Abstract
Molecular determinants underlying the production of siderophores in the human and animal pathogen Staphylococcus aureus and the contribution of siderophore production to the virulence of this bacterium have, until now, remained undefined. Here, we show that S. aureus strains RN6390 and Newman produce siderophore when the cells are starved for iron. We further identified and characterized a nine-gene, iron-regulated operon, designated sbn and situated between sirABC and galE on the S. aureus chromosome, that is involved in the production of a siderophore. Mutation of the sbnE gene, in both RN6390 and Newman, eliminates the ability of these strains to produce a siderophore under iron-limited growth conditions, while introduction of multicopy sbnE into sbnE mutants complemented the inability of the mutants to produce the siderophore. sbnE mutants, in both the RN6390 and Newman backgrounds, displayed a drastic growth deficiency, compared to the wild type, in iron-restricted growth medium, whereas no such deficiency was observed during growth in iron-replete medium. Complemented mutants showed a restored ability to grow under iron restriction. We further showed that an sbnE mutant was compromised in a murine kidney abscess model of S. aureus infection, illustrating the importance of siderophore production to the pathogenicity of S. aureus. sbn genes were present in all S. aureus strains tested (and all S. aureus genome sequences) but were undetectable in any of the 13 coagulase-negative staphylococci tested, including Staphylococcus epidermidis.
Iron is an absolute requirement for the growth of most microorganisms, with the possible exceptions of lactobacilli (2) and Borrelia burgdorferi (24). Despite being the fourth most abundant element in the Earth's crust, iron is frequently a growth-limiting nutrient. In aerobic environments and at physiological pH, iron is present in the ferric (Fe3+) state and forms insoluble hydroxide and oxyhydroxide precipitates. Mammals overcome iron restriction by possessing high-affinity iron-binding glycoproteins such as transferrin and lactoferrin that serve to solubilize and deliver iron to host cells (33). This results in a further restriction of free extracellular iron, and, accordingly, the concentration of free iron in the human body is estimated to be 10−18 M, a concentration that is several orders lower than that required to support a productive bacterial infection (3).
To overcome iron restriction, bacteria have evolved several different mechanisms to acquire this essential nutrient. For example, members of the family Pasteurellaceae may express receptors for the recognition of iron-loaded forms of transferrin and lactoferrin (9). One of the most common iron acquisition mechanisms, though, is the use of low-molecular-weight, high-affinity iron chelators, termed siderophores, and cognate cell envelope receptors that serve to actively internalize ferric-siderophore complexes. Many siderophores are able to successfully compete with transferrin and lactoferrin for host iron. Indeed, the ferric-siderophore uptake systems are critical virulence factors in bacteria such as septicemic E. coli (34), Vibrio anguillarum (6), Erwinia chrysanthemi (8), and Pseudomonas aeruginosa (19).
Staphylococcus aureus may cause numerous syndromes in humans, ranging from minor skin and wound infections to more serious sequelae such as endocarditis, osteomyelitis, and septicemia (1). The ability of S. aureus to invade and colonize many tissues may be ascribed to its capacity to express several virulence factors such as fibronectin-, elastin-, and collagen-binding proteins that aid in tissue adherence and multiple exotoxins and proteases that result in tissue destruction and bacterial dissemination. The ability of this bacterium to acquire iron during in vivo growth is also likely important to its pathogenesis, and several research groups have characterized several different genes whose products are involved in the binding and/or transport of host iron compounds (17, 20, 30).
S. aureus possesses several different iron-regulated ABC transporters, including those encoded by the sstABCD (21), sirABC (12), and fhuCBG (28) operons. While the transported substrates are unknown for the sst and sir systems, the fhuCBG genes, in concert with fhuD1 and fhuD2 (27), are involved in the acquisition of iron(III)-hydroxamate complexes. Several members of the staphylococci, including numerous coagulase-negative staphylococci (CoNS) and strains of S. aureus, produce siderophores. Two of these siderophores, staphyloferrin A (14, 18) and staphyloferrin B (7, 11), are of the polycarboxylate class, while the third, aureochelin (5), is chemically uncharacterized. Leading into this study, no molecular-genetic information was known about the synthesis of any of the staphylococcal siderophores.
The objectives of this study were to identify genetic loci involved in siderophore production in S. aureus, to generate siderophore-deficient strains of S. aureus, and to utilize the mutants so as to investigate the importance of siderophore production to the iron-limited growth of this bacterium. Finally, we sought to investigate the importance of siderophore production to the virulence of S. aureus. We report here the identification and characterization of an iron-regulated, nine-gene operon (designated sbn) whose products are involved in the biosynthesis of a siderophore in S. aureus. We show that the operon is present in both laboratory strains and clinical isolates of S. aureus but is not present in CoNS. Finally, we demonstrate that the expression of this operon not only is important for iron-restricted growth of S. aureus in laboratory culture but also prolongs the ability of S. aureus to survive in vivo.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth media.
Bacterial strains and plasmids used in this study are described in Table 1. Escherichia coli and S. aureus strains were routinely cultured in Luria-Bertani broth (Difco) and tryptic soy broth (Difco), respectively. Iron-restricted bacterial growth was performed in Tris-minimal succinate medium (TMS), the composition of which has been described previously (28). Residual free iron was chelated from TMS medium by the addition of ethylenediamine-di(o-hydroxyphenylacetic acid) (EDDHA) (1 μM unless otherwise stated), or TMS was made iron replete by the addition of 50 μM FeCl3. The following antibiotics were used at the indicated concentrations: erythromycin (5 μg/ml), lincomycin (20 μg/ml), neomycin (50 μg/ml), kanamycin (50 μg/ml), and tetracycline (4 μg/ml) for S. aureus selection and ampicillin (100 μg/ml), tetracycline (10 μg/ml), and erythromycin (300 μg/ml) for E. coli selection. All reagents were made with water purified through a Milli-Q water purification system (Millipore, Mississauga, Ontario, Canada).
TABLE 1.
Bacterial strains, plasmids, and oligonucleotides used in this studya
| Bacterial strain, plasmid, or oligonucleotide application | Description or sequences | Source or reference |
|---|---|---|
| Bacteria | ||
| E. coli DH5α | φ80dlacZΔM15 recA1 endA1 gyrA96 thi-1 hsdR17(rK− mK+) supE44 relA1 deoR Δ(lacZYA-argF)U169 | Promega |
| S. aureus | ||
| RN4220 | rK− mK+; accepts foreign DNA | 15 |
| RN6390 | Prophage-cured wild-type strain | 23 |
| Newman | Wild-type strain | O. Schneewind |
| SA113 | T. Foster | |
| ATCC 25923 | ATCC | |
| MJH010 | 8325-4 fur::Tet; Tetr | S. Foster |
| H295 | RN6390 fur::Km; Kmr | 28 |
| H706 | Newman fur::Km (fur::Km marker from H295 was transduced into Newman); Kmr | This study |
| H438 | RN4220 sbnF::pMUTIN4; Emr | This study |
| H479 | H295 sbnF::pMUTIN4; Emr Kmr | This study |
| H520 | RN4220 SA0121::pMUTIN4; Emr | This study |
| H521 | RN4220 galE::pMUTIN4; Emr | This study |
| H551 | RN4220 sbnI::pMUTIN4; Emr | This study |
| H557 | RN4220 sbnH::pMUTIN4; Emr | This study |
| H572 | RN4220 sbnA::pMUTIN4; Emr | This study |
| H672 | RN6390 sbnE::Km; Kmr | This study |
| H675 | RN6390 sbnE::Km fur::Tet; Kmr Tetr | This study |
| H686 | Newman sbnE::Km; Kmr | This study |
| H16 | Clinical isolate | LHSC |
| H50 | Clinical isolate | LHSC |
| H51 | Clinical isolate | LHSC |
| Coagulase-negative staphylococci (CoNS) | ||
| S. auricularis ATCC 33753 | ATCC | |
| S. capitis ATCC 35661 | ATCC | |
| S. caprae ATCC 35538 | ATCC | |
| S. chromogenes ATCC 43764 | ATCC | |
| S. cohnii ATCC 29973 | ATCC | |
| S. epidermidis LK819 | M. Valvano | |
| S. haemolyticus ATCC 29970 | ATCC | |
| S. intermedius ATCC 29663 | ATCC | |
| S. hominis ATCC 27846 | ATCC | |
| S. sciuri ATCC 29062 | ATCC | |
| S. simulans ATCC 27851 | ATCC | |
| S. warneri ATCC 27836 | ATCC | |
| S. xylosus ATCC 35663 | ATCC | |
| Burkholderia cepacia CEP024 | Genomovar III isolate from cystic fibrosis patient | M. Valvano |
| Plasmids | ||
| pAUL-A | Temperature-sensitive S. aureus suicide vector; Emr Lcr | 4 |
| pAW8 | E. coli-S. aureus shuttle vector; Tetr | 32 |
| pBC SK(+) | E. coli cloning vector; Cmr | Stratagene |
| pDG782 | pMLT22 derivative that carries a kanamycin resistance cassette; Apr Kmr | 10 |
| pMUTIN4 | lacZ fusion vector; Apr (E. coli) Emr (S. aureus) | 31 |
| pSED12 | pBC SK+ derivative carrying sbnE; Cmr | This study |
| pSED17 | pSED12 derivative containing sbnE::Km; Cmr Kmr | This study |
| pSED18 | pAUL-A derivative containing sbnE::Km; Kmr Emr | This study |
| pSED32 | pAW8 derivative carrying sbnE; Tetr | This study |
| Oligonucleotidesb | ||
| Generation of sbnA-lacZ fusion | TTGGATCCAGTATATGAATCCTGGAGGC (forward) | |
| TTGGATCCAAAAATGACTGACCCTTTCGCATC (reverse) | ||
| Generation of sbnF-lacZ fusion | TGGATCCCATCACCAATTGAGCGTGTCGTAGGAGAT (forward) | |
| TGGATCCTTTCAATTGTATGAGGCGCCAACACTCGT (reverse) | ||
| Generation of sbnH-lacZ fusion | TTGCGGCCGCGATAGATAGAGATATCATTA (forward) | |
| TTGGATCCTAGTTAACGCCTATGCCACC (reverse) | ||
| Generation of sbnI-lacZ fusion | TTGCGGCCGCCCCAACACAATTTGGTATTTCTGAA (forward) | |
| TTGGATCCTACTTGAAAATGTGCTTCGC (reverse) | ||
| Generation of SA0121-lacZ fusion | TTGCGGCCGCAAGTTCCATTTGGTGTGTGG (forward) | |
| TTGGATCCGGTAAAACAGTGAAAAGAGC (reverse) | ||
| Generation of galE-lacZ fusion | TTGCGGCCGCTATTATCGCTTTAGTATTAT (forward) | |
| TTGGATCCTCAACGCCTGCTTGAGATGTT (reverse) | ||
| Cloning of sbnE gene | TTGGATCCATTAGCAGACATAGATATAT (forward) | |
| TTGGATCCTAGTGTCTCATCATTAATCG (reverse) |
Abbreviations: Apr, Cmr, Kmr, Lcr, and Tetr, resistance to ampicillin, chloramphenicol, kanamycin, lincomycin, and tetracycline, respectively; LHSC, London Health Sciences Centre; ATCC, American Type Culture Collection.
Data under the heading “Oligonucleotides” are descriptions. Restriction sites for subsequent cloning of the PCR products are underlined.
Recombinant DNA methodology.
Plasmid DNA was isolated from E. coli using Qiaprep mini-spin kits (Qiagen). DNA manipulations, including restriction enzyme digestion and DNA ligation, were performed according to standard procedures (25). Restriction enzymes were purchased from Life Technologies, MBI Fermentas, New England Biolabs, or Roche Diagnostics, and DNA ligations were performed using the Roche Rapid DNA Ligation kit. PwoI (Roche) was used for all PCRs. Oligonucleotides were obtained from Life Technologies and are described in Table 1.
Chromosomal DNA isolation and Southern blotting.
Chromosomal DNA was isolated from various staphylococcal strains using procedures as previously described (28). Briefly, cells were lysed at 37°C using 10 μg of lysostaphin (Sigma) in STE (0.1 M NaCl, 10 mM Tris-HCl [pH 8.0], 1 mM EDTA [pH 8.0]), or for CoNS, lysozyme (1 μg) was added to STE. Sodium dodecyl sulfate (0.1%) and proteinase K (0.5 mg) were added to the preparations and incubated for 2 h at 55°C. Southern blotting techniques were performed essentially as previously described (25), and hybridization was performed with digoxigenin (DIG) (Roche Diagnostics)-labeled probes, prepared and used according to the manufacturer's instructions. Light emission was detected by exposing blots to Hyperfilm ECL (Amersham Biosciences).
Construction of an sbnE mutant.
A 3,037-bp DNA fragment carrying sbnE was PCR amplified from the chromosome of S. aureus RN6390 and cloned into pBCSK+ (BamHI), generating pSED12. The sbnE coding region was interrupted at a unique NcoI site (end polished with Klenow enzyme) by the insertion of a kanamycin resistance cassette, derived from plasmid pDG782, to create pSED17. A BamHI fragment containing the disrupted sbnE gene was removed from pSED17 and cloned into the temperature-sensitive S. aureus suicide plasmid, pAUL-A, to generate pSED18. Plasmid pSED18 was introduced into S. aureus RN4220 before being transduced into S. aureus RN6390 using bacteriophage 80α, using methods previously described (28). S. aureus RN6390 carrying pSED18 was grown to mid-log phase at 30°C before the growth temperature was shifted to 42°C. After 4 h of incubation at 42°C, the culture was plated onto medium containing kanamycin and neomycin and incubated at 42°C overnight. The sbnE mutant, resistant to kanamycin and neomycin and sensitive to erythromycin and lincomycin, was isolated as a result of allelic exchange between chromosomal sbnE and the insertionally inactivated copy. The chromosomal insertion of the Kmr cassette into sbnE was confirmed by PCR.
Creation of transcriptional lacZ fusions and β-galactosidase assays.
Internal fragments of individual genes were cloned into the multiple-cloning site of pMUTIN4 (31), a vector that does not replicate in gram-positive bacteria. S. aureus RN4220 was then transformed with recombinant pMUTIN4 plasmids, and homologous recombination between the cloned DNA sequences and those present on the chromosome resulted in the integration of recombinant plasmids into the chromosome. Chromosomal integrations were confirmed by PCR amplification of pMUTIN4-specific DNA sequences.
S. aureus strains bearing transcriptional fusions to lacZ were assayed for β-galactosidase activity using previously described methods (30). Briefly, cultures were grown in TMS supplemented with 1 μM EDDHA or FeCl3 to an optical density at 600 nm of 0.8. Cells (5 × 108) were lysed in a solution containing 10 mM potassium phosphate buffer (pH 7.8), 15 mM EDTA, 1% Triton X-100, and 10 μg of lysostaphin at 37°C. After centrifugation of cell debris, 5 μl of supernatant was assayed for β-galactosidase activity using the Galacto-Light Plus Chemluminescent reporter gene kit (Tropix) in a Berthold luminometer. The background was set at 50 relative light units/s and the data presented are mean relative light units per second for three independent samples ± standard error.
Siderophore production assays and isolation of siderophore.
Siderophore activity in spent culture supernatants was assayed using chrome azurol S (CAS) by procedures previously described (26). Dilutions of culture supernatants were mixed with equal volumes of CAS shuttle solution and allowed to interact for 30 min at room temperature. With TMS medium serving as the blank, and Desferal serving as the reference standard, the absorbance at 630 nm was determined. Siderophore units were calculated as follows: (A630 of TMS − A630 of sample)/A630 of TMS × 100%.
For siderophore isolations, S. aureus strains were vigorously shaken in TMS for 48 h at 37°C. Culture supernatants were recovered by centrifugation and lyophilized. The concentrated supernatant was resuspended in 100% methanol to 1/10 of the volume of the original culture supernatant and passed through Whatman no. 1 filter paper to remove particulate material. Rotary evaporation was used to reduce the volume before application to an LH-20 column (Amersham Biosciences). Fractions were collected, and those testing positive with CAS shuttle solution and for biological activity in siderophore plate bioassays were dried, resuspended in water, and examined by high-performance liquid chromatography (HPLC). Analytical reversed-phase HPLC was used for final purification of siderophore. The column utilized was a Waters ODS2 Spherisorb column (4.6 by 150 mm). Trifluoroacetic acid (0.1%) in water represented solvent A, whereas 0.1% trifluoroacetic acid in acetonitrile was used as solvent B. The chromatographic method used was as follows: at a flow rate of 0.75 ml/min, 6% B for 3.5 min, followed by a gradient of 6 to 60% B over 20 min. Staphylobactin was detected at 210 nm and had a retention time of approximately 17 min. Staphylobactin was collected, dried, and rechromatographed to check for purity and activity before being analyzed by electrospray ionization-mass spectrometry (ESI-MS).
ESI-MS.
ESI-MS and MS-mass spectrography analyses were performed on a quadrupole-time-of-flight (Q-TOF2) mass spectrometer fitted with a Z-spray source (Micromass, Manchester, United Kingdom). The detector was calibrated using an MS-mass spectrography spectrum of [Glu]-fibrinopeptide B. The molecular mass of the siderophore sample was determined by flow injection analysis using a Waters CapLC system with a carrier solvent of 1:1 HPLC-grade methanol-HPLC-grade water at a flow rate of 30 μl/min. Spectra were acquired in positive-ion mode with an m/z range of 50 to 1,800 using the following parameters: capillary voltage, 3.2 kV; cone voltage, 30 to 40 V; desolvation temperature, 200°C; source temperature, 80°C. Tandem mass spectra were acquired for the parent ion of interest using argon as the collision gas and collision energies ranging from 10 to 30 eV. All spectra were acquired and processed using MassLynx 3.5 (Micromass).
Siderophore plate bioassays.
The ability of siderophores to promote the iron-restricted growth of S. aureus was assessed using siderophore plate bioassays, performed as previously described (28). Briefly, S. aureus RN6390 was incorporated into solid TMS medium (1.4 × 104 cells/ml) containing 20 μΜ EDDHA. The ability of purified siderophores to promote growth of S. aureus was assessed after incubation of plates for 36 h at 37°C.
Mouse kidney abscess experiments.
Female Swiss-Webster mice, weighing 25 g, were purchased from Charles River Laboratories Canada, Inc., and housed in microisolator cages. Bacteria were grown overnight in tryptic soy broth (TSB), harvested, and washed three times in sterile saline. Pilot experiments demonstrated that S. aureus Newman colonized mice better in this model than did RN6390 and that the optimal amount of S. aureus Newman to inject into the tail vein to obtain an acute but nonlethal kidney infection was 107 CFU. Bacteria, suspended in sterile saline, were administered intravenously via the tail vein. The number of viable bacteria injected was confirmed by plating serial dilutions of the inoculum on TSB-agar. On days 5 and 6 postinjection, mice were sacrificed and kidneys were aseptically removed. Using a PowerGen 700 Homogenizer, kidneys were homogenized for 45 s in sterile phosphate-buffered saline containing 0.1% Triton X-100, and homogenate dilutions were plated on TSB-agar to enumerate recovered bacteria. Data presented are the log CFU recovered per mouse.
Computer analyses.
DNA sequence analysis, oligonucleotide primer design and nucleotide sequence alignments were performed using the Vector NTI Suite software package (Informax Inc., Bethesda, Md.).
Nucleotide sequence accession number.
The nucleotide sequence for the sbn operon has been deposited in GenBank and has been assigned accession no. AY251022.
RESULTS
S. aureus RN6390 and Newman produce siderophore.
The objectives of this study were to characterize the role that siderophore production plays in the iron-restricted growth of S. aureus in culture and also to examine its importance to in vivo growth and pathogenicity of this bacterium. To accomplish this, our goal was to generate genetically defined siderophore-deficient mutants from siderophore-producing strains of S. aureus.
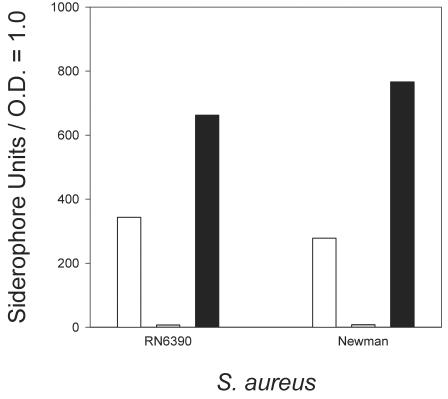
Previous studies have shown that various different isolates of S. aureus have the potential to produce multiple siderophores, including staphyloferrin A and staphyloferrin B (18), and that the genetically characterized strain 8325-4 produced siderophore(s), but of undetermined identity (12, 13). We have demonstrated that two additional S. aureus strains that are used in our laboratory, strain RN6390 and strain Newman, produce readily detectable quantities of siderophore activity when the cells are grown under conditions of iron starvation but produce very little siderophore during growth in iron-replete medium (Fig. 1). Noting that high-affinity iron acquisition systems, including siderophore production and iron(III)-siderophore uptake, are typically regulated by Fur in many different bacteria, we further showed that, indeed, in strains RN6390 and Newman, siderophore production was regulated by exogenous iron concentrations via the Fur protein, since fur derivatives of both RN6390 (H295) and Newman (H706) produced high levels of siderophore activity even when grown in iron-replete medium (Fig. 1). These findings are consistent with published results of Horsburgh et al., who used S. aureus 8325-4 (13).
FIG. 1.
Siderophore levels in spent culture supernatants of RN6390 and Newman and their respective fur derivatives, H295 and H706. Bacteria were grown in an iron-deficient (open bars) or an iron-replete (iron-deficient medium supplemented with 50 μM iron chloride) (gray bars) medium, while the fur::Km derivatives of both RN6390 and Newman (solid bars) were grown in an iron-replete medium. Siderophore units were calculated as described in Materials and Methods. O.D., optical density.
Isolation of siderophore from S. aureus.
We wanted to identify which siderophore(s) was produced by S. aureus RN6390 and related strains. Given that siderophore production was derepressed in fur backgrounds, we isolated siderophore from culture supernatants of strain H295 (RN6390 fur::Km). Our initial experiments focused on the isolation of staphyloferrin A and staphyloferrin B using published procedures (11, 18). However, these purifications yielded extremely little CAS-positive material, suggesting that strain RN6390 produces no, or extremely little, staphyloferrin A or staphyloferrin B. Extraction of culture supernatants using a procedure that has previously been used to isolate ornibactins (29) did, however, result in the isolation of significant quantities of CAS-positive material. Chromatography of methanol-extracted culture supernatant through an LH-20 column yielded discrete fractions that both were CAS positive and promoted the iron-restricted growth of S. aureus in siderophore plate bioassays. Further purification by reversed-phase HPLC yielded an isolated peak of material that retained biological activity. ESI-MS analysis of the isolated material showed that it contained an abundance of a molecule with an m/z = 822, which is significantly greater than that of previously characterized staphylococcal siderophores (staphyloferrin A m/z = 480; staphyloferrin B m/z = 448). We were unable to detect the presence of compounds in the active LH-20 fractions that matched the masses of either staphyloferrin A or staphyloferrin B. Taken together, these results strongly suggest that we have isolated a siderophore that has not previously been identified in the staphylococci. Efforts are ongoing to elucidate the structure of the molecule. Until such time as we determine that this is not a siderophore previously identified from another organism, we have tentatively named the molecule staphylobactin.
Identification and analysis of a siderophore biosynthetic gene cluster in S. aureus.
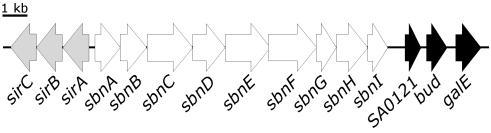
At the time we initiated these studies, genetic information underlying siderophore biosynthesis in the staphylococci had yet to be resolved. We searched S. aureus genome sequences from several strains and identified several open reading frames (ORFs) whose products shared significant similarity with enzymes with demonstrated roles in siderophore biosynthesis. In particular, we identified an 11.5-kb gene cluster, situated between the sirABC operon and galE on the staphylococcal chromosome (Fig. 2), whose products share significant similarity with known or predicted siderophore biosynthetic enzymes in other bacteria (Table 2). While the SirABC proteins share a high degree of similarity to iron(III)-siderophore transport proteins (12), galE (encoding UDP-galactose-4-epimerase) is involved in nucleotide-sugar precursor formation. Hypothesizing that the 11.5-kb gene cluster was involved in siderophore biosynthesis, we designated the coding regions sbn, for siderophore biosynthesis.
FIG. 2.
Schematic representation of the sir-galE region of the S. aureus chromosome. Arrows are representative of individual coding regions. The coding regions within the sbn operon are shown with open arrows, the sir coding regions are shown with gray arrows, and coding regions likely not involved in iron uptake are shown with black arrows. SA0121, hypothetical ORF with nomenclature that is derived from N315 genome sequence; bud, butanediol dehydrogenase.
TABLE 2.
Amino acid identity and similarity to proteins expressed from the sbn operon
| Protein | Closest match or function | Bacterium(a) | Identity (%) | Similarity (%) |
|---|---|---|---|---|
| SbnA | O-Acetyl serine sulfhydrylase | Streptomyces ayermitilis | 42 | 62 |
| O-Acetyl serine sulfhydrylase | E. coli | 29 | 45 | |
| SbnB | Ornithine cyclodeaminase | Archaeoglobus fulgidis | 32 | 53 |
| SbnC | AcsA (achromobactin biosynthesis) | Pectobacterium chrysanthemi | 32 | 50 |
| PvsB (vibrioferrin biosynthesis) | Vibrio parahaemolyticus | 23 | 42 | |
| IucC (aerobactin biosynthesis) | E. coli | 24 | 40 | |
| SbnD | Multidrug efflux | Listeria spp. | 26 | 47 |
| SbnE | RhbC (rhizobactin 1021 biosynthesis) | Sinorhizobium meliloti | 26 | 45 |
| PvsD (vibrioferrin biosynthesis) | Vibrio parahaemolyticus | 25 | 45 | |
| AcsD (achromobactin biosynthesis) | Pectobacterium chrysanthemi | 25 | 43 | |
| IucA (aerobactin biosynthesis) | E. coli | 24 | 42 | |
| SbnF | AcsC (achromobactin biosynthesis) | Pectobacterium chrysanthemi | 45 | 63 |
| RhbF (rhizobactin 1021 biosynthesis) | Sinorhizobium meliloti | 28 | 48 | |
| AlcC (alcaligin biosynthesis) | Bordetella bronchiseptica | 25 | 47 | |
| IucC (aerobactin biosynthesis) | E. coli | 25 | 44 | |
| SbnG | AcsB (achromobactin biosynthesis) | Pectobacterium chrysanthemi | 47 | 67 |
| 4-Hydroxy-2-oxovalerate aldolase | Xanthomonas campestris | 35 | 51 | |
| 2-Dehydro-3-deoxyglucarate aldolase | E. coli | 29 | 51 | |
| SbnH | PvsE (vibrioferrin biosynthesis) | Vibrio parahaemolyticus | 42 | 59 |
| Diaminopimelate decarboxylase | Xanthomonas campestris | 39 | 57 | |
| SbnI | Unknown | NDa | ND |
ND, not determined.
To confirm that the sbn gene cluster was involved in siderophore biosynthesis in S. aureus, we insertionally inactivated the fifth ORF (sbnE) with a kanamycin resistance cassette in S. aureus RN6390, thus creating strain H672. Methanol extracts of spent culture supernatant from iron-restricted H672 contained no trace of material that promoted S. aureus growth in siderophore plate bioassays. Biologically active siderophore was, however, consistently isolated from methanol extracts of iron-restricted supernatants of both the wild-type strain (RN6390) and strain H672 complemented with pSED32, a plasmid carrying sbnE, where expression of sbnE was driven by the plac promoter present on the vector. The staphylobactin molecule isolated from iron-restricted wild-type cultures was completely absent in iron-restricted supernatants of H672 and H675 (RN6390 fur sbnE). These results implicated sbnE as a key gene involved in the production of a siderophore and, more specifically, staphylobactin. The sbnE::Km mutation was also transduced into S. aureus Newman, to create strain H686. Whereas staphylobactin was undetectable in supernatants of iron-starved H686, it was readily detectable in culture supernatants of iron-starved Newman. These results were confirmed by ESI-MS.
The sbnABCDEFGHI genes comprise an operon, and iron, via Fur, regulates its transcription.
Predicted coding regions of the first nine ORFs of the sbn locus either overlap or have very short noncoding segments separating them from one another, whereas approximately 600 bp exists between the 3′ end of the 9th coding region and the 5′ end of the 10th coding region. This suggested that the operon may be comprised of nine ORFs. The 10th coding region encodes a predicted protein of unknown function, the product of the 11th coding region displays significant similarity to butanediol dehydrogenases (acetoin reductases), and the 12th coding region is galE, encoding UDP-galactose-4-epimerase, which is involved in sugar-nucleotide precursor formation in polysaccharide biosynthesis. In an effort to characterize the transcriptional regulation of the sbn operon, and to delineate the limits of the operon, chromosomal lacZ reporter gene fusions targeted to several coding regions, both within and beyond the putative sbn operon, were created. β-Galactosidase expression was then monitored in strains bearing lacZ fusions when the cells were grown in either iron-replete or iron-deficient growth medium. When grown in the presence of 50 μM FeCl3, expression of β-galactosidase in strains bearing fusions to sbnA, sbnF, sbnH, and sbnI was at low, background levels, whereas expression was well above background in strains bearing fusions to SA0121 and galE (Table 3). When grown in iron-deficient medium, however, all strains showed high levels of β-galactosidase expression. These results indicate that transcription of the sbn operon is iron regulated through the 9th coding region (sbnI) and that expression of the 10th coding region and galE is not iron regulated and likely plays no role in the production of siderophore. The observation that sbnA and sbnF were transcribed to the highest levels under iron-deficient growth conditions, while sbn genes further downstream appeared to be transcribed to lesser amounts under similar growth conditions, suggests that expression of the operon is controlled by one iron-regulated promoter element present upstream of the sbnA coding region.
TABLE 3.
β-Galactosidase expression from sbn-lacZ fusions
| Bacterial strain | Presence of Fe | Mean β-galactosidase activity ± SD (RLU/s) |
|---|---|---|
| RN4220 | + | 0 ± 0 |
| RN4220 | − | 0 ± 0 |
| RN4220 sbnA::pMUTIN4 | + | 0 ± 0 |
| RN4220 sbnA::pMUTIN4 | − | 144,763 ± 6,080 |
| RN4220 sbnF::pMUTIN4 | + | 0 ± 0 |
| RN4220 sbnF::pMUTIN4 | − | 193,944 ± 3,398 |
| RN4220 sbnH::pMUTIN4 | + | 0 ± 0 |
| RN4220 sbnH::pMUTIN4 | − | 4,660 ± 209 |
| RN4220 sbnI::pMUTIN4 | + | 0 ± 0 |
| RN4220 sbnI::pMUTIN4 | − | 3,330 ± 188 |
| RN4220 SA0121::pMUTIN4 | + | 106 ± 3 |
| RN4220 SA0121::pMUTIN4 | − | 89 ± 10 |
| RN4220 galE::pMUTIN4 | + | 3,046 ± 525 |
| RN4220 galE::pMUTIN4 | − | 2,146 ± 76 |
| RN4220 fur sbnF::pMUTIN4 | + | 264,425 ± 6,581 |
| RN4220 fur sbnF::pMUTIN4 | − | 231,425 ± 5,720 |
The putative sbnA start codon is preceded by a sequence which resembles a staphylococcal Shine-Dalgarno sequence (AGGAAGA) (Fig. 3) (22). Approximately 50 bp further upstream, a 19-bp sequence (TGAGAATCATTATCAATTA) that bears a striking resemblance to consensus Fur boxes was found, suggesting that expression of the sbn operon is regulated by exogenous iron concentrations via the S. aureus Fur homolog. This would be consistent with our earlier observations (see above) that siderophore production was derepressed in a fur background. Indeed, in a fur-deficient background, β-galactosidase expression from the strain bearing an sbnF-lacZ fusion was extremely high when the cells were grown in iron-replete medium, indicating that the Fur protein represses transcription of the sbn operon under iron-rich growth conditions.
FIG. 3.
Promoter region for the sirABC and sbn operons. Putative Fur box sequences are boxed. Also shown are the predicted start codons for the sirA and sbnA genes, along with predicted Shine-Dalgarno sequences.
An sbnE mutant demonstrates a growth defect in iron-deficient medium.
To assess the contribution of siderophore production to in vitro growth of S. aureus, RN6390 and Newman, their isogenic sbnE::Km mutants (H672 and H686, respectively) and the complemented mutants were grown in defined minimal medium. When grown in TMS medium supplemented with 10 μM EDDHA and 50 μM FeCl3 (iron-replete medium), the growth yields of all of the strains were not appreciably different from one another (Fig. 4A). However, the growth of both H672 and H686 (sbnE mutants) was severely impaired, relative to that of their isogenic parents and the sbnE mutants carrying plasmid pSED32 (carrying multicopy sbnE gene), in the identical medium but lacking FeCl3 (Fig. 4B). Given that the iron-sufficient versus the iron-deficient medium differed only by the presence or absence of FeCl3, the suggestion that the poor growth phenotype of the sbnE mutants was due to the possible chelation of other essential elements by EDDHA can be ruled out. Thus, the sbnE mutants are impaired solely in iron acquisition.
FIG. 4.
Effect of an sbnE mutation on the growth of S. aureus. Shown are growth curves of S. aureus RN6390 (•), Newman (○), H672 (RN6390 sbnE::Km) (▾), H686 (Newman sbnE::Km) (▿), H672 plus pSED32 (▪), and H686 plus pSED32 (□) grown in TMS medium supplemented with 10 μM EDDHA in the presence (A) or absence (B) of 50 μM FeCl3. Bacteria were grown in sidearm flasks with vigorous shaking, and growth was monitored using a Klett meter. Growth experiments were performed in duplicate in three separate experiments. The results of a typical experiment are shown.
Siderophore production enhances the virulence of S. aureus.
S. aureus can survive and replicate in blood to cause infection despite the fact that this environment is iron restrictive. Moreover, recent reports have demonstrated that S. aureus can express proteins with the ability to bind to host iron sources such as heme and hemoglobin (17). Thus, in an effort to determine whether siderophore production in S. aureus is involved in the pathogenesis of this bacterium, the ability of the sbnE mutant to colonize mice was compared to that of its isogenic parent. Swiss-Webster mice were used in a murine kidney abscess model of S. aureus infection. The kidneys of individual mice injected with S. aureus Newman contained an average of more than 108 bacteria at both 5 and 6 days postinjection (Fig. 5). Kidneys from these mice possessed multiple cortical and medullar abscesses. In contrast, the kidneys from mice injected with H686 (Newman sbnE::Km) lacked observable abscesses, average numbers of bacteria recovered from the kidneys were below 107 at day 5 postinjection, and no bacteria were recoverable at day 6 postinjection (Fig. 5), illustrating that the sbnE mutant bacteria were significantly attenuated in this model. These data implicate siderophore production as an important factor in the ability of S. aureus to survive in vivo.
FIG. 5.
An sbnE mutant is compromised in a murine kidney abscess model. Two groups of 12 mice were injected in the tail vein with 107 bacteria. One group received S. aureus Newman, while the second group was infected with H686 (Newman sbnE::Km). CFU recovered from the kidneys of mice at both 5 (eight mice) and 6 (four mice) days postinfection are plotted. Each symbol represents the staphylococcal count in the kidneys of one animal, and the dotted line represents the limit of detection for staphylococci in this assay system. Data are representative of three independent experiments. Statistical significance was determined using the Student unpaired t test, and differences were found to be highly significant (P < 0.003).
The sbn operon is present in S. aureus but not in the CoNS.
Given the demonstrated importance of siderophore production to the pathogenicity of S. aureus, we wished to determine whether the sbn genes were specific to S. aureus or whether they were also present in other staphylococci. Dot blotting experiments, performed under low-stringency hybridization conditions, were performed in efforts to detect sbnA, sbnC, sbnE, and sbnH homologs in several other members of the staphylococci. Whereas sbn genes were readily detected in all laboratory and clinical strains of S. aureus tested (see Table 1 for a complete list of strains used), we were unable to detect the presence of these genes in any of 13 different species of CoNS (Table 1). Homologs of these genes are also not present in the genome sequences of Staphylococcus epidermidis ATCC 12228 or RP62A. Thus, the sbn operon appears to be specific to S. aureus among the staphylococci.
The sbn operon is found in Ralstonia solanacearum.
Interestingly, searches of the databases did reveal a similarly sized operon, present on a megaplasmid in the completed genome sequence of the phytopathogen R. (formerly Pseudomonas) solanacearum, whose products bear striking similarity to Sbn proteins (Table 4). Indeed, it is highly likely that the two operons evolved from the same ancestor since the Ralstonia homologs are present in the same order as the sbn genes in S. aureus. The sbnE homolog in Ralstonia, however, is present on the complementary strand compared with the rest of the coding regions in the Ralstonia operon. Another minor difference between the regions in S. aureus and R. solanacearum is that the R. solanacearum sbnC and sbnD homologs appear to be fused into one coding region. A striking dissimilarity between the sbn operon in S. aureus and the homologous region of DNA in R. solanacearum is the G+C content (in moles percent) of the respective operons. Whereas the operon in R. solanacearum has a G+C content of 72 mol%, the S. aureus sbn operon has a G+C content of 37 mol%. The G+C content of the S. aureus genome is approximately 32 mol%.
TABLE 4.
Homolog of the sbn operon in R. solanacearum
| Sbn protein |
R. solanacearum homologa
|
|
|---|---|---|
| Identity (%) | Similarity (%) | |
| SbnA | 56 | 75 |
| SbnB | 58 | 75 |
| SbnC | 29 | 44 |
| SbnD | 28 | 42 |
| SbnE | 32 | 52 |
| SbnF | 36 | 54 |
| SbnG | 42 | 59 |
| SbnH | 47 | 63 |
| SbnI | NDb | ND |
Identities and similarities are between the predicted protein products.
ND, not determined.
DISCUSSION
S. aureus possesses both siderophore-mediated and nonsiderophore iron uptake systems, and the relative role that each system plays during pathogenesis needs to be resolved. In this study, we characterized a genetic locus (sbn) that is involved in the biosynthesis of a siderophore in S. aureus. We showed that an sbnE mutation correlates with impaired growth in an iron-deficient medium and decreased virulence of S. aureus in a murine kidney abscess model.
Mutation of the sbnE coding region resulted in the inability of S. aureus strains to synthesize an as yet structurally uncharacterized molecule that we have named staphylobactin. Introduction of multicopy sbnE into sbnE mutants resulted in the restored ability of S. aureus to produce staphylobactin, a molecule which when purified by HPLC promoted the iron-restricted growth of S. aureus. It would appear, then, that the staphylobactin molecule is a product synthesized from expression of the sbn operon; however, in order to validate this hypothesis, an elucidation of the structure of this molecule as well as a detailed functional characterization of the remainder of the sbn genes will be required.
The sbnE mutant derivatives of RN6390 and Newman, H672 and H686, respectively, grew equivalently to their isogenic wild-type parents in iron-rich medium. In contrast, the sbnE mutants were severely compromised in their ability to grow, relative to the wild type, under conditions of severe iron starvation (i.e., TMS supplemented with 10 μM EDDHA). We did observe, however, that at moderate levels of iron restriction (i.e., TMS supplemented with 1 μM EDDHA), H672 and H686 grew nearly as well as the wild type. The supernatants of mutants grown under these conditions did react positively in CAS assays, but we were unable to detect staphylobactin in culture supernatants. As well, S. aureus RN6390 grew significantly better under severe iron restriction than S. aureus Newman did, and the former seemed to produce higher levels of siderophore activity as measured by CAS assays. It is plausible that S. aureus RN6390 produces an additional siderophore(s) that Newman lacks and that they are produced under moderate levels of iron restriction. The significantly longer lag period of Newman versus RN6390 in growth assays under conditions of severe iron restriction (Fig. 4B) supports this argument. Alternatively, there may be differences in the regulation of staphylobactin production between the two strains. For example, the levels of iron restriction needed for expression of sbn genes or the amount of staphylobactin produced, may be different in Newman than in RN6390. Other research groups have reported differences in the levels of siderophore produced by different members of the staphylococci (5, 16). Our results support the suggestion that expression of the sbn genes is important in situations of severe iron starvation in culture and, most certainly, is important in vivo since an sbnE mutant was attenuated in mice. Although the sbnE mutant was unable to survive for any period beyond 5 days in vivo, it is interesting that an average of 6 × 106 bacteria could still be recovered from kidneys at day 5 of the infection. Although it appears that the bacteria were actively being cleared, this observation supports the notion that siderophore production may not be required for the establishment of an infection but most certainly appears to be required for prolonged survival of S. aureus in vivo. S. aureus may be able to scavenge various different sources of host iron (e.g., heme and hemoglobin) during the establishment of an infection, and indeed, S. aureus does possess the ability to bind heme and hemoglobin (17).
Attempts to isolate readily detectable quantities of staphyloferrin A and staphyloferrin B from S. aureus RN6390 and Newman were unsuccessful even when the culture media were supplemented with dl-ornithine and 2,3-diaminopropionic acid, precursors in the synthesis of staphyloferrin A and staphyloferrin B, respectively. This was surprising since previous studies have demonstrated staphyloferrin A and staphyloferrin B production by several different strains of S. aureus and several CoNS. Staphylobactin, with an m/z of 822—significantly larger than that of staphyloferrin A (m/z = 480), staphyloferrin B (m/z = 448), or the structurally uncharacterized aureochelin (m/z = 577)—may represent a novel structure. It is also feasible that one of the staphyloferrin molecules may comprise a part of the structure of staphylobactin. Further characterization of this molecule is obviously required to address this question.
The sbnABCDEFGHI operon (SA0112 to SA0120 in S. aureus N315 and SAV0116 to SAV0124 in S. aureus Mu50) was identified in searches of several S. aureus genomes for coding regions whose products shared significant similarity with siderophore biosynthetic genes in other bacteria. The operon is present in all completed and ongoing genome sequencing projects, and we could detect sbn genes in all S. aureus strains used in this study. We were unable to detect the presence of sbn genes, using low-stringency hybridization techniques, in CoNS species. We also could not identify homologs of any of the sbn genes in the genome sequences of S. epidermidis RP62A and S. epidermidis ATCC 12228. Since a previous investigation demonstrated the presence of the staphyloferrins in S. epidermidis strains (18), this lends further support to the idea that the sbn operon is responsible for the production of a siderophore not previously identified in the staphylococci. Our results suggest that the CoNS, generally less pathogenic than S. aureus due in large part to a relative lack of virulence factors, would appear to lack the ability to produce staphylobactin. As noted in this study, the ability to produce this siderophore, synthesized via expression of the sbn operon, correlates with enhanced virulence of S. aureus in a murine kidney abscess model and may, therefore, represent another key determinant that dictates differences in the virulence of CoNS versus S. aureus.
An operon homologous to the S. aureus sbnABCDEFGHI locus was identified in the genome sequence of R. solanacearum, specifically on the 2.1-Mb megaplasmid present in this bacterium. While the G+C contents of the two operons are quite different, reflecting the codon bias of the two organisms (72 mol% for the R. solanacearum locus; 37 mol% for sbn in S. aureus), the organization of predicted genes within the two loci and the similarities between the respective predicted proteins are striking. Given the different niches occupied by the two bacteria (primarily humans for S. aureus versus the soil for R. solanacearum), it is quite interesting to speculate as to how this locus might have been acquired by, or how it evolved in, these two different bacteria.
A consensus Fur box is centered approximately 60 bp upstream of sbnA, and our data support a role for Fur in the transcriptional regulation of the sbn operon. The Fur box sequence shares 17 of 19 bp with the S. aureus consensus Fur box as described previously (13). Divergently transcribed from the sbn operon is sirABC, an iron-regulated operon whose products are similar to ABC transporters involved in the uptake of iron(III) siderophores (12). The sir and sbn operons are separated by 230 bp that possesses two putative Fur box sequences for the transcriptional control of each operon. The predicted sirA gene product is a 36-kDa lipoprotein that shares similarity with a periplasmic iron(III) siderophore-binding protein in E. chrysanthemi, while SirB and SirC, sharing significant similarity with CbrB and CbrC, respectively, are likely to encode transmembrane permease components of an ABC transporter. The proximity of the sbn and sir operons lead us to speculate that the SirABC proteins are involved in the transport of staphylobactin, a hypothesis we are currently testing.
Acknowledgments
This work was supported by operating grant MOP-38002 from the Canadian Institutes of Health Research (CIHR) to D.E.H.
S.E.D. is the recipient of a Natural Sciences and Engineering Research Council PGS-A scholarship, and D.E.H. is a CIHR New Investigator.
We thank James Henderson for excellent technical assistance and O. Schneewind, S. Foster, T. Foster, and M. Valvano for reagents.
Editor: V. J. DiRita
REFERENCES
- 1.Archer, G. L. 1998. Staphylococcus aureus: a well-armed pathogen. Clin. Infect. Dis. 26:1179-1181. [DOI] [PubMed] [Google Scholar]
- 2.Archibald, F. 1983. Lactobacillus plantarum, an organism not requiring iron. FEMS Microbiol. Lett. 19:29-32. [Google Scholar]
- 3.Braun, V., K. Hantke, and W. Köster. 1998. Bacterial iron transport: mechanisms, genetics, and regulation. Metal Ions Biol. Syst. 35:67-145. [PubMed] [Google Scholar]
- 4.Chakraborty, T., M. Leimeister-Wächter, E. Domann, M. Hartl, W. Goebel, T. Nichterlein, and S. Notermans. 1992. Coordinate regulation of virulence genes in Listeria monocytogenes requires the product of the prfA gene. J. Bacteriol. 174:568-574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Courcol, R. J., D. Trivier, M.-C. Bissinger, G. R. Martin, and M. R. W. Brown. 1997. Siderophore production by Staphylococcus aureus and identification of iron-regulated proteins. Infect. Immun. 65:1944-1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Crosa, J. H., L. L. Hodges, and M. H. Schiewe. 1980. Curing of a plasmid is correlated with an attenuation of virulence in the marine fish pathogen Vibrio anguillarum. Infect. Immun. 27:897-902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dreschel, H., S. Freund, G. Nicholson, H. Haag, O. Jung, H. Zähner, and G. Jung. 1993. Purification and chemical characterization of staphyloferrin B, a hydrophilic siderophore from staphylococci. BioMetals 6:185-192. [DOI] [PubMed] [Google Scholar]
- 8.Enard, C., A. Diolez, and D. Expert. 1988. Systemic virulence of Erwinia chrysanthemi 3937 requires a functional iron assimilation system. J. Bacteriol. 170:2419-2426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gray-Owen, S. D., and A. B. Schryvers. 1996. Bacterial transferrin and lactoferrin receptors. Trends Microbiol. 4:185-191. [DOI] [PubMed] [Google Scholar]
- 10.Guérout-Fleury, A. M., K. Shazand, N. Frandsen, and P. Stragier. 1995. Antibiotic-resistance cassettes for Bacillus subtilis. Gene 167:335-336. [DOI] [PubMed] [Google Scholar]
- 11.Haag, H., H. P. Fiedler, J. Meiwes, H. Drechsel, G. Jung, and H. Zähner. 1994. Isolation and biological characterization of staphyloferrin B, a compound with siderophore activity from staphylococci. FEMS Microbiol. Lett. 115:125-130. [DOI] [PubMed] [Google Scholar]
- 12.Heinrichs, J. H., L. E. Gatlin, C. Kunsch, G. H. Choi, and M. S. Hanson. 1999. Identification and characterization of SirA, an iron-regulated protein from Staphylococcus aureus. J. Bacteriol. 181:1436-1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Horsburgh, M. J., E. Ingham, and S. J. Foster. 2001. In Staphylococcus aureus, Fur is an interactive regulator with PerR, contributes to virulence, and is necessary for oxidative stress resistance through positive regulation of catalase and iron homeostasis. J. Bacteriol. 183:468-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Konetschny-Rapp, S., G. Jung, J. Meiwes, and H. Zähner. 1990. Staphyloferrin A: a structurally new siderophore from staphylococci. Eur. J. Biochem. 191:65-74. [DOI] [PubMed] [Google Scholar]
- 15.Kreiswirth, B. N., S. Lofdahl, M. J. Bentley, M. O'Reilly, P. M. Schlievert, M. S. Bergdoll, and R. P. Novick. 1983. The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305:680-685. [DOI] [PubMed] [Google Scholar]
- 16.Lindsay, J. A., and T. V. Riley. 1994. Staphylococcal iron requirements, siderophore production, and iron-regulated protein expression. Infect. Immun. 62:2309-2314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mazmanian, S. K., E. P. Skaar, A. H. Gaspar, M. Humayun, P. Gornicki, J. Jelenska, A. Joachmiak, D. M. Missiakas, and O. Schneewind. 2003. Passage of heme-iron across the envelope of Staphylococcus aureus. Science 299:906-909. [DOI] [PubMed] [Google Scholar]
- 18.Meiwes, J., H.-P. Fiedler, H. Haag, H. Zähner, S. Konetschny-Rapp, and G. Jung. 1990. Isolation and characterization of staphyloferrin A, a compound with siderophore activity from Staphylococcus hyicus DSM 20459. FEMS Microbiol. Lett. 67:201-206. [DOI] [PubMed] [Google Scholar]
- 19.Meyer, J. M., A. Neely, A. Stintzi, C. Georges, and I. A. Holder. 1996. Pyoverdin is essential for virulence of Pseudomonas aeruginosa. Infect. Immun. 64:518-523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Modun, B., R. W. Evans, C. L. Joannou, and P. Williams. 1998. Receptor-mediated recognition and uptake of iron and human transferrin by Staphylococcus aureus and Staphylococcus epidermidis. Infect. Immun. 66:3591-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Morrissey, J. A., A. Cockayne, P. J. Hill, and P. Williams. 2000. Molecular cloning and analysis of a putative siderophore ABC transporter from Staphylococcus aureus. Infect. Immun. 68:6281-6288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Novick, R. P. 1991. Genetic systems in staphylococci. Methods Enzymol. 204:587-636. [DOI] [PubMed] [Google Scholar]
- 23.Peng, H. L., R. P. Novick, B. Kreiswirth, J. Kornblum, and P. Schlievert. 1988. Cloning, characterization, and sequencing of an accessory gene regulator (agr) in Staphylococcus aureus. J. Bacteriol. 170:4365-4372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Posey, J. E., and F. C. Gherardini. 2000. Lack of a role for iron in the Lyme disease pathogen. Science 288:1651-1653. [DOI] [PubMed] [Google Scholar]
- 25.Sambrook, J., E. F. Fritsch, and T. Maniatis. 1989. Molecular cloning: a laboratory manual, 2nd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 26.Schwyn, B., and J. B. Neilands. 1987. Universal chemical assay for the detection and determination of siderophores. Anal. Biochem. 160:47-56. [DOI] [PubMed] [Google Scholar]
- 27.Sebulsky, M. T., and D. E. Heinrichs. 2001. Identification and characterization of fhuD1 and fhuD2, two genes involved in iron-hydroxamate uptake in Staphylococcus aureus. J. Bacteriol. 183:4994-5000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sebulsky, M. T., D. Hohnstein, M. D. Hunter, and D. E. Heinrichs. 2000. Identification and characterization of a membrane permease involved in iron-hydroxamate transport in Staphylococcus aureus. J. Bacteriol. 182:4394-4400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sokol, P. A., P. Darling, D. E. Woods, E. Mahenthiralingam, and C. Kooi. 1999. Role of ornibactin biosynthesis in the virulence of Burkholderia cepacia: characterization of pvdA, the gene encoding l-ornithine N5-oxygenase. Infect. Immun. 67:4443-4455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Taylor, J. M., and D. E. Heinrichs. 2002. Transferrin binding in Staphylococcus aureus: Involvement of a cell wall anchored protein. Mol. Microbiol. 43:1603-1614. [DOI] [PubMed] [Google Scholar]
- 31.Vagner, V., E. Dervyn, and S. D. Ehrlich. 1998. A vector for systematic gene inactivation in Bacillus subtilis. Microbiology 144:3097-3104. [DOI] [PubMed] [Google Scholar]
- 32.Wada, A., and H. Watanabe. 1998. Penicillin-binding protein 1 of Stapylococcus aureus is essential for growth. J. Bacteriol. 180:2759-2765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Weinberg, E. D. 1999. Iron loading and disease surveillance. Emerg. Infect. Dis. 5:346-352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Williams, P. H. 1979. Novel iron uptake system specified by ColV plasmids: an important component in the virulence of invasive strains of Escherichia coli. Infect. Immun. 26:925-932. [DOI] [PMC free article] [PubMed] [Google Scholar]