Abstract
Both anti- and proapoptotic activities have been reported to occur during chlamydial infection. To reconcile the apparent controversy, we compared host cell apoptotic responses to infection with 17 different chlamydial serovars and strains. None of the serovars caused any biologically significant apoptosis in the infected host cells. Host cells in chlamydia-infected cultures can continue to undergo DNA synthesis and mitosis. Chlamydia-infected cells are resistant to apoptosis induction, although the extent of the antiapoptotic ability varied between serovars. These observations have demonstrated that an anti- but not proapoptotic activity is the prevailing event in chlamydia-infected cultures.
Chlamydiae are obligate intracellular bacterial pathogens consisting of three major species with each causing unique health problems for humans (6, 8, 9, 11, 32). The Chlamydia trachomatis species has more than 15 different serovars, including serovars A to K, serovars L1 to L3, and a murine strain known as the mouse pneumonitis agent, which has recently been designated chlamydia muridarum. In general, serovars A to C cause ocular infection that can lead to blinding trachoma in developing countries (48), while serovars D to K are the leading cause of sexually transmitted bacterial diseases in the United States and other developed nations (44). In some cases, the lymphogranuloma venereum (LGV) serovars (L1, L2, and L3) can cause infections leading to LGV (2). The species Chlamydia pneumoniae causes human respiratory infection that has recently been linked to atherosclerosis (9), while the species Chlamydia psittaci is primarily an animal pathogen, although humans can acquire infection from C. psittaci-infected animals (11). Many different C. psittaci strains have been identified, including the strains GPIC (25) and 6BC (14).
Despite the diversity in tissue tropism and pathogenic phenotypes, all chlamydial serovars and strains must replicate in the cytoplasmic vacuoles of eukaryotic cells (21). A typical chlamydial infection starts with the entry of an infectious elementary body (EB) into eukaryotic cells via endocytosis. The internalized EB then develops into a noninfectious but metabolically active reticular body for replication. The progeny reticular bodies finally differentiate back into EBs for exiting the infected cell and infecting new target cells. In an in vitro cell culture system, the entire replication process, occurring within a modified cytoplasmic vacuole (also called an inclusion), can take one to several days to complete, depending on the serovars or strains. In general, at an infection load of 50%, most organisms are differentiated back to EBs by ∼30 h after infection for chlamydia muridarum, GPIC, and 6BC; by ∼40 h for L1 to L3; by 60 to 70 h for A to K; and by 3 to 5 days for C. pneumoniae strains. However, the timeline for organism differentiation during natural infection is still unknown, although it is thought that chlamydiae can persist in the infected host for long periods of time, which may be a major contributor to the pathogenesis of chlamydial diseases in humans (4, 28).
The fact that chlamydiae have been able to survive and replicate intracellularly so successfully suggests that chlamydiae have evolved the ability to manipulate host cells. It is known that chlamydia-laden vacuoles can actively avoid fusion with lysosomes, although the molecular mechanism involved remains to be elucidated (21, 42). We have previously shown that chlamydiae possess various molecular means for protecting the infected cells from being destroyed by host defense mechanisms, including suppression of host cell major histocompatibility complex antigen expression (52-54) and blockade of host cell apoptosis pathways (16). However, due to the complex interactions between chlamydiae and host cells, different labs have reported varied outcomes in regard to chlamydial effects on the host apoptosis program. Since our initial description of chlamydial interactions with host cell apoptosis (16), there have been more than 30 publications on the subject. Both anti- and proapoptotic activities have been described for different serovars and strains and at different times after infection (12, 13, 16, 19, 34, 35, 37). To reconcile the apparent conflicting observations, we compared 17 different chlamydial serovars and strains from both the C. trachomatis and C. psittaci species for their effects on host cell apoptosis. Since an antiapoptotic activity has been consistently assigned to the C. pneumoniae species under various culture conditions (1, 18, 37), this species was not included in the comparison. When the 17 serovars and strains from both the C. trachomatis and C. psittaci species were analyzed, we found that although statistically significant apoptosis events were observed in cultures infected with some serovars and strains, especially at the late stages of infection, none caused biologically significant apoptosis, since <15% of cells were apoptotic even in the samples with the highest apoptosis rate. In fact, host cells in chlamydia-infected culture can continue to undergo active DNA synthesis and form mitotic spindles. More importantly, all chlamydial serovars exhibited a clear antiapoptotic activity throughout the experiments, although the extent of the antiapoptotic ability varied between serovars. These observations have extended our previous finding and demonstrated that an anti- and not a proapoptotic activity is the biologically significant event in cultures infected with chlamydiae.
MATERIALS AND METHODS
Chlamydial organisms and infection.
The chlamydial serovars and strains used for the present study include A, B, C, D, E, F, G, H, I, K, L1, L3, and Ba (obtained from Harlan Caldwell at the Rocky Mountain Laboratory, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Hamilton, Mont.); 6BC (kindly provided by Thomas Hatch, University of Tennessee, Memphis) (14); chlamydia muridarum (kindly provided by Louis De La Maza, University of California, Irvine); and L2 and GPIC (our stocks). These organisms were grown, purified, and titrated as previously described (55). Aliquots of the organisms were stored at −80°C until use. HeLa cells (American Type Culture Collection, Manassas, Va.) and MS74 cells (endometrial epithelial cells; kindly provided by John Alderete, University of Texas Health Science Center at San Antonio) were maintained in Dulbecco's modified Eagle's medium (GIBCO BRL, Rockville, Md.) with 10% fetal calf serum (GIBCO BRL), while human umbilical vein endothelial cells (HUVEC) (Cambrex Bio Science Rockland, Inc., East Rutherford, N.J.) were maintained in complete EGM medium (Cambrex). For infection, cells were grown on glass coverslips in 24-well plates overnight prior to chlamydial inoculation. Chlamydial organisms diluted in DMEM with 10% fetal calf serum were directly inoculated onto the cell monolayers. The infection dose was pretitrated for individual serovars, and an infection rate of ∼50% was used for all serovars in all infection experiments. The corresponding multiplicities of infection (MOIs) were ∼0.5 for chlamydia muridarum, GPIC, and 6BC; 1 for LGV serovars, D, E, I, and K; and 2 for A, B, C, F, and G in HeLa cells. MOIs of 1 for L2 in MS74 cells and 5 in HUVEC were used. HUVEC were least susceptible to chlamydial infection. The variations in MOIs between chlamydial strains for achieving the same infection rate in different cells were largely caused by the differences in chlamydial growth dependence on cycloheximide. Our stocks were titrated in the presence of cycloheximide in HeLa cells, and no cycloheximide was used in the infection experiments. Our intention was to avoid high infection dose-induced toxicity and to also allow sufficient numbers of both infected and uninfected cells to be counted from the same cultures. The cell samples were cultured at 37°C in a CO2 incubator and processed at various time points after infection, as indicated for individual experiments, for microscopic observations as described below. For apoptosis induction, a proapoptotic stimulus, staurosporine (Sigma, St. Louis, Mo.), was added to the cultures at a final concentration of ∼1 μg/ml and left for 4 h. For bromodeoxyuridine (BrDu) incorporation experiments, BrDu (catalog no. 550891; PharMingen, San Diego, Calif.) was added to cultures at a final concentration of 1 μg/ml and left for 4 h prior to the termination of the culture experiments.
Immunofluorescence staining.
Cells grown on coverslips were fixed with 2% paraformaldehyde dissolved in phosphate-buffered saline for 30 min at room temperature, followed by permeabilization with 1% saponin for an additional 30 min. To avoid detaching the apoptotic cells, all solutions were prewarmed to room temperature and added to each well gently. After washing and blocking, the cell samples were subjected to various combinations of antibody and chemical labeling. For visualizing apoptotic cells, three different combinations of triple labeling were used: Hoechst stain (blue for binding to DNA) (Sigma) plus anti-chlamydial lipopolysaccharide (LPS) (clone M5B9, murine immunoglobulin G3 [mIgG3]) (unpublished data) plus anti-cytochrome c (6H2.B4, mIgG1) (PharMingen), Hoechst stain plus anti-chlamydial LPS plus terminal deoxynucleotidyltransferase-mediated dUTP-biotin nick end labeling (TUNEL) stain (labeling free DNA ends with fluorescein isothiocyanate-dUTP as instructed by the manufacturer) (Promega, Madison, Wis.), or Hoechst stain plus anti-cytochrome c plus TUNEL stain. The bound primary antibodies were visualized with either Cy3 (red)- or Cy2 (green)-conjugated goat anti-mouse IgG1 or IgG3 (Molecular Probes, Eugene, Oreg.) so that each of the triple stainings was in a different color. For detecting spindle microtubules, the cells were similarly processed and stained with Hoechst stain plus anti-chlamydial LPS plus anti-acetylated tubulin (611b1, mIgG2b) (Sigma). The primary antibodies were visualized as described above except that the goat anti-mouse IgG3 was replaced by a goat anti-mouse IgG2b. For visualizing BrDu incorporation, the cell samples were similarly processed except that a base denaturation step (HCl at a final concentration of 1 N for 1 h at 37°C) was used to break double-stranded DNA. The cell samples were then stained with Hoechst stain plus an antichlamydia rabbit antiserum (R1L2, raised with purified EBs of C. trachomatis serovar L2) (unpublished data) plus anti-BrDu (BU33, mIgG1) (Sigma). The primary antibody labeling was visualized with a goat anti-rabbit IgG conjugated with Cy2 and a goat anti-mouse IgG conjugated with Cy3 (Caltag, Burlingame, Calif.).
Fluorescence microscopy.
The cell samples after the appropriate immunolabeling were used for both image acquisition and cell counting with an AX-70 fluorescence microscope equipped with multiple filter sets (Olympus, San Antonio, Tex.). For acquiring images, the multicolor-labeled samples were exposed under a given filter set at a time, and the single-color images were acquired with a Hamamatsu digital camera. The single-color images were then superimposed with the software SimplePCI to display multicolors. For counting cells with particular staining phenotypes, five random views per coverslip were counted under the appropriate objective lenses. In each experiment, the number of cells or nuclei per view was calculated from 10 random views of duplicate coverslips.
Statistical analysis.
A two-tailed Student t test from http://faculty.vassar.edu/lowry/tu.html or in the software SigmaPlot was used for statistical analysis of data.
RESULTS
Chlamydial infection induces a low level of apoptosis, and most apoptosis is detected in uninfected cells.
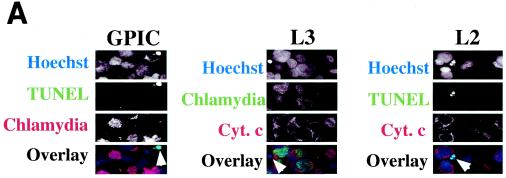
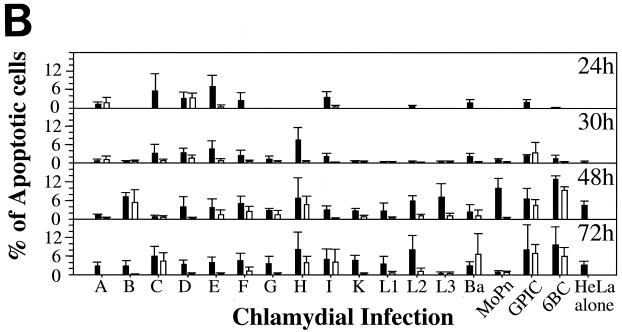
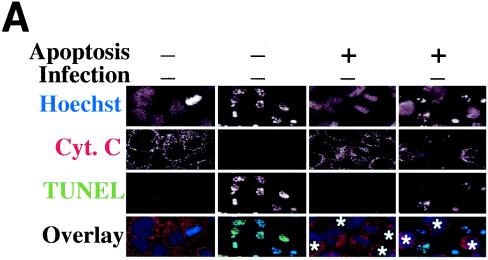
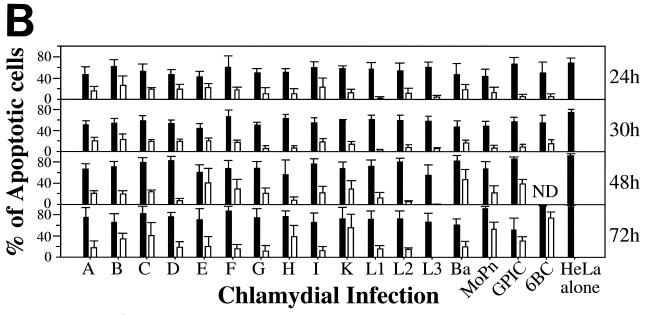
Since DNA fragmentation or nuclear condensation and mitochondrial cytochrome c release into cytosol are two major hallmarks for most apoptosis events, we used these apoptotic features to evaluate the effects of chlamydial infection on host cell apoptosis (Fig. 1). A comparison of three triple-labeling combinations revealed that simultaneous visualization of DNA together with chlamydial inclusion and cytochrome c or DNA fragmentation allows us to accurately monitor the apoptosis events in cultures infected with different strains of chlamydiae. Shown in Fig. 1A are representative examples of three different combinations of labeling for cells infected with GPIC, L3, and L2 for 30 h. Hoechst staining clearly revealed both host cell nuclei and chlamydial inclusions. Furthermore, the condensed host cell nuclei exhibit extremely intense Hoechst staining, which has been conventionally used as an indicator of nuclear apoptosis (16). Indeed, TUNEL staining revealed DNA fragmentation in the condensed nuclei, and anti-cytochrome c staining showed mitochondrial cytochrome c release into cytosol of the cells with condensed nuclei. Based on these staining patterns, we quantitated the number of apoptotic cells in cultures with or without infection with 17 different chlamydial serovars at 24, 30, 48, and 72 h after infection (Fig. 1B). To differentiate the effect of chlamydial infection on the infected cells (bearing chlamydial inclusions) from that on uninfected cells (bearing no inclusions) in the same infected cultures, apoptotic events in these two cell populations were quantitated separately. At 24 and 30 h after infection, a level of <5% apoptotic cells was detected in either the infected or uninfected cell populations from most culture samples, with the exception of the uninfected cells of samples from cultures infected with serovars C (5.5% apoptosis at 24 h), E (8% at 24 h and 5% at 30 h), and H (8% at 30 h). By 48 and 72 h, the overall apoptotic events increased in all cultures, including the noninfection control culture with ∼5% (48 h) or ∼4% (72 h) of apoptosis. However, most of the infected cultures maintained <10% apoptotic cells, with the exception that the chlamydia muridarum-infected culture reached ∼10% apoptosis at 48 h and 6BC-infected cells reached ∼14% at 48 h and ∼10% at 72 h. These apoptotic events occurred in cells bearing no inclusions, although these cells were in the infected cultures. In fact, the level of apoptosis was generally higher in cells without inclusions than in those with inclusions in the same infected cultures. For example, there were statistically significant differences in the percentage of apoptosis between cells with (1%) or without (10%) inclusions in chlamydia muridarum-infected cultures at 48 h (P < 0.01) and between cells with (2%) or without (8%) inclusions in L2-infected culture at 72 h (P < 0.05). However, the cultures infected with either GPIC (72 h after infection) or 6BC (48 and 72 h) displayed a high apoptosis rate in both infected and uninfected cell populations. Nevertheless, the overall level of apoptosis even in cultures with the highest apoptosis rate was still relatively low (<15%), which suggests that the apoptotic events in chlamydia-infected cultures are not likely to be biologically significant.
FIG. 1.
Effects of chlamydial infection on host cell apoptosis. HeLa cells with or without infection with 17 different chlamydial serovars were processed at various times after infection and subjected to triple labeling by using an immunofluorescence staining assay as described in Materials and Methods. (A) Representative images of three different combinations of triple labeling, using cells infected with chlamydial GPIC, L3, and L2 serovars for 30 h. DNA was labeled blue with Hoechst stain, fragmented DNA was labeled green by a TUNEL assay, and chlamydiae were labeled either red or green and cytochrome c (Cyt. c) was labeled either red or green by antibody-directed staining. The images were acquired a single color at a time (each in gray; first three rows) and assembled into tricolor via computer software-mediated overlay (bottom row). The single apoptotic cell in each column (from each culture) is indicated with an arrowhead in the overlay panels. Note that apoptotic cells can be easily identified with each of the three staining combinations. (B) Cells in the triply labeled cell samples were counted under a fluorescence microscope as described in Materials and Methods. Although all three combinations of triple staining described above were used, only one combination was used for a given experiment. Cells with or without chlamydial inclusion and with or without apoptotic nuclei were counted independently from the same view, and five random views were counted from each coverslip. The results (means ± standard deviations) are from three to eight experiments, each with duplicates and were compared for cells bearing inclusions (open bars) and those bearing no inclusions (solid bars) from the same infected cultures or the cells from the cultures of HeLa cells alone (last solid bar). MoPn, chlamydia muridarum.
Chlamydia-infected cells are able to undergo DNA synthesis and mitosis.
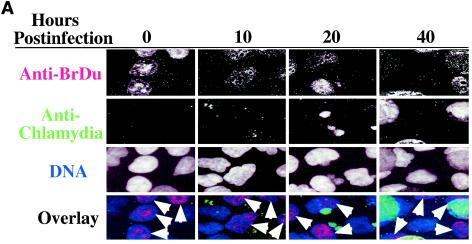
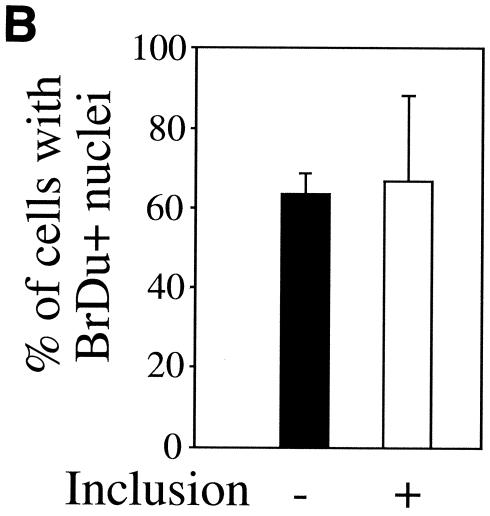
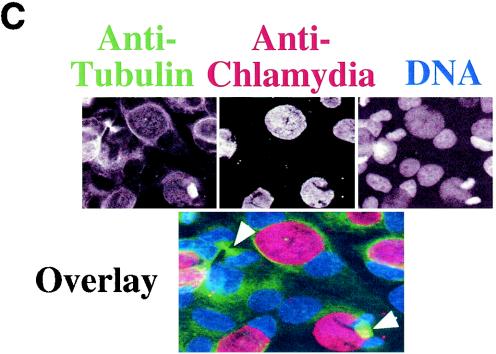
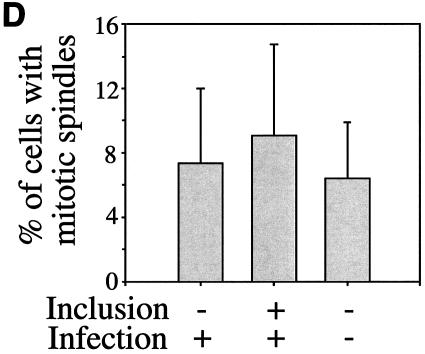
Since most cells in chlamydia-infected cultures are not apoptotic, we next evaluated whether the cells are at a quiescent stage or can undergo normal proliferation. By definition, resting cells not only fail to undergo apoptosis but also lack the ability to proliferate unless stimulated. We then measured the host DNA synthesis and mitotic spindle formation, both of which are indicative of cell proliferation, in chlamydia L2-infected cultures (Fig. 2). Active BrDu incorporation was detected in nuclei of cells regardless of L2 infection and infection stage, including 0, 10, 20, and 40 h after infection (Fig. 2A). The percentage of host cells positive for BrDu incorporation was similar for cells with (65%) or without (62%) L2 inclusions in the same infected cultures (P > 0.05) (Fig. 2B), suggesting that chlamydial infection did not affect the ability of either cell population to carry out DNA synthesis. We further compared the rate of mitotic spindle formation between various cell populations 40 h after infection with L2. Obvious spindle microtubules were detected in cells regardless of chlamydia L2 infection (Fig. 2C). There was no significant difference in the percentage of cells positive for mitotic spindles between the infected (9%) and uninfected (7.5%) cells in the same infected culture or the cells (7%) in the noninfection control culture (P > 0.05 for both comparisons) (Fig. 2D), demonstrating that chlamydial infection does not prevent the infected cells from entering mitosis. Due to the fact that BrDu incorporation is an irreversible process and thus accumulates, while mitotic spindle formation is a transient process, we have observed an obvious difference in the percentage of cells positive for DNA synthesis versus positive for mitotic spindles.
FIG. 2.
Effects of chlamydial infection on host cell DNA synthesis (A and B) and mitosis (C and D). HeLa cells with or without infection with chlamydial L2 serovar were processed for staining and counting. (A) The infected cell cultures were processed at 0, 10, 20, or 40 h after infection for staining. Nuclei incorporating BrDu were labeled red (indicated with arrowheads in the overlay panels), chlamydial inclusions were labeled green, and DNA was labeled blue. Note that host cell nuclei incorporated BrDu regardless of L2 infection, and BrDu can also be taken up by chlamydial organisms (last panel). (B) Samples stained 40 h after infection were subjected to counting as described in the legend to Fig. 1B. The percentages of cells with BrDu-positive nuclei (means ± standard deviations) were compared for cells with or without chlamydial inclusions in the same infected cultures (P > 0.05). The data are from five independent experiments. (C) HeLa cells with or without L2 infection for 40 h were labeled for acetylated tubulin (green), chlamydial inclusion (red), and DNA (blue). Mitotic spindles are indicated by arrowheads in the composite tricolor panel. (D) The samples stained as described for panel C were counted as described for Fig. 1B. The results (means ± standard deviations) are from five independent experiments and were compared between cells with chlamydial inclusions (middle bar) and without inclusions (left bar) (P > 0.05) from the same infected cultures or the cells from the noninfection cultures (right bar) (P > 0.05). Note that chlamydial infection had no significant effects on host cell DNA synthesis and mitosis.
Chlamydia-infected cells are resistant to apoptosis induction.
Normally, when mammalian cells are not arrested and are competent for mitosis, these cells should be able to undergo apoptosis in response to stress signals. The question is why most chlamydia-infected cells fail to proceed with apoptosis in response to chlamydial infection. We have previously demonstrated that chlamydia-infected cells are profoundly resistant to apoptosis induction due to the chlamydial ability to block host cell apoptosis pathways. Since these earlier studies were based on only a few serovars of C. trachomatis species, we decided to extend our previous studies by comparing the antiapoptotic activities among 17 different chlamydial serovars from both the C. trachomatis and C. psittaci species (Fig. 3). A triple staining for DNA, cytochrome c, and DNA fragmentation was used to reveal the apoptosis induced by staurosporine in cell cultures with or without chlamydial infection. Shown in Fig. 3A are images from an L2-infected culture. Clearly, the staurosporine-induced apoptosis can be accurately determined based on this triple staining. The apoptotic cells detected either in the noninfection control culture or in the L2-infected culture all showed striking nuclear condensation with obvious DNA fragmentation and clear mitochondrial cytochrome c release. It should be noted that the chlamydial infection rate was intentionally adjusted to ∼50% so that the apoptosis in cells with or without chlamydial inclusions from the same infected cultures can be conveniently quantitated and the rates of apoptosis in these two cell populations can be compared (Fig. 3B). Overall, all chlamydia-infected cells exhibited a significant antiapoptotic activity during the entire infection course. This was true when the rate of apoptosis was compared either between cells with or without chlamydial inclusions in the same infected cultures or between the inclusion-positive cells in the infected cultures and cells in the noninfection cultures. It should be emphasized that the comparison between two cell populations in the same cultures may lead to more reliable conclusions, since both cell populations compared are exposed to the exact same culture conditions. The level of antiapoptotic activity varied between different serovars and different times after infection. The lowest antiapoptotic activity was detected in cells infected with 6BC, followed by GPIC, K, and chlamydia muridarum at 72 h postinfection. It is apparent that a higher apoptosis rate was always detected at the late stages of infection, suggesting that older cells are more susceptible to apoptosis induction with staurosporine. Nevertheless, chlamydia-infected cells still maintained a significantly high level of antiapoptotic activity even at the late stages of infection.
FIG. 3.
Chlamydia-infected cells are resistant to apoptosis induction. HeLa cells were infected and processed way as described in the Fig. 1 legend except that the cultures were induced to undergo apoptosis with staurosporine at 4 h prior to the termination of the cultures. (A) Representative images showing that both the cells in the noninfection control cultures and the cells without chlamydial inclusions in the serovar L2-infected cultures were induced to undergo apoptosis by staurosporine. The DNA were labeled blue (first row), cytochrome c were labeled red (second row), and fragmented DNA were labeled green (third row). Chlamydial inclusions identified based on Hoechst staining (last two panels of the first row) are marked with asterisks (last two panels of bottom row). Note that although the uninfected cells were induced to undergo apoptosis, the infected cells (cells with inclusions) were resistant to apoptosis induction. Other combinations of triple staining as described in the Fig. 1A legend were also used in this experiment. (B) HeLa cells with or without infection with 17 different chlamydial serovars were induced to undergo apoptosis and processed at various times after infection. The cell samples were stained as described in the Fig. 1A and 3A legends. The cells with or without inclusions and with or without apoptotic nuclei were counted independently as described in the Fig. 1B legend. The results (means ± standard deviations) are from three to eight experiments, each with duplicates, and were compared between cells bearing inclusions (open bars) and those bearing no inclusions (solid bars) from the same infected cultures or the cells from the cultures of HeLa cells alone (last solid bar). Note that cells with inclusions from each of the 17 infected cultures all showed significant antiapoptotic activity. ND, not detected. MoPn, chlamydia muridarum.
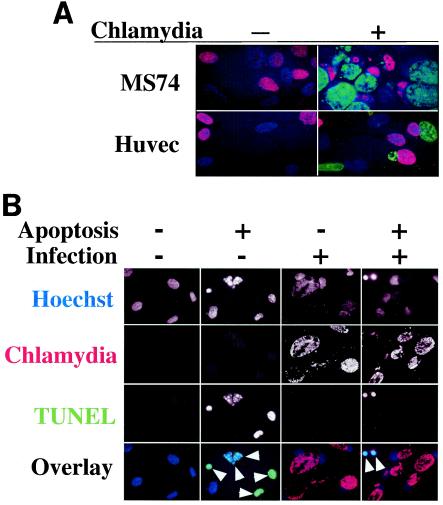
Human primary cells infected with chlamydia L2 can undergo DNA synthesis and resist apoptosis induction.
To test whether the chlamydial effects observed in HeLa cells are cell line dependent, we evaluated the effects of chlamydial infection on host cell DNA synthesis and apoptosis programs in human primary cells. Both endometrial epithelial cells (MS74) and HUVEC efficiently incorporated BrDu regardless of L2 infection (Fig. 4A). The BrDu-positive cells were counted in three separate experiments, and the BrDu incorporation rate was between 40 and 60% for each of the three cell populations (all cells in noninfection control cultures and uninfected cells or infected cells [with obvious chlamydial inclusion bodies] in the infected cultures) (data not shown). Furthermore, HUVEC with L2 infection for 40 h did not show any significant apoptosis, and instead host cell apoptosis induced by staurosporine was profoundly inhibited (Fig. 4B). When the apoptotic events were quantitated by counting the apoptotic cells in two independent experiments, about 80% of either the normal HUVEC in the noninfection culture or the uninfected cells in the infected culture were induced to undergo apoptosis, while fewer than 5% of the infected cells were apoptotic regardless of staurosporine induction (data not shown), demonstrating a strong antiapoptotic activity by chlamydia in human primary cells. These observations have confirmed our findings in HeLa cells.
FIG. 4.
Human primary cells infected with L2 can also undergo DNA synthesis and resist apoptosis induction. Either MS74 cells or HUVEC were infected with L2 as described in Materials and Methods and processed as described in the Fig. 2A and 3A legends. (A) Representative images of either MS74 cells or HUVEC infected with L2 for 40 h were labeled with BrDu and processed as described in Materials and Methods. The samples were stained for nuclei incorporating BrDu (red), chlamydial inclusions (green), and DNA (blue). Note that nuclei of either cell type incorporated BrDu regardless of L2 infection, indicating that both of the primary cells can undergo DNA synthesis in the presence of chlamydial infection. (B) Representative images of HUVEC with or without L2 infection for 40 h and with or without apoptosis induction by staurosporine. The DNA was labeled blue (first row), chlamydial inclusions were labeled red (second row), and fragmented DNA was labeled green (third row). Apoptotic cells are indicated with arrowheads in the tricolor overlay images. Note that although most of the uninfected cells (in either the noninfection control culture or the infected culture) were induced to undergo apoptosis, the infected cells (cells with inclusions) were resistant to apoptosis induction.
DISCUSSION
We have demonstrated that although apoptosis can be detected in chlamydia-infected cultures, antiapoptotic activity is the prevailing event in chlamydia-infected cells during the entire course of chlamydial infection. This finding is consistent with the following observations: (i) chlamydia-infected cells can continue to undergo DNA synthesis and mitosis during the entire course of infection (Fig. 2) (7); (ii) chlamydia-infected cells are known to be able to synthesize and secrete cytokines even at very late stages of infection (24, 31, 38); (iii) an up-regulated metabolic activity was detected in chlamydia-infected cells (23, 33); (iv) active phosphorylation of both host and chlamydial proteins, likely by host kinases, was identified (3, 17, 43); (v) chlamydiae can actively import nutrients, including lipids and energy, from host cell cytosol into chlamydial vacuoles for chlamydial biosynthesis (10, 22, 49, 51), which requires host cells to be viable; and (v) preventing the infected cells from undergoing apoptosis may help the intracellular chlamydial organisms to evade host defense mechanisms (16), which is consistent with the observation that animals with or without deficiency in perforin-mediated apoptosis showed a similar susceptibility to chlamydial infection (36). All of these observations support a central concept that what chlamydiae really want from their host cells is a safe and rich niche to live in, which inevitably requires chlamydiae to manipulate host cells and to prevent but not to induce host cell apoptosis. However, the role of chlamydial antiapoptotic activity in chlamydial evasion of cytotoxic T lymphocyte (CTL)-induced apoptosis requires further investigation, since it has been shown that chlamydial antigen-specific CD8+ T cells can be induced and these CD8+ T cells can cause cell lysis in chlamydia-infected cultures (29, 30, 45, 50). The apparent discrepancies between the chlamydial antiapoptotic activity and CTL-induced cell lysis may be explained from the following two aspects. First, the chlamydial antiapoptotic activity peaks at middle cycle of chlamydial growth, while many of the CTL assays were carried out with cultures infected with chlamydia at early stages of chlamydial infection. Second, the chlamydial antiapoptotic activity was detected in the infected cells at the single-cell level (16) (see below for details), while the CTL-induced cell lysis was measured based on Cr51 release from the entire infected culture. It is known that the adjacent uninfected cells in the infected cultures are more susceptible to apoptosis induction by factors secreted in the same infected culture (41). It is not clear how much the nonspecific cell lysis can contribute to the Cr51 lysis release after the chlamydia-infected cultures are mixed with CTL preparations. The challenge will be to visualize the apoptosis events at the single-cell level in chlamydia-infected cultures after induction with chlamydia-specific CTLs.
The observation that apoptosis was detected in chlamydia-infected cultures and even increased at the late stages of infection (Fig. 1B and 3B) may be due to the following reasons. First, the increased apoptosis late in the infection course may reflect the elevated susceptibility of older cells to stress signals, since the noninfection control cells also displayed a higher rate of apoptosis 48 h after culture (Fig. 1B). Second, the fact that apoptosis of the uninfected cells from the infected cultures was significantly higher than that of the cells from the noninfection cultures (Fig. 1B) suggests that there are proapoptotic factors in the infected cultures to promote apoptosis. It is possible that either chlamydia-derived toxins or toxic materials released by chlamydia-infected cells play some role in promoting apoptosis. Several chlamydial toxin homologue genes (including chlamydia muridarum and GPIC) encode a homologue of the complete clostridium large toxin (39, 40, 46). It has been demonstrated that the chlamydial toxin homologue genes are expressed late in the growth cycle, and chlamydia muridarum is more toxic to cultured cells than other C. trachomatis strains, which contain truncated toxin genes or no toxin genes at all (5). These observations are consistent with our finding in the present study that cultures infected with chlamydia muridarum or either of the two C. psittaci strains (GPIC and 6BC) displayed higher levels of apoptosis at the late stages of infection (Fig. 1B). Finally, many host cell-derived cytokines, such as tumor necrosis factor alpha, can have proapoptotic activity (26), and chlamydia-infected cells are known to secrete these inflammatory cytokines (38). It has been observed that supernatants from live chlamydia-infected macrophage cultures were toxic to lymphocytes (reference 47 and unpublished data). Although these proapoptotic conditions induced up to 15% of the uninfected cells to undergo apoptosis, most infected cells from the same infected cultures showed no significant apoptosis (Fig. 1B). This finding is consistent with a previous study showing that in chlamydia-infected mouse cell cultures, significant apoptosis was detected in the uninfected cells but not in the infected cells (41). It has been suggested that chlamydiae may induce apoptosis of leukocytes recruited to the site of infection for evading host defense (27). These observations together support the concept that chlamydia-induced apoptosis occurs mainly in the nearby uninfected cells, while the infected cells are resistant to apoptosis induction.
The increased apoptosis rate in chlamydia-infected cultures at the late stages of infection has led some to hypothesize that chlamydiae may exit the infected cells via host cell apoptosis. Although there is evidence supporting this idea (12, 35), our observations suggest that the situation is complex and requires more studies. For example, although more cells in chlamydia-infected cultures became apoptotic at the late stages of infection, the overall apoptosis levels were not high enough (involving only 15% of the cells) (Fig. 1B) to consider apoptosis an active or effective process for chlamydial spread. More importantly, most of the apoptosis occurred in the uninfected cells (Fig. 1B), and chlamydia-infected cells were actually resistant to apoptosis induction throughout the entire infection course (Fig. 3B). Since chlamydiae reside within cytoplasmic vacuoles, active lysis of both vacuolar and cytoplasmic membranes or activation of unknown EB excretion pathways is required for chlamydial organisms to reach the extracellular space, which cannot be achieved by apoptosis. On the contrary, apoptotic cells not only maintain the cytoplasmic membrane integrity but also provide signals for phagocytosis, which serves no advantage for chlamydial survival in the infected host cells. Therefore, more experimentation is required to clarify the role of host cell apoptosis in chlamydial release and to understand the mechanisms by which chlamydiae achieve intercellular transmission.
We have noticed that the methods used for detecting apoptosis in chlamydia-infected cultures can affect the results (data not shown). Although in general, apoptosis can be detected at the cytoplasmic membrane, cytosol, or nuclear level by many well-established methods, detecting apoptosis in chlamydia-infected cultures represents a unique situation, and not all methods can be used for accurately quantitating apoptosis in chlamydia-infected cultures. This is because the massive chlamydial organism structures and chlamydial infection-induced host cell alterations that are independent of host cell apoptosis can all contribute to final readouts of the apoptosis measurements. For example, chlamydial DNA can interfere with apoptosis measurements based on DNA quantitation, such as flow cytometry or enzyme-linked immunosorbent assay. Unfortunately, many of the studies showing chlamydial proapoptotic activities have used the flow cytometry-based assay for measuring apoptosis events in chlamydia-infected cultures. The fact that chlamydiae can induce transient exposure of phosphatidylserine (20) makes annexin V-based measurements unreliable for identifying apoptosis in chlamydia-infected cultures, although the transiently exposed phosphatidylserine may be used as signal for phagocytosis (15). Similarly, it is not appropriate to use the chlamydia-induced activation of caspase-1 for identifying apoptosis (31), since caspase-1 has many other functions, such as processing interleukin-1β and -18, and activation of capase-1 does not necessarily drive the cells into apoptosis. Due to the complexity of the chlamydia-host cell interaction, visualization of apoptosis at the single-cell level, although tedious, may represent one of the most reliable methods for detecting apoptosis, since it avoids the cell population-based interference caused by the presence of chlamydial infection. This microscopic approach may introduce subjectivity, and it could underestimate the apoptosis events when apoptotic cells are detached from the coverslips prior to counting. In our experience, the cell detachment can be minimized by careful sample processing, and the extent of detachment can be monitored by comparing the cell number or density between normal and proapoptotic staurosporine-treated cell samples. Caution was taken in the present study to ensure that there was no significant detachment of apoptotic cells.
Finally, the most interesting question is, of course, how chlamydiae manage to prevent the infected host cells from undergoing apoptosis. Based on both our previous studies (16) and the present work, we hypothesize that chlamydiae may secrete factors into host cells for actively blocking host apoptosis pathways. Efforts are under way to identify the potential antiapoptotic factor(s) by using a combination of genetic selection and biochemical purification approaches (52) and to further understand the mechanisms of chlamydial antiapoptotic activity.
Acknowledgments
This work was supported in part by grants (to G. Zhong) from the U.S. Department of Health and Human Services (R01 AI47997 and R01 HL64883).
Editor: F. C. Fang
REFERENCES
- 1.Airenne, S., H. M. Surcel, J. Tuukkanen, M. Leinonen, and P. Saikku. 2002. Chlamydia pneumoniae inhibits apoptosis in human epithelial and monocyte cell lines. Scand. J. Immunol. 55:390-398. [DOI] [PubMed] [Google Scholar]
- 2.Bauwens, J. E., H. Orlander, M. P. Gomez, M. Lampe, S. Morse, W. E. Stamm, R. Cone, R. Ashley, P. Swenson, and K. K. Holmes. 2002. Epidemic Lymphogranuloma venereum during epidemics of crack cocaine use and HIV infection in the Bahamas. Sex. Transm. Dis. 29:253-259. [DOI] [PubMed] [Google Scholar]
- 3.Bea, F., M. H. Puolakkainen, T. McMillen, F. N. Hudson, N. Mackman, C. C. Kuo, L. A. Campbell, and M. E. Rosenfeld. 2003. Chlamydia pneumoniae induces tissue factor expression in mouse macrophages via activation of Egr-1 and the MEK-ERK1/2 pathway. Circ Res. 92:394-401. [DOI] [PubMed] [Google Scholar]
- 4.Beatty, W. L., R. P. Morrison, and G. I. Byrne. 1994. Persistent chlamydiae: from cell culture to a paradigm for chlamydial pathogenesis. Microbiol. Rev. 58:686-699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Belland, R. J., M. A. Scidmore, D. D. Crane, D. M. Hogan, W. Whitmire, G. McClarty, and H. D. Caldwell. 2001. Chlamydia trachomatis cytotoxicity associated with complete and partial cytotoxin genes. Proc. Natl. Acad. Sci. USA 98:13984-13989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bentsi, C., C. A. Klufio, P. L. Perine, T. A. Bell, L. D. Cles, C. M. Koester, and S. P. Wang. 1985. Genital infections with Chlamydia trachomatis and Neisseria gonorrhoeae in Ghanaian women. Genitourin. Med. 61:48-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bose, S. K., and H. Liebhaber. 1979. Deoxyribonucleic acid synthesis, cell cycle progression, and division of Chlamydia-infected HeLa 229 cells. Infect. Immun. 24:953-957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brunham, R. C., M. Laga, J. N. Simonsen, D. W. Cameron, R. Peeling, J. McDowell, H. Pamba, J. O. Ndinya-Achola, G. Maitha, and F. A. Plummer. 1990. The prevalence of Chlamydia trachomatis infection among mothers of children with trachoma. Am. J. Epidemiol. 132:946-952. [DOI] [PubMed] [Google Scholar]
- 9.Campbell, L. A., and C. C. Kuo. 2003. Chlamydia pneumoniae and atherosclerosis. Semin. Respir. Infect. 18:48-54. [DOI] [PubMed] [Google Scholar]
- 10.Carabeo, R. A., D. J. Mead, and T. Hackstadt. 2003. Golgi-dependent transport of cholesterol to the Chlamydia trachomatis inclusion. Proc. Natl. Acad. Sci. USA 12:12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Centers for Disease Control and Prevention. 2000. Compendium of measures to control Chlamydia psittaci infection among humans (psittacosis) and pet birds (avian chlamydiosis), 2000. Morb. Mortal. Wkly. Rep. Recomm. Rep. 49:3-17. [PubMed] [Google Scholar]
- 12.Coutinho-Silva, R., J. L. Perfettini, P. M. Persechini, A. Dautry-Varsat, and D. M. Ojcius. 2001. Modulation of P2Z/P2X(7) receptor activity in macrophages infected with Chlamydia psittaci. Am. J. Physiol. Cell Physiol. 280:C81-C819. [DOI] [PubMed] [Google Scholar]
- 13.Dean, D., and V. C. Powers. 2001. Persistent Chlamydia trachomatis infections resist apoptotic stimuli. Infect. Immun. 69:2442-2447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Everett, K. D., A. A. Andersen, M. Plaunt, and T. P. Hatch. 1991. Cloning and sequence analysis of the major outer membrane protein gene of Chlamydia psittaci 6BC. Infect. Immun. 59:2853-2855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fadok, V. A., D. L. Bratton, D. M. Rose, A. Pearson, R. A. Ezekewitz, and P. M. Henson. 2000. A receptor for phosphatidylserine-specific clearance of apoptotic cells. Nature 405:85-90. [DOI] [PubMed] [Google Scholar]
- 16.Fan, T., H. Lu, H. Hu, L. Shi, G. A. McClarty, D. M. Nance, A. H. Greenberg, and G. Zhong. 1998. Inhibition of apoptosis in chlamydia-infected cells: blockade of mitochondrial cytochrome c release and caspase activation. J. Exp. Med. 187:487-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fawaz, F. S., C. van Ooij, E. Homola, S. C. Mutka, and J. N. Engel. 1997. Infection with Chlamydia trachomatis alters the tyrosine phosphorylation and/or localization of several host cell proteins, including cortactin. Infect. Immun. 65:5301-5308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Geng, Y., R. B. Shane, K. Berencsi, E. Gonczol, M. H. Zaki, D. J. Margolis, G. Trinchieri, and A. H. Rook. 2000. Chlamydia pneumoniae inhibits apoptosis in human peripheral blood mononuclear cells through induction of IL-10. J. Immunol. 164:5522-5529. [DOI] [PubMed] [Google Scholar]
- 19.Gibellini, D., R. Panaya, and F. Rumpianesi. 1998. Induction of apoptosis by Chlamydia psittaci and Chlamydia trachomatis infection in tissue culture cells. Zentralbl. Bakteriol. 288:35-43. [DOI] [PubMed] [Google Scholar]
- 20.Goth, S. R., and R. S. Stephens. 2001. Rapid, transient phosphatidylserine externalization induced in host cells by infection with Chlamydia spp. Infect. Immun. 69:1109-1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hackstadt, T. 1998. The diverse habitats of obligate intracellular parasites. Curr. Opin. Microbiol. 1:82-87. [DOI] [PubMed] [Google Scholar]
- 22.Hackstadt, T., D. D. Rockey, R. A. Heinzen, and M. A. Scidmore. 1996. Chlamydia trachomatis interrupts an exocytic pathway to acquire endogenously synthesized sphingomyelin in transit from the Golgi apparatus to the plasma membrane. EMBO J. 15:964-977. [PMC free article] [PubMed] [Google Scholar]
- 23.Hatch, G. M., and G. McClarty. 1998. Cardiolipin remodeling in eukaryotic cells infected with Chlamydia trachomatis is linked to elevated mitochondrial metabolism. Biochem. Biophys. Res. Commun. 243:356-360. [DOI] [PubMed] [Google Scholar]
- 24.Hess, S., C. Rheinheimer, F. Tidow, G. Bartling, C. Kaps, J. Lauber, J. Buer, and A. Klos. 2001. The reprogrammed host: Chlamydia trachomatis-induced up-regulation of glycoprotein 130 cytokines, transcription factors, and antiapoptotic genes. Arthritis Rheum. 44:2392-2401. [DOI] [PubMed] [Google Scholar]
- 25.Hsia, R. C., and P. M. Bavoil. 1996. Sequence analysis of the omp2 region of Chlamydia psittaci strain GPIC: structural and functional implications. Gene 176:155-162. [DOI] [PubMed] [Google Scholar]
- 26.Inada, H., I. Izawa, M. Nishizawa, E. Fujita, T. Kiyono, T. Takahashi, T. Momoi, and M. Inagaki. 2001. Keratin attenuates tumor necrosis factor-induced cytotoxicity through association with TRADD. J. Cell Biol. 155:415-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jendro, M. C., T. Deutsch, B. Korber, L. Kohler, J. G. Kuipers, B. Krausse-Opatz, J. Westermann, E. Raum, and H. Zeidler. 2000. Infection of human monocyte-derived macrophages with Chlamydia trachomatis induces apoptosis of T cells: a potential mechanism for persistent infection. Infect. Immun. 68:6704-6711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joyner, J. L., J. M. Douglas, Jr., M. Foster, and F. N. Judson. 2002. Persistence of Chlamydia trachomatis infection detected by polymerase chain reaction in untreated patients. Sex. Transm. Dis. 29:196-200. [DOI] [PubMed] [Google Scholar]
- 29.Kim, S. K., M. Angevine, K. Demick, L. Ortiz, R. Rudersdorf, D. Watkins, and R. DeMars. 1999. Induction of HLA class I-restricted CD8+ CTLs specific for the major outer membrane protein of Chlamydia trachomatis in human genital tract infections. J. Immunol. 162:6855-6866. [PubMed] [Google Scholar]
- 30.Kim, S. K., L. Devine, M. Angevine, R. DeMars, and P. B. Kavathas. 2000. Direct detection and magnetic isolation of Chlamydia trachomatis major outer membrane protein-specific CD8+ CTLs with HLA class I tetramers. J. Immunol. 165:7285-7292. [DOI] [PubMed] [Google Scholar]
- 31.Lu, H., C. Shen, and R. C. Brunham. 2000. Chlamydia trachomatis infection of epithelial cells induces the activation of caspase-1 and release of mature IL-18. J. Immunol. 165:1463-1469. [DOI] [PubMed] [Google Scholar]
- 32.Morrison, R. P., K. Lyng, and H. D. Caldwell. 1989. Chlamydial disease pathogenesis. Ocular hypersensitivity elicited by a genus-specific 57-kD protein J. Exp. Med. 169:663-675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ojcius, D. M., H. Degani, J. Mispelter, and A. Dautry-Varsat. 1998. Enhancement of ATP levels and glucose metabolism during an infection by Chlamydia. NMR studies of living cells J. Biol. Chem. 273:7052-7058. [DOI] [PubMed] [Google Scholar]
- 34.Ojcius, D. M., P. Souque, J. L. Perfettini, and A. Dautry-Varsat. 1998. Apoptosis of epithelial cells and macrophages due to infection with the obligate intracellular pathogen Chlamydia psittaci. J. Immunol. 161:4220-4226. [PubMed] [Google Scholar]
- 35.Perfettini, J. L., D. M. Ojcius, C. W. Andrews, Jr., S. J. Korsmeyer, R. G. Rank, and T. Darville. 2003. Role of proapoptotic BAX in propagation of Chlamydia muridarum (the mouse pneumonitis strain of Chlamydia trachomatis) and the host inflammatory response. J. Biol. Chem. 278:9496-9502. [DOI] [PubMed] [Google Scholar]
- 36.Perry, L. L., K. Feilzer, S. Hughes, and H. D. Caldwell. 1999. Clearance of Chlamydia trachomatis from the murine genital mucosa does not require perforin-mediated cytolysis or Fas-mediated apoptosis. Infect. Immun. 67:1379-1385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rajalingam, K., H. Al-Younes, A. Muller, T. F. Meyer, A. J. Szczepek, and T. Rudel. 2001. Epithelial cells infected with Chlamydophila pneumoniae (Chlamydia pneumoniae) are resistant to apoptosis. Infect. Immun. 69:7880-7888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Rasmussen, S. J., L. Eckmann, A. J. Quayle, L. Shen, Y. X. Zhang, D. J. Anderson, J. Fierer, R. S. Stephens, and M. F. Kagnoff. 1997. Secretion of proinflammatory cytokines by epithelial cells in response to Chlamydia infection suggests a central role for epithelial cells in chlamydial pathogenesis. J. Clin. Invest. 99:77-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Read, T. D., R. C. Brunham, C. Shen, S. R. Gill, J. F. Heidelberg, O. White, E. K. Hickey, J. Peterson, T. Utterback, K. Berry, S. Bass, K. Linher, J. Weidman, H. Khouri, B. Craven, C. Bowman, R. Dodson, M. Gwinn, W. Nelson, R. DeBoy, J. Kolonay, G. McClarty, S. L. Salzberg, J. Eisen, and C. M. Fraser. 2000. Genome sequences of Chlamydia trachomatis MoPn and Chlamydia pneumoniae AR39. Nucleic Acids Res. 28:1397-1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Read, T. D., G. S. Myers, R. C. Brunham, W. C. Nelson, I. T. Paulsen, J. Heidelberg, E. Holtzapple, H. Khouri, N. B. Federova, H. A. Carty, L. A. Umayam, D. H. Haft, J. Peterson, M. J. Beanan, O. White, S. L. Salzberg, R. C. Hsia, G. McClarty, R. G. Rank, P. M. Bavoil, and C. M. Fraser. 2003. Genome sequence of Chlamydophila caviae (Chlamydia psittaci GPIC): examining the role of niche-specific genes in the evolution of the Chlamydiaceae. Nucleic Acids Res. 31:2134-2147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schoier, J., K. Ollinger, M. Kvarnstrom, G. Soderlund, and E. Kihlstrom. 2001. Chlamydia trachomatis-induced apoptosis occurs in uninfected McCoy cells late in the developmental cycle and is regulated by the intracellular redox state. Microb. Pathog. 31:173-184. [DOI] [PubMed] [Google Scholar]
- 42.Scidmore, M. A., E. R. Fischer, and T. Hackstadt. 2003. Restricted fusion of Chlamydia trachomatis vesicles with endocytic compartments during the initial stages of infection. Infect. Immun. 71:973-984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Scidmore, M. A., and T. Hackstadt. 2001. Mammalian 14-3-3beta associates with the Chlamydia trachomatis inclusion membrane via its interaction with IncG. Mol. Microbiol. 39:1638-1650. [DOI] [PubMed] [Google Scholar]
- 44.Sherman, K. J., J. R. Daling, A. Stergachis, N. S. Weiss, H. M. Foy, S. P. Wang, and J. T. Grayston. 1990. Sexually transmitted diseases and tubal pregnancy. Sex. Transm. Dis. 17:115-121. [DOI] [PubMed] [Google Scholar]
- 45.Starnbach, M. N., M. J. Bevan, and M. F. Lampe. 1995. Murine cytotoxic T lymphocytes induced following Chlamydia trachomatis intraperitoneal or genital tract infection respond to cells infected with multiple serovars. Infect. Immun. 63:3527-3530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Stephens, R. S., S. Kalman, C. Lammel, J. Fan, R. Marathe, L. Aravind, W. Mitchell, L. Olinger, R. L. Tatusov, Q. Zhao, E. V. Koonin, and R. W. Davis. 1998. Genome sequence of an obligate intracellular pathogen of humans: Chlamydia trachomatis. Science 282:754-759. [DOI] [PubMed] [Google Scholar]
- 47.Su, H., and H. D. Caldwell. 1995. Kinetics of chlamydial antigen processing and presentation to T cells by paraformaldehyde-fixed murine bone marrow-derived macrophages. Infect. Immun. 63:946-953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Taylor, H. R., S. L. Johnson, J. Schachter, H. D. Caldwell, and R. A. Prendergast. 1987. Pathogenesis of trachoma: the stimulus for inflammation. J. Immunol. 138:3023-3027. [PubMed] [Google Scholar]
- 49.Tipples, G., and G. McClarty. 1993. The obligate intracellular bacterium Chlamydia trachomatis is auxotrophic for three of the four ribonucleoside triphosphates. Mol. Microbiol. 8:1105-1114. [DOI] [PubMed] [Google Scholar]
- 50.Wizel, B., B. C. Starcher, B. Samten, Z. Chroneos, P. F. Barnes, J. Dzuris, Y. Higashimoto, E. Appella, and A. Sette. 2002. Multiple Chlamydia pneumoniae antigens prime CD8+ Tc1 responses that inhibit intracellular growth of this vacuolar pathogen. J. Immunol. 169:2524-2535. [DOI] [PubMed] [Google Scholar]
- 51.Wylie, J. L., G. M. Hatch, and G. McClarty. 1997. Host cell phospholipids are trafficked to and then modified by Chlamydia trachomatis. J. Bacteriol. 179:7233-7242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Zhong, G., P. Fan, H. Ji, F. Dong, and Y. Huang. 2001. Identification of a chlamydial protease-like activity factor responsible for the degradation of host transcription factors. J. Exp. Med. 193:935-942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Zhong, G., T. Fan, and L. Liu. 1999. Chlamydia inhibits interferon gamma-inducible major histocompatibility complex class II expression by degradation of upstream stimulatory factor 1. J. Exp. Med. 189:1931-1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhong, G., L. Liu, T. Fan, P. Fan, and H. Ji. 2000. Degradation of transcription factor RFX5 during the inhibition of both constitutive and interferon gamma-inducible major histocompatibility complex class I expression in chlamydia-infected cells. J. Exp. Med. 191:1525-1534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Zhong, G. M., R. E. Reid, and R. C. Brunham. 1990. Mapping antigenic sites on the major outer membrane protein of Chlamydia trachomatis with synthetic peptides. Infect. Immun. 58:1450-1455. [DOI] [PMC free article] [PubMed] [Google Scholar]