain was toxic to yeast.
The individual wild-type and mutant GAP and ADPRT domains were then cloned into an expression plasmid under control of a galactose-inducible/glucose repressible promoter, transformed into yeast, and selected for growth on media lacking tryptophan and containing glucose. Following overnight growth under noninducing conditions, serial 10-fold dilutions were plated onto glucose-containing (noninducing) and galactose-containing (inducing) conditions. Figure 7 shows that yeast strains expressing either the wild-type or mutant GAP or ADPRT domains plated under noninducing conditions grew equally well as yeast containing the vector only. However, when plated under inducing conditions, yeast transformants expressing the wild-type GAP and wild-type ADPRT domains grew substantially less well than yeast expressing the mutant domains or yeast expressing only the vector. Of note, even the vector-only-containing strain grew less well on galactose-containing media compared to its growth on glucose-containing media, a less efficient carbon source. Together these results demonstrate that ExoT is functional in yeast and that intact GAP or ADPRT domains are required to inhibit growth. Furthermore, it suggests that S. cerevisiae can be used as a model system to study the targets of ExoT.
FIG. 7.
Growth of yeast is inhibited by expression of the GAP or ADPRT domains of ExoT. S. cerevisiae strain L4852 carrying a high-copy-number plasmid encoding galactose-inducible expression of the indicated domains of ExoT were grown overnight in noninducing synthetic selective broth containing glucose as a carbon source. The cultures were washed twice in water and normalized to an OD600 of 1.0. Tenfold serial dilutions were then spotted onto selective medium plates containing 1% glucose (noninducing) or 1% galactose (inducing).
DISCUSSION
P. aeruginosa possesses an impressive array of cell-associated and secreted factors to subvert host cell functions and resist host cell defenses. A key protein in this host pathogen interplay is the type III secreted protein ExoT. Upon translocation into the host cell, this toxin contributes to disruption of the epithelial cell barrier in several ways. It causes disruption of the host cell actin cytoskeleton, leading to cell rounding and loss of cell-cell junctions. It prevents epithelial cell migration in the context of wound healing. Finally, it inhibits bacterial internalization into epithelial cells and macrophages. Avoiding uptake by macrophages may further contribute to bacterial evasion of host cell defenses.
In previous studies, we have shown that the GAP activity of ExoT towards Rho, Rac, and Cdc42 contributes to each of these processes but that mutations in the GAP domain, despite their complete loss of GAP activity, only partially eliminated cell rounding and anti-internalization activity (19). In this report, we definitively demonstrate by several assays that the ADPRT domain clearly contributes to the biological functions of ExoT. After first eliminating the possibility that other portions of the GAP domain contribute to its biological activity, we found that deletions in the ADPRT domain diminished ExoT activity as did mutating the two key catalytic glutamic acid residues in the ADPRT domain (which results in loss of in vivo ADPRT activity (48). Inactivation of both domains led to a complete loss of activity in the wound healing, cell rounding, and internalization assays. Expression of either an active GAP or ADPRT domain in yeast inhibited growth, suggesting that both domains are catalytically active and that there are yeast targets for these two enzymatic activities. While other bacterial GAPs have been shown to be toxic to yeast (34), to the best of our knowledge, this is the first demonstration that a bacterial ADPRT inhibits yeast growth.
The role of the ADPRT domain was most striking in the wound healing experiments. Indeed, inactivation of the ADPRT domain, even in the context of an active GAP domain, reduced its ability to inhibit epithelial cell migration and wound closure. The activity of the GAP mutant in this assay was also diminished to a similar extent, suggesting that both domains contribute equally to this physiologically important activity. This loss of function was observed as early as 4 h after infection, though it became more accentuated at later time points. Interestingly, we have previously observed that the subcellular localization of paxillin and focal adhesion kinase was differentially affected by different mutants of ExoT (21). These two proteins are components of focal adhesions and their activity and/or phosphorylation states are modulated as cells migrate (37). When scrape-wounded cells were exposed to wild-type ExoT, these proteins, along with actin, redistributed from focal adhesions to a glob-like structure. In contrast, exposure to the GAP mutant [ExoT(R149K)] resulted in a cytoplasmic staining pattern of these two proteins. This observation suggests that the ADPRT activity of ExoT affects focal adhesions.
In contrast, in the cell rounding experiments, the GAP activity was most important for the early changes in cell morphology. This observation is consistent with our previously published findings that ExoT-mediated cell rounding could be blocked by overexpression of constitutively active RhoA (29). However, video microscopy revealed the effect exerted by the ADPRT domain. Consistent with the static pictures, the GAP activity caused extensive cell rounding, with loss of cell-cell contacts. In contrast, the ADPRT domain appeared primarily to affect cell-substrate adhesion and while leaving cell-cell contacts unaffected. The effect of wild-type ExoT appeared to be a sum of the GAP and ADPRT activities.
As suggested by our previous demonstration that P. aeruginosa enters cells through a RhoA-dependent pathway (30), the GAP activity of ExoT accounted for most of the anti-internalization activity. However, in both the cell rounding and anti-internalization assays, the double mutant had a more severe loss of function phenotype than either single mutant, again underscoring the contribution of the ADPRT domain.
While this work was under review, Sun and Barbieri identified three to five targets of ExoT, depending upon the cell type (48). Besides auto-ADP ribosylation of itself, ExoT ADP ribosylates two isoforms of Crk. This SH2- and SH3-containing adaptor protein links integrin-mediated signaling through focal adhesions via Rac and Rap1 to multiple host cell responses, including cell migration and adhesion (reviewed in reference 9). Our results, together with theirs, provide an explanation as to why the ADPRT activity of ExoT is prominent in wound healing and cell migration as well as a mechanism for its effects on host cell morphology. We would suggest that the anti-internalization activity of ExoT on epithelial cells reflects the action of its GAP domain on RhoA-modulated processes. The effect on epithelial cell morphology again is primarily through the inactivation of Rho by the GAP activity of ExoT, but there is also a contribution by the ADPRT domain. The loss of cell substrate adhesion seen with the ADPRT domain alone may reflect the disruption of focal adhesions via an effect on Crk. Finally, the inhibition of wound migration involves the inactivation of Rho, Rac, and Cdc42 by the GAP activity as well as the modulation of Crk activity by the ADPRT domain of ExoT. There are likely other targets of the ADPRT activity of ExoT which may affect additional processes. Consistent with this prediction is our observation that expression of the ExoT ADPRT domain in yeast is toxic, despite the fact that S. cerevisiae lacks a Crk homolog.
ExoT thus joins a growing family of bifunctional proteins that are translocated by the type III secretion system from gram-negative bacteria into eukaryotic cells. There are now several examples in which a GAP domain is fused to an ADPRT domain, including the closely related protein ExoS as well as AexT, a recently identified type III secreted protein in Aeromonas salmonicida (3, 4). In the case of SptP of Salmonella enterica serovar Typhimurium, the GAP domain is fused to a domain with tyrosine phosphatase activity (15, 16). It is interesting to speculate on the evolutionary advantages of having a protein that encodes two different enzymatic activities. It may simply be a more cost-efficient way to introduce bacterial proteins into host cells (“two for the price of one”). Alternatively, both domains may share a substrate or benefit from being colocalized in the host cell. For example, Rac is apparently a substrate for both the GAP and ADPRT domains of ExoS (14, 33) and the ExoS ADPRT domain preferentially modifies membrane-bound over cytoplasmic Ras (41). Finally, the activity of one domain may modify the activity of the second domain. In fact, it has recently been shown that the ADPRT domain of ExoS modifies the catalytic arginine in its own GAP domain and thus has the potential to down-regulate its own GAP activity (42). Likewise, ExoT exhibits auto-ADP ribosylation activity in vivo (48). It will be interesting to determine if ExoT exhibits a similar regulatory mechanism. We have observed that the when the GAP domain is expressed in the absence of a functional ADPRT domain, cell rounding occurs faster and the in vivo GAP activity is greater (P. Balachandran, S. Shafikhani, and J. Engel, unpublished data). Together, these observations suggest that ExoT may similarly downregulate its GAP activity by ADP ribosylation of the critical catalytic arginine residue.
In summary, our work suggests that the ADPRT domain of ExoT is functional in vivo and that the modulation of target proteins synergizes with the GAP activity to disrupt the actin cytoskeleton. These activities likely contribute to the role of ExoT in the pathogenesis of P. aeruginosa infections.
Acknowledgments
We thank Keith Mostov, Eric Brown, Andrew Finch, and members of the Engel laboratory for continuing discussions and thoughtful comments on the paper. We thank Hitem Madhani and Joachim Li for yeast strains and plasmids.
This work was supported by grants from the NIH (AI 42806 and HL55980 to J.N.E., HL51854 to M.A.M., and AI053194 to J.N.E. and to M.A.M). L.K.G.-R. was supported by the Bank of American Gianinni Foundation and the American Lung Association.
Editor: D. L. Burns
REFERENCES
- 1.Alm, R. A., and J. A. Mattick. 1996. Identification of two genes with prepilin-like leader sequences required for type 4 fimbrial biogenesis in Pseudomonas aeruginosa. J. Bacteriol. 178:3809-3817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Barbieri, A. M., Q. Sha, P. Bette-Bobillo, P. D. Stahl, and M. Vidal. 2001. ADP-ribosylation of Rab5 by ExoS of Pseudomonas aeruginosa affects endocytosis. Infect. Immun. 69:5329-5334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Braun, M., K. Stuber, Y. Schlatter, T. Wahli, P. Kuhnert, and J. Frey. 2002. Characterization of an ADP-ribosyltransferase toxin (AexT) from Aeromonas salmonicida subsp. salmonicida. J. Bacteriol. 184:1851-1858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Burr, S. E., K. Stuber, T. Wahli, and J. Frey. 2002. Evidence for a type III secretion system in Aeromonas salmonicida subsp. salmonicida. J. Bacteriol. 184:5966-5970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cowell, B. A., D. Y. Chen, D. W. Frank, A. J. Vallis, and S. M. J. Fleiszig. 2000. ExoT of cytotoxic Pseudomonas aeruginosa prevents uptake by corneal epithelial cells. Infect. Immun. 68:403-406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.de Bentzmann, S., M. Polette, J. M. Zahm, J. Hinnrasky, C. Kileztky, O. Bajolet, J. M. Klossek, A. Filloux, A. Lazdunski, and E. Puchelle. 2000. Pseudomonas aeruginosa virulence factors delay airway epithelial wound repair by altering the actin cytoskeleton and inducing overactivation of epithelial matrix metalloproteinase-2. Lab. Investig. 80:209-219. [DOI] [PubMed] [Google Scholar]
- 7.de Bentzmann, S., P. Roger, and E. Puchelle. 1996. Pseudomonas aeruginosa adherence to remodelling respiratory epithelium. Eur. Resp. J. 10:2145-2150. [DOI] [PubMed] [Google Scholar]
- 8.Engel, J. N. (ed.). 2002. Molecular pathogenesis of acute Pseudomonas aeruginosa infections. Kluwer Academic/Plenum Press, New York, N.Y.
- 9.Feller, S. M. 2001. Crk family adaptors-signalling complex formation and biological roles. Oncogene 20:6348-6371. [DOI] [PubMed] [Google Scholar]
- 10.Feltman, H., G. Schulert, S. Khan, M. Jain, L. Peterson, and A. R. Hauser. 2001. Prevalence of type III secretion genes in clinical and environmental isolates of Pseudomonas aeruginosa. Microbiology 147:2659-2669. [DOI] [PubMed] [Google Scholar]
- 11.Finck-Barbancon, V., J. Goranson, L. Zhu, T. Sawa, J. P. Wiener-Kronish, S. M. J. Fleiszig, C. Wu, L. Mende-Mueller, and D. Frank. 1997. ExoU expression by Pseudomonas aeruginosa correlates with acute cytotoxicity and epithelial injury. Mol. Microbiol. 25:547-557. [DOI] [PubMed] [Google Scholar]
- 12.Frank, D. W. 1997. The exoenzyme S regulon of Pseudomonas aeruginosa. Mol. Microbiol. 4:621-629. [DOI] [PubMed] [Google Scholar]
- 13.Fraylick, J. E., J. R. La Rocque, T. S. Vincent, and J. C. Olson. 2001. Independent and coordinate effects of ADP-ribosyltransferase and GTPase-activating activities of exoenzyme S on HT-29 epithelial cell function. Infect. Immun. 69:5318-5328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fraylick, J. E., E. A. Rucks, D. M. Greene, T. S. Vincent, and J. C. Olson. 2002. Eukaryotic cell determination of ExoS ADP-ribosyltransferase substrate specificity. Biochem. Biophys. Res. Commun. 291:91-100. [DOI] [PubMed] [Google Scholar]
- 15.Fu, Y., and J. E. Galan. 1999. A Salmonella protein antagonizes Rac-1 and Cdc42 to mediate host-cell recovery after bacterial invasion. Nature 401:293-297. [DOI] [PubMed] [Google Scholar]
- 16.Fu, Y., and J. E. Galan. 1998. The Salmonella typhimurium tyrosine phosphatase SptP is translocated into host cells and disrupts the actin cytoskeleton. Mol. Microbiol. 27:359-368. [DOI] [PubMed] [Google Scholar]
- 17.Ganesan, A. K., D. W. Frank, R. P. Misra, G. Schmidt, and J. T. Barbieri. 1998. Pseudomonas aeruginosa exoenzyme S ADP-ribosylates Ras at multiple sites. J. Biol. Chem. 273:7332-7337. [DOI] [PubMed] [Google Scholar]
- 18.Ganesan, A. K., T. S. Vincent, J. C. Olson, and J. T. Barbieri. 1999. Pseudomonas aeruginosa exoenzyme S disrupts Ras-mediated signal transduction by inhibiting guanine nucleotide exchange factor-catalyzed nucleotide exchange. J. Biol. Chem. 274:21823-21829. [DOI] [PubMed] [Google Scholar]
- 19.Garrity-Ryan, L., B. Kazmierczak, R. Kowal, J. Commolli, A. Hauser, and J. Engel. 2000. The arginine finger domain of ExoT is required for actin cytoskeleton disruption and inhibition of internalization of Pseudomonas aeruginosa by epithelial cells and macrophages. Infect. Immun. 68:7100-7113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Geiser, T., P. H. Jarreau, K. Atabai, and M. A. Matthay. 2000. Interleukin-1β augments in vitro alveolar epithelial repair. Am. J. Physiol. Lung Cell. Mol. Physiol. 279:L1184-L1190. [DOI] [PubMed] [Google Scholar]
- 21.Geiser, T., B. Kazmierczak, L. Garrity-Ryan, M. Matthay, and J. Engel. 2001. Pseudomonas aeruginosa ExoT inhibits in vitro lung epithelial wound repair. Cell. Microbiol. 3:223-236. [DOI] [PubMed] [Google Scholar]
- 22.Goehring, U. M., G. Schmidt, K. J. Pederson, K. Aktories, and J. T. Barbieri. 1999. The N-terminal domain of Pseudomonas aeruginosa exoenzyme S is a GTPase-activating protein for Rho GTPases. J. Biol. Chem. 274:36369-36372. [DOI] [PubMed] [Google Scholar]
- 23.Hauser, A., E. Cobb, M. Bodi, D. Mariscal, J. Valles, J. Engel, and J. Rello. 2002. Impact of Pseudomonas aeruginosa type III secretion on clinical outcomes in patients with ventilator-associated pneumonia. Crit. Care Med. 30:521-528. [DOI] [PubMed] [Google Scholar]
- 24.Hauser, A. R., P. J. Kang, and J. Engel. 1998. PepA, a novel secreted protein of Pseudomonas aeruginosa, is necessary for cytotoxicity and virulence. Mol. Microbiol. 27:807-818. [DOI] [PubMed] [Google Scholar]
- 25.Hauser, A. R., P. J. Kang, S. J. M. Fleiszig, K. Mostov, and J. Engel. 1998. Defects in type III secretion correlate with internalization of Pseudomonas aeruginosa by epithelial cells. Infect. Immun. 66:1413-1420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hueck, C. 1998. Type III protein secretion systems in bacterial pathogens of animals and plants. Microbiol. Mol. Biol. Rev. 62:379-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.James, P., J. Halladay, and E. A. Craig. 1996. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics 144:1425-1436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kang, P. J., A. R. Hauser, G. Apodaca, S. Fleiszig, J. Wiener-Kronish, K. Mostov, and J. N. Engel. 1997. Identification of Pseudomonas aeruginosa genes required for epithelial cell injury. Mol. Microbiol. 24:1249-1262. [DOI] [PubMed] [Google Scholar]
- 29.Kazmierczak, B., and J. Engel. 2002. Pseudomonas aeruginosa ExoT acts in vivo as a GTPase activating protein for RhoA, Rac1, and Cdc42. Infect. Immun. 70:2198-2205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kazmierczak, B. I., T.-S. Jou, K. Mostov, and J. Engel. 2001. Rho-GTPase activity modulates Pseudomonas aeruginosa internalization by epithelial cells. Cell. Microbiol. 3:85-98. [DOI] [PubMed] [Google Scholar]
- 31.Kheradmand, F., H. G. Folkesson, L. Shum, R. Derynk, R. Pytela, and M. A. Matthay. 1994. Transforming growth factor enhances alveolar epithelial repair in a new in vitro model. Am. J. Physiol. 267:728-738. [DOI] [PubMed] [Google Scholar]
- 32.Krall, R., G. Schmidt, K. Aktories, and J. T. Barbieri. 2000. Pseudomonas aeruginosa ExoT is a Rho GTPase-activating protein. Infect. Immun. 68:6066-6068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Krall, R., J. Sun, K. Pederson, and J. Barbieri. 2002. In vivo Rho GTPase-activating protein activity of Pseudomonas aeruginosa cytotoxin ExoS. Infect. Immun. 70:360-367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lesser, C. F., and S. I. Miller. 2001. Expression of microbial virulence proteins in Saccharomyces cerevisiae models mammalian infection. EMBO J. 20:1840-1849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu, S., T. L. Yahr, D. W. Frank, and J. T. Barbieri. 1997. Biochemical relationships between the 53-kilodalton (Exo53) and 49-kilodalton (ExoS) forms of exoenzyme S of Pseudomonas aeruginosa. J. Bacteriol. 179:1609-1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mumberg, D., R. Muller, and M. Funk. 1994. Regulatable promoters of Saccharomyces cerevisiae: comparison of transcriptional activity and their use for heterologous expression. Nucleic Acids Res. 22:5767-5768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Panetti, T. S. 2002. Tyrosine phosphorylation of paxillin, FAK, and p130CAS: effects on cell spreading and migration. Front. Biosci. 7:d143-d150. [DOI] [PubMed] [Google Scholar]
- 38.Pederson, K. J., and J. T. Barbieri. 1998. Intracellular expression of the ADP-ribosyltransferase domain of Pseudomonas exoenzyme S is cytotoxic to eukaryotic cells. Mol. Microbiol. 30:751-760. [DOI] [PubMed] [Google Scholar]
- 39.Pederson, K. J., A. J. Vallis, K. Aktories, D. W. Frank, and J. T. Barbieri. 1999. The amino-terminal domain of Pseudomonas aeruginosa ExoS disrupts actin filaments via small-molecular-weight GTP-binding proteins. Mol. Microbiol. 32:393-401. [DOI] [PubMed] [Google Scholar]
- 40.Radke, J., K. J. Pederson, and J. T. Barbieri. 1999. Pseudomonas aeruginosa exoenzyme S is a biglutamic acid ADP-ribosyltransferase. Infect. Immun. 67:1508-1510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Riese, M. J., and J. T. Barbieri. 2002. Membrane localization contributes to the in vivo ADP-ribosylation of Ras by Pseudomonas aeruginosa ExoS. Infect. Immun. 70:2230-2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Riese, M. J., U. M. Goehring, M. E. Ehrmantraut, J. Moss, J. T. Barbieri, K. Aktories, and G. Schmidt. 2002. Auto-ADP-ribosylation of Pseudomonas aeruginosa ExoS. J. Biol. Chem. 277:12082-12088. [DOI] [PubMed] [Google Scholar]
- 43.Riese, M. J., A. Wittinghofer, and J. T. Barbieri. 2001. ADP ribosylation of Arg41 of Rap by ExoS inhibits the ability of Rap to interact with its guanine nucleotide exchange factor, C3G. Biochemistry 40:3289-3294. [DOI] [PubMed] [Google Scholar]
- 44.Roy-Burman, A., R. Savel, S. Racine, B. Swanson, N. Revadigar, J. Fijimoto, T. Sawa, D. Frank, and J. Wiener-Kronish. 2001. Type III protein secretion is associated with death in lower respiratory and systemic Pseudomonas aeruginosa infections. J. Infect. Dis. 183:1767-1774. [DOI] [PubMed] [Google Scholar]
- 45.Sato, H., D. W. Frank, C. J. Hillard, J. B. Feix, R. R. Pankhaniya, K. Moriyama, V. Finck-Barbancon, A. Buchaklian, M. Lei, R. M. Long, J. Wiener-Kronish, and T. Sawa. 2003. The mechanism of action of the Pseudomonas aeruginosa-encoded type III cytotoxin, ExoU. EMBO J. 22:2959-2969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Simon, R., U. Priefer, and A. Puhler. 1983. A broad host range mobilization system for in vivo genetic engineering: transposon mutagenesis in gram negative bacteria. Bio/Technology 1:784-791. [Google Scholar]
- 47.Stebbins, C. E., and J. E. Galan. 2001. Structural mimicry in bacterial virulence. Nature 412:701-705. [DOI] [PubMed] [Google Scholar]
- 48.Sun, J., and J. T. Barbieri. 2003. Pseudomonas aeruginosa ExoT ADP-ribosylates CT10 regulator of kinase (Crk) proteins. J. Biol. Chem. 278:32794-32800. [DOI] [PubMed]
- 49.Vallis, A. J., V. Finck-Barbançon, T. L. Yahr, and D. W. Frank. 1999. Biological effects of Pseudomonas aeruginosa type III-secreted proteins on CHO cells. Infect. Immun. 67:2040-2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.von Pawel-Rammingen, U. M., V. Telpnev, G. Schmidt, K. Aktories, H. Wolf-Watz, and R. Rosqvist. 2000. GAP activity of the Yersinia YopE cytotoxin specifically targets the Rho pathway: a mechanism for disruption of actin microfilament structure. Mol. Microbiol. 36:737-748. [DOI] [PubMed] [Google Scholar]
- 51.West, S. E., S. A. Kaye, A. N. Hamood, and B. H. Iglewski. 1994. Characterization of Pseudomonas aeruginosa mutants that are deficient in exotoxin A synthesis and are altered in expression of regA, a positive regulator of exotoxin A. Infect. Immun. 62:897-903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yahr, T. L., J. T. Barbieri, and D. W. Frank. 1996. Genetic relationship between the 53- and 49-kilodalton forms of exoenzyme S from Pseudomonas aeruginosa. J. Bacteriol. 178:1412-1419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Yahr, T. L., A. J. Vallis, M. K. Hancock, J. T. Barbieri, and D. W. Frank. 1998. ExoY, an adenylate cyclase secreted by the Pseudomonas aeruginosa type III system. Proc. Natl. Acad. Sci. USA 95:13899-13904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang, L., H. Wang, S. C. Masters, B. Wang, J. T. Barbieri, and H. Fu. 1999. Residues of 14-3-3 zeta required for activation of exoenzyme S of Pseudomonas aeruginosa. Biochemistry 38:12159-12164. [DOI] [PubMed] [Google Scholar]