Abstract
Chronic lung infections with Pseudomonas aeruginosa biofilms are associated with refractory and fatal pneumonia in cystic fibrosis (CF). In this study, a group of genomically diverse P. aeruginosa isolates were compared with the reference strain PAO1 to assess the roles of motility, twitching, growth rate, and overproduction of a capsular polysaccharide (alginate) in biofilm formation. In an in vitro biofilm assay system, P. aeruginosa displayed strain-specific biofilm formation that was not solely dependent on these parameters. Compared with non-CF isolates, CF isolates expressed two opposing growth modes: reduced planktonic growth versus efficient biofilm formation. Planktonic cells of CF isolates showed elevated sensitivity to hydrogen peroxide, a reactive oxygen intermediate, and decreased lung colonization in an aerosol infection mouse model. Despite having identical genomic profiles, CF sequential isolates produced different amounts of biofilm. While P. aeruginosa isolates exhibited genomic diversity, the genome size of these isolates was estimated to be 0.4 to 19% (27 to 1,184 kb) larger than that of PAO1. To identify these extra genetic materials, random amplification of polymorphic DNA was coupled with PAO1-subtractive hybridization. Three loci were found within the genomes of two CF isolates encoding one novel homolog involved in retaining a Shigella virulence plasmid (mvpTA) and two divergent genes that function in removing negative supercoiling (topA) and biosynthesis of pyoverdine (PA2402). Together, P. aeruginosa biodiversity could provide one cause for the variation of morbidity and mortality in CF. P. aeruginosa may possess undefined biofilm adhesins that are important to the development of an antibiofilm therapeutic target.
Pseudomonas aeruginosa, a gram-negative environmental bacterium, is responsible for the majority of morbidity and mortality in cystic fibrosis (CF) (27). Lung infections with this bacterium manifest with varied disease severity. Particularly, in CF chronic lung infections, P. aeruginosa develops a characteristic phenotype, surrounded by an overproduced capsular polysaccharide called alginate (17) and associated into large bacterial conglomerates known as biofilms (9). It is this formation that allows the bacteria to survive within the CF lung and elicit unproductive immune responses that ultimately result in the demise of the patient (19).
Biofilms are formed from individual free-floating (planktonic) cells and are defined as exopolysaccharide-surrounded bacteria, or microcolonies, growing on biotic or abiotic surfaces (37). Biofilms are ubiquitous in nature and are also associated with numerous chronic or recurrent bacterial infections and diseases (13). Formation of biofilms by this bacterium can be viewed as a developmental process (31) that is roughly divided into four steps: (i) adhesion, (ii) monolayer, (iii) microcolony, and (iv) mature biofilms (10). In recent years, considerable progress has been made with regard to the molecular components that promote biofilm formation. Several surface-associated factors, such as flagella and type IV pili, have been shown to be essential for adhesion and microcolony formation, respectively (32, 49), as well as other undefined adhesins (12, 35, 52, 53). Flagella act to overcome repulsive forces between the bacterium and surface to allow the initial contact (37). Following adhesion of single bacteria into the formation of a monolayer, twitching motility is used to initiate the grouping of cells into microcolonies (32). Further development of biofilms leads to the emergence of mature biofilms, which are characterized by alginate-encased bacterial aggregates (17).
The genome of the reference strain P. aeruginosa PAO1 is available (50). However, the functions of the majority of the genes within this genome remain elusive. Furthermore, P. aeruginosa isolates are known to possess extensive genome diversity (40), the origin of which is yet to be completely defined. There are, however, many isolates that carry additional genetic materials that are missing in PAO1 (25, 47). Two such examples include the identification of P. aeruginosa genomic islands 1 (26) and 2 (2) (PAGI-1 and -2, respectively). While the functions of PAGI-1 are proposed to play a role in immune evasion (26), PAGI-2 is involved in glycosylation of the flagellin protein, a function that is not present in PAO1 (2). Therefore, genomic diversity may allow this bacterium to expand its pathogenic potentials.
While CF isolates are known to have the ability to attach to a surface and form biofilms (46), it is not clear whether there is an intrinsic difference in biofilm formation among genomically diverse environmental and clinical isolates of P. aeruginosa. In this report, we will examine the roles of four biofilm-related factors in biofilm formation: motility, twitching, growth rate, and overproduction of alginate. Our studies have shown that while the activities of flagella and type IV pili are required for the initiation of biofilm formation, they play no quantitative role in the progressive development of the initial biofilm microcolony. Also, we demonstrate increased H2O2 sensitivity and decreased lung colonization in an aerosol infection mouse model with CF isolates relative to non-CF isolates. Two opposing modes of growth are indicative of CF isolates: efficient biofilm formation and reduced planktonic growth rate. The genomic diversity found throughout many P. aeruginosa isolates appears to be due considerably to the presence of large portions of DNA not found in PAO1. We also examined the cause of genomic diversity as a function of novel DNA inclusion. Rapid amplification of polymorphic DNA (RAPD) coupled with subtractive hybridization was used to identify genes novel to or existing in a highly varied form in PAO1. Our results lead us to conclude that CF isolates of P. aeruginosa differ greatly in many aspects of biofilm formation among each other and also when compared with non-CF isolates.
MATERIALS AND METHODS
Bacterial strains.
The strains used in this study are listed in Table 1. P. aeruginosa and Escherichia coli strains were grown at 37°C in Lennox broth (LB), on LB agar, or Pseudomonas isolation agar (PIA; DIFCO) plates unless otherwise noted. For the flagellar activity assay, P. aeruginosa was grown on motility test medium (MTM; Becton Dickinson).
TABLE 1.
Phenotypic characteristics of P. aeruginosa isolates used in this study
| Isolatea | Increase in biofilm formationb | Motilityc | Type IV pilus twitching activity (mm)d | Doubling time (h)e | Mucoidy (μg of uronic acid/mg of wet cell massf | Origin | Sourceg |
|---|---|---|---|---|---|---|---|
| CFS1 | 3.1 | 0.8 | 23 | 1.3 | 1.2 | CF-94 | Speert |
| CFS2 | 4.7 | 0.3 | 11 | 1.9 | 0.9 | CF-97 | Speert |
| CFS3 | 1.6 | 0.3 | — | 1.0 | 0.5 | CF-98 | Speert |
| CFS4 | 4.6 | 0.6 | — | 1.8 | 1.5 | CF-98 | Speert |
| CF001 | 4.0 | 0.2 | — | 2.3 | 0.5 | CF | Speert |
| CF005 | 5.5 | 0.3 | — | 3.0 | 1.7 | CF | Speert |
| CF017 | 5.5 | 0.1 | 29 | 2.7 | 2.4 | CF | Hoiby |
| CF040 | 4.6 | 0.8 | 9 | 1.3 | 2.0 | CF | Phibbs |
| CF049 | 3.2 | 0.5 | — | 1.8 | 1.0 | CF | Speert |
| CF5 | 2.2 | 0.3 | — | 1.4 | 0.8 | CF | Govan |
| CF15 | 3.9 | 0.4 | 7 | 1.3 | 0.8 | CF | Govan |
| CF25 | 2.2 | 0.4 | 22 | 2.6 | 4.0 | CF | Govan |
| CF29 | 1.7 | 0.1 | — | 1.8 | 5.5 (M) | CF | Govan |
| CF37 | 3.2 | 0.3 | — | 1.0 | 5.0 | CF | Govan |
| CF46 | 1.8 | 0.8 | 31 | 1.1 | 1.5 | CF | Govan |
| CF149 | 3.8 | 0.7 | 22 | 0.8 | 1.2 | CF | Pier |
| 104035 | 1.3 | 0.9 | — | 1.1 | 1.8 | Biopsy | Guymon |
| 114199 | 2.9 | 0.6 | 11 | 1.1 | 2.1 | Sputum | Guymon |
| 203084 | 3.5 | 0.5 | 15 | 0.8 | 1.0 | Sputum | Guymon |
| 203097 | 6.6 | 0.6 | 25 | 1.2 | 1.0 | Wound | Guymon |
| 226281 | 6.0 | 0.6 | 25 | 1.3 | 0.6 | Eye | Guymon |
| 311058 | 1.8 | 0.9 | 19 | 1.5 | 1.9 | Sputum | Guymon |
| 822026 | 2.9 | 0.9 | 30 | 1.4 | 3.4 | Blood | Guymon |
| ERC-1 | 3.8 | 0.9 | 22 | 0.8 | 0.8 | Env.h | Stoodley |
| ENV42 | 2.2 | 0.6 | 16 | 0.8 | 2.3 | Env. | Speert |
| OR | 2.8 | 0.3 | 26 | 1.5 | 1.7 | Env. | Somerville |
| PAO1 | 1.0 | 1.0 | — | 0.8 | 1.7 | Wound | Phibbs |
| CF032 | 0.6 | 0.6 | — | 1.7 | 0.6 | CF | Hoiby |
| CF9 | 0.1 | 0.3 | 16 | 2.5 | 0.6 | CF | Govan |
| CF011 | < | 0.1 | — | 1.2 | 1.9 | CF | Hoiby |
| CF029 | < | 0.0 | — | 4.8 | 0.8 | CF | Hoiby |
| PA14 | < | 0.8 | 26 | 1.0 | 0.7 | Wound | Ausubel |
A group of 32 isolates were used. Except for the sequential isolates (CFS1 to CFS4), the genomic profiles of these isolates were different when examined by SpeI digestion of genomes monitored by PFGE separation.
Shown is the relative fold increase in biofilm formation in comparison to PAO1. <, the level of biofilm formation was much lower than the level for PAO1.
Motility is expressed as a relative ratio of the swimming zone between the isolate and PAO1.
Type IV pilus-mediated twitching activity was measured as described by Alm and Mattick (1). Shown are the actual twitching zones in millimeters. —, measurement was below the limit of detection.
Doubling time represents the time required to double the bacterial population in LB medium within the 96-well plate at 37°C, as determined during the log phase of bacterial growth.
Alginate production was measured based on growth on LB plates at 37°C for 48 h. Mucoidy was expressed as micrograms of uronic acid per milligram of wet cell mass. The mucoid (M) phenotype was determined after 48 h of growth on LB agar at 37°C.
The P. aeruginosa isolates used in this study were obtained from the following individuals: D. Speert, University of British Columbia, British Columbia, Canada; N. Hoiby, University of Copenhagen, Copenhagen, Denmark; P. Phibbs, East Carolina University Pseudomonas Genetic Stock Center, Greenville, N.C.; J. Govan, University of Edinburgh, Edinburgh, Scotland; G. Pier, Harvard Medical School, Boston, Mass.; C. Guymon, U.S. Army Institute of Surgical Research, Fort Sam Houston, Tex.; P. Stoodley, Montana State University, Bozeman; C. Somerville, Marshall University, Huntington, W.V.; and F. Ausubel, Harvard Medical School, Boston, Mass.
Env., environmental.
Biofilm assay.
The assay for biofilm formation was adapted from the procedure previously described (33). Briefly, P. aeruginosa overnight culture was diluted 1:100 in fresh LB medium, dispensed (125 μl) to wells of a 96-well polyvinyl chloride (PVC) microtiter plate and grown for 15 h at 37°C (unless otherwise noted) with no aeration. Wells were stained with 100 μl of 0.25% crystal violet (CV) for 30 min at room temperature. Stain was discarded, and the plate was rinsed three to five times in standing water and allowed to dry. Stained biofilm was solubilized with 200 μl of 95% ethanol for 10 min and read according to the optical density at 570 nm (OD570). For light microscopy, biofilm samples were removed from the stained biofilm assay 96-well microtiter plates with a razor blade and wet-mounted on a specimen slide. Samples were viewed under normal light microscopy.
Determination of motility and twitching activity.
Equal concentrations of P. aeruginosa isolates, as adjusted by OD (corresponding to CFU per milliliter) from an overnight culture, were used to inoculate MTM or LB plates. An inoculation needle was placed in the culture and stabbed vertically into the medium. Plates were incubated overnight at 37°C for the motility assay, and the motility-mediated swimming zone was measured in millimeters. After incubation, the agar was removed from the twitching activity plate, and the plate was stained with 0.25% (wt/vol) Coomassie blue for 30 min. Stain was removed, and twitching activity was measured in millimeters.
Determination of planktonic and biofilm growth rate.
For static planktonic growth rate, OD570 readings of biofilm assay plates were taken every 30 min starting at t = 0 until the density reached 0.07, at which point, readings were taken every 15 min for a total time of 15 h. For biofilm growth rate, duplicate 96-well plates were tested for biofilm formation at t = 0, 1, 3, 4.5, 6, 8, 10, and 15 h. Furthermore, a shaken 200-ml culture was assayed every 30 min for a total time of 6.5 h. For determining the viable cell count from biofilm, 96-well plates were tested for biofilm formation after 15 h. Duplicate plates were washed gently with sterile water, and the biofilm growth was removed by sterile swab and plated, and the number of CFU per 16 wells was determined. To calculate the specific growth rate for all growth modes, the exponential growth phase was selected on a graph of ln OD versus time (24). The specific growth rate was defined as the slope of the regression line (1/h).
H2O2 sensitivity assay.
This assay was based on a previously published method (58), except paper disks were impregnated with 10 μl of 12% H2O2.
Catalase assay.
P. aeruginosa cell suspensions in phosphate buffer (pH 7.4) were bead beated for 60 s three times while on ice. The supernatant was measured for protein concentration by using the Bio-Rad DC protein assay. Catalase activity was determined by catalase assay kit (Cayman Chemical, Ann Arbor, Mich.).
PFGE.
Pulsed-field gel electrophoresis (PFGE) analysis of genomic profiles of P. aeruginosa isolates was adapted from the standard enterobacterial procedure developed by the Centers for Disease Control and Prevention (PulseNet, section 5, “Preparation of PFGE plugs from agar cultures” [www.cdc.gov]). Briefly, the P. aeruginosa overnight culture (5 ml) was pelleted and resuspended in 1 ml of cell suspension buffer (100 mM Tris, 100 mM EDTA [pH 8.0]) to 10% transmittance as indicated by the BioMerieux Vitek colorimeter. The cell suspension (200 μl) was combined with 200 μg of proteinase K and inverted six times. A mixture of 1.6% InCert agarose (FMC BioProducts, Rockland, Maine) and 1% sodium dodecyl sulfate (SDS) (0.8 g of InCert agarose, 2.5 ml of 20% SDS, 46.7 ml of Tris-EDTA buffer [TE]) (200 μl) was mixed with the cell suspension-proteinase K mixture and dispensed in an agarose plug mold (Bio-Rad). After solidifying, the cells were lysed within the plug using 1.5 ml of cell lysis buffer (50 mM Tris, 50 mM EDTA [pH 8.0], 1% N-lauroylsarcosine) and 800 μg of proteinase K for 1.5 h in a 54°C water bath with agitation. Plugs were then washed twice with 10 ml of preheated, sterile, reagent-grade H2O for 15 min and twice with 10 ml of preheated TE buffer for 15 min, with all washes performed in a 48°C water bath with agitation. Portions of the plugs (1.5 mm) were digested with the restriction enzyme SpeI (20 U), DpnI (14 U), or XbaI (50 U) for 1 h at 37°C. Portions of the plugs (6 mm) were equilibrated with reaction buffer for 3 h at 4°C and digested with I-CeuI (0.8 U) for 2 h at 37°C. Digested portions were loaded into an agarose gel of a specific concentration and subjected to certain separation conditions (both noted in the relevant figure). The running buffer was 0.5× Tris-borate EDTA (TBE). After PFGE, the gel was stained with ethidium bromide (1 μg/ml) for 30 min and photographed under UV light.
Alginate assay.
The alginate assay was based on a previously published method (23) with the following modifications. Isolates were inoculated onto LB plates and incubated for 48 h at 37°C. Bacteria were scraped off the plates and suspended in 10 ml of phosphate-buffered saline (PBS; pH 7.4) and centrifuged at 1,500 × g for 20 min. The supernatant was used for the alginate assay.
RAPD.
RAPD reactions were carried out with purified genomic DNA of P. aeruginosa isolates by using standard primers as previously described (28). The conditions that were selected as the optimal conditions for obtaining accurate amplified band profiles with the eight primers are as follows: assays were performed in 50 μl of EasyStart PCR tubes (Molecular BioProducts) with 2.5 U of MasterAmp Taq DNA polymerase (Epicentre) and 0.25 μM each primer. The following temperature cycling was used with a Bio-Rad iCycler system: 94°C for 4 min; 4 cycles consisting of 94°C for 2 min, 45°C for 5 min, and 72°C for 5 min, followed by 30 cycles consisting of 94°C for 1 min, 45°C for 1 min, and 72°C for 2 min; and a final extension step consisting of 72°C for 10 min.
Southern hybridization.
Agarose gels were soaked in 0.25 N HCl for 30 min, rinsed in H2O, and soaked in 1.5 M NaCl-0.5 M NaOH for 30 min and 1.5 M NaCl-0.5 M Tris-Cl (pH 8.0) for 30 min. A blotting apparatus was constructed with a filter paper wick and a Hybond-N+ membrane (Amersham Pharmacia Biotech) and transferred with 20× SSC (1× SSC is 0.15 M NaCl plus 0.015 M sodium citrate) transfer buffer for 24 h. After transfer, the membrane was rinsed in transfer buffer and UV cross-linked. Hybridization and digoxigenin probe labeling were carried out as described by the manufacturer (Roche Molecular Biochemicals). PAO1 DNA was fully digested with Sau3A, separated by electrophoresis on a 1% agarose gel, and stained with ethidium bromide. A particular section of the digested DNA gel corresponding to the size of 0.5 to 2 kb was removed, extracted by QIAquick gel extraction (Qiagen) according to the specifications of the manufacturer, and used for labeling.
Transmission electron microscopy.
P. aeruginosa overnight culture (2 ml) was pelleted, washed with 500 μl of PBS, pelleted, and resuspended in 100 μl of PBS. The cell suspension (10 μl) and 2% phosphotungstic acid (PTA; 10 μl) were combined on a glass slide. A Formvar-coated grid was placed on top of the mixture for 30 s. The grid was allowed to dry overnight at room temperature. Grids were viewed by transmission electron microscopy (Hitachi).
Examination of genomic islands by PCR.
Amplification reactions of PAGI-1 and PAGI-2 were carried out with purified genomic DNA by using the following primers: for PAGI-1, orf18 forward (5′TAAGGGGTTCTAGCGGC) and reverse (5′AATCGGTGCAAGGGAGTA), orf22 forward (5′GACTTGCATGGGGCTT) and reverse (5′TGCCGAACACGATCAA), and PAO1-specific (PA2221) forward (5′TATCAGTGTCGGGCAAGA) and reverse (5′AGCTCCGGCAACCACTA); and for PAGI-2, orfA forward (5′TATGTTCCGCAAGGTCT) and reverse 5′AATGGTACATGGGGAAGT) and PAO1-specific (PA1089) forward (5′TGTGCGCACTGCCTAC) and reverse (5′GCAAGGTATTGGTTCGG). All primers were made at the Marshall DNA Core Facility. Assays were performed in 50-μl EasyStart PCR tubes (Molecular BioProducts) with 2.5 U of MasterAmp Taq DNA polymerase (Epicentre) and 0.25 μM each primer set. The following temperature cycling was done with the Bio-Rad iCycler system: 94°C for 1 min; 34 cycles consisting of 94°C for 1 min, 54°C for 2 min, and 72°C for 2 min; and a final extension step consisting of 72°C for 8 min.
P. aeruginosa aerosol infection mouse model.
The P. aeruginosa overnight culture (1 ml) was used to inoculate 100 ml of LB and was grown for 14 to 16 h with aeration at 37°C. The cells were pelleted and resuspended in 10 ml of P-PBS (1% Proteose Peptone-PBS). The cell suspension (5 ml) was dispensed into the nebulizer-Venturi unit of the inhalation exposure system (Glas-Col, Terre Haute, Ind.) and aerosolized for 30 min (compressed air control = 20, vacuum control = 50), cloud decayed for 25 min, and UV decontaminated for 5 min. A total of 10 mice were exposed for each strain, with 5 mice terminated at t = 0 and t = 6 h. Mice were sacrificed by carbon dioxide. The right lung was removed, homogenized in 1 ml of P-PBS, and plated via serial dilution on LB agar.
DNA sequencing and bioinformatic analysis of novel genomic sequences.
Purified plasmids were sequenced by using universal M13 forward and reverse primers. Sequencing was performed with 11 μl (approximately 2 μg) of purified plasmid, 1 μl (3.2 pmol) of unlabeled sequencing primer, and 8 μl of Big Dye terminator reaction mix (PE Applied Biosystems). DNA sequencing was performed on a LI-COR 4200 DNA sequencer (Lincoln, Neb.) at the Core Facility at Marshall University School of Medicine. Once the DNA sequences were available, they were compared by BLAST to GenBank and to the annotation of the PAO1 genome (www.pseudomonas.com).
RESULTS
P. aeruginosa isolates demonstrate different abilities of in vitro biofilm formation.
While P. aeruginosa strains possess complex, diversified genomes (22) and readily form biofilms (9), it is unclear whether there are any variations in biofilm formation among clonally diverse clinical and environmental isolates of P. aeruginosa. A total of 151 isolates (101 CF and 50 non-CF) collected from geographically diversified areas were first examined for their clonal relationship by macrorestriction digestions of chromosomal DNA with SpeI, XbaI, and/or DpnI, followed by separation by PFGE. The PFGE pattern of each isolate was compared with that of the reference strain PAO1 (50) and the patterns were also compared between these isolates. All PFGE patterns were unique except for 16 sequential CF isolates, which is consistent with previous reports that CF pulmonary infections are mainly associated with a predominant strain (6, 28, 39). Next, all isolates were tested for biofilm-forming ability by using an adaptation of a PVC 96-well microtiter dish method (33). Since some CF isolates are auxotrophic and often require the presence of several amino acids for growth in minimal media (3, 4), we chose to grow these isolates in complex LB medium. The strain PAO1 was included as a standard in every microtiter dish used to determine biofilm formation. A significant variation in biofilm formation was found among all isolates tested (Fig. 1A). To confirm that the crystal violet-stained samples at the air-liquid interface on the dish were true biofilm, a sample was prepared and visualized microscopically, which was consistent with characteristics of initial biofilms (Fig. 1B). Furthermore, the majority of these isolates showed increased amounts of biofilm compared with PAO1 (Fig. 1A). A group of 32 phenotypically diverse isolates were selected for further analysis. As shown in Table 1, biofilm formation of these samples varied greatly in relation to PAO1. Only five of the samples (CF032, CF9, CF011, CF029, and PA14) formed less biofilm than PAO1. Sequential isolates (CFS1 to CFS4) (57) from the same patient over a period of 4 years expressed different levels of biofilm formation. Furthermore, these isolates were also tested for biofilm formation in a 12-well polystyrene plate, which was consistent with the PVC plate results (data not shown). To test whether the observed variations could be due to the relative hydrophobicity of these isolates, the biofilm formation was also tested on a glass surface. The dynamics and trend of biofilm formation on this surface were identical to those seen with PVC (data not shown), suggesting that the relative hydrophobicity of these isolates is not involved in the differences in biofilm formation. Since some isolates are from the environment, biofilm formation was compared between 30 and 37°C. No difference was seen among these isolates (data not shown).
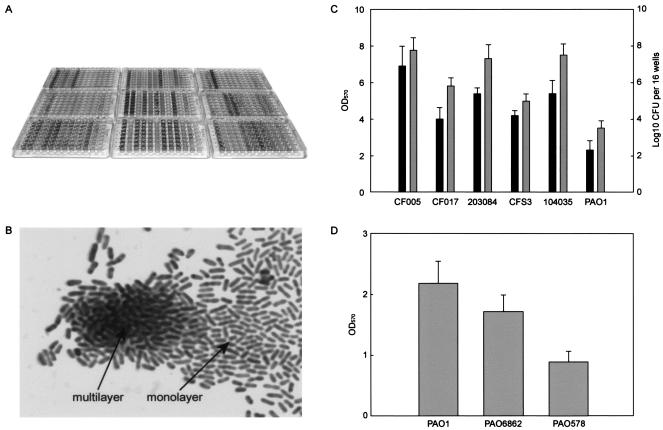
FIG. 1.
Variations in (A) and light microscopy (B) of biofilm formation produced by genomically diversified clinical and environmental isolates of P. aeruginosa in an in vitro biofilm assay system. For each isolate within each 96-well plate in panel A, the assay was repeated for a total of eight times (vertically). Biofilm formation was analyzed along with PAO1 (far-right vertical lane of each plate). (C) Relationship between CV-stained materials (black bars) and biofilm viable counts (grey bars). (D) Relationship between alginate production in PAO1 and its derivatives and biofilm formation.
However, this variation, as observed with CV staining (Fig. 1A), could be due to the different amounts of CV-absorbing extracellular materials. To test this, viable counts of biofilms were measured and compared with the OD of CV-stained materials. There was a correlation between biofilm CFU and OD (Fig. 1C), suggesting that the optical intensity is representative of the cell density in biofilms. Furthermore, three isogenic strains, PAO1, PAO6862 (PAO1 algD::Gmr) (55), and PAO578 (PAO1 mucA22), a hypermucoid strain known to maintain mucoid status on LB plates (11), were assayed for the contribution of mucoidy on biofilm formation. When stained with CV, PAO1 showed the largest amount of biofilm followed by PAO6862 and the producer of the least biofilm, PAO578 (Fig. 1D). These results are consistent with the recent observation that alginate is probably not required for the initial phase of biofilm formation (18, 54) and also conclude that CV staining is not indicative of the extracellular matrix but the viable cells inside biofilms.
Flagellum- and type IV pilus-mediated activities are not proportional to biofilm formation.
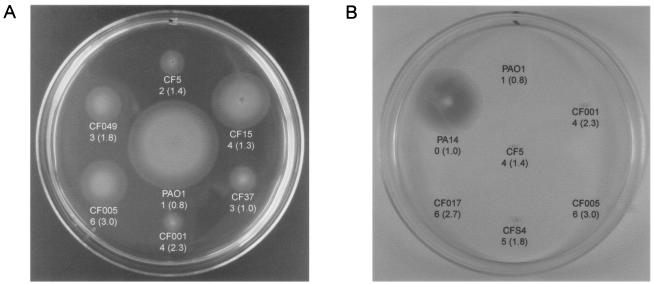
The formation of biofilms is a multistep process that requires participation of structural appendages, such as flagella and type IV pili (32). Variations in biofilm formation, as seen in Fig. 1, could be due to altered activities of these structural appendages. To test this, we measured the motility and twitching abilities of the isolates in Table 1. The flagellum activity of PAO1 was the greatest among all strains tested (Table 1 and Fig. 2A). However, PAO1 formed the smallest amount of biofilm compared to the other six CF isolates shown in Fig. 2A. Furthermore, other isolates with increased motility displayed elevated biofilm formation (Table 1, CF040 and ERC-1). Thus, no quantitative correlation was seen between the ability of the bacteria to assume a biofilm mode and motility. In addition, when samples were negatively stained with 2% PTA and examined by electron microscopy, all isolates showed the presence of flagella (data not shown). We also assayed the twitching activity of the 32 isolates. While twitching was not visible in all samples, one that displayed such activity was strain PA14, which formed the smallest amount of biofilm compared with other isolates (Fig. 2B). There were, however, isolates forming more biofilm coupled with increased twitching activity (CF017 and 203097, Table 1). Therefore, no quantitative correlation was seen between twitching and biofilm formation.
FIG. 2.
Motility and twitching activity of P. aeruginosa and the relationship with biofilm formation and planktonic growth rate. Shown are motility (A) and twitching (B) zones between six isolates and PAO1. The first number under the strain designation indicates the relative fold increase in biofilm formation versus PAO1; the number in parentheses indicates the doubling time in hours.
CF isolates express slow planktonic growth in contrast to efficient biofilm formation.
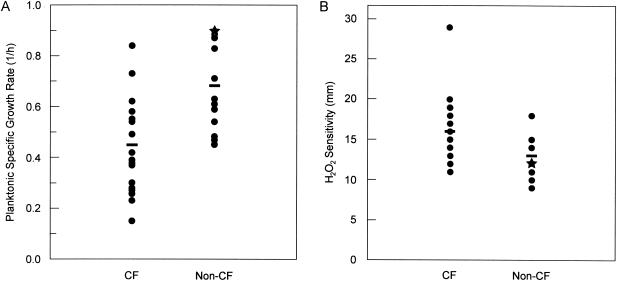
Biofilm formation could be affected by inherent differences in the planktonic growth rate of each isolate (34). To measure this growth property, we developed a method to simultaneously measure the specific growth rate of multiple samples utilizing the same 96-well microtiter plates used for biofilm formation. P. aeruginosa isolates grown in this system displayed a typical bacterial growth curve. While the generation time of CF029 was the lowest and equivalent to 4.8 h (0.15 h−1), PAO1 had the highest growth rate of about 0.8 h (0.9 h−1) under this condition (Table 1). The CF isolates as a whole were found to grow significantly slower than non-CF isolates (mean, 0.45 versus 0.7 h−1, respectively; P = 0.008, t test) (Fig. 3). When grouped, the CF isolates were found to form an amount of biofilm similar to that formed by non-CF isolates (P = 0.615, t test; Table 1). Comparatively, the biofilm-specific growth of these isolates at different time points also mimics this trend (data not shown). Furthermore, in comparison with PAO1, the majority of CF isolates displayed slower planktonic growth but efficient biofilm formation. For example, the planktonic growth rates of CF005 and CF001 were 3 and 2.3 h−1, respectively, but they generated three- and sixfold more biofilm than PAO1, respectively (Table 1). To eliminate the possibility of cell partitioning that could be derived from the static culture method, which may affect the measurement of the planktonic growth rate, a shaken culture method was also performed. Again, the majority of CF isolates showed the same trend of reduced planktonic growth rate compared with the environmental isolates, non-CF isolates, and reference strains, such as PAO1 and PA14. Furthermore, CF isolates such as CF149, CF37, and CF46 showed a pattern of fast planktonic growth, while CF005, CF029, and CF25 had slow planktonic growth, consistent with the results obtained with the static culture method (Table 1). Thus, there was no correlation between planktonic growth rate and biofilm formation.
FIG. 3.
Comparison of planktonic growth rate (A) and sensitivity to hydrogen peroxide (B) between CF and non-CF isolates. Specific growth rates and sensitivity to H2O2 of each isolate in comparison with PAO1 (stars) were determined as described in Materials and Methods. The horizontal bar in each group represents the median values of growth rates (P = 0.008, t test) and growth inhibition zones in millimeters (P = 0.008, t test).
No correlation between mucoidy and initial biofilm formation.
Mucoidy (overproduction of alginate) is an important virulence factor in chronic P. aeruginosa lung infections in CF (17). Recently, it has been shown that alginate may not play a key role in initial biofilm formation (18, 54). A correlation between uronic acid (an alginate precusor) production and initial biofilm formation was tested among different isolates. As seen in Table 1, the highest alginate producers, CF25, CF29, and CF37, did not lead to the largest amounts of biofilm formed in this study. Conversely, some strains with low alginate production generated large (CFS2, CF005, and 226281) and small amounts of biofilms (CF9 and CF029). Therefore, no correlation was seen between alginate production and initial biofilm formation.
Planktonic cells of CF isolates are more sensitive to H2O2.
P. aeruginosa biofilms are more resistant to reactive oxygen intermediates (ROI) than their planktonic counterparts (21). However, it is not clear whether individual planktonic cells of genomically diverse isolates have altered sensitivity to H2O2, an abundant ROI species within neutrophils. When compared, CF isolates were found to be more sensitive to this particular ROI species than those of non-CF origin (Fig. 3B, P = 0.008, t test).
Nonmucoid CF isolates are associated with reduced lung colonization in an acute aerosol infection mouse model.
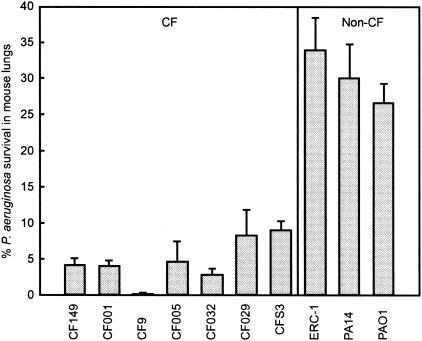
To test whether there is a correlation between increased in vitro biofilm formation and in vivo lung colonization, we utilized an aerogenic infection mouse model (57). A group of age- and sex-matched C57BL/6 mice were exposed to aerosols of seven CF and three non-CF isolates (Fig. 4). Only nonmucoid isolates were aerosolized, because previous work had indicated that aerosolized mucoid isolates retained more effectively in the lungs than isogenic nonmucoid strains (56). Bacterial survival in the lungs was measured for each isolate 6 h after aerosol exposure in comparison with the initial deposition dose. CF isolates were cleared more effectively (up to 10% retention) from the murine lungs than non-CF isolates (25 to 35% retention, Fig. 4). PAO1 was markedly less capable of being cleared than the CF isolates tested.
FIG. 4.
Nonmucoid CF isolates of P. aeruginosa are cleared more efficiently from the mouse lungs than non-CF isolates in an acute aerosol infection mouse model. A group of C57BL/6 mice were exposed to aerosols of different P. aeruginosa isolates as labeled on the x axis. For each exposure, five mice each were included for determination of bacterial deposition to the lungs at t = 0 and 6 h.
CF isolates have increased genome sizes with genomic insertions.
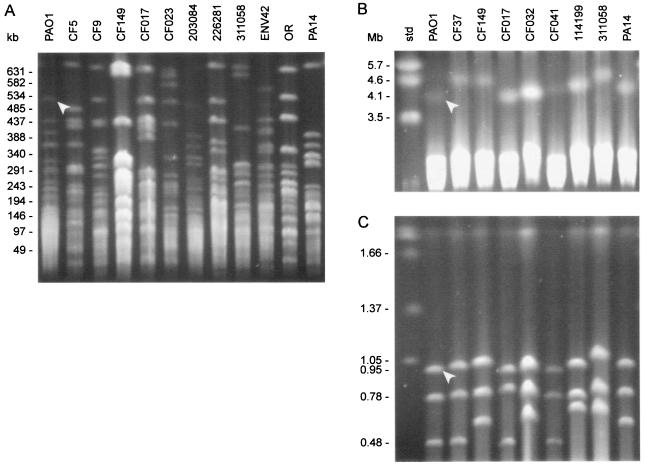
Horizontal gene transfer is a cause of genomic diversity in P. aeruginosa (2, 25, 26, 47). When the genomes of the isolates in Table 1 were examined for genomic diversity by SpeI macrorestriction digestion coupled with PFGE separation (Fig. 5A), the isolates possessed at least one band larger than that of the largest PAO1 fragment (SpeA, Fig. 5A). Also, the PFGE banding pattern was highly diverse among these isolates (Fig. 5A). This suggests that genome size of these isolates could be hypervariable in relation to PAO1. To determine whether intrinsic diversity was related to genome size, the chromosomes of these isolates were digested by the homing endonuclease I-CeuI, which specifically cuts at the rrn operons. A selected group of isolates were analyzed for genome size (Table 2). As shown in Fig. 5B and C, all isolates examined had an equal number of four rrn operons, just like PAO1. The genome sizes of these isolates ranged from 0.4 to 18.9% larger than that of PAO1 (Table 2). Since many clinical isolates carry specific genomic sequences called PAGI-1 and -2 (2, 26), we examined the presence or absence of PAGI-1 and -2 by PCR. As shown in Table 2, PAGI-1 and -2 were present within the genomes of CF149 and 311058, corresponding with the largest genomes identified.
FIG. 5.
Relationship between genome diversity and genome size among clinical and nonclinical isolates of P. aeruginosa. (A) PFGE separation of SpeI-digested chromosomal DNA from a selected group of P. aeruginosa isolates. An arrowhead indicates the largest SpeI fragment of PAO1 (SpeA) (57). (B and C) PFGE separation of genomic DNA digested with I-CeuI. Arrowheads indicate the largest PAO1 fragments, 4,064 and 950 kb, respectively. The running conditions were 18 h, 6 V/cm, 120° angle, 1% agarose, and a switch time of 15 to 40 s with a gradient of a = 0.35741 in panel A; 48 h, 2 V/cm, 106° angle, 0.8% agarose, and a switch time of 20 s to 30 s with a linear gradient in panel B; and 22 h, 6 V/cm, 120° angle, 0.8% agarose, and a switch time of 90 s in panel C. The molecular size standards were Schizosaccharomyces pombe (B) and Hansenula wingei (C) (Bio-Rad).
TABLE 2.
Genomic characteristics of a selected group of P. aeruginosa isolates used in this study
| Strain | Genome size (kb) | Difference from estimated size (kb)a | % Difference | Presence ofb:
|
|
|---|---|---|---|---|---|
| PAGI-1 | PAGI-2 | ||||
| PAO1 | 6,264 | 0 | 0.0 | − | − |
| PA14 | 6,618 | 354 | 5.7 | − | − |
| CF37 | 6,862 | 598 | 9.6 | + | − |
| CF149 | 6,990 | 726 | 11.6 | + | + |
| CF017 | 6,291 | 27 | 0.4 | − | + |
| CF032 | 6,659 | 395 | 6.3 | − | + |
| CF041 | 6,386 | 122 | 2.0 | + | − |
| 114199 | 6,875 | 611 | 9.8 | − | − |
| 311058 | 7,448 | 1,184 | 18.9 | + | + |
Genome sizes were estimated as shown in Fig. 5B and C.
The presence or absence of PAGI-1 and -2 was determined by PCR amplification of the PAGI-specific genes. For PAGI-1 (GenBank accession no. AF241171), three sets of primers were used for PCR. The first set amplified the gene coding for phytoene dehydrogenase (orf22; 1.5 kb), and the second set amplified the gene coding for 1,3-propanediol dehydrogenase (orf22; 1.4 kb). Both genes are present only on PAGI-1. The third set of primers produced a PAO1-specific transposase gene (PA2221; 1.5 kb). +, orf18/orf22 present and PA2221 absent; −, PA2221 present, but orf18/orf22 missing. For PAGI-2 (GenBank accession no. AF332547), two sets of primers were used. The first set amplified orfA (1.2 kb), and the second set produced a partial PAO1-specific gene (PA1089; 0.8 kb).
Identification of novel DNA sequences in CF isolates.
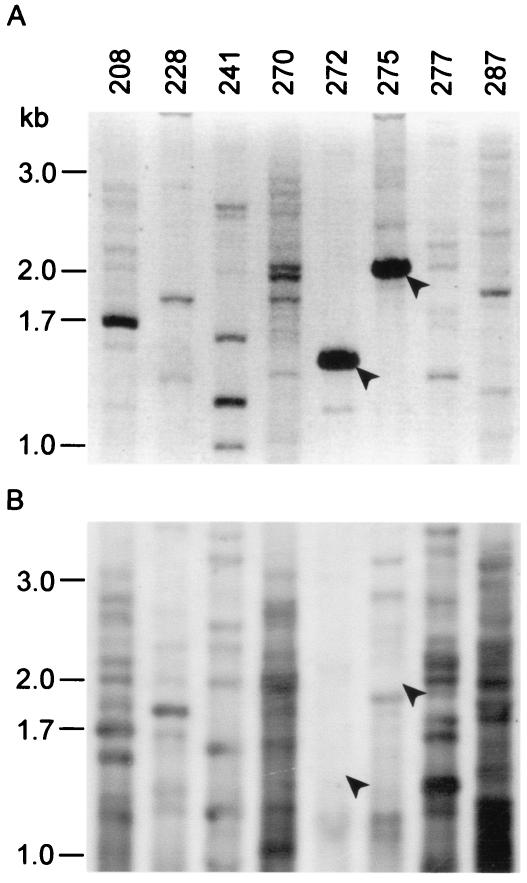
To identify novel DNA sequences in these isolates, we developed a simple method based on RAPD coupled with subtractive hybridization using PAO1 DNA as a probe. While the resolving power of RAPD mainly allows for strain differentiation (28), it is not clear what genes are amplified by this PCR-based method. We hypothesized that some of the randomly amplified PCR products from the larger genomes could be missing from the PAO1 genome and may represent unknown genomic islands. A Southern hybridization of a RAPD membrane with randomly labeled PAO1 genomic DNA yielded two fragments (1.4 and 2.0 kb) that failed to be hybridized (Fig. 6B). Using this method, we identified three novel genomic sequences from two CF isolates, CF005 and CF023 (Table 3). The sequence CF023-228 was matched to a probable nonribosomal peptide synthetase (PA2402) that had homology with the recently released highly divergent pyoverdine biosynthetic locus from two CF isolates (47). The CF005-275 and -272 sequences were not present in the GenBank database, with the first one encoding a variant of topoisomerase I from P. aeruginosa and the second encoding a homolog of mvpTA from Shigella flexneri (42) that has not been identified in P. aeruginosa.
FIG. 6.
Identification of novel DNA sequences from CF isolates of P. aeruginosa. (A) Randomly amplified PCR products using CF005 as a template with primers as indicated above the gel were separated in a 1% agarose gel. After blotting, filters were hybridized with random primer-labeled total PAO1 genomic DNA. (B) Southern blot of the gel shown in panel A with arrows indicating two major bands amplified by primers 272 and 275, respectively, but which failed to be bound by PAO1 DNA.
TABLE 3.
Novel DNA sequences identified from CF isolates of P. aeruginosa
| Strain | Primera (insert size in kb) | GenBank accession no. | Gene | Homolog product (accession no.) | E value (BLAST search) |
|---|---|---|---|---|---|
| CF005 | 272 (1.5) | AY258908 | mvpTA | Plasmid maintenance protein (S. flexneri; NP_490590.1) | 9E-48 |
| 275 (2.1) | AY265810 | topA | Topoisomerase I (X. fastidiosa; NP_061659.1) | 4E-50 | |
| CF023 | 228 (1.6) | AY261781 | PA2402 | Probable nonribosomal peptide synthetase (AE004667_1) | 2E-59 |
Primers were used as previously described (28). The number in parentheses represents the fragment amplified by this primer, which was cloned into pCR4-TOPO (Invitrogen) and sequenced with universal M13 primers.
DISCUSSION
Four primary conclusions can be drawn from this cross-sectional analysis of clinical and environmental isolates of P. aeruginosa. First, various P. aeruginosa strains have different capacities of in vitro biofilm formation. Second, the majority of CF isolates in comparison with non-CF isolates demonstrate two opposing growth modes: reduced planktonic growth versus efficient biofilm formation. Third, nonmucoid CF isolates have a reduced ability of lung colonization compared with PAO1, PA14, and environmental isolates. Fourth, the presence of novel DNA is common in CF isolates and represents a major cause of genomic diversity in P. aeruginosa.
Biofilm formation is an important phenotype associated with chronic P. aeruginosa pulmonary infections in CF (10). In this study, we evaluated the effect of motility, twitching, growth rate, and mucoidy on biofilm formation using a group of 32 P. aeruginosa isolates. A standard method based on bacterial growth within 96-well microtiter plates (33) was used to assess the biofilm-forming ability of these strains. This simple method allows for a high throughput and reliable analysis of biofilm formation on an abiotic surface. This surface is by no means indicative of CF lungs; however, it allows for analysis of strain-dependent biofilm-forming ability (Fig. 1A). Furthermore, the biofilms formed in this system possess the characteristics of mono- and multilayer community structures (Fig. 1B). Laboratory strains such as PAO1 and PA14, although originally isolated from clinical settings (20, 38), have reduced biofilm formation compared with fresh isolates from clinical and environmental sources. While PAO1 and PA14 grow optimally as planktonic cells (0.8 and 1.0 h, respectively), it appears that the capacity of biofilm formation in these two strains is at a basal level when compared with other isolates (Table 1). This is perhaps related to laboratory attenuation of these two strains. Again, CF isolates form much more biofilm than PAO1 (Table 1). This suggests that PAO1 may be equipped with endogenous biofilm suppression mechanisms, or, conversely, CF isolates possess mechanisms to promote biofilm formation. Also, most approaches in studying biofilm formation focus on screening mutants defective in biofilm formation (32). However, a complementary approach that has not been actively pursued is to use mutants with increased biofilm formation to identify biofilm repressors in these two strains. Furthermore, CF isolates are proficient biofilm formers, since they exhibit efficient biofilm formation and despite the fact that they are deficient in planktonic growth (Fig. 3). It is known that P. aeruginosa is capable of making the transition from an environmental state to a chronic colonizing state in CF (41). However, such an adjustment comes at a price. For example, most CF isolates are auxotrophic mutants and are incapable of synthesizing methionine, leucine, arginine, and/or ornithine (3, 4). Intriguingly, this auxotrophic defect is linked with conversion to mucoidy, known to confer increased resistance to antibiotics and host defenses (17). The production of alginate in this chronic state is a major consumer of cellular energy in return for securing the survival of the bacteria. This adjustment and rerouting of cellular resources to adequately suit the new environment compromise the planktonic growth of the bacterium (51).
The activities of flagella and type IV pili have been shown to play a role in biofilm formation (37). Since all isolates examined in this study had flagella, our results about the relationship between motility and biofilm formation suggest that the flagellar activity of CF isolates, as manifested by the observed swimming zones, is not the predominant factor involving the progressive development of biofilms in vitro. When grouping all samples in Table 1, a slight correlation was seen between planktonic generation time and motility (R2 = 0.30). This trend is expected in that motility, as measured in this study, may be a reflection of how fast the bacteria are dividing planktonically. This is particularly evident with PAO1, which has the fastest growth rate and is the most motile (Fig. 2A). Furthermore, twitching, as mediated by type IV pili, was not observed in all of the samples tested (Table 1); however, many of those without this activity formed more biofilm than PAO1. This is assuming the twitching assay is a reflection of the role type IV pili play in microcolony formation. Since both assays were inoculated with the same number of bacteria, the observed variations in these activities could not have been caused by a difference in the initial inoculum size. Since some CF isolates grow primarily for surface attachment (Table 1) independent of these two known biofilm-related factors, these results suggest the presence of a novel biofilm adhesin. To explain why the activities of flagella and type IV pili do not correlate with the amount of biofilm formed, these structural appendages may only act as biofilm adhesins qualitatively. To support this, a report showed that increased initial biofilm formation was linked with isogenic variants deficient in flagellum and type IV pilus activities (7, 12). Also, an additional surface structure called curli has been implicated with increased biofilm formation in an E. coli K12 mutant (53). However, mucoidy status of these isolates did not affect initial biofilm formation in vitro (Table 1), which is in agreement with previous reports that mucoid exopolysaccharide is needed mainly for maintaining the altered architecture of mature biofilms (18, 29). Furthermore, although the specific adhesins that promote biofilm formation on a PVC surface are unknown, our studies showed that the same type of adhesins are also involved in binding to another abiotic surface, polystyrene. While four sequential isolates have identical genomic profiles, as revealed by PFGE analysis, there exists a significant variation in biofilm formation, motility, type IV pilus activity, growth rate, and alginate production among them (Table 1). These isolates may constantly adapt themselves to changes in the CF lung environment over the course of chronic colonization. In support of this conjecture, sequential isolates can express different phenotypes, such as untypeable O-antigen (47), and can be either prototrophic or auxotrophic (3).
When the specific planktonic growth rates of CF and non-CF isolates were compared, the growth rates of CF isolates were significantly lower (Fig. 3). These results suggest that particular responses taken by CF isolates to evade environmental stresses begin to tax the cellular resources, at least to a greater extent than those of the non-CF clinical isolates, thereby expressing a reduced planktonic growth rate. The CF lung may be a more extreme environment to adapt to than other clinical environments, such as burn wounds, therefore resulting in a reduced planktonic growth rate. The present work presents a model that while the majority of CF isolates retain their biofilm-forming ability during their transition from an in vivo to in vitro environment, the shift from planktonic to biofilm growth may be beneficial to bacterial survival (9). Association into a biofilm offers a selective advantage allowing the collective unit to operate as one, protected from the external environment, associated with increased resistance to antibiotics and host defenses (14), cooperating metabolically and evolving as a community, potentially by horizontal gene transfer (16).
By inducing a bacterial aerosol-induced lung infection (BAILI) using an inhalation exposure system (57), a clear distinction in colonization is displayed between the CF and non-CF isolates (Fig. 4). An environmental sample (ERC-1) (48) and burn patient isolates PA14 (38) and PAO1 (20) all showed increased retention in the mouse lung compared to the CF isolates. While the biofilm assay determines initial attachment to a PVC surface, BAILI verifies not only attachment to lung tissue, but also host defense intervention. The explanation of bacterial clearance from the lung can be approached in two stages. First, the inherent adhesive ability of a bacterial strain determines whether and how efficiently an initial colonization will take place. This may be due to the presence of different bacterial surface structures. For example, CF isolates can express a novel lipid A portion of lipopolysaccharide (15). Also, fresh clinical isolates fail to bind to an asialo-GM1, generally considered a lung epithelial receptor for a laboratory strain (44), indicating CF isolates may have novel surface proteins that mediate attachment to lung tissue. Once this colonization exists, the host defenses are the next consideration as the ability of the isolate to evade clearance is determined. The innate pulmonary defenses include antimicrobial defensins and neutrophils that carry an abundance of reactive oxygen species, like hydrogen peroxide. Our results suggest that CF isolates, when in the planktonic phase, are significantly more sensitive to H2O2 than are non-CF isolates (Fig. 3B), which showed increased mouse lung retention (Fig. 4). However, a direct catalase enzymatic activity assay on the total cellular extracts of all these isolates indicated no difference between the CF and non-CF isolates. While this is inconclusive, the first assay appears to be quantitative and explains the clearance data. Incidentally, the aerosol mouse model simulates an acute infection condition caused by planktonic cells, not biofilms. Using this model, bacteria deposited to the alveoli were in a single-cell form (data not shown). Also, neutrophil migration from the bloodstream to the lung occurs within the first 4 h following aerosol exposure (56). This may in part explain why in vivo mouse lung clearance does not correlate with the in vitro microtiter dish biofilm assay. On the other hand, CFTR knockout mice have recently been shown to develop lung infections when pretreated with oral antibiotics (8). It would be interesting to test the response of this orally treated CFTR mouse model to aerosolized bacteria to determine whether the modified CFTR-deficient lung environment affects colonization.
The inherent genomic diversity in P. aeruginosa may be caused by four means of novel DNA introduction into the bacterial genome: nucleotide substitution, insertions of transposons and bacterial phages, horizontal gene transfer, and preexisting novel genomic sequences. First, CF isolates of P. aeruginosa often display a high frequency of mutation after long-term colonization in CF (30), possibly due to single-nucleotide polymorphisms (SNPs). One example of CF-related SNPs involves mutations in mucA, giving rise to overproduction of alginate (5). It is known that PAO1 carries transposons and a bacteriophage (50). Two genomic islands, PAGI-1 and -2, have been identified in isolates of P. aeruginosa (2, 26). Another consequence of horizontal gene transfer is the increase in genome size. In this study, we used a low-throughput PFGE technique to estimate the genome size of various isolates of P. aeruginosa coupled with highly specific digestion using I-CeuI. This method allowed accurate determination of the genome size of P. aeruginosa clone C, which was 6.52 Mb, about 256 kb larger (4%) than the PAO1 genome (43). Using this approach, we determined that a selected group of isolates in Table 1 had much larger genomes than that of PAO1 (Fig. 5). These results agree with data showing the presence of PAGI-1 and -2 in the largest genomes analyzed (Table 2, CF149 and 311058). It has been shown recently that although extensive novel sequences are present in the genomes of CF isolates, the backbone of the PAO1 genome is preserved in several P. aeruginosa isolates (25, 47). In combination with our lung clearance data, this suggests that PAO1 remains an excellent model for studying virulence, even though its genome size is not representative of most CF isolates (Table 2). Since bacteria in biofilms have an increased incidence of horizontal gene transfer (16), we also note that while genome size was increased, the lung colonization ability was reduced in P. aeruginosa CF isolates (Fig. 4). This is in agreement with the trend that a CF isolate had an overall reduction in virulence when examined in a highly sensitive plant model (45). However, in enterohemorrhagic E. coli O157:H7, the increase in genome size (19.2%) is associated with increased virulence when compared with standard laboratory strain K12 (36). Variations in biofilm formation in different isolates lead us to speculate whether there are some biofilm-specific genomic islands in CF isolates. However, the PAO1 genome can be viewed as a mutant. It could be possible that the ancestor of PAO1 had slowly deleted genes that were not required for specific environments, thus maintaining only a minimal number of genes (backbone) for P. aeruginosa survival (47). This view contradicts the concept of novel gene acquisition by horizontal gene transfer. However, this view can explain why many strains of P. aeruginosa, including those studied here, have genes not present in PAO1 (25, 47). The predecessor of PAO1 could have previously possessed many of the genes found throughout P. aeruginosa isolates, as opposed to the introduction of many novel genes into PAO1 giving rise to these isolates. Together, what may actually exist could be a combination of both mechanisms.
Genomic comparison between clinical and environmental isolates can yield useful information on the inherent virulence properties. While shotgun sequencing of multiple bacterial genomes is an ideal choice, the costs and time associated are often high. An alternative is the use of microarray gene expression profiles in conjunction with subtractive genomic hybridization. However, this high-throughput method relies on the correct annotation of the existing genomes, which may carry misannotated genes. To complement these methods, we developed a simple method that allows for a quick comparison between two genomes using RAPD in combination with PAO1 DNA subtractive hybridization (Fig. 6). This subtractive method has two key features: (i) RAPD-based amplification of an unknown genome minus (ii) the existing PAO1 genome. Three novel genetic loci were identified in two clinical isolates (Table 3). The mvpAT locus is located on a large Shigella virulence plasmid and is involved in plasmid retention (42). However, the order is reversed (mvpTA) in a CF isolate (CF005). In addition, we isolated a topA homolog that encodes a topoisomerase I variant. Two topA genes were found in CF005: one with 100% homology to PA3011 in PAO1 and the other topA variant that failed to be hybridized by PAO1 genomic DNA. Furthermore, this approach led us to identify a variant of PA2402 that has been recently hypothesized to be involved in synthesis of pyoverdine (47). One limitation of this method is the inability to pinpoint the exact location of these novel genes within the PAO1 backbone; however, this can be resolved by performing inverse PCR. Altogether, RAPD coupled with subtractive hybridization is an effective way to examine the genomic difference between isolates. This method also allows identification of smaller DNA segments absent in the PAO1 genome. Variations of this approach with random degenerate primers coupled with various amplification conditions are expected to generate more novel DNA sequences.
In addition to the improved understanding of the phenotypic diversity with relation to biofilm formation in P. aeruginosa, this report suggests that the extensive genomic diversity in this species may become an unexplored cause for the variation in morbidity and mortality in CF. Furthermore, it demonstrates the presence of multiple mechanisms of biofilm formation, which is aided by novel biofilm adhesins that could become antibiofilm therapeutic targets.
Acknowledgments
This work was supported by a student traineeship from the Cystic Fibrosis Foundation (HEAD01H0) and by Public Health Service grant DK58128 from the National Institutes of Health.
Editor: V. J. DiRita
REFERENCES
- 1.Alm, R. A., and J. S. Mattick. 1995. Identification of a gene, pilV, required for type 4 fimbrial biogenesis in Pseudomonas aeruginosa, whose product possesses a pre-pilin-like leader sequence. Mol. Microbiol. 16:485-496. [DOI] [PubMed] [Google Scholar]
- 2.Arora, S. K., M. Bangera, S. Lory, and R. Ramphal. 2001. A genomic island in Pseudomonas aeruginosa carries the determinants of flagellin glycosylation. Proc. Natl. Acad. Sci. USA 98:9342-9347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Barth, A. L., and T. L. Pitt. 1995. Auxotrophic variants of Pseudomonas aeruginosa are selected from prototrophic wild-type strains in respiratory infections in patients with cystic fibrosis. J. Clin. Microbiol. 33:37-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Barth, A. L., and T. L. Pitt. 1996. The high amino-acid content of sputum from cystic fibrosis patients promotes growth of auxotrophic Pseudomonas aeruginosa. J. Med. Microbiol. 45:110-119. [DOI] [PubMed] [Google Scholar]
- 5.Boucher, J. C., H. Yu, M. H. Mudd, and V. Deretic. 1997. Mucoid Pseudomonas aeruginosa in cystic fibrosis: characterization of muc mutations in clinical isolates and analysis of clearance in a mouse model of respiratory infection. Infect. Immun. 65:3838-3846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Burns, J. L., R. L. Gibson, S. McNamara, D. Yim, J. Emerson, M. Rosenfeld, P. Hiatt, K. McCoy, R. Castile, A. L. Smith, and B. W. Ramsey. 2001. Longitudinal assessment of Pseudomonas aeruginosa in young children with cystic fibrosis. J. Infect. Dis. 183:444-452. [DOI] [PubMed] [Google Scholar]
- 7.Chiang, P., and L. L. Burrows. 2003. Biofilm formation by hyperpiliated mutants of Pseudomonas aeruginosa. J. Bacteriol. 185:2374-2378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Coleman, F. T., S. Mueschenborn, G. Meluleni, C. Ray, V. J. Carey, S. O. Vargas, C. L. Cannon, F. M. Ausubel, and G. B. Pier. 2003. Hypersusceptibility of cystic fibrosis mice to chronic Pseudomonas aeruginosa oropharyngeal colonization and lung infection. Proc. Natl. Acad. Sci. USA 100:1949-1954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costerton, J. W. 2001. Cystic fibrosis pathogenesis and the role of biofilms in persistent infection. Trends Microbiol. 9:50-52. [DOI] [PubMed] [Google Scholar]
- 10.Costerton, J. W., P. S. Stewart, and E. P. Greenberg. 1999. Bacterial biofilms: a common cause of persistent infections. Science 284:1318-1322. [DOI] [PubMed] [Google Scholar]
- 11.Deretic, V., J. R. Govan, W. M. Konyecsni, and D. W. Martin. 1990. Mucoid Pseudomonas aeruginosa in cystic fibrosis: mutations in the muc loci affect transcription of the algR and algD genes in response to environmental stimuli. Mol. Microbiol. 4:189-196. [DOI] [PubMed] [Google Scholar]
- 12.Deziel, E., Y. Comeau, and R. Villemur. 2001. Initiation of biofilm formation by Pseudomonas aeruginosa 57RP correlates with emergence of hyperpiliated and highly adherent phenotypic variants deficient in swimming, swarming, and twitching motilities. J. Bacteriol. 183:1195-1204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Donlan, R. M., and J. W. Costerton. 2002. Biofilms: survival mechanisms of clinically relevant microorganisms. Clin. Microbiol. Rev. 15:167-193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Drenkard, E., and F. M. Ausubel. 2002. Pseudomonas biofilm formation and antibiotic resistance are linked to phenotypic variation. Nature 416:740-743. [DOI] [PubMed] [Google Scholar]
- 15.Ernst, R. K., E. C. Yi, L. Guo, K. B. Lim, J. L. Burns, M. Hackett, and S. I. Miller. 1999. Specific lipopolysaccharide found in cystic fibrosis airway Pseudomonas aeruginosa. Science 286:1561-1565. [DOI] [PubMed] [Google Scholar]
- 16.Ghigo, J. M. 2001. Natural conjugative plasmids induce bacterial biofilm development. Nature 412:442-445. [DOI] [PubMed] [Google Scholar]
- 17.Govan, J. R., and V. Deretic. 1996. Microbial pathogenesis in cystic fibrosis: mucoid Pseudomonas aeruginosa and Burkholderia cepacia. Microbiol. Rev. 60:539-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hentzer, M., G. M. Teitzel, G. J. Balzer, A. Heydorn, S. Molin, M. Givskov, and M. R. Parsek. 2001. Alginate overproduction affects Pseudomonas aeruginosa biofilm structure and function. J. Bacteriol. 183:5395-5401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hoiby, N., A. Fomsgaard, E. T. Jensen, H. K. Johansen, G. Kronborg, S. S. Pedersen, T. Pressler, and A. Kharazmi. 1995. The immune response to bacterial biofilms, p. 233-250. In H. M. Lappin-Scott and J. W. Costerton (ed.), Microbial biofilms. Cambridge University Press, Cambridge, United Kingdom.
- 20.Holloway, B. W. 1955. Genetic recombination in P. aeruginosa. J. Gen. Microbiol. 13:572-581. [DOI] [PubMed] [Google Scholar]
- 21.Jensen, E. T., A. Kharazmi, N. Hoiby, and J. W. Costerton. 1992. Some bacterial parameters influencing the neutrophil oxidative burst response to Pseudomonas aeruginosa biofilms. APMIS 100:727-733. [PubMed] [Google Scholar]
- 22.Kiewitz, C., and B. Tummler. 2000. Sequence diversity of Pseudomonas aeruginosa: impact on population structure and genome evolution. J. Bacteriol. 182:3125-3135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Knutson, C. A., and A. Jeanes. 1968. A new modification of the carbazole reaction: application to heteropolysaccharides. Anal. Biochem. 24:470-481. [DOI] [PubMed] [Google Scholar]
- 24.Koch, A. L. 1994. Growth measurement, p. 248-277. In P. Gerhardt (ed.), General and molecular bacteriology. ASM Press, Washington, D.C.
- 25.Larbig, K. D., A. Christmann, A. Johann, J. Klockgether, T. Hartsch, R. Merkl, L. Wiehlmann, H. J. Fritz, and B. Tummler. 2002. Gene islands integrated into tRNAGly genes confer genome diversity on a Pseudomonas aeruginosa clone. J. Bacteriol. 184:6665-6680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liang, X., X. Q. Pham, M. V. Olson, and S. Lory. 2001. Identification of a genomic island present in the majority of pathogenic isolates of Pseudomonas aeruginosa. J. Bacteriol. 183:843-853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lyczak, J. B., C. L. Cannon, and G. B. Pier. 2002. Lung infections associated with cystic fibrosis. Clin. Microbiol. Rev. 15:194-222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mahenthiralingam, E., M. E. Campbell, J. Foster, J. S. Lam, and D. P. Speert. 1996. Random amplified polymorphic DNA typing of Pseudomonas aeruginosa isolates recovered from patients with cystic fibrosis. J. Clin. Microbiol. 34:1129-1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Nivens, D. E., D. E. Ohman, J. Williams, and M. J. Franklin. 2001. Role of alginate and its O acetylation in formation of Pseudomonas aeruginosa microcolonies and biofilms. J. Bacteriol. 183:1047-1057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Oliver, A., R. Canton, P. Campo, F. Baquero, and J. Blazquez. 2000. High frequency of hypermutable Pseudomonas aeruginosa in cystic fibrosis lung infection. Science 288:1251-1254. [DOI] [PubMed] [Google Scholar]
- 31.O'Toole, G., H. B. Kaplan, and R. Kolter. 2000. Biofilm formation as microbial development. Annu. Rev. Microbiol. 54:49-79. [DOI] [PubMed] [Google Scholar]
- 32.O'Toole, G. A., and R. Kolter. 1998. Flagellar and twitching motility are necessary for Pseudomonas aeruginosa biofilm development. Mol. Microbiol. 30:295-304. [DOI] [PubMed] [Google Scholar]
- 33.O'Toole, G. A., and R. Kolter. 1998. Initiation of biofilm formation in Pseudomonas fluorescens WCS365 proceeds via multiple, convergent signalling pathways: a genetic analysis. Mol. Microbiol. 28:449-461. [DOI] [PubMed] [Google Scholar]
- 34.O'Toole, G. A., L. A. Pratt, P. I. Watnick, D. K. Newman, V. B. Weaver, and R. Kolter. 1999. Genetic approaches to study of biofilms. Methods Enzymol. 310:91-109. [DOI] [PubMed] [Google Scholar]
- 35.Parkins, M. D., H. Ceri, and D. G. Storey. 2001. Pseudomonas aeruginosa GacA, a factor in multihost virulence, is also essential for biofilm formation. Mol. Microbiol. 40:1215-1226. [DOI] [PubMed] [Google Scholar]
- 36.Perna, N. T., G. Plunkett III, V. Burland, B. Mau, J. D. Glasner, D. J. Rose, G. F. Mayhew, P. S. Evans, J. Gregor, H. A. Kirkpatrick, G. Posfai, J. Hackett, S. Klink, A. Boutin, Y. Shao, L. Miller, E. J. Grotbeck, N. W. Davis, A. Lim, E. T. Dimalanta, K. D. Potamousis, J. Apodaca, T. S. Anantharaman, J. Lin, G. Yen, D. C. Schwartz, R. A. Welch, and F. R. Blattner. 2001. Genome sequence of enterohaemorrhagic Escherichia coli O157:H7. Nature 409:529-533. [DOI] [PubMed] [Google Scholar]
- 37.Pratt, L. A., and R. Kolter. 1999. Genetic analyses of bacterial biofilm formation. Curr. Opin. Microbiol. 2:598-603. [DOI] [PubMed] [Google Scholar]
- 38.Rahme, L. G., E. J. Stevens, S. F. Wolfort, J. Shao, R. G. Tompkins, and F. M. Ausubel. 1995. Common virulence factors for bacterial pathogenicity in plants and animals. Science 268:1899-1902. [DOI] [PubMed] [Google Scholar]
- 39.Romling, U., B. Fiedler, J. Bosshammer, D. Grothues, J. Greipel, H. von der Hardt, and B. Tummler. 1994. Epidemiology of chronic Pseudomonas aeruginosa infections in cystic fibrosis. J. Infect. Dis. 170:1616-1621. [DOI] [PubMed] [Google Scholar]
- 40.Romling, U., J. Greipel, and B. Tummler. 1995. Gradient of genomic diversity in the Pseudomonas aeruginosa chromosome. Mol. Microbiol. 17:323-332. [DOI] [PubMed] [Google Scholar]
- 41.Romling, U., J. Wingender, H. Muller, and B. Tummler. 1994. A major Pseudomonas aeruginosa clone common to patients and aquatic habitats. Appl. Environ. Microbiol. 60:1734-1738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Sayeed, S., L. Reaves, L. Radnedge, and S. Austin. 2000. The stability region of the large virulence plasmid of Shigella flexneri encodes an efficient postsegregational killing system. J. Bacteriol. 182:2416-2421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schmidt, K. D., B. Tummler, and U. Romling. 1996. Comparative genome mapping of Pseudomonas aeruginosa PAO with P. aeruginosa C, which belongs to a major clone in cystic fibrosis patients and aquatic habitats. J. Bacteriol. 178:85-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Schroeder, T. H., T. Zaidi, and G. B. Pier. 2001. Lack of adherence of clinical isolates of Pseudomonas aeruginosa to asialo-GM1 on epithelial cells. Infect. Immun. 69:719-729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Silo-Suh, L., S. J. Suh, P. A. Sokol, and D. E. Ohman. 2002. A simple alfalfa seedling infection model for Pseudomonas aeruginosa strains associated with cystic fibrosis shows AlgT (sigma-22) and RhlR contribute to pathogenesis. Proc. Natl. Acad. Sci. USA 99:15699-15704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Singh, P. K., A. L. Schaefer, M. R. Parsek, T. O. Moninger, M. J. Welsh, and E. P. Greenberg. 2000. Quorum-sensing signals indicate that cystic fibrosis lungs are infected with bacterial biofilms. Nature 407:762-764. [DOI] [PubMed] [Google Scholar]
- 47.Spencer, D. H., A. Kas, E. E. Smith, C. K. Raymond, E. H. Sims, M. Hastings, J. L. Burns, R. Kaul, and M. V. Olson. 2003. Whole-genome sequence variation among multiple isolates of Pseudomonas aeruginosa. J. Bacteriol. 185:1316-1325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stewart, P. S., A. K. Camper, S. D. Handran, C. Huang, and M. Warnecke. 1997. Spatial distribution and coexistence of Klebsiella pneumoniae and Pseudomonas aeruginosa in biofilms. Microb. Ecol. 33:2-10. [DOI] [PubMed] [Google Scholar]
- 49.Stickler, D. 1999. Biofilms. Curr. Opin. Microbiol. 2:270-275. [DOI] [PubMed] [Google Scholar]
- 50.Stover, C. K., X. Q. Pham, A. L. Erwin, S. D. Mizoguchi, P. Warrener, M. J. Hickey, F. S. Brinkman, W. O. Hufnagle, D. J. Kowalik, M. Lagrou, R. L. Garber, L. Goltry, E. Tolentino, S. Westbrock-Wadman, Y. Yuan, L. L. Brody, S. N. Coulter, K. R. Folger, A. Kas, K. Larbig, R. Lim, K. Smith, D. Spencer, G. K. Wong, Z. Wu, and I. T. Paulsen. 2000. Complete genome sequence of Pseudomonas aeruginosa PA01, an opportunistic pathogen. Nature 406:959-964. [DOI] [PubMed] [Google Scholar]
- 51.Terry, J. M., S. E. Pina, and S. J. Mattingly. 1991. Environmental conditions which influence mucoid conversion Pseudomonas aeruginosa PAO1. Infect. Immun. 59:471-477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Vallet, I., J. W. Olson, S. Lory, A. Lazdunski, and A. Filloux. 2001. The chaperone/usher pathways of Pseudomonas aeruginosa: identification of fimbrial gene clusters (cup) and their involvement in biofilm formation. Proc. Natl. Acad. Sci. USA 98:6911-6916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Vidal, O., R. Longin, C. Prigent-Combaret, C. Dorel, M. Hooreman, and P. Lejeune. 1998. Isolation of an Escherichia coli K-12 mutant strain able to form biofilms on inert surfaces: involvement of a new ompR allele that increases curli expression. J. Bacteriol. 180:2442-2449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Wozniak, D. J., T. J. Wyckoff, M. Starkey, R. Keyser, P. Azadi, G. A. O'Toole, and M. R. Parsek. 2003. Alginate is not a significant component of the extracellular polysaccharide matrix of PA14 and PAO1 Pseudomonas aeruginosa biofilms. Proc. Natl. Acad. Sci. USA 100:7907-7912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu, H., J. C. Boucher, N. S. Hibler, and V. Deretic. 1996. Virulence properties of Pseudomonas aeruginosa lacking the extreme-stress sigma factor AlgU (σE). Infect. Immun. 64:2774-2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Yu, H., M. Hanes, C. E. Chrisp, J. C. Boucher, and V. Deretic. 1998. Microbial pathogenesis in cystic fibrosis: pulmonary clearance of mucoid Pseudomonas aeruginosa and inflammation in a mouse model of repeated respiratory challenge. Infect. Immun. 66:280-288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Yu, H., and N. E. Head. 2002. Persistent infections and immunity in cystic fibrosis. Front. Biosci. 7:D442-D457. [DOI] [PubMed] [Google Scholar]
- 58.Yu, H., M. J. Schurr, and V. Deretic. 1995. Functional equivalence of Escherichia coli σE and Pseudomonas aeruginosa AlgU: E. coli rpoE restores mucoidy and reduces sensitivity to reactive oxygen intermediates in algU mutants of P. aeruginosa. J. Bacteriol. 177:3259-3268. [DOI] [PMC free article] [PubMed] [Google Scholar]