SUMMARY
Reactive oxygen species (ROS), particularly H2O2, act as intracellular second messengers in many signaling pathways. Protein-tyrosine phosphatases (PTPs) are now believed to be important targets of ROS. PTPs contain a conserved catalytic cysteine with an unusually low pKa. This property allows PTPs to execute nucleophilic attack on substrate phosphotyrosyl residues, but also renders them highly susceptible to oxidation. Reversible oxidation, which inactivates PTPs, is emerging as an important cellular regulatory mechanism and might contribute to human diseases, including cancer. Given their potential toxicity, it seems likely that ROS generation is highly controlled within cells to restrict oxidation to those PTPs that must be inactivated for signaling to proceed. Thus, identifying ROS-inactivated PTPs could be tantamount to finding the PTP(s) that critically regulate a specific signaling pathway. This article provides an overview of the methods currently available to identify and quantify PTP oxidation and outlines future challenges in redox signaling.
Keywords: Protein-tyrosine phosphatases, Tyrosyl phosphorylation, Signal transduction, Oxidation, Reactive oxygen species, Activity-based probes, Mass spectrometry, Dimedone
INTRODUCTION
The phosphorylation of proteins on tyrosyl residues is a critical post-translational modification that regulates several fundamental cellular processes, including cell growth, proliferation and migration [1, 2]. The levels of phosphotyrosine are regulated by protein-tyrosine kinases (PTKs) [1] and protein-tyrosine phosphatases (PTPs) [2, 3]. Dysregulation of either protein family leads to abnormal levels of phosphorylation and can contribute to several human diseases, including cancer (reviewed in [4, 5]).
The PTP superfamily consists of 107 genes, which are divisible into four families [2]. The class I cysteine-based PTPs comprise the largest PTP family and can be subdivided into classical and dual-specificity PTPs (DUSPs) [2]. The classical PTPs include 21 receptor and 17 non-receptor PTPs, which exclusively hydrolyze phosphotyrosyl residues [2]. All classical PTPs contain a highly conserved “signature motif”, [I/V]HCSXGXGR[S/T]G, wherein the invariant cysteine is essential for catalysis [6]. In most classical PTPs, this cysteinyl residue has a pKa between 4.5–5.5 due to its close contact with several nearby main-chain amide groups and a hydrogen bond with the side chain of the serine residue in the signature motif [7]. This property allows the catalytic cysteine to remain in the thiolate (S−) state at physiological pH, facilitating nucleophilic attack on substrate phosphotyrosines, but it also renders PTPs highly susceptible to oxidation [8–10].
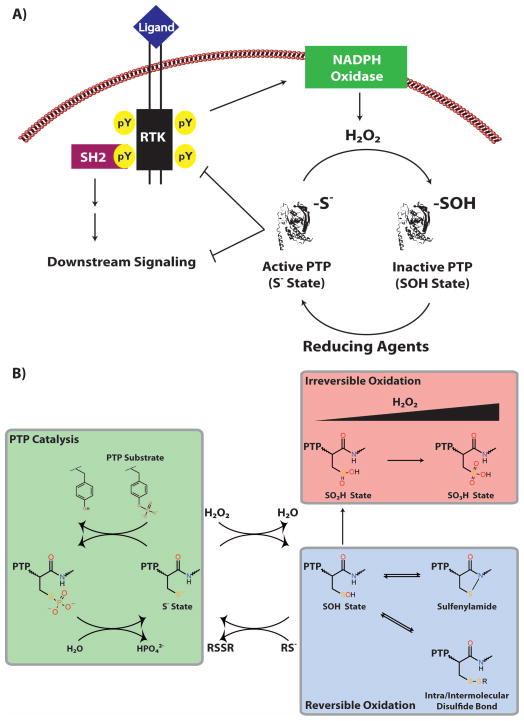
Recent studies suggest that cells capitalize on the exquisite sensitivity of PTPs to oxidation by employing reactive oxygen species (ROS), particularly hydrogen peroxide (H2O2), as intracellular second messengers in many signaling pathways (reviewed in [11–14]). ROS are transiently and locally generated within cells by NADPH oxidases (NOXs) following growth factor stimulation (reviewed in [15]) and are required for full receptor phosphorylation and activation of downstream signaling (Figure 1A) [16, 17]. Growth factor-induced ROS production leads to the reversible oxidation and inactivation of PTPs to the sulfenic acid (SOH) state (Figure 1B) [10]. The SOH state is labile and, in different PTP family members, rapidly rearranges to form a sulfenylamide [18–20] with the adjacent main chain nitrogen or a disulfide bond [21, 22] with a nearby cysteinyl residue. The sulfenylamide and disulfide states help to prevent “hyper-oxidation” to the biologically irreversible sulfinic (SO2H) and sulfonic (SO3H) acid states (Figure 1B) [10, 18, 19]. Several studies have reported the oxidation of specific PTPs in response to different types of cell stimuli, including PTPN1 (PTP1B) in EGFR signaling [23], PTPN11 (SHP2) in PDGF signaling [24], PTP1B and PTPN2 (TC-PTP) in insulin signaling [25] and PTPN6 (SHP1) in B cell receptor signaling [26–28] (reviewed in [14]). In contrast to normal cells, cancer cells often produce high levels of ROS, leading to decreased basal PTP activity and enhanced tyrosyl phosphorylation [29–34]. Readers are directed to a review in this issue by Ostman et al. for a detailed discussion of the role of PTP oxidation in pathological cell signaling.
Figure 1. Redox regulation of PTPs.
(A) Model for PTP redox regulation. RTK activation results in the tranisent and localized production of H2O2 by NOX enzymes. Due to the intrinsic sensitivity of their catalytic cysteinyl residues, PTPs are reversibly oxidized (PTP-SOH) and inactivated in the presence of H2O2, leading to increased tyrosyl phosphorylation and downstream signaling. As signaling continues, NOX enzymes are inactivated, resulting in decreased H2O2 levels, PTP re-activation (PTP-S−) by reducing agents (i.e., glutathione peroxidases) and a reduction in tyrosyl phosphorylation/signal transmission. (B) Schematic depicting PTP catalysis and oxidation. In the active (PTP-S−) state, PTPs can dephosphorylate phosphotyrosyl substrates; however, in the presence of physiological levels of H2O2, PTPs are reversibly oxidized to the sulfenic acid (PTP-SOH) state and thereby inactivated. This state is labile and, in different PTP family members, rapidly rearranges to form a intramolecular sulfenylamide or a disulfide bond with a nearby cysteine residue. The sulfenylamide and disulfide states help to prevent hyper-oxidation to the biologically irreversible sulfinic (PTP-SO2H) and sulfonic (PTP-SO3H) acid states. Figure adapted from [87].
Given the prominent role of ROS in regulating normal and pathological cell signaling, the identification of ROS-inactivated PTPs might be tantamount to finding the PTP(s) that critically regulate a specific signaling pathway. Several different approaches are available to monitor classical PTP oxidation, each of which exploits the biochemical properties of PTP catalysis or oxidation. This article provides a critical analysis for each of these methods with a particular emphasis on their applicability to a global proteomic approach. We also outline future challenges in improving the identification of redox- regulated PTPs.
DETECTION OF REVERSIBLE PTP OXIDATION
Basal or ligand-induced PTP oxidation results in two pools of PTPs: oxidized (SOH; inactive) and reduced (S−; active). Numerous techniques have been developed to identify reversibly oxidized PTPs; these can be classified as “indirect” or “direct.” Indirect methods are the most common and, because they exploit conserved biochemical properties of PTP catalysis, also can be applied to detect PTP expression. Direct approaches rely on detecting the oxidized form of PTPs (or structural changes that arise due to oxidation).
Indirect approaches
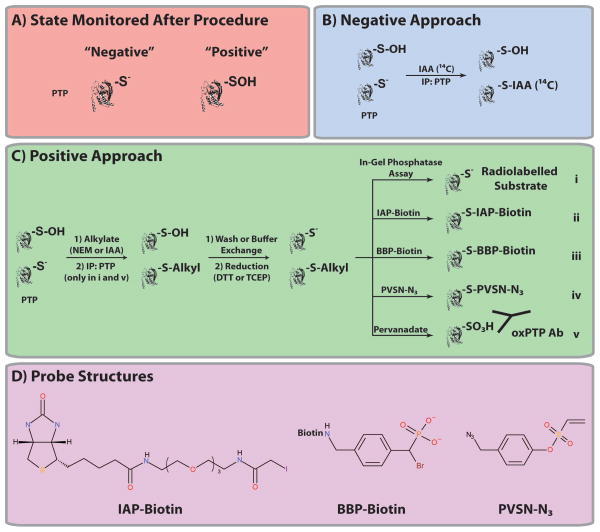
Indirect approaches share a similar experimental workflow, but they can be divided into two general categories based on whether they detect a decrease in active PTPs (S−; “negative”) or an increase in oxidized PTPs (SOH; “positive”). Both rely on the ability of active PTPs to react stoichiometrically and irreversibly with alkylating agents (PTP-S-Alkyl; i.e., iodoacetic acid [IAA] or N-ethylmaleimide [NEM]) (see Introduction) and the resistance of oxidized PTPs to alkylation. In negative techniques, cells are lysed in the presence of a “labelled” (e.g., radioactive or biotin-tagged IAA) alkylating agent, and PTP oxidation is measured based on decreased detection of that probe (Figure 2A). Cells also are lysed in the presence of an alkylating agent in positive approaches; however, oxidized PTPs are then converted to the active state (using a reducing agent) and captured and detected using a PTP-reactive probe (e.g., biotin-tagged IAA; Figure 2B).
Figure 2. Indirect methods to monitor PTP oxidation.
(A) The state (S− or SOH) monitored after the indicated method is shown. Summary of the two general categories of indirect approaches to quantify PTP oxidation. In negative techniques (B), cells are lysed in the presence of a labelled (e.g., radioactive) alkylating agent, and PTP oxidation is reflected by decreased detection of the probe. In positive approaches (C), cells also are lysed in the presence of an alkylating agent; however, oxidized PTPs are then converted to the active state (using a reducing agent) and reacted with a radiolabelled substrated (modified in-gel PTPase assay; i), IAP-Biotin (ii), BBP- Biotin (iii), PVSN-N3 (iv) or pervanadate (v). (C) Structures of IAP-Biotin, BBP-Biotin and PVSN-N3 are shown.
Indirect approaches can be “targeted” or “global.” In targeted methods, the PTP of interest is immunoprecipitated; by contrast, with global approaches, oxidation of the entire PTP family can be assessed.
Negative approaches
Negative approaches were the first methods developed to detect and quantify PTP oxidation. Lee et al. used radioactively labelled IAA (14C) to monitor oxidation of the non-receptor PTP PTP1B (Figure 2A and Table 1) [23]. In their experiments, oxidation was assessed by first lysing cells in the presence of radiolabelled IAA to alkylate and irreversibly label the catalytic cysteinyl residue of active PTPs, while oxidized PTPs remained unaffected [23]. PTP1B was then immunoprecipitated, resolved on an SDS-PAGE gel and the radioactivity incorporated (into PTP1B) was quantified [23]. A decrease in incorporation (compared with no treatment) indicated PTP1B oxidation [23]. Using this procedure, Lee et al. demonstrated that PTP1B is oxidized reversibly following H2O2 treatment of recombinant PTP1B and EGF stimulation of A431 cells [23]. The latter result was particularly important, because it provided the first direct evidence that PTPs are physiologically relevant targets of growth factor-evoked ROS, consistent with the earlier suggestion that PTPs must be inhibited by ROS to allow for full receptor tyrosine kinase (RTK) phosphorylation and activation of downstream signaling [16, 17].
Table 1.
Comparison of the current methods to detect oxidized PTPs.
| Method | Positive or Negative | Applied to MS | Potential for MS | PTP Specific | Benefits | Limitations | Selected References | |
|---|---|---|---|---|---|---|---|---|
| Indirect | Radiolabelled Assay | Negative | No | Yes | No |
|
|
[23, 35] |
| Modified In-Gel PTPase Assay | Positive | No | No | Yes |
|
|
[24, 38, 39] | |
| IAP-Biotin | Positive | No | Yes | No |
|
|
[40, 43, 44] | |
| ABPs | Positive | No | Yes | Yes |
|
|
[40, 53, 54, 56] | |
| qPTPome/ q-oxPTPome | Positive | Yes | Yes | Yes |
|
|
[62–66] | |
| Direct | Dimedone-based chemical probes | Positive | Yes | Yes | No |
|
|
[69–74, 77–82] |
| Dimedone antibodies | Positive | No | Yes | No |
|
|
[83, 84] | |
| Conformation- sensing antibodies | Positive | No | Yes | Yes |
|
|
[86] |
Lee et al. also provided a framework to begin to experimentally measure the level of oxidized PTPs following ligand stimulation [23]. But despite its success in identifying PTP1B as a negative regulator of EGFR signaling, this method is inherently insensitive because it relies on measuring decreases in labelling. Although Lee et al. reported a 40% increase in PTP1B oxidation following EGF stimulation, this is likely an overestimate based on recent measurements of growth factor-induced oxidation (see below). Furthermore, because this is a targeted method, each PTP must be tested individually, provided antibodies are available. Several variations of this method have been used to measure PTP oxidation in different contexts [35, 36]. Rather than relying on radiolabelled IAA, these assays used a biotin- or fluorescently-tagged IAA probe [35, 36]. Such approaches also are amenable to measuring the oxidation of any reactive cysteine-containing proteins [37].
Modified in-gel phosphatase (PTPase) assay
The modified in-gel PTPase assay was the first technique used to detect an increase in PTP oxidation (i.e., positive signal) [24, 25, 38]. It is based on the in-gel PTPase assay, developed in the mid-1990s by Burridge and Nelson to monitor PTP expression. In the in-gel PTPase assay, total cell lysates are resolved on a denaturing SDS-PAGE gel, and proteins are then renatured and reacted with a radioactively labelled PTP substrate (i.e., 32P-labelled poly(GluTyr)) that is incorporated into the gel prior to polymerization [39]. PTPs are detected by autoradiography as regions where 32P has been selectively removed (i.e., negative bands) [39]. By applying this approach, Burridge and Nelson detected PTP expression in several cell lines and tissues [39].
The modified in-gel PTPase assay permits the identification of oxidized PTPs (Figure 2B and Table 1): lysates are first alkylated with IAA followed by the in-gel PTPase assay, which enables reactivation of formerly oxidized PTPs and their detection based on reactivity with the radiolabelled substrate [24, 25, 38]. This assay, developed by Meng et al., was first used to detect increases in PTP oxidation in Rat1 cells following H2O2 stimulation, and also identified PTPN11 as a negative regulator of PDGF signaling [24]. It was subsequently applied to demonstrate reversible oxidation of PTP1B and PTPN2 during insulin signaling [25]. By detecting oxidation of different PTPs in these signaling pathways, Meng et al. added to the growing body of evidence demonstrating that PTPs are critical ROS targets and that different PTPs may be oxidized following activation of different RTKs.
The development and application of the modified in-gel PTPase assay was an important step forward because it enabled the global, positive identification of PTP oxidation, which could not be assessed previously, and opened the door for the development of more sensitive assays (see below). Yet despite its success, the modified in-gel PTPase assay has several limitations. It is biased toward non-transmembrane PTPs, because most RPTPs do not renature well [39]. Furthermore, although this assay can detect whether a PTP is oxidized, this approach still requires identification of the oxidized PTP by depletion experiments, and due to the differential ability of PTPs to renature, is not quantitative [39].
Modified cysteinyl-labelling assay: Iodoacetylpolyethylene oxide biotin (IAP-Biotin)
The modified cysteinyl-labelling assay, a clear improvement from the modified in-gel PTPase assay, detects oxidized proteins based on their reactivity with the biotin-tagged alkylating agent, IAP-Biotin (Figures 2B and C and Table 1) [40], which can react with any reactive cysteinyl residue [40]. In this assay, cells are lysed in the presence of IAA, followed by a desalting column/buffer exchange to remove IAA [40]. Eluates are treated with DTT (to reduce reversibly oxidized proteins) and subsequently incubated with the IAP-Biotin probe to capture reduced (formerly oxidized) proteins [40]. These can then be purified with streptavidin-Sepharose beads and detected by immunoblotting with streptavidin or specific antibodies [40]. To limit the background of non-PTP proteins, the assay can be performed under mildly acidic conditions (pH 5.5), which ensures that the majority of cysteine-containing proteins are in the thiol (SH) state and cannot react with the probe [40]. At this pH, the catalytic cysteine of most PTPs (including classical PTPs and DUSPs) along with other highly reactive cysteine-containing proteins, such as hydroxylases, peroxidases and thiol proteases, remains in the active thiolate (S−) state and are reactive [40].
The concept underlying the modified cysteinyl-labelling assay was conceived in the late-1990s to assess the oxidation of non-PTP proteins, such as p53 [41]. PTPs were first assessed by Li et al., who used NEM-Biotin (3-(N-maleimido-propionnyl)biocytin; MPB) probe, to measure the nitrosylation of PTP1B in response to the treatment of A431 and Jurkat cells with various nitrosylation agents (i.e., SNAP and GSNO) [42]. Boivin et al. developed the modified cysteinyl-labelling assay to detect increased oxidation of a number of proteins in PDGF-transformed angiomyolipoma cells (compared with non-transformed cells), which was prevented when the cells were treated with the NOX inhibitor diphenyliodonium (DPI) or the antioxidant N-acetyl cysteine (NAC) [40]. Boivin et al. also demonstrated that this assay could detect oxidation of PTP1B and the RPTPs PTPRF (LAR) and PTPRA (RPTPα), along with the lipid phosphatase PTEN and the MAPK phosphatase MKP-1 in the same cells [40]. Subsequently, the authors published several papers to outline the method to the redox signaling field [43, 44], which will facilitate its adaptation to a global MS-based approach. In fact, a modified IAP-Biotin probe was recently used to identify reactive cysteinyl residues by MS. Although >1000 cysteine-containing peptides were identified, only three were PTP-derived, and these did not include active site peptides [37]. Notably, these assays were performed at physiological pH (7.5), which as previously discussed, would increase the background of abundant (non-PTP) peptides [37, 40]. By performing these experiments under mildly acidic conditions, it is possible that this approach could assess PTP expression (and oxidation) by MS, along with other highly reactive cysteinyl-containing proteins. Without using MS though, this assay is limited by its inability to identify the oxidized proteins. Furthermore, in its current format, unlike the modified in-gel PTPase assay, where only PTPs are measured, a large panel of non-PTP proteins would be detected. The identification of other proteins maybe beneficial depending on the intended experimental aim (i.e., assessment of all redox-regulated proteins or exclusively classical PTPs). However, probe reactivity with non-PTP proteins might reduce the assay’s sensitivity and result in decreased detection of classical PTPs, as was evident in the aforementioned study [37].
Although global proteomic applications of the modified cysteinyl-labelling assay are currently unavailable to assess PTP oxidation, targeted MS approaches have been reported. For example, Lou et al. developed a MS-based technique whereby all PTP oxidation states can be measured [31]. In their method, cell lysates prepared in the presence of iodoacetamide (IAM) are applied to a gel filtration column, and the eluate is then reduced. PTPs of interest are immunoprecipitated, resolved on an SDS-PAGE gel and stained with Coomassie [31]. The region corresponding to the PTP is excised and subjected to in-gel trypsinization [31]. Tryptic peptides are then detected by MALDI-MS, permitting the identification (but not the quantification) of all PTP oxidation states (i.e., S−, SOH, SO2H, SO3H) [31]. The authors applied this procedure to show that PTP1B is reversibly (SOH) and irreversibly (SO2H and SO3H) oxidized in several cancer cells [31]. For this PTP, quantification was possible because its S− and SOH forms migrate differently on SDS-PAGE compared with the SO2H and SO3H forms [31]. These experiments demonstrated that >25% of PTP1B can be reversibly oxidized basally in cancer cells [31]. This study provided a seminal contribution to the field by providing the first quantification of all PTP oxidation states; however, it is unlikely to be applicable to a global proteomic approach since the oxidized forms of most, in not all PTPs are unlikely to migrate differently.
Subsequently, Held et al. developed a targeted proteomic approach that relies on selected reaction monitoring (SRM, also known as multiple reaction monitoring or MRM) to quantify protein oxidation [45]. SRM is a label-free, tandem MS method that allows the relative abundance of characterized peptide ions to be quantified (reviewed in [46, 47]). In their assay, which they term oxidized SRM (OxSRM), cells are lysed in the presence of trichloroacetic acid (TCA) to quench all redox reactions [45]. Proteins are then resuspended in a denaturing buffer containing unlabelled (d0) NEM to alkylate free cysteinyl residues, precipitated with TCA and resuspended in denaturing buffer containing TCEP and a stable isotopic version of NEM with 5 deuteriums (d5) to label formerly oxidized proteins [45]. The protein of interest is then immunoprecipitated, digested with trypsin and the peptides corresponding to the alkylated d5 NEM (oxidized) and d0 NEM (reduced) cysteines, which differ by 5 Da are then monitored by SRM to determine their ratio (oxidized/reduced [d5/d0]) [45]. Similar to the modified in-gel PTPase assay, this method is not PTP specific and can be applied to monitor the oxidation of any protein, provided antibodies are available [45]. The authors applied this method to demonstrate that PTP1B is oxidized following H2O2 and diamide (a ROS- producing agent) stimulation of MCF7 cells and that p53 is oxidized following diamide treatment [45].
The OxSRM assay provides a reliable general procedure to assess protein oxidation by SRM [45]. By using SRM for quantification, the authors can monitor the oxidation of any reactive cysteinyl residue provided the peptide can be initially detected by LC-MS/MS [45]. One of the limitations of the method, as the authors discuss, is the overestimation of the fraction of reversibly oxidized proteins if a high proportion are in the SO2H or SO3H state. However, this is a limitation shared by most approaches to assess oxidation [45]. Overall, this is a promising technique to monitor oxidation, particularly since the fraction of each protein oxidized can be quantified using different isotopes of NEM for alkylation.
Modified cysteinyl-labelling assay: Activity-based PTP probes
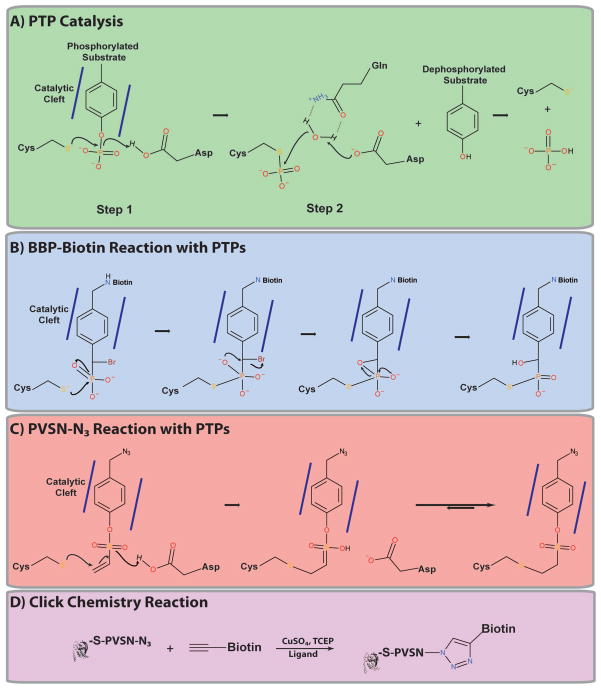
Activity-based probes (ABPs) are reagents designed to react with mechanistically-related enzymes [48, 49]. ABPs have been developed to target several protein families, including caspases [50], papains [51], glycosidases [52] and hydrolases [53]. By exploiting the properties of PTP catalysis, Zhang and coworkers have developed ABPs targeting classical PTPs (Figure 3A). All classical PTPs contain two residues that are critical for the catalytic mechanism: the catalytic cysteine and an aspartic acid (D) residue in the WPD loop [6–8]. Phosphotyrosine substrates bind to, and are stabilized in, the non-polar PTP catalytic cleft. The WPD loop then undergoes a dramatic conformational change, closing over the phenol ring of the phosphotyrosine, thereby holding the substrate in place and positioning it for dephosphorylation [6–8]. The catalytic thiolate then executes a nucleophilic attack on the phosphate moiety of the phosphotyrosine substrate [6–8]. The aspartic acid residue acts as the general acid, donating a proton (H+) to the tyrosine, facilitating release of the dephosphorylated substrate and formation of a PTP-S-PO42− intermediate [6–8]. The same aspartic acid residue then acts as a general base, removing a proton from water, and promotes nucleophilic attack by the resulting hydroxyl ion (OH−) on the PTP-S-PO42− intermediate, releasing the reactivated PTP and PO42− [6–8].
Figure 3. Activity-based PTP probes.
(A) Schematic of PTP catalysis. Mechanism for the reaction of a PTP with the activity-based probes BBP-Biotin (B) and PVSN-N3 (C). Figures adapted from [54, 55]. (D) Example of a Cu-catalyzed azide-alkyne cycloaddition click chemistry reaction.
Zhang et al. developed ABPs targeting the classical PTP family by synthesizing molecules that: (1) resemble phosphotyrosine, allowing ABP binding to the PTP catalytic cleft; (2) contain a reactive group, permitting the reaction and subsequent trapping of the PTP; and (3) have a linker connecting the ABP to a reporter/affinity tag, enabling purification of the PTP-ABP adduct [54–56]. Two of these ABPs are α-bromobenzylphosphonate (BBP)-Biotin (Figure 2C) [54] and 4-(azidomethyl)phenyl ethenesulfonate azide (PVSN-N3; Figure 2C) [55]. Both contain a phenyl ring to promote binding to the PTP catalytic cleft and initiation of a normal substrate reaction. Each, however, traps PTPs differently (Figures 3B and C) [54, 55]. The phosphonate moiety of BBP-Biotin reacts with the PTP catalytic cysteine, generating a transient phosphorane-like intermediate, which leads to the formation and subsequent opening of a three-membered epoxide ring, release of bromide, and rearrangement to yield a PTP-S- BBP-Biotin adduct (Figure 3B) [54]. PVSN-N3 reacts with the catalytic cysteine via a 1,4-conjugate addition, forming a stable thioether bond (PTP-S-PVSN-N3; Figure 3C) [55]. In experiments using BBP-Biotin, PTPs are isolated by binding to streptavidin- Sepharose [54], whereas PVSN-N3 adducts must first undergo a click chemistry reaction with an alkyne-conjugated reporter/affinity tag (Figure 3D) [55]. Click chemistry is a modular reaction that permits the reaction of two stable and otherwise inert reactants (i.e., azide and alkyne) in an efficient and irreversible reaction [57, 58], most often Cu-catalyzed azide-alkyne cycloaddition [59, 60] or Staudinger ligation [61]. PVSN-N3 lacks the bulky biotin group associated with BBP-Biotin, which might increase PVSN-N3 reactivity with PTPs by avoiding steric hindrance by the biotin moiety. Furthermore, PVSN-N3 can readily cross cell membranes in cells and react with PTPs, unlike BBP- Biotin, which is too bulky [55]. Zhang et al. demonstrated that both of these probes can react with PTPs, first using the Yersinia PTP, YopH, and then the human PTPs: PTP1B, PTPN11 and PTPRJ (DEP1) [54, 55]. These ABPs also react with DUSPs (i.e., Cdc14, VHR, PRL-3) and the low molecular weight protein-tyrosine phosphatase (LMW-PTP) [54, 55].
Boivin et al. demonstrated that BBP-Biotin can be used to monitor PTP oxidation [40]. Briefly, cells are lysed in the presence of IAA, applied to a gel filtration column, reduced and then reacted with the probe, allowing purification of formerly oxidized PTPs (Figure 2B and Table 1) [40]. BBP-Biotin detected oxidation of several PTPs in PDGF-transformed angiomyolipoma cells, including PTP1B, PTPRF and PTPRA [40]. PVSN-N3 has not yet been applied to detect PTP oxidation; however, based on its ability to detect PTP expression, this should be possible. Furthermore, it might have improved reactivity compared with BBP-Biotin, due to the absence of the bulky biotin group (see above).
A benefit of using ABPs to monitor PTP oxidation is the reduction of background (non-PTP) proteins due to the mechanism-based specificity of these probes [54, 55]. As yet, neither of these ABPs have been applied to MS experiments. Furthermore, a complicated and lengthy series of reactions are needed to generate BBP-Biotin, which could restrict its use by the general redox signaling field [54]. By contrast, PVSN-N3 requires only a few synthetic steps, making it an ideal candidate to globally detect PTP expression and oxidation [55].
Oxidized PTP active site antibody (oxPTP Ab)
The Ostman group employed an antibody-based strategy to detect oxidation of classical PTPs by immunizing rabbits with a peptide comprising a portion of the signature motif of PTP1B hyper-oxidized to the SO3H state (VHCSO3HSAG) [62, 63].
The high conservation of the signature motif ([I/V]HCSXGXGR[S/T]G; see Introduction) allows this antibody to cross-react with other classical PTPs (see below). Persson et al. used this antibody in targeted experiments to show reactivity with several classical PTPs, including PTP1B, PTPN2, PTPN6, PTPN11, PTPRA and PTPRJ [62, 64]. Using an approach analogous to the modified cysteinyl-labelling assay, the oxPTP Ab also could be used to measure PTP oxidation (Figure 2B). In this procedure, cells are lysed in the presence of NEM, and the PTP of interest is immunoprecipitated, reduced, washed, and then hyper-oxidized to the SO3H state using pervanadate (PV). The wash step between the reduction and oxidation reaction is required to prevent the conversion of PV to vanadate (VO43−), a reversible PTP inhibitor. Using this targeted protocol, Persson et al. showed that PTPN11 is oxidized following PDGF stimulation [62], the D2 domain of PTPRA is highly sensitive to oxidation [62, 64], and that PTPN6 and PTPN11 are susceptible to H2O2-induced oxidation [65].
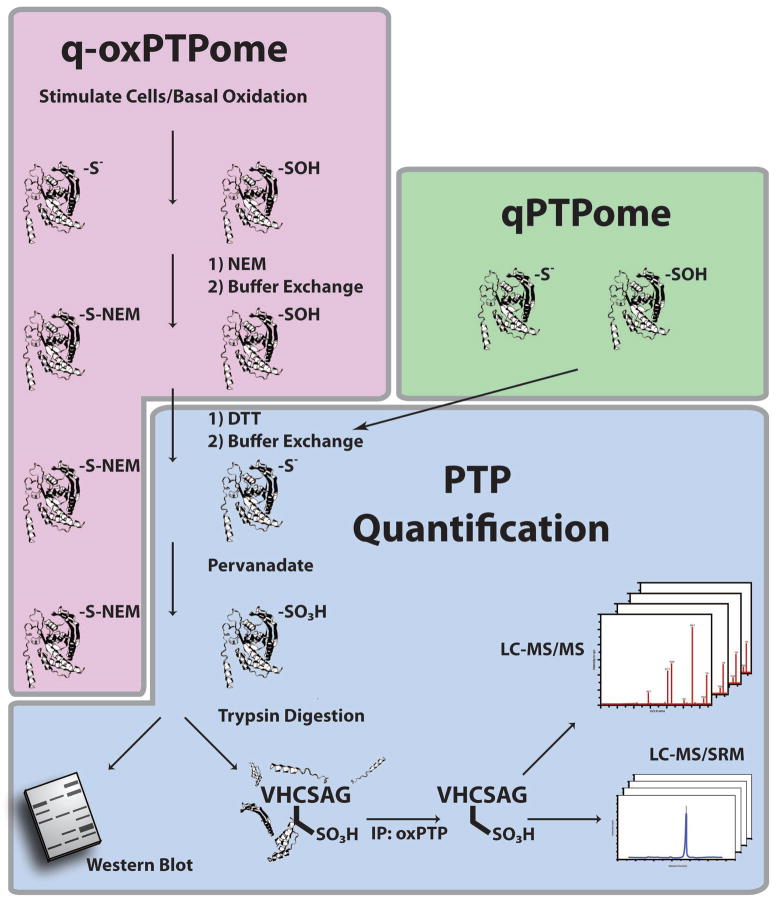
Our group modified the antibody-based approach to enable global detection of classical PTP expression and oxidation. To measure PTP expression, we applied DTT-treated lysates to a gel filtration column and then treated with PV (Figure 4) [66]. Combined with a newly available monoclonal oxPTP Ab, we used this method, termed qPTPome, to detect several PTPs in PV-treated lysates following immunoprecipitation and immunoblotting with the oxPTP Ab. Efficient PTP purification can be achieved only if lysates are denatured prior to immunopurification, suggesting that the oxPTP Ab cannot bind PTPs in their native PTP-SO3H conformation. We further modified this assay to an MS-based approach by generating tryptic digests of PV-treated lysates, immunoprecipitating PTP-SO3H active site peptides with the oxPTP Ab, and subsequently analyzing the peptides by LC-MS/MS. These modifications allowed us to identify 17 PTPs in NIH3T3 cells, including the D1 and D2 domains for several RPTPs. In subsequent experiments, we demonstrated the oxPTP Ab can isolate PTPs from mouse, human or rat cells provided that their active sites match the consensus sequence [I/V]HCS[A/S]G, permitting the detection of all catalytically active classical PTPs (36 of 38 classical PTPs). Notably, the oxPTP Ab cannot react with other phosphatases, such as DUSPs or LMW-PTP.
Figure 4. qPTPome and q-oxPTPome.
Schematic of qPTPome and q-oxPTPome, respectively. For q-oxPTPome, basal or ligand stimulated PTP oxidation (PTP-SOH) is monitored by lysing cells in the presence of NEM, which irreversibly alkylates active PTPs (PTP-S-NEM). Excess NEM is removed by gel filtration, and oxidized PTPs are reduced with DTT (PTP-S−). A second buffer exchange is applied to remove DTT, and reduced PTPs (representing PTPs that were initially oxidized) are hyper-oxidized to the sulfonic acid (PTP-SO3H) state by PV treatment. For qPTPome, the alkylation step is omitted, cells are lysed in the presence of DTT and all PTPs initially present in cells are hyper-oxidized to the sulfonic acid state by PV treatment. In both methods, hyper-oxidized PTPs can then be monitored by immunoblotting or processed for MS. For MS analysis, PV-treated lysates are digested with trypsin, and the hyper-oxidized active site peptides of classical PTPs are immunoprecipitated with the oxPTP Ab and detected using either “discovery”-based proteomics by LC-MS/MS or in “monitoring”-mode by LC-MS/SRM. Note that PTP catalytic domains are represented as ribbon diagrams. Figure adapted from [66].
We then developed a multiplexed SRM (mSRM) protocol to enable, in a single MS assay, comprehensive quantification of PTP expression (and oxidation) of mouse and human classical PTPs using qPTPome-processed samples (Figure 4). Applying this method, we demonstrated that both relative and absolute (when combined with absolute quantification [AQUA]) levels of PTP expression can be measured.
To measure PTP oxidation, we developed q-oxPTPome, wherein cells are lysed in the presence of NEM, applied to a gel filtration column, reduced, applied to a second gel filtration column, and then treated with PV (Figure 4 and Table 1). PV-treated lysates are treated with trypsin, and PTP-SO3H peptides are immunopurified by the oxPTP Ab and quantified by mSRM. By combining q-oxPTPome with qPTPome, the fraction of each PTP oxidized (q-oxPTPome/qPTPome) can be determined. Because the oxPTP Ab detects all RPTP D1 domains and most D2 domains, this approach can be used to monitor the differential oxidation of these two domains. Furthermore, modifications to the procedure allow quantification of all PTP oxidation states (i.e., S−, SOH, SO2H, SO3H). By applying this approach, we showed that q-oxPTPome could monitor differential changes in PTP oxidation: (1) following H2O2 stimulation of NIH3T3 cells; (2) following the alteration of the intracellular redox environment by depleting glutathione; (3) in several different cancer cell lines; and (4) following inhibition of a driver oncogene. Taken together, these data suggested that PTP oxidation adds an additional layer of complexity to the cancer phenotype.
Although q-oxPTPome can quantify the above changes in PTP oxidation, we have been unable to identify the subset of PTPs oxidized in response to growth factor stimulation of normal cells. This likely reflects the complex procedure involved in q-oxPTPome and the small and localized subset of PTPs that are inactivated by normal levels of growth factor-evoked ROS. Because the assay is quantitative, it provides an upper bound on growth factor-induced increases in PTP oxidation (<5%). Studies are underway in our laboratory to increase the sensitivity of q-oxPTPome to circumvent this limitation. Without these improvements, the assay will be unable to begin to interrogate the critical questions confronting the redox signaling field. Furthermore, owing to the divergence of some of the PTP active site sequences, the D2 domains of a few RPTPs (Ptprc, Ptprk, Ptprm, Ptprt and Ptpru) escape detection by the oxPTP Ab. However, the oxPTP Ab can detect their D1 domains, allowing quantification of their expression (and oxidation).
Overall, qPTPome and q-oxPTPome provide a general workflow to begin to study PTP signaling at a systems level. For example, qPTPome can be combined with phosphotyrosine proteomics to suggest PTP substrates. A similar protocol could be used in other systems to identify key PTP substrates. It might also be possible to combine q-oxPTPome with phosphotyrosine proteomics to identify PTP substrates following stimulation with different levels of H2O2, by correlating the levels of PTP oxidation with increases in phosphorylation of specific proteins. Modifications of qPTPome might be developed to facilitate the simultaneous identification of all PTP post-translational modifications: classical PTPs could be isolated at the protein level using the oxPTP Ab, followed by secondary enrichment of trypsinized peptides based on a specific post-translational modification (i.e., anti-phosphotyrosine antibodies to detect tyrosyl phosphorylation) and MS analysis. Such approaches could help to bridge the gap in our understanding of PTP and PTK signaling.
Summary of indirect approaches
Indirect approaches to assess PTP expression and oxidation have improved since the development of the radiolabelled assay by Lee et al. [23]. Although there currently is no “gold standard” assay, the redox-signaling field is rapidly moving towards the development of more robust methods to monitor PTP oxidation. Indirect approaches provide the unique opportunity to probe both PTP expression and oxidation by exploiting the properties of PTP catalysis. Because these approaches can monitor the entire classical PTP family, it will soon be possible for the signaling field to begin to study the global involvement of PTPs in signaling, rather than its current targeted format. Despite the potential of each of these assays, particularly the modified cysteinyl-labelling assay and qPTPome/q-oxPTPome, additional improvements are required to begin to interrogate the role of PTPs in regulating growth factor signaling at a systems level.
Direct approaches
Unlike indirect approaches, which rely on the reactivity of active PTPs, direct methods monitor PTP oxidation by exploiting the properties of oxidized PTPs. Currently, there are two methods to directly assess PTP oxidation. The first relies on dimedone (5,5-dimethyl-1,3-cyclohexanedione), a cell-permeable small molecule that selectively and irreversibly reacts with sulfenic acids (SOH; Figure 5A). Because dimedone reacts with the oxidized thiol, it targets the entire redoxome (not just PTPs), in principle, making it an ideal approach to directly and globally assess protein oxidation. The second approach relies on conformation-sensing antibodies that recognize structural modifications that occur in oxidized PTPs (compared with the active enzyme) that can trap, purify and inhibit PTP reactivation.
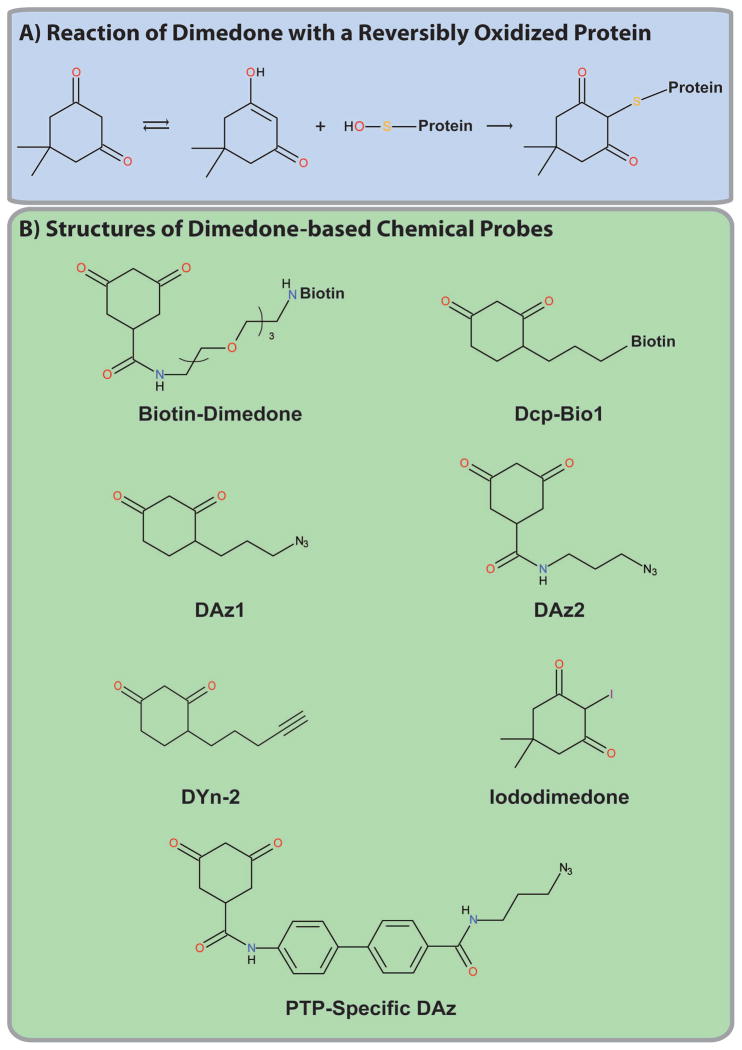
Figure 5. Dimedone-based probes.
(A) Reaction of dimedone with a reversibly oxidized protein. (B) Structures of the indicated dimedone-based probes.
Dimedone-based probes
Dimedone-based probes provided the first direct method used to assess protein oxidation. Their reactivity is based on the 1,3-cyclohexadione backbone, which undergoes ketoenol tautomerism and, in the enol state, can react with the electrophilic sulfur atom in protein-sulfenic acids, releasing water and forming a thioether (C-S-C) bond with the oxidized protein (Figure 5A). Dimedone was used to measure protein oxidation as early as the late 1960s, when it was shown to react with oxidized papain [67] and glyceraldehyde-3-phosphate dehydrogenase (GAPDH) [68]. However, its application was limited to targeted approaches due to the absence of a reporter/affinity tag.
Poole and colleagues synthesized several dimedone-based probes consisting of the dimedone reactive group, a linker and a reporter/affinity tag or an azide group (Figure 5B and Table 1) [69–74]. Direct conjugation of dimedone to a fluorophore or biotin tag allows the detection of oxidized proteins using a single reagent, but, as for ABPs, might limit reactivity with proteins due to steric constraints [69–74]. By contrast, dimedone probes with an azide moiety require click chemistry to connect the probe to an alkyne-linked reporter/affinity tag [69–74]. Poole et al. used AhpC, a cysteine-based peroxidase in Salmonella typhimurium, to test these probes [69–71], showing by MS that dimedone selectively labels oxidized AhpC, but not active AhpC or a Cys46Ser mutant [75, 76]. Both the direct and “clickable” probes also could react with oxidized AhpC using a targeted in vitro approach [69–71]. Subsequently, the same group demonstrated that these probes could detect an overall increase in Biotin-Dimedone (Dcp-Bio1; Figure 5B) labelling following T cell activation and that both PTPN6 and PTPN11 were oxidized reversibly [77]. This probe was also used to detect PTP1B and PTPRJ (DEP1) oxidation following VEGF stimulation of HUVECs, demonstrating that Dcp-Bio1 can react with both non-receptor and receptor PTPs [78]. Recently, Carrroll and colleagues also demonstrated that DYn-2, a click chemistry-based dimedone probe could detect oxidation of PTP1B, PTPN11 and PTEN in response to EGF stimulation of A431 cells [79].
Dimedone probes were first applied to measure protein oxidation globally by Charles et al., who compared H2O2-stimulated and control rat ventricular mycocyte lysates treated with Biotin-Dimedone [80]. Oxidized proteins were purified on streptavidin beads, and resolved by SDS-PAGE, subjected to in-gel trypsinization (in regions that showed increased labelling after H2O2 stimulation), followed by LC-MS/MS [80]. Twenty-two proteins were oxidized inducibly; however, these corresponded to highly abundant proteins, such as myosin or ATP synthase [80]. It is possible that the bulky nature of the biotin moiety prevented the probes from reacting with signaling proteins, such as PTPs, whose oxidized cysteines are located within sterically restrictive “pockets” or “clefts.”
Using a click chemistry-based dimedone probe (DAz1; Figure 5B), Leonard et al. identified 189 reversibly oxidized proteins in HeLa cells using a similar MS-based approach. Unlike the previous study, however, several signaling proteins were identified suggesting that these probes have increased reactivity [81]. Despite this improvement, though, PTPs were not identified. A recent publication by Leonard et al. suggested that previous dimedone-based probes have limited reactivity with classical PTPs [81]. To circumvent this limitation, the authors synthesized several dimedone-based probes containing a “PTP-binding module,” such as a phenyl group, which increased probe binding within the PTP catalytic cleft and consequently increased its reactivity (Figure 5B) [82]. These probes showed significantly increased reactivity with oxidized YopH, a Yersinia PTP, compared with previous dimedone probes [82]. Although these probes have yet to be applied to monitor PTP oxidation, they are promising candidates to apply to develop a global proteomic approach to monitor PTP oxidation.
Dimedone-specific antibodies
Dimedone-specific antibodies also are available to directly monitor protein oxidation (Table 1). To generate these antibodies, Seo et al. synthesized a keynote limpet hemocyanin (KLH)-conjugated dimedone hapten by joining the dimedone moiety with cysteamine via a thioether bond followed by a five carbon linker [83]. KLH-conjugated dimedone was injected into rabbits to produce polyclonal dimedone-specific antibodies [83]. In principle, these antibodies could be used to detect oxidized proteins by treating cells or lysates with dimedone, followed by immunoprecipitation and/or immunoblotting [83]. Indeed, in a targeted approach, dimedone-specific antibodies reacted with oxidized, but not reduced GAPDH, peroxiredoxin I (Prx1) and actin [83]. Oxidized proteins also could be detected in cell lysates by immunoblotting or in cells using immunofluorescence [83]. Accordingly, the authors applied these dimedone-specific antibodies to measure the differential protein oxidation in a panel of breast cancer cell lines by treating the cells with dimedone for 2 hours, followed by cell lysis and immunoblotting [83]. Cells that were not treated with dimedone showed very low reactivity with the antibody, in contrast to dimedone-labelled cells, from which several immunoreactive bands were detected [83]. The intensity and pattern of bands detected across the panel of breast cancer lines differed, suggesting each breast cancer line has a unique profile of oxidized proteins [83].
One limitation of the approach used by Seo et al. is that oxidation was assessed after 2 hours of dimedone treatment [83]. As dimedone irreversibly reacts with sulfenic acids, incubating cells with dimedone for such an extended period of time, probably alters the intracellular redox state, leading to increases of protein oxidation. It would be best to treat cells with dimedone for as short a period of time as possible or to treat lysates rather than cells with dimedone to limit the alteration of redox homeostasis. The later approach might lead to post-lysis oxidation, though, making it crucial that the experiment is performed under anaerobic conditions.
A second dimedone-specific antibody was developed by Maller et al., who used KLH that was reacted directly with dimedone as the immunogen [84]. Unfortunately, these antibodies only detected oxidized GAPDH in H2O2-stimulated rat myocytes, with very few additional immunoreactive bands, unlike the previous antibodies [84]. The inability of this antibody to detect several immunoreactive bands will limit its application.
Although the antibodies developed by Seo et al. appears to specifically react with a large panel of reversibly oxidized proteins, it is unclear whether it can detect growth factor-induced changes in protein oxidation. Furthermore, it will be essential to identify these dimedone-immunoreactive bands by MS. Given dimedone’s low reactivity with PTPs, it may be difficult to apply these antibodies to monitor PTP oxidation; however, this will be unclear until targeted approaches are used to directly assess PTP oxidation or it is applied to MS. Seo et al. also recently developed an iododimedone (d0) derivative (Figure 5B) that can react with any reactive cysteinyl-containing protein, permitting the quantification of the fraction of a protein oxidized when combined with a stable isotope version of dimedone (d6; d6/do) [85]. It will be interesting to see whether this can be combined with the dimedone-specific antibodies to develop a quantitative global proteomic approach to assess protein oxidation.
Conformation-sensing antibodies of oxidized PTP1B (PTP1B-OX)
Recently, the Tonks group developed conformation-sensing antibodies targeting the oxidized form of PTP1B (PTP1B-OX; Table 1) [86]. Earlier crystallographic studies showed that in the presence of ROS, PTP1B is oxidized to the sulfenic acid (SOH) state and rapidly rearranges to form a sulfenylamide (see Introduction) [18, 19]. Sulfenylamide formation dramatically alters the PTP1B active site, exposing Tyr46 [18, 19]. Haque et al. identified a mutant of PTP1B (CA/SA) that adopts a similar conformation to PTP1B-OX [86]. The authors then employed phage display combined with subtractive panning to develop antibodies that specifically detected PTP1B-CA/SA [86]. The resulting antibodies also reacted with PTP1B-OX in vitro and could prevent its reactivation in the prescence of the reducing agent TCEP [86]. Remarkably, this antibody did not bind to closely related PTPN2, which shares ~50% identity with PTP1B [86]. When expressed in cells as an “intrabody,” it could be used to immunoprecipitate and image PTP1B-OX in response to H2O2 or insulin stimulation, demonstrating for the first time the existence of the sulfenylamide state in vivo [86]. Using immunofluoerescence, the authors reported that ~40% of PTP1B co-localized with PTP1B-OX (suggesting that upto 40% of PTP1B might be oxidized) following insulin stimulation, although, based on immunoprecipitation experiments, it appeared that only 5–10% was oxidized. This discrepancy might reflect the limited spatial resolution of the immunofluorescence measurements. The immunoprecipitation data probably represents the “true” level of PTP1B oxidation. This might be an over-estimate of the fraction “instantaneously” oxidized, because the conformation-specific antibodies bind and prevent reactivation of PTP1B. Consequently, PTP1B-OX accumulates over time, leading to an “integral” measurement of PTP1B-OX, rather than the instantaneous value measured by other methods. These data suggest that the fraction of PTP1B oxidized following insulin stimulation is <5%, demonstrating the importance of localized PTP oxidation in growth factor signal transduction. It will be interesting to apply these antibodies to monitor PTP1B-OX in other signaling pathways.
Summary of direct approaches
Direct approaches to monitor PTP oxidation provide a unique opportunity to assess oxidation, as they omit the complicated series of steps required by indirect methods. Although these steps are generally considered to go to completion, each step probably results in some losses that lead to an overall decrease in assay sensitivity. In principle then, direct approaches are preferable for quantifying PTP oxidation, but it has been difficult to develop direct methods that monitor oxidation of the entire classical PTP family. Dimedone-based approaches provide a promising technique to directly and globally measure PTP oxidation. Recent improvements, such as the PTP-specific probes developed by Seo et al [81] might allow this approach to be applied to the entire PTP family.
Summary and future perspectives
Oxidation has emerged as an important mechanism for regulating PTP activity in both normal and oncogenic cell signaling. ROS levels are increased following growth factor stimulation and in several diseases, including cancer. Elevated levels of ROS lead to an increased fraction of oxidized PTPs, shifting the dynamic equilibrium between PTPs and PTKs in favour of increased tyrosyl phosphorylation and downstream signaling. Detecting ROS-inactivated PTPs should help to identify PTP(s) that regulate specific signaling pathways and to delineate the physiological roles of redox regulation. Ideally identification of these oxidized PTPs would involve the application of global approaches that provide a systems-level understanding of PTP signaling.
We have outlined several currently available methods to assess PTP expression and oxidation. Initially, these techniques were exclusively targeted methods, which could only monitor oxidation of a specific PTP at any given time. However, our increased understanding of PTP catalysis and oxidation, combined with improvements to current proteomic technologies, now provide the framework for beginning to monitor PTPs at a systems-level. Despite the success of recent approaches, improvements to these methods are required to routinely quantify changes in PTP oxidation. These changes will not only allow the quantification of PTP oxidation and expression, but will likely provide the opportunity to interrogate other PTP regulatory mechanisms, such as tyrosyl phosphorylation. Only time will tell if new innovations can help to bridge the gap in our understanding between PTP and PTK signaling.
Acknowledgments
Work in the Neel lab is supported by National Institutes of Health Grant R37CA49152 (B.G.N.) and the International Human Frontiers Science Program Organization Grant RGP0039/2009C102 (B.G.N.). Additional support was provided by the Ontario Ministry of Health and Long Term Care and the Princess Margaret Hospital Foundation. B.G.N. is a Canada Research Chair, Tier 1. R.K. is the recipient of a graduate fellowship from the Canadian Institutes of Health Research.
Abbreviations
- ABP
Activity-based probe
- AQUA
Absolute quantification
- BBP-Biotin
α-bromobenzylphosphonate biotin
- DPI
Diphenyliodonium
- DTT
Dithiothreitol
- DUSP
Dual-specificity phosphatase
- EGFR
Epidermal growth factor receptor
- GAPDH
Glyceraldehyde-3-phosphate dehydrogenase
- GSNO
S-nitrosoglutathione
- H2O2
Hydrogen peroxide
- HUVEC
Human umbilical vein endothelial cells
- IAA
Iodoacetic acid
- IAM
Iodoacetamide
- IAP-Biotin
Iodoacetylpolyethylene oxide biotin
- KLH
Keynote limpet hemocyanin
- LC-MS/MS
Liquid chromatography-tandem mass spectrometry
- LMW-PTP
Low molecular weight protein-tyrosine phosphatase
- MRM
Multiple reaction monitoring
- mSRM
Multiplexed selected reaction monitoring
- NAC
N-acetyl cysteine
- NEM
N-ethylmaleimide
- NOX
NADPH oxidase
- oxPTP Ab
Oxidized PTP active site antibody
- OxSRM
Oxidized SRM
- q-oxPTPome
Quantitative analysis of the oxidized classical PTPome
- qPTPome
Quantitative analysis of the classical PTPome
- PDGF
Platelet-derived growth factor
- PrxI
Peroxiredoxin I
- PTK
Protein-tyrosine kinase
- PTP
Protein-tyrosine phosphatase
- PTP1B-OX
Oxidized form of PTP1B
- PV
Pervanadate
- PVSN-N3
4-(azidomethyl)phenyl ethenesulfonate azide
- ROS
Reactive oxygen species
- RPTP
Receptor protein-tyrosine phosphatase
- RTK
Receptor tyrosine kinase
- S−
Thiolate
- SDS-PAGE
Sodium dodecyl sulfate polyacrylamide electrophoresis
- SH
Thiol
- SNAP
S-nitroso-N-acetylpenicillamine
- SO2H
Sulfinic acid
- SO3H
Sulfonic acid
- SOH
Sulfenic acid
- SRM
Selected reaction monitoring
- TCA
Trichloroacetic acid
- TCEP
Tris(2-carboxyethyl)phosphine
- VEGF
Vascular endothelial growth factor
References
- 1.Lemmon MA, Schlessinger J. Cell signaling by receptor tyrosine kinases. Cell. 2010;141:1117–34. doi: 10.1016/j.cell.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Alonso A, Sasin J, Bottini N, Friedberg I, Friedberg I, Osterman A, Godzik A, Hunter T, Dixon J, Mustelin T. Protein tyrosine phosphatases in the human genome. Cell. 2004;117:699–711. doi: 10.1016/j.cell.2004.05.018. [DOI] [PubMed] [Google Scholar]
- 3.Tonks NK. Protein tyrosine phosphatases: from genes, to function, to disease. Nat Rev Mol Cell Biol. 2006;7:833–46. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- 4.Julien SG, Dube N, Hardy S, Tremblay ML. Inside the human cancer tyrosine phosphatome. Nat Rev Cancer. 2011;11:35–49. doi: 10.1038/nrc2980. [DOI] [PubMed] [Google Scholar]
- 5.Lahiry P, Torkamani A, Schork NJ, Hegele RA. Kinase mutations in human disease: interpreting genotype-phenotype relationships. Nat Rev Genet. 2010;11:60–74. doi: 10.1038/nrg2707. [DOI] [PubMed] [Google Scholar]
- 6.Andersen JN, Mortensen OH, Peters GH, Drake PG, Iversen LF, Olsen OH, Jansen PG, Andersen HS, Tonks NK, Moller NP. Structural and evolutionary relationships among protein tyrosine phosphatase domains. Mol Cell Biol. 2001;21 :7117–36. doi: 10.1128/MCB.21.21.7117-7136.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang ZY. Mechanistic studies on protein tyrosine phosphatases. Prog Nucleic Acid Res Mol Biol. 2003;73:171–220. doi: 10.1016/s0079-6603(03)01006-7. [DOI] [PubMed] [Google Scholar]
- 8.Denu JM, Dixon JE. Protein tyrosine phosphatases: mechanisms of catalysis and regulation. Current opinion in chemical biology. 1998;2:633–41. doi: 10.1016/s1367-5931(98)80095-1. [DOI] [PubMed] [Google Scholar]
- 9.Salmeen A, Barford D. Functions and mechanisms of redox regulation of cysteine-based phosphatases. Antioxidants & redox signaling. 2005;7:560–77. doi: 10.1089/ars.2005.7.560. [DOI] [PubMed] [Google Scholar]
- 10.Tanner JJ, Parsons ZD, Cummings AH, Zhou H, Gates KS. Redox regulation of protein tyrosine phosphatases: structural and chemical aspects. Antioxidants & redox signaling. 2010;15:77–97. doi: 10.1089/ars.2010.3611. [DOI] [PubMed] [Google Scholar]
- 11.den Hertog J, Groen A, van der Wijk T. Redox regulation of protein-tyrosine phosphatases. Arch Biochem Biophys. 2005;434:11–5. doi: 10.1016/j.abb.2004.05.024. [DOI] [PubMed] [Google Scholar]
- 12.Rhee SG, Bae YS, Lee SR, Kwon J. Hydrogen peroxide: a key messenger that modulates protein phosphorylation through cysteine oxidation. Sci STKE. 2000;2000:pe1. doi: 10.1126/stke.2000.53.pe1. [DOI] [PubMed] [Google Scholar]
- 13.Rhee SG, Kang SW, Jeong W, Chang TS, Yang KS, Woo HA. Intracellular messenger function of hydrogen peroxide and its regulation by peroxiredoxins. Curr Opin Cell Biol. 2005;17:183–9. doi: 10.1016/j.ceb.2005.02.004. [DOI] [PubMed] [Google Scholar]
- 14.Tonks NK. Redox redux: revisiting PTPs and the control of cell signaling. Cell. 2005;121:667–70. doi: 10.1016/j.cell.2005.05.016. [DOI] [PubMed] [Google Scholar]
- 15.Lambeth JD. NOX enzymes and the biology of reactive oxygen. Nature reviews Immunology. 2004;4:181–9. doi: 10.1038/nri1312. [DOI] [PubMed] [Google Scholar]
- 16.Sundaresan M, Yu ZX, Ferrans VJ, Irani K, Finkel T. Requirement for generation of H2O2 for platelet-derived growth factor signal transduction. Science. 1995;270:296–9. doi: 10.1126/science.270.5234.296. [DOI] [PubMed] [Google Scholar]
- 17.Finkel T. Oxygen radicals and signaling. Curr Opin Cell Biol. 1998;10:248–53. doi: 10.1016/s0955-0674(98)80147-6. [DOI] [PubMed] [Google Scholar]
- 18.Salmeen A, Andersen JN, Myers MP, Meng TC, Hinks JA, Tonks NK, Barford D. Redox regulation of protein tyrosine phosphatase 1B involves a sulphenyl-amide intermediate. Nature. 2003;423:769–73. doi: 10.1038/nature01680. [DOI] [PubMed] [Google Scholar]
- 19.van Montfort RL, Congreve M, Tisi D, Carr R, Jhoti H. Oxidation state of the active-site cysteine in protein tyrosine phosphatase 1B. Nature. 2003;423:773–7. doi: 10.1038/nature01681. [DOI] [PubMed] [Google Scholar]
- 20.Yang J, Groen A, Lemeer S, Jans A, Slijper M, Roe SM, den Hertog J, Barford D. Reversible oxidation of the membrane distal domain of receptor PTPalpha is mediated by a cyclic sulfenamide. Biochemistry. 2007;46:709–19. doi: 10.1021/bi061546m. [DOI] [PubMed] [Google Scholar]
- 21.Chen CY, Willard D, Rudolph J. Redox regulation of SH2-domain-containing protein tyrosine phosphatases by two backdoor cysteines. Biochemistry. 2009;48:1399–409. doi: 10.1021/bi801973z. [DOI] [PubMed] [Google Scholar]
- 22.van der Wijk T, Overvoorde J, den Hertog J. H2O2-induced intermolecular disulfide bond formation between receptor protein-tyrosine phosphatases. The Journal of biological chemistry. 2004;279:44355–61. doi: 10.1074/jbc.M407483200. [DOI] [PubMed] [Google Scholar]
- 23.Lee SR, Kwon KS, Kim SR, Rhee SG. Reversible inactivation of protein-tyrosine phosphatase 1B in A431 cells stimulated with epidermal growth factor. The Journal of biological chemistry. 1998;273:15366–72. doi: 10.1074/jbc.273.25.15366. [DOI] [PubMed] [Google Scholar]
- 24.Meng TC, Fukada T, Tonks NK. Reversible oxidation and inactivation of protein tyrosine phosphatases in vivo. Molecular cell. 2002;9:387–99. doi: 10.1016/s1097-2765(02)00445-8. [DOI] [PubMed] [Google Scholar]
- 25.Meng TC, Buckley DA, Galic S, Tiganis T, Tonks NK. Regulation of insulin signaling through reversible oxidation of the protein-tyrosine phosphatases TC45 and PTP1B. The Journal of biological chemistry. 2004;279:37716–25. doi: 10.1074/jbc.M404606200. [DOI] [PubMed] [Google Scholar]
- 26.Capasso M, Bhamrah MK, Henley T, Boyd RS, Langlais C, Cain K, Dinsdale D, Pulford K, Khan M, Musset B, Cherny VV, Morgan D, Gascoyne RD, Vigorito E, DeCoursey TE, MacLennan IC, Dyer MJ. HVCN1 modulates BCR signal strength via regulation of BCR-dependent generation of reactive oxygen species. Nature immunology. 2010;11:265–72. doi: 10.1038/ni.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Rolli V, Gallwitz M, Wossning T, Flemming A, Schamel WW, Zurn C, Reth M. Amplification of B cell antigen receptor signaling by a Syk/ITAM positive feedback loop. Molecular cell. 2002;10:1057–69. doi: 10.1016/s1097-2765(02)00739-6. [DOI] [PubMed] [Google Scholar]
- 28.Reth M. Hydrogen peroxide as second messenger in lymphocyte activation. Nature immunology. 2002;3:1129–34. doi: 10.1038/ni1202-1129. [DOI] [PubMed] [Google Scholar]
- 29.Cairns RA, Harris IS, Mak TW. Regulation of cancer cell metabolism. Nat Rev Cancer. 2011;11:85–95. doi: 10.1038/nrc2981. [DOI] [PubMed] [Google Scholar]
- 30.Liou GY, Storz P. Reactive oxygen species in cancer. Free Radic Res. 2010;44 :479–96. doi: 10.3109/10715761003667554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lou YW, Chen YY, Hsu SF, Chen RK, Lee CL, Khoo KH, Tonks NK, Meng TC. Redox regulation of the protein tyrosine phosphatase PTP1B in cancer cells. Febs J. 2008;275:69–88. doi: 10.1111/j.1742-4658.2007.06173.x. [DOI] [PubMed] [Google Scholar]
- 32.Sattler M, Verma S, Shrikhande G, Byrne CH, Pride YB, Winkler T, Greenfield EA, Salgia R, Griffin JD. The BCR/ABL tyrosine kinase induces production of reactive oxygen species in hematopoietic cells. The Journal of biological chemistry. 2000;275:24273–8. doi: 10.1074/jbc.M002094200. [DOI] [PubMed] [Google Scholar]
- 33.Gianni D, Bohl B, Courtneidge SA, Bokoch GM. The involvement of the tyrosine kinase c-Src in the regulation of reactive oxygen species generation mediated by NADPH oxidase-1. Mol Biol Cell. 2008;19:2984–94. doi: 10.1091/mbc.E08-02-0138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ostman A, Hellberg C, Bohmer FD. Protein-tyrosine phosphatases and cancer. Nat Rev Cancer. 2006;6:307–20. doi: 10.1038/nrc1837. [DOI] [PubMed] [Google Scholar]
- 35.Wu RF, Terada LS. Oxidative modification of protein tyrosine phosphatases. Sci STKE. 2006;2006:pl2. doi: 10.1126/stke.3322006pl2. [DOI] [PubMed] [Google Scholar]
- 36.Kwon J, Qu CK, Maeng JS, Falahati R, Lee C, Williams MS. Receptor-stimulated oxidation of SHP-2 promotes T-cell adhesion through SLP-76-ADAP. The EMBO journal. 2005;24:2331–41. doi: 10.1038/sj.emboj.7600706. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Weerapana E, Wang C, Simon GM, Richter F, Khare S, Dillon MB, Bachovchin DA, Mowen K, Baker D, Cravatt BF. Quantitative reactivity profiling predicts functional cysteines in proteomes. Nature. 2010;468:790–5. doi: 10.1038/nature09472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Meng TC, Hsu SF, Tonks NK. Development of a modified in-gel assay to identify protein tyrosine phosphatases that are oxidized and inactivated in vivo. Methods (San Diego, Calif. 2005;35:28–36. doi: 10.1016/j.ymeth.2004.07.005. [DOI] [PubMed] [Google Scholar]
- 39.Burridge K, Nelson A. An in-gel assay for protein tyrosine phosphatase activity: detection of widespread distribution in cells and tissues. Anal Biochem. 1995;232:56–64. doi: 10.1006/abio.1995.9961. [DOI] [PubMed] [Google Scholar]
- 40.Boivin B, Zhang S, Arbiser JL, Zhang ZY, Tonks NK. A modified cysteinyl-labeling assay reveals reversible oxidation of protein tyrosine phosphatases in angiomyolipoma cells. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:9959–64. doi: 10.1073/pnas.0804336105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu HH, Momand J. Pyrrolidine dithiocarbamate prevents p53 activation and promotes p53 cysteine residue oxidation. The Journal of biological chemistry. 1998;273:18898–905. doi: 10.1074/jbc.273.30.18898. [DOI] [PubMed] [Google Scholar]
- 42.Li S, Whorton AR. Regulation of protein tyrosine phosphatase 1B in intact cells by S-nitrosothiols. Arch Biochem Biophys. 2003;410:269–79. doi: 10.1016/s0003-9861(02)00696-3. [DOI] [PubMed] [Google Scholar]
- 43.Boivin B, Yang M, Tonks NK. Targeting the reversibly oxidized protein tyrosine phosphatase superfamily. Sci Signal. 2010;3:pl2. doi: 10.1126/scisignal.3137pl2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Boivin B, Tonks NK. Analysis of the redox regulation of protein tyrosine phosphatase superfamily members utilizing a cysteinyl-labeling assay. Methods in enzymology. 2010;474:35–50. doi: 10.1016/S0076-6879(10)74003-9. [DOI] [PubMed] [Google Scholar]
- 45.Held JM, Danielson SR, Behring JB, Atsriku C, Britton DJ, Puckett RL, Schilling B, Campisi J, Benz CC, Gibson BW. Targeted quantitation of site-specific cysteine oxidation in endogenous proteins using a differential alkylation and multiple reaction monitoring mass spectrometry approach. Mol Cell Proteomics. 2010;9:1400–10. doi: 10.1074/mcp.M900643-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Elschenbroich S, Kislinger T. Targeted proteomics by selected reaction monitoring mass spectrometry: applications to systems biology and biomarker discovery. Molecular bioSystems. 2010;7:292–303. doi: 10.1039/c0mb00159g. [DOI] [PubMed] [Google Scholar]
- 47.Picotti P, Bodenmiller B, Mueller LN, Domon B, Aebersold R. Full dynamic range proteome analysis of S. cerevisiae by targeted proteomics. Cell. 2009;138:795–806. doi: 10.1016/j.cell.2009.05.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cravatt BF, Wright AT, Kozarich JW. Activity-based protein profiling: from enzyme chemistry to proteomic chemistry. Annual review of biochemistry. 2008;77:383–414. doi: 10.1146/annurev.biochem.75.101304.124125. [DOI] [PubMed] [Google Scholar]
- 49.Jessani N, Cravatt BF. The development and application of methods for activity-based protein profiling. Current opinion in chemical biology. 2004;8:54–9. doi: 10.1016/j.cbpa.2003.11.004. [DOI] [PubMed] [Google Scholar]
- 50.Kato D, Boatright KM, Berger AB, Nazif T, Blum G, Ryan C, Chehade KA, Salvesen GS, Bogyo M. Activity-based probes that target diverse cysteine protease families. Nature chemical biology. 2005;1:33–8. doi: 10.1038/nchembio707. [DOI] [PubMed] [Google Scholar]
- 51.Yuan F, Verhelst SH, Blum G, Coussens LM, Bogyo M. A selective activity-based probe for the papain family cysteine protease dipeptidyl peptidase I/cathepsin C. Journal of the American Chemical Society. 2006;128:5616–7. doi: 10.1021/ja060835v. [DOI] [PubMed] [Google Scholar]
- 52.Saghatelian A, Jessani N, Joseph A, Humphrey M, Cravatt BF. Activity-based probes for the proteomic profiling of metalloproteases. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:10000–5. doi: 10.1073/pnas.0402784101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Liu Y, Patricelli MP, Cravatt BF. Activity-based protein profiling: the serine hydrolases. Proceedings of the National Academy of Sciences of the United States of America. 1999;96:14694–9. doi: 10.1073/pnas.96.26.14694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Kumar S, Zhou B, Liang F, Wang WQ, Huang Z, Zhang ZY. Activity-based probes for protein tyrosine phosphatases. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:7943–8. doi: 10.1073/pnas.0402323101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Liu S, Zhou B, Yang H, He Y, Jiang ZX, Kumar S, Wu L, Zhang ZY. Aryl vinyl sulfonates and sulfones as active site-directed and mechanism-based probes for protein tyrosine phosphatases. Journal of the American Chemical Society. 2008;130:8251–60. doi: 10.1021/ja711125p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang ZY. Functional studies of protein tyrosine phosphatases with chemical approaches. Biochimica et biophysica acta. 2005;1754:100–7. doi: 10.1016/j.bbapap.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 57.Kolb HC, Finn MG, Sharpless KB. Click Chemistry: Diverse Chemical Function from a Few Good Reactions. Angewandte Chemie (International ed) 2001;40 :2004–2021. doi: 10.1002/1521-3773(20010601)40:11<2004::AID-ANIE2004>3.0.CO;2-5. [DOI] [PubMed] [Google Scholar]
- 58.Sletten EM, Bertozzi CR. Bioorthogonal chemistry: fishing for selectivity in a sea of functionality. Angewandte Chemie (International ed) 2009;48:6974–98. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Meldal M, Tornoe CW. Cu-catalyzed azide-alkyne cycloaddition. Chemical reviews. 2008;108:2952–3015. doi: 10.1021/cr0783479. [DOI] [PubMed] [Google Scholar]
- 60.Rodionov VO, Presolski SI, Diaz DD, Fokin VV, Finn MG. Ligand-accelerated Cu-catalyzed azide-alkyne cycloaddition: a mechanistic report. Journal of the American Chemical Society. 2007;129:12705–12. doi: 10.1021/ja072679d. [DOI] [PubMed] [Google Scholar]
- 61.Kiick KL, Saxon E, Tirrell DA, Bertozzi CR. Incorporation of azides into recombinant proteins for chemoselective modification by the Staudinger ligation. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:19–24. doi: 10.1073/pnas.012583299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Persson C, Sjoblom T, Groen A, Kappert K, Engstrom U, Hellman U, Heldin CH, den Hertog J, Ostman A. Preferential oxidation of the second phosphatase domain of receptor-like PTP-alpha revealed by an antibody against oxidized protein tyrosine phosphatases. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:1886–91. doi: 10.1073/pnas.0304403101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Persson C, Kappert K, Engstrom U, Ostman A, Sjoblom T. An antibody-based method for monitoring in vivo oxidation of protein tyrosine phosphatases. Methods (San Diego, Calif. 2005;35:37–43. doi: 10.1016/j.ymeth.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 64.Groen A, Lemeer S, van der Wijk T, Overvoorde J, Heck AJ, Ostman A, Barford D, Slijper M, den Hertog J. Differential oxidation of protein-tyrosine phosphatases. The Journal of biological chemistry. 2005;280:10298–304. doi: 10.1074/jbc.M412424200. [DOI] [PubMed] [Google Scholar]
- 65.Weibrecht I, Bohmer SA, Dagnell M, Kappert K, Ostman A, Bohmer FD. Oxidation sensitivity of the catalytic cysteine of the protein-tyrosine phosphatases SHP-1 and SHP-2. Free radical biology & medicine. 2007;43:100–10. doi: 10.1016/j.freeradbiomed.2007.03.021. [DOI] [PubMed] [Google Scholar]
- 66.Karisch R, Fernandez M, Taylor P, Virtanen C, St-Germain JR, Jin LL, Harris IS, Mori J, Mak TW, Senis YA, Ostman A, Moran MF, Neel BG. Global proteomic assessment of the classical protein-tyrosine phosphatome and "Redoxome". Cell. 2011;146:826–40. doi: 10.1016/j.cell.2011.07.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Morihara K. Chemical nature of the active site of papain. I. Inhibition by carbonyl reagents with active methylene groups. Journal of biochemistry. 1967;62:250–62. doi: 10.1093/oxfordjournals.jbchem.a128655. [DOI] [PubMed] [Google Scholar]
- 68.Benitez LV, Allison WS. The inactivation of the acyl phosphatase activity catalyzed by the sulfenic acid form of glyceraldehyde 3-phosphate dehydrogenase by dimedone and olefins. The Journal of biological chemistry. 1974;249:6234–43. [PubMed] [Google Scholar]
- 69.Poole LB, Zeng BB, Knaggs SA, Yakubu M, King SB. Synthesis of chemical probes to map sulfenic acid modifications on proteins. Bioconjugate chemistry. 2005;16:1624–8. doi: 10.1021/bc050257s. [DOI] [PubMed] [Google Scholar]
- 70.Poole LB, Klomsiri C, Knaggs SA, Furdui CM, Nelson KJ, Thomas MJ, Fetrow JS, Daniel LW, King SB. Fluorescent and affinity-based tools to detect cysteine sulfenic acid formation in proteins. Bioconjugate chemistry. 2007;18:2004–17. doi: 10.1021/bc700257a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Poole LB, Nelson KJ. Discovering mechanisms of signaling-mediated cysteine oxidation. Current opinion in chemical biology. 2008;12:18–24. doi: 10.1016/j.cbpa.2008.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Nelson KJ, Klomsiri C, Codreanu SG, Soito L, Liebler DC, Rogers LC, Daniel LW, Poole LB. Use of dimedone-based chemical probes for sulfenic acid detection methods to visualize and identify labeled proteins. Methods in enzymology. 2010;473:95–115. doi: 10.1016/S0076-6879(10)73004-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wani R, Qian J, Yin L, Bechtold E, King SB, Poole LB, Paek E, Tsang AW, Furdui CM. Isoform-specific regulation of Akt by PDGF-induced reactive oxygen species. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:10550–5. doi: 10.1073/pnas.1011665108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Reddie KG, Seo YH, Muse WB, III, Leonard SE, Carroll KS. A chemical approach for detecting sulfenic acid-modified proteins in living cells. Molecular bioSystems. 2008;4:521–31. doi: 10.1039/b719986d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ellis HR, Poole LB. Novel application of 7-chloro-4-nitrobenzo-2-oxa-1,3-diazole to identify cysteine sulfenic acid in the AhpC component of alkyl hydroperoxide reductase. Biochemistry. 1997;36:15013–8. doi: 10.1021/bi972191x. [DOI] [PubMed] [Google Scholar]
- 76.Poole LB, Ellis HR. Identification of cysteine sulfenic acid in AhpC of alkyl hydroperoxide reductase. Methods in enzymology. 2002;348:122–36. doi: 10.1016/s0076-6879(02)48632-6. [DOI] [PubMed] [Google Scholar]
- 77.Michalek RD, Nelson KJ, Holbrook BC, Yi JS, Stridiron D, Daniel LW, Fetrow JS, King SB, Poole LB, Grayson JM. The requirement of reversible cysteine sulfenic acid formation for T cell activation and function. J Immunol. 2007;179:6456–67. doi: 10.4049/jimmunol.179.10.6456. [DOI] [PubMed] [Google Scholar]
- 78.Oshikawa J, Urao N, Kim HW, Kaplan N, Razvi M, McKinney R, Poole LB, Fukai T, Ushio-Fukai M. Extracellular SOD-derived H2O2 promotes VEGF signaling in caveolae/lipid rafts and post-ischemic angiogenesis in mice. PloS one. 2010;5:e10189. doi: 10.1371/journal.pone.0010189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Paulsen CE, Truong TH, Garcia FJ, Homann A, Gupta V, Leonard SE, Carroll KS. Peroxide-dependent sulfenylation of the EGFR catalytic site enhances kinase activity. Nature chemical biology. 2012;8:57–64. doi: 10.1038/nchembio.736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Charles RL, Schroder E, May G, Free P, Gaffney PR, Wait R, Begum S, Heads RJ, Eaton P. Protein sulfenation as a redox sensor: proteomics studies using a novel biotinylated dimedone analogue. Mol Cell Proteomics. 2007;6:1473–84. doi: 10.1074/mcp.M700065-MCP200. [DOI] [PubMed] [Google Scholar]
- 81.Leonard SE, Reddie KG, Carroll KS. Mining the thiol proteome for sulfenic acid modifications reveals new targets for oxidation in cells. ACS chemical biology. 2009;4:783–99. doi: 10.1021/cb900105q. [DOI] [PubMed] [Google Scholar]
- 82.Leonard SE, Garcia FJ, Goodsell DS, Carroll KS. Redox-based probes for protein tyrosine phosphatases. Angewandte Chemie (International ed) 2011;50:4423–7. doi: 10.1002/anie.201007871. [DOI] [PubMed] [Google Scholar]
- 83.Seo YH, Carroll KS. Profiling protein thiol oxidation in tumor cells using sulfenic acid-specific antibodies. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:16163–8. doi: 10.1073/pnas.0903015106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Maller C, Schroder E, Eaton P. Glyceraldehyde 3-phosphate dehydrogenase is unlikely to mediate hydrogen peroxide signaling: studies with a novel anti-dimedone sulfenic acid antibody. Antioxidants & redox signaling. 2011;14:49–60. doi: 10.1089/ars.2010.3149. [DOI] [PubMed] [Google Scholar]
- 85.Seo YH, Carroll KS. Quantification of Protein Sulfenic Acid Modifications Using Isotope-Coded Dimedone and Iododimedone. Angewandte Chemie (International ed) 2011 doi: 10.1002/anie.201007175. [DOI] [PubMed] [Google Scholar]
- 86.Haque A, Andersen JN, Salmeen A, Barford D, Tonks NK. Conformation-Sensing Antibodies Stabilize the Oxidized Form of PTP1B and Inhibit Its Phosphatase Activity. Cell. 2011;147:185–98. doi: 10.1016/j.cell.2011.08.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Sarma BK, Mugesh G. Redox regulation of protein tyrosine phosphatase 1B (PTP1B): a biomimetic study on the unexpected formation of a sulfenyl amide intermediate. Journal of the American Chemical Society. 2007;129:8872–81. doi: 10.1021/ja070410o. [DOI] [PubMed] [Google Scholar]