Abstract
Arsenic (+3 oxidation state) methyltransferase (AS3MT) is the key enzyme in the pathway for methylation of arsenicals. A common polymorphism in the AS3MT gene that replaces a threonyl residue in position 287 with a methionyl residue (AS3MT/M287T) occurs at a frequency of about 10% among populations worldwide. Here, we compared catalytic properties of recombinant human wild-type (wt) AS3MT and AS3MT/M287T in reaction mixtures containing S-adenosylmethionine, arsenite (iAsIII) or methylarsonous acid (MAsIII) as substrates and endogenous or synthetic reductants, including glutathione (GSH), a thioredoxin reductase (TR)/thioredoxin (Trx)/NADPH reducing system, or tris (2-carboxyethyl) phosphine hydrochloride (TCEP). With either TR/Trx/NADPH or TCEP, wtAS3MT or AS3MT/M287T catalyzed conversion of iAsIII to MAsIII, methylarsonic acid (MAsV), dimethylarsinous acid (DMAsIII), and dimethylarsinic acid (DMAsV); MAsIII was converted to DMAsIII and DMAsV. Although neither enzyme required GSH to support methylation of iAsIII or MAsIII, addition of 1 mM GSH decreased Km and increased Vmax estimates for either substrate in reaction mixtures containing TR/Trx/NADPH. Without GSH, Vmax and Km values were significantly lower for AS3MT/M287T than for wtAS3MT. In the presence of 1 mM GSH, significantly more DMAsIII was produced from iAsIII in reactions catalyzed by the M287T variant than in wtAS3MT-catalyzed reactions. Thus, 1 mM GSH modulates AS3MT activity, increasing both methylation rates and yield of DMAsIII. AS3MT genotype exemplified by differences in regulation of wtAS3MT and AS3MT/M287T-catalyzed reactions by GSH may contribute to differences in the phenotype for arsenic methylation and, ultimately, to differences in the disease susceptibility in individuals chronically exposed to inorganic arsenic.
Keywords: Arsenic methylation, AS3MT polymorphism, thioredoxin reductase, glutathione
INTRODUCTION
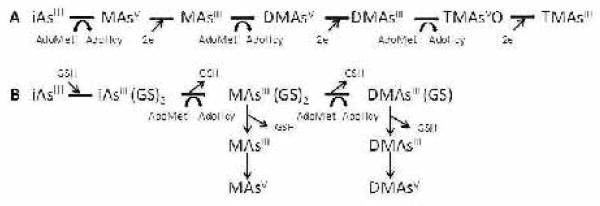
Enzymatically catalyzed methylation of inorganic arsenic (iAs) is the main pathway for the metabolism of iAs (Styblo et al., 1995; Vahter 1999). Conversion of iAs to methylated metabolites affects the distribution and retention of arsenic (As) and also produces As species that mediate some of the toxic effects associated with iAs exposure (Chen et al., 2011; Hughes et al., 2010; Thomas et al,. 2001). Enzymatically catalyzed methylation transfers methyl groups from S-adenosylmethionine (AdoMet) to As to produce monomethylarsenic (MAs), dimethylarsenic (DMAs), and trimethylarsenic (TMAs) metabolites that contain either trivalent As (AsIII) or pentavalent As (AsV) (Cullen et al., 1984; Challenger 1951). Strong evidence suggests that arsenic (+3 oxidation state) methyltransferase (As3mt, EC 2.1.1.137) is the key enzyme catalyzing reactions that form all known methylated oxyarsenical metabolites of iAs (Lin et al., 2002; Thomas et al., 2007). Two pathways have been proposed for As3mt-catalyzed methylation of iAs (Figure 1). The oxidative methylation pathway proposed by Challenger involves oxidative addition of a methyl group to a trivalent arsenical to yield a methylated product containing pentavalent As (Challenger 1951). The pentavalent arsenical is then reduced to trivalency allowing repeated cycles of oxidative methylation. Coupled rounds of oxidative methylation of trivalent As and reduction of pentavalent As converts arsenite (iAsIII) to methylarsonic acid (MAsV), methylarsonous acid (MAsIII), dimethylarsinic acid (DMAsV), dimethylarsinous acid (DMAsIII), trimethylarsine oxide (TMAsVO), and finally, trimethylarsine (TMAsIII). Hirano and associates have proposed a reductive methylation pathway in which trivalent arsenicals bound to glutathione (GSH) or a protein thiol are substrates for repeated rounds of enzymatically catalyzed methylation (Hayakawa et al., 2005). In this pathway, methylated trivalent arsenicals remain bound to thiols throughout the cycle and pentavalent methylated arsenicals are produced by oxidation of AsIII-thiol complexes. Consistent with either mechanism, both trivalent and pentavalent inorganic and methylated arsenicals have been detected in urine of individuals chronically exposed to iAs, in cells and medium following in vitro exposure to arsenicals, and in reaction systems that contain recombinant As3mt (Del Razo et al., 2001, 2011; Devesa et al., 2004; Hernández-Zavala et al., 2008; Valenzuela et al., 2005).
Figure 1.
Two postulated pathways for methylation of inorganic arsenic by AS3MT: A. Oxidative methylation pathway; B. Reductive methylation pathway; iAsIII, arsenite; MAsV, methylarsonic acid; MAsIII, methylarsonous acid, DMAsV, dimethylarsinic acid; DMAsIII, dimethylarsinous acid; TMAsV, trimethylarsine oxide; TMAsIII, trimethylarsine; AdoMet, S-adenosylmethionine; AdoHcy, S-adenosylhomocysteine; GSH, glutathione; e, electron.
Previous studies of kinetic aspects and cofactor requirements for methylation reactions catalyzed by recombinant rat As3mt (rAs3mt) found that catalysis was supported by endogenous dithiol reductants, thioredoxin (Trx), glutaredoxin, or lipoic acid (Waters et al. 2004a,b). Thus, incorporation of coupled systems consisting of thioredoxin reductase (TR), thioredoxin (Trx), and NADPH, or glutaredoxin/GSH/GSH reductase/NADPH, or lipoic acid/TR/NADPH into reaction mixtures supported rAs3mt-catalyzed methylation of iAsIII to yield mono-, di-and trimethylated arsenicals, including MAsV, MAsIII, DMAsV, DMAsIII, TMAsVO and TMAsIII. Synthetic reductants, dithiothreitol (DTT) and tris(2-carboxylethyl)-phosphine hydrochloride (TCEP), also supported rAs3mt-catalyzed methylation. Although the monothiol reductant GSH did not support rAs3mt-catalyzed methylation of iAsIII and inhibited conversion of DMAs to TMAsVO and trimethylarsine, its addition to reaction mixtures that also contained endogenous dithiol reductants or synthetic reductants increased methylation rates and DMAs yield. Studies with recombinant human AS3MT (hAS3MT) have also examined kinetic aspects of As methylation and the role of reductants in catalysis (Hayakawa et al. 2005; Song et al., 2009, 2010; Wood et al., 2006). Reductants evaluated with hAS3MT include monothiols (mercaptoethanol, cysteine, GSH) and a dithiol DTT. Reactions catalyzed by hAS3MT in the presence of DTT yielded high DMAs/MAs ratios that were similar to the ratios of these metabolites found in urine of humans ingesting iAs or in primary cultures of human hepatocytes exposed to iAs. In contrast, reaction mixtures containing hAS3MT and GSH as the sole reductant displayed low methylation rates, high MAs levels, and low DMAs/MAs ratios. Thus, hAS3MT and rAs3mt are absolutely dependent on a dithiol reductant for optimal activity. For both rAs3mt and hAS3MT, it is likely that GSH acts as modulator, not a primary determinant, of catalytic activity.
Because the catalytic activity of hAS3MT in presence of endogenous dithiol reductants has not been systemically examined, we characterized the function of two commonly occurring forms of hAS3MT. These are wildtype form (wtAS3MT) which contains a methionyl residue in position 287 and a variant form AS3MT/M287T in which this methionyl residue is replaced by a threonyl residue. This change arises from a single nucleotide C14458T polymorphism (rs11191439) in the coding region of AS3MT (NM_020682.3: c.860T>C) and occurs in ~10% of Caucasian, African American and Latino populations (Del Razo, et al., 2011; Wood et al., 2006). Individuals with wtAS3MT and AS3MT/M287T genotypes display different urinary profiles of methylated metabolites of iAs, including changes in the DMAs/MAs ratio (Agusa et al., 2009; Chung et al., 2009; Engström et al., 2007, 2011; Fujihara et al., 2009; Hernández et al., 2008a,b; Hwang et al., 2010; Valenzuela et al., 2009), suggesting that AS3MT genotype and As methylation phenotype can be linked. Therefore, we have compared the kinetics of As methylation in reactions catalyzed by recombinant wtAS3MT and AS3MT/M287T using physiological and synthetic reductants and examined the role of GSH as a modulator of catalytic activities of wtAS3MT and AS3MT/M287T. Differences in kinetic behavior of the wildtype protein and its common variant, particularly modulation of activity by GSH, may account for interindividual variation in capacity to methylate iAs.
MATERIALS AND METHODS
Arsenicals
Arsenicals used as substrates for kinetic studies or as analytical standards included iAsIII (NaAsIIIO2), sodium arsenate (Na HAsV 2 O4) (Sigma-Aldrich, St. Louis, MO), MAsV [CH AsV 3 O(ONa)2] and DMAsV [(CH ) V 3 2As O(OH)] (Chem Service, West Chester, PA). These commercial reagents were stated to be greater than 98% pure. MAsIII [(CH AsIII 3 O)4], DMAsIII [(CH ) III 3 2As I], and TMAsVO were provided by Dr. William Cullen (University of British Columbia, Vancouver, Canada). Arsenic content of each arsenical was determined by graphite furnace-atomic absorption spectrometry (AAS) (Matoušek et al., 2008). To minimize oxidation of unstable MAsIII and DMAsIII, fresh solutions of these arsenicals were prepared shortly before each experiment.
Expression and Purification of Human Recombinant wtAS3MT and AS3MT/M287T
Human wtAS3MT DNA was PCR amplified from normal human liver cDNA (Invitrogen, Carlsbad, CA). The PCR product was cloned into the pRSET expression vector (Invitrogen). The AS3MT/T860C mutant that encodes the AS3MT/M287T variant was generated using the QuikChange II Site-Directed Mutagenesis Kit (Agilent Technologies, Inc., Santa Clara, CA) and the following primers: sense 5′AATTACAGGACATGAAAAAGAACTAACGTTTGATGCCAATTTTACATTTAAGG3′ and antisense 5′CCTTAAATGTAAAATTGGCATCAAACGTTAGTTCTTTTTCATGTCCTGTAATT3′. Identity and orientation of the wtAS3MT and AS3MT/T860C inserts in expression vectors were confirmed by sequencing. His-tagged wtAS3MT and AS3MT/M287T proteins were expressed in BL21(DE3)pLysS E. coli strain (Bioline, Tauton, MA) and purified by Ni-NTA agarose following the manufacturer’s instructions (Qiagen, Valencia, CA). Identity and purity of purified proteins were confirmed by gel electrophoresis with Coomassie Blue staining and by immunoblots using a mouse monoclonal anti-His antibody (Qiagen, Valencia, CA) and a rabbit polyclonal anti-human AS3MT antibody (Santa Cruz Biotechnology, Inc., Santa Cruz, CA) (Figure S1, Supplementary Data). Protein concentrations were measured using a BCA kit from Sigma-Aldrich with bovine serum albumin as a standard. Purified proteins were aliquoted and stored at −80°C until use.
In Vitro Methylation Assay
Catalytic activities of wtAS3MT and AS3MT/M287T were examined in reaction mixtures that contained 100 mM Tris-HCl buffer (pH 7.4), a recombinant enzymes (0.5 – 5 μg), 1 mM AdoMet (Sigma), an arsenical substrate, and one or several of the following reductants: GSH (Sigma-Aldrich), TCEP (Sigma-Aldrich), recombinant rat TR (American Diagnostica, Greenwich, CT) or TR purified from rat liver (Sigma-Aldrich), Trx from E. coli (Sigma-Aldrich), and NADPH (Sigma-Aldrich). The final volume of the mixture was 100 μl. The mixtures were incubated in capped 1.5-ml Eppendorf tubes at 25°C or 37°C for up to 2 hours. Michaelis-Menten constants (Km and Vmax) were calculated to determine capacity of AS3MT variants to methylate arsenicals and their affinity for arsenical substrates.
Analysis of Products of In Vitro Methylation Reactions
Concentrations of As species in reaction mixtures were determined by oxidation state specific hydride generation (HG)-cryotrapping (CT)- AAS as previously described (Hernández-Zavala et al., 2008; Matoušek et al., 2008). Briefly, HG-CT-AAS measured As contents in arsine and methyl-substituted arsines (methyl- and dimethylarsine) generated in reactions of iAs, MAs and DMAs with sodium borohydride (EM Science, Gibbstown, NJ) at pH 6. Arsines from trivalent arsenicals (iAsIII, MAsIII and DMAsIII) were generated directly. For generation of arsines from both tri- and pentavalent arsenicals, the latter were reduced with 2% cysteine hydrochloride (EMD Chemicals Inc., Gibbstown, NJ). Concentrations of AsV-species were then calculated as a difference between the two measurements. To prevent oxidation of unstable DMAsIII, one-half of an in vitro reaction mixture (50 μl) was analyzed for trivalent arsenicals immediately after incubation. The remaining sample was snap frozen in dry ice and stored at -80°C for analysis of AsIII+V species. Limits of detection for analysis of trivalent and pentavalent iAs, MAs and DMAs species by HG-CT-AAS ranged from 8 to 20 pg As. Percentage As recovery from a reaction mixture was calculated by comparing sum of the tri- and pentavalent arsenicals determined by HG-CT-AAS after incubation and the amount of substrate added into the reaction mixture before incubation. Recoveries of As in this study ranged from 89 to 105%.
Statistical Analysis
All statistical analyses used the SAS software package (SAS Institute Inc., Cary, North Carolina). A general linear model or one-way analysis of variance (ANOVA) with Student-Newman-Keuls or Bonferoni correction for multiple comparisons was used to identify statistically significant differences (p < 0.05) between rates and yields of in vitro methylation reactions catalyzed by wtAS3MT or AS3MT/M287T. These methods were also used to characterize differences between corresponding Km and Vmax values.
RESULTS
Effects of Reductants on Methylation of iAsIII by wtAS3MT and AS3MT/M287T
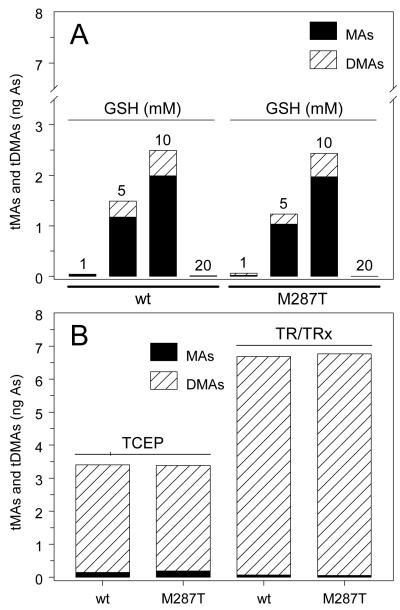
The function of GSH as a reductant was evaluated in reaction mixtures containing recombinant wtAS3MT or AS3MT/M287T (5 μg), 1 mM AdoMet, 1 μM iAsIII and 1 to 20 mM GSH. The mixtures were incubated for 2 hours at 37°C, treated with 2% cysteine and analyzed by HG-CT-AAS for presence of total iAs (iAsIII + iAsV), total MAs (tMAs = MAsIII + MAsV), total DMAs (tDMAs = DMAsIII + DMAsV). In the presence of 1 mM GSH as the sole reductant, both wtAS3MT and AS3MT/M287T catalyzed conversion of less than 1% of substrate to tMAs and tDMAs (Figure 2A). Although methylation yields increased proportionally in the presence of 5 and 10 mM GSH, tMAs, not tDMAs, was the major metabolite. In contrast, addition of 20 mM GSH sharply inhibited methylation catalyzed by either protein. Addition of 1 mM TCEP or coupled system consisting of 0.2 μM TR, 10 μM Trx and 300 μM NADPH to reaction mixtures containing wtAS3MT or AS3MT/M287T supported iAsIII methylation (Figure 2B). For either protein, about 45% and 90% of iAsIII was methylated over the 2-hour incubation period in the presence of TCEP and TR/Trx/NADPH, respectively. For either reductant and either protein, tDMAs accounted for more than 95% of methylated product.
Figure 2.
Effects of reductants on methylation of arsenite (iAsIII) by wtAS3MT and AS3MT/M287T. Reaction mixtures contained 100 mM Tris-HCl buffer (pH 7.4), recombinant enzyme (5 μg), 1 mM AdoMet, 1 μM iAsIII and one of the following reductants or reducing systems: A. GSH (1, 5, 10 or 20 mM); B. 1 mM TCEP or a coupled enzymatic system consisting of 0.2 μM TR, 10 μM Trx and 300 μM NADPH. Reaction mixtures were incubated at 37°C for 2 hours. Cumulative yields of tMAs (MAsIII + MAsV) and tDMAs (DMAsIII + DMAsV) shown (Mean ± SD, n = 3).
Modulation by GSH of iAsIII Methylation Catalyzed by wtAS3MT or AS3MT/M287T
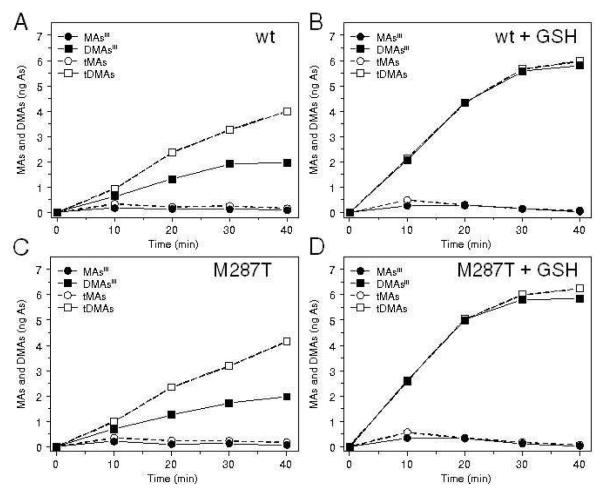
The effect of GSH on methylation of iAsIII was examined in reaction mixtures that contained 1 μM iAsIII, wtAS3MT or AS3MT/M287T (5 μg), and TR/Trx/NADPH coupled system. Reaction mixtures were incubated at 37°C for up to 40 minutes and yields of tri-and pentavalent methylated metabolites were determined by HG-CT-AAS. Without added GSH, profiles of metabolites were similar in reactions catalyzed by wtAS3MT or AS3MT/M287T. Methylated arsenicals containing either AsIII or AsV; MAsIII, MAsV, DMAsIII, and DMAsV were detected in these reaction mixtures. tDMAs yields increased in proportion to incubation time, accounting for ~55% of total substrate at 40 minutes; DMAsIII accounted for ~40-60% of tDMAs (Figure 3A,C). Addition of 1 mM GSH to reaction mixtures approximately doubled yields of methylated meta bolites (Figure 3B,D). At all time points, addition of GSH produced significantly higher yields of tDMAs in reactions catalyzed by AS3MT/M287T than in reactions catalyzed by wtAS3MT (p < 0.05). In reactions catalyzed by either protein in the presence of GSH, DMAsIII was the predominant metabolite, accounting for >90% tDMAs. The 10-, 20-and 30-minute DMAsIII yields were significantly higher in reactions catalyzed by AS3MT/M287T as compared to wtAS3MT-catalyzed reactions (p < 0.05). In reaction mixtures that contained TCEP as reductant, addition of GSH also increased methylation rates and DMAsIII yields (Figure S2, Supplementary Data). Notably, methylation rates in TCEP-supported reactions were lower than methylation rates in reaction mixtures containing TR/Trx/NADPH.
Figure 3.
Effects of glutathione (GSH) on methylation of arsenite (iAsIII) catalyzed by wtAS3MT or AS3MT/M287T in the presence of TR/Trx/NADPH. Reaction mixtures contained 100 mM Tris-HCl buffer (pH 7.4), recombinant enzyme (5 μg), 1 mM AdoMet, 1 μM iAsIII, 0.2 μM TR, 10 μM Trx, 300 μM NADPH, and either 0 mM GSH (A,C) or 1 mM GSH (B,D). Reactions mixtures were incubated at 37°C for up to 40 minutes. Yields of MAsIII tMAs, DMAsIII and tDMAs are shown as mean ± SD for n = 3. (Note that the SD bars for all time points are smaller than the symbols marking the corresponding mean values.)
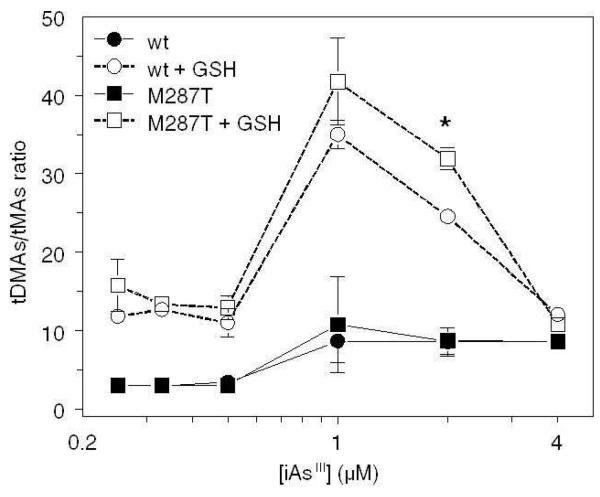
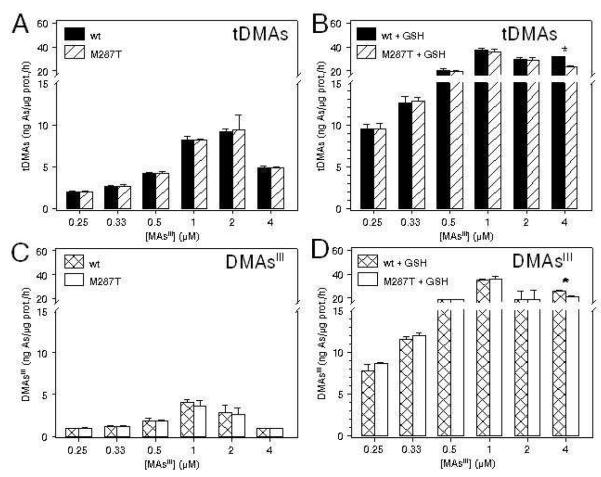
Effect of iAsIII Concentration on wtAS3MT and AS3MT/M287T Activities
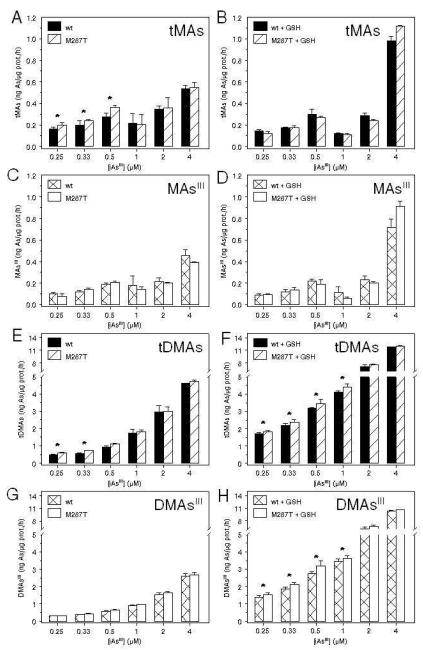
Rates and yields of wtAS3MT-or AS3MT/M287T-catalyzed reactions were examined with or without GSH in reaction mixtures that contained 0.25 to 4 μM iAsIII and TR/Trx/NADPH (Figure 4). Methylation rates increased proportionally with iAsIII concentration for both wtAS3MT-and AS3MT/M287T-catalyzed reactions. In the absence of GSH, DMAsIII and DMAsV were major metabolites; DMAsIII accounted for ~40-60% of tDMAs. At all substrate concentrations, MAsIII and MAsV were minor metabolites. At 0.25 to 0.5 μM iAsIII, the AS3MT/M287T-catalyzed reactions generated significantly more tMAs than reactions catalyzed by wtAS3MT. Addition of 1 mM GSH to reaction mixtures with either enzyme increased methylation rates 2-to 3-fold and increased the fraction of tDMAs present as DMAsIII. Addition of 1 mM GSH to reaction mixtures with either enzyme significantly increased tMAs production in reaction mixtures at low substrate (≤ 1 μM) concentrations. However, with added GSH, AS3MT/M287T-catalyzed reactions at low substrate (≤ 1 μM) concentrations yielded tDMAs and DMAsIII at significantly higher rates than did wtAS3MT-catalyzed reactions. Catalytic efficiencies of wtAS3MT and AS3MT/M287T were compared using tDMAs/tMAs ratios for either enzyme in the absence or presence of GSH (Figure 5). Without added GSH, tDMAs/tMAs ratios for both enzymes remained low and varied only about 2-fold over the range of substrate concentrations. For both enzymes, addition of GSH to reaction mixtures increased tDMAs/tMAs ratios over the range of substrate concentrations. The stimulatory effect of GSH was greatest for 1 and 2 μM iAsIII. At 2 μM iAsIII, tDMAs/tMAs ratios for the enzymes differed significantly (p < 0.05). Similar concentration-dependent increases in tDMA and DMAsIII production were found in wtAS3MT- or AS3MT/M287T-catalyzed reactions that contained 1 mM TCEP as reductant (Figure S3, Supplementary Data). Although tDMA and DMAsIII production rates were lower in the presence of TCEP than of TR/Trx/NADPH, addition of 1 mM GSH stimulated both tMAs and tDMAs production that resulted in decreased tDMAs/tMAs ratios in the methylation reaction when TCEP as reductant (results not shown). In the absence and presence of GSH rates of tMAs and tDMAs production were generally higher in AS3MT/M287T-catalyzed reactions than in wtAS3MT -catalyzed reactions.
Figure 4.
Effect of glutathione (GSH) on rates of wtAS3MT and AS3MT/M287T-catalyzed methylation of arsenite (iAsIII). Reaction mixtures contained 100 mM Tris-HCl buffer (pH 7.4), recombinant enzyme (5 μg), 1 mM AdoMet, 0.2 μM TR, 10 μM Trx, 300 μM NADPH, iAsIII (0.25, 0.33, 0.5, 1, 2 or 4 μM), and either 0 mM GSH (A, C, E, G) or 1 mM GSH (B, D, F, H). Reaction mixtures were incubated at 37°C for 20 minutes. The rates of tMAs, MAsIII, tDMAs and DMAsIII production (ng As/μg protein/hour) shown (Mean ± SD, n = 3). *Rates of methylation by wtAS3MT and AS3MT/M287T are significantly different (p < 0.05).
Figure 5.
Effects of arsenite (iAsIII) concentration and glutathione (GSH) on tDMAs/tMAs ratio. Reaction mixtures contained 100 mM Tris-HCl buffer (pH 7.4), recombinant enzyme (5 μg), 1 mM AdoMet, 0.2 μM TR, 10 μM Trx, 300 μM NADPH, iAsIII (0.25, 0.33, 0.5, 1, 2 or 4 μM) and either 0 or 1 mM GSH. Reaction mixtures were incubated at 37°C for up to 20 minutes. tDMAs/tMAs ratios shown (Mean ± SD, n = 3). *Denotes a significant difference (p < 0.05) between tDMAs/tMAs ratios for reactions catalyzed by wtAS3MT and AS3MT/M287T.
Methylation of MAsIII by wtAS3MT and AS3MT/M287T
Methylation of MAsIII (0.25 – 4 μM) was examined in reaction mixtures containing recombinant wtAS3MT or AS3MT/M287T and a TR/Trx/NADPH coupled system as reductant. The mixtures were incubated at 37°C. Because rates of MAsIII methylation catalyzed by either AS3MT protein much exceeded iAsIII methylation rates, assay conditions were modified to use less protein (1 μg). Both DMAsIII and DMAsV were products of MAsIII methylation (Figure 6). Rates of wtAS3MT- and AS3MT/M287T-catalyzed reactions increased over the 0.25 to 1 μM MAsIII concentration range but remained unchanged or declined at higher concentrations suggesting substrate inhibition. Addition of 1 mM GSH to reaction mixtures increased rates of MAsIII methylation and DMAsIII yields for both proteins (p < 0.05). At MAsIII concentrations ≤ 2 μM, reaction rates in wtAS3MT- or AS3MT/M287T-catalyzed reactions were equally stimulated by GSH. In reaction mixtures that contained 4 μM MAsIII, addition of GSH resulted in significantly higher (p < 0.05) rates of both tDMAs and DMAsIII formation in wtAS3MT-catalyzed reactions than in AS3MT/M287T-catalyzed reactions. With 0.25 to 4 μM MAsIII in TCEP-containing reaction mixtures, methylation profiles with or without GSH addition resembled those found in reaction mixtures containing TR/Trx/NADPH (Figure S4, Supplementary Data). For both proteins, tDMAs production rates in TCEP-containing reaction mixtures were much lower than those in reaction mixtures containing TR/Trx/NADPH but were stimulated by addition of GSH. Notably, inhibition of wtAS3MT-or AS3MT/M287T-catalyzed methylation by 2 and 4 μM MAsIII was more pronounced in the presence of TCEP than in the reaction mixtures containing TR/Trx/NADPH.
Figure 6.
Effect of glutathione (GSH) on methylation of methylarsonous acid (MAsIII) by wtAS3MT and AS3MT/M287T. Reaction mixtures contained 100 mM Tris-HCl buffer (pH 7.4), recombinant enzyme (1 μg), 1 mM AdoMet, 0.2 μM TR, 10 μM Trx, 300 μM NADPH, MAsIII (0.25, 0.33, 0.5, 1, 2 or 4 μM) and either 0 or 1 mM GSH. Reaction mixtures were incubated at 37°C for up to 60 minutes in the absence of GSH (A,C) and for up to 30 minutes in the presence of GSH (B,D). Rates of tDMAs (A,B) and DMAsIII (C,D) production (ng As/μg protein/hour) shown (Mean ± SD, n = 3). *wtAS3MT-and AS3MT/M287T-catalyzed rates of methylation are significantly different (p < 0.05).
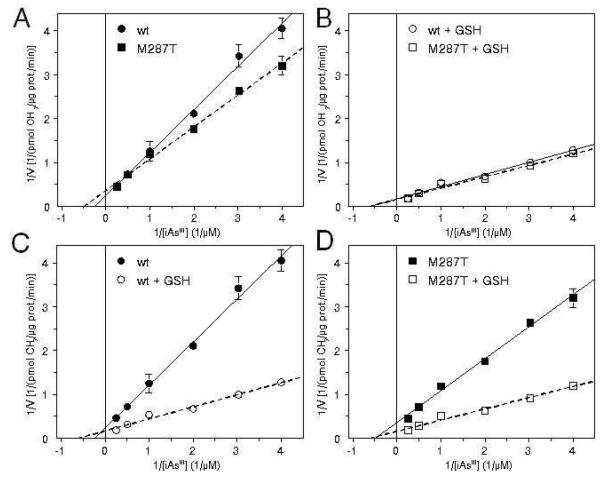
Michaelis-Menten Kinetics for Methylation of iAsIII and MAsIII by wtAS3MT and AS3MT/M287T
Michaelis-Menten constants, Km and Vmax, were calculated for wtAS3MT-and AS3MT/M287T-catalyzed methylation of iAsIII from equations characterizing double-reciprocal plots of 1/v vs 1/[iAsIII] for iAsIII concentrations of 0.25, to 4.0 μM (Figure 7). The reactions were carried out with 5 μg recombinant protein at 37°C. The reaction rate (v) was expressed as pmol of CH3 groups transferred to substrate by 1 μg of recombinant enzyme in 1 minute. Double-reciprocal plots were also used to calculate Km and Vmax for methylation of MAsIII (plots not shown). However, because MAsIII concentrations > 1.0 μM inhibited methylation catalyzed by either enzyme, these plots were calculated from data obtained at lower MAsIII concentrations. In the presence of TR/Trx/NADPH and the absence of GSH, both Km and Vmax values for methylation of iAsIII were significantly lower (p<0.05) for reactions catalyzed by AS3MT/M287T than for reactions catalyzed by wtAS3MT (Table 1). Addition of 1 mM GSH increased Vmax values and decreased Km values for reactions catalyzed by either enzyme. However, in the presence of GSH, Km and Vmax values for wtAS3MT and AS3MT/M287T were not statistically different. Km and Vmax values were also determined for wtAS3MT-and AS3MT/M287T-catalyzed reactions using MAsIII as substrate and 0.5 μg of recombinant protein per assay. The rate of MAsIII methylation at 37°C was significantly higher than the rate of iAsIII methylation under the same conditions. To accurately measure the rate of MAsIII methylation during the linear phase of the reaction, the reaction mixtures were incubated at 25°C. For MAsIII, AS3MT/M287T-catalyzed reactions yielded a significantly higher Km value and a significantly higher Vmax value than did a wtAS3MT-catalyzed reaction (both p < 0.05). Addition of 1 mM GSH to reaction mixtures containing either protein increased Vmax and decreased Km values and minimized differences between these constants in wtAS3MT-and AS3MT/M287T-catalyzed reactions. Notably, Vmax values for MAsIII methylation catalyzed by wtAS3MT and AS3MT/M287T both in the presence and absence of GSH were several-fold higher than corresponding Vmax values for iAsIII methylation. The K value characterizing wtAS3MT-catalyzed activity in the absence of GSH was higher for iAsIII (4.1 μM) than for MAsIII (1.5 μM). In contrast, the AS3MT/M287T-catalyzed activity was characterized by a low Km for iAsIII (2.0 μM) and a high Km for MAsIII (3.0 μM). In the presence of GSH, Km value was significantly lower for MAsIII than for iAsIII for reactions catalyzed by wtAS3MT but not for reactions catalyzed by AS3MT/M287T. Km and Vmax values were also calculated for iAsIII and MAsIII methylation catalyzed by wtAS3MT and AS3MT/M287T with TCEP as reductant (Table S1, Supplementary Data). Here, Km and Vmax values for wtAS3MT-and AS3MT/M287T-catalyzed methylation of iAsIII in the absence of GSH were similar. Addition of 1 mM GSH increased Vmax and decreased Km value by similar margins for both variants. Without addition of GSH, Vmax and Km values for MAsIII methylation were higher for AS3MT/M287T than for wtAS3MT. Addition of GSH decreased Km values for MAsIII methylation by both AS3MT variants, but had little or no effects on Vmax values.
Figure 7.
Double-reciprocal plots of 1/v vs. 1/[iAsIII] for the methylation of arsenite (iAsIII) by wtAS3MT and AS3MT/M287T: Effects of polymorphism on double-reciprocal plots for the methylation reactions in the presence (A) or absence (B) of 1 mM GSH, and effects of 1 mM GSH on double-reciprocal plots for the methylation reactions catalyzed by wtAS3MT (C) and AS3MT/M287T (D) (Mean ± SD, n = 3). For reaction conditions see Figure 4.
Table 1.
Effect of glutathione (GSH) on kinetic constants for methylation of iAsIII and MAsIII catalyzed by recombinant human wild type AS3MT (wt) and its polymorph AS3MT/M287T (M287T).
| Substrate | |||||
|---|---|---|---|---|---|
| iAsIII* | MAsIII** | ||||
| Genotype | GSH | Vmax (pmol CH3/μg prot./min) |
Km (μM) |
Vmax (pmol CH3/μg prot./min) |
Km (μM) |
| wt | 0 | 4.1 ± 0.56b | 4.1 ± 0.74a | 7.3 + 1.33f,§ | 1.5 ± 0.27e,§ |
| M287T | 0 | 2.8 ± 0.16c | 2.0 ± 0.17b | 15.0 ± 1.14d,e,§ | 3.0 ± 0.31d,§ |
| wt | 1 mM | 5.8 ± 0.14a | 1.6 ± 0.05c | 14.3 ± 0.84e,§ | 0.7 ± 0.05f, |
| M287T | 1 mM | 5.9 ± 0.59a | 1.6 ± 0.12c | 16.8 ± 0.43d,§ | 0.9 ± 0.05f |
Reaction mixtures with iAsIII contained 5 μg of recombinant protein, 1 μM AdoMet, and a TR/Trx/NAPDH coupled system and were incubated at 37°C for up to 20 minutes. Mean ± SD values for n = 3 shown.
Reaction mixtures with MAsIII contained 0.5 μg of recombinant protein, 1 μM AdoMet, and a TR/Trx/NAPDH coupled system and were incubated at 25°C for up to 15 minutes. Mean ± SD values for n = 3 are shown.
Different letters indicate statistically significant differences (p <0.05) in Vmax or Km values between wtAS3MT- and AS3MT/M287T-catalyzed methylation of iAsIII in the presence or absence of GSH.
Different letters indicate statistically significant differences (p <0.05) in Vmax or Km values between wtAS3MT- and AS3MT/M287T-catalyzed methylation of MAsIII in the presence or absence of GSH.
Vmax or Km value for methylation of MAsIII is significantly different from the corresponding values for the methylation of iAsIII.
Examination of wtAS3MT and AS3MT/M287T Activities with Pentavalent Arsenicals
Here, the reaction mixtures containing 5 μg of recombinant protein, 1 mM AdoMet, the TR/Trx/NADPH reducing system, and 1 μM iAsV, MAsV or DMAsV were incubated in the presence or absence of 1 mM GSH at 37°C for 2 hours. The reaction mixtures were then analyzed for tri- and pentavalent As species by HG-CT-AAS. None of the pentavalent arsenicals was methylated under these conditions. However, ~0.6% of DMAsV was reduced to DMAsIII in the absence and 7.2% in the presence of GSH. About 0.1-0.2% of MAsV was reduced to MAsIII regardless of GSH presence.
DISCUSSION
The coupled TR/Trx/NADPH system supported methylation by two most common variants of hAS3MT resulting in high methylation rates and high DMAs/MAs ratios in presence of GSH. Both hAS3MT forms catalyzed reactions that converted iAsIII to MAsIII, MAsV, DMAsIII, and DMAsV, and MAsIII to DMAsIII, and DMAsV. The arrays of metabolites produced in reaction mixtures were consistent with those found in urine of individuals exposed to iAs (Valenzuela et al. 2005; Del Razo et al, 2011). Unlike rAs3mt, human wtAS3MT and AS3MT/M287T did not catalyze formation of trimethylated arsenicals.
Catalytic functions of both rAs3mt and hAS3MT are absolutely dependent on a reductant (Hayakawa et al., 2005; Song et al., 2010; Waters et al., 2004a,b). A number of synthetic and physiologically relevant compounds have been shown to support the catalytic activity of both rAs3mt and hAS3MT. The TR/Trx/NADPH system has been most effective in supporting the rAs3mt activity (Waters et al., 2004a,b), but has never been tested as a cofactor of hAS3MT. Although GSH, the most abundant low-molecular weight reductant in mammalian cells, did not support catalysis by rAs3mt (Waters et al., 2004a,b), it did support methylation of iAsIII catalyzed by hAS3MT (Hayakawa et al., 2005; Song et al., 2010). However, with GSH as the sole reductant, rates of hAS3MT-catalyzed methylation were low and MAs, not DMAs, was the prime metabolite. The current study examined reductant requirements of the most common variants of hAS3MT and found that Trx fully supported hAS3MT catalysis. In contrast, GSH was neither necessary nor sufficient to support catalysis by hAS3MT but did modulate the amount and pattern of metabolites. Notably, the in vitro assay conditions used in the present study (the enzyme:substrate ratio, AdoMet and GSH concentrations, pH and temperature) were comparable to those used in previous studies examining iAs methylation by either rAs3mt (Waters et al., 2004a,b) or hAS3MT (Hayakawa et al., 2005; Song et al., 2010).
Catalytically active vicinal cysteines in TR that reduce protein disulfides may reduce highly conserved cysteinyl residues in rAs3mt and hAS3MT that are required for catalytic activity (Sun et al., 1999; Thomas et al, 2007). The targets for reduction by TR could be disulfides generated during oxidative methylation of trivalent As or reduction of pentavalent As to trivalency. TR-coupled arsenate reductases have been identified in several prokaryotes (Messens and Silver 2006). For example, TR is oxidized in the reduction of a disulfide between critical cysteinyl residues in a TR-dependent ArsC arsenate reductase (Roos et al., 2009). A possible linkage between As methylation and the role of TR in AS3MT catalysis occurs through production of MAsIII, a competitive inhibitor of Trx (Ki ~ 100 nM) (Lin et al., 1999). Exposure of cultured primary rat hepatocytes to MAsIII inhibits TR activity and reduces capacity to methylate iAs (Lin et al, 2001). Thus, in the cellular environment, production of methylated metabolites of iAs catalyzed by AS3MT could be modulated by the availability of TRx which is regulated by the TR activity.
In reaction mixtures containing TR/Trx/NADPH or TCEP, GSH modulated hAS3MT activity. The physiological concentration of 1 mM GSH increased overall methylation rates and DMAs/MAs ratio. With added 1 mM GSH, metabolites formed from iAsIII and MAsIII were almost exclusively trivalent methylated arsenicals (MAsIII and DMAsIII). The predominance of trivalent arsenicals could reflect GSH-dependent reduction of pentavalent As to trivalency and formation of stable complexes of GSH and trivalent arsenicals. In addition, complexes of trivalent arsenicals and GSH may serve as substrates for reactions catalyzed by rat As3mt (Waters et al., 2004b) and human AS3MT (Hayakawa et al., 2005). However, the HG-CT-AAS method used in this study cannot differentiate between free trivalent arsenicals and arsenical-GSH complexes and thus, cannot inform on the mechanism (oxidative vs. reductive) of the methylation reactions. Studies with an AsIII S-adenosylmethionine methyltransferase from the thermophilic eukaryotic alga Cyanidioschyzon sp. 5508 (CmArsM) that is orthologous with hAS3MT show that the substrate-GSH complexes iAsIII(GS)3 and MAsIII-(GS)2 are preferred substrates for methylation (Marapakala et al., 2012). Notably, the present study found that even in the presence of a one-thousand fold molar excess of GSH, iAsV, MAsV, or DMAsV were not substrates for methylation by either hAS3MT form. This suggests that neither GSH-dependent reduction of pentavalent arsenicals nor complexation of resulting trivalent arsenicals by GSH was sufficient to provide substrates used for methylation reactions catalyzed by this enzyme. Alternatively, GSH could modulate the activity of hAS3MT by glutathionylation, the formation of mixed disulfides between GSH and protein cysteinyl residues. Glutathionylation of critical cysteinyls in hAS3MT could regulate protein function (Gallogly and Mieyal 2007) by affecting interactions between these residues that contribute to the protein’s catalytic function. Thus, an extensive glutathionylation could be also responsible for the inhibition of AS3MT activity in the reaction assays containing 20 mM GSH (Figure 1).
This study characterized catalysis of As methylation by two major variants of human AS3MT, wtAS3MT and AS3MT/M287T. Differences between the catalytic properties of these two variants depended specifically on presence of GSH in reaction mixtures. Thus, in the presence of TR/Trx/NADPH and GSH, AS3MT/M287T produced more DMAsIII and tDMAs from iAsIII than did wtAS3MT. Although this difference was small, it was statistically significant over the substrate range of 0.25 to 1 μM iAsIII. No differences in DMAsIII and tDMAs yields were found for higher iAsIII concentrations (2 and 4 μM), indicating that the catalytic capacities of both variants were saturated. Consistent with saturation of the second methylation step (i.e., the conversion of MAs to DMAs) was also the sharp increase in MAs yield found in reaction mixture containing 4 μM iAsIII (Figure 4B,D). In contrast, when MAsIII was used as the substrate, tDMAs or DMAsIII production by the AS3MT variants did not differ except at 4 μM MAsIII. This suggests that the two methylation reactions catalyzed by the enzyme are not equivalent in terms of modulation by GSH. The non-equivalency of mono-and dimethylation reactions was also partly reflected by differences in Km and Vmax values obtained with different arsenical substrates. Comparison of wtAS3MT and AS3MT/M287T in reaction mixtures with iAsIII and TR/Trx/NADPH found lower Km and Vmax values for AS3MT/M287T; this is consistent with a higher affinity but a lower capacity for methylation of substrate in AS3MT/M287T-catalyzed reactions in absence of GSH. In reaction mixtures with MAsIII and TR/Trx/NADPH in absence of GSH, the affinity of wtAS3MT was higher than that of AS3MT/M287T, but the capacity to methylate MAsIII was lower. Addition of GSH to reaction mixtures containing TR/Trx/NADPH increased Vmax and decreased Km values and minimized differences between Km and Vmax values in wtAS3MT-and AS3MT/M287T-catalyzed reactions.
The ratio of tDMAs/tMAs in urine has been used as a heuristic indicator of methylation capacity of individuals exposed to iAs and in studies of interindividual variability in susceptibility to adverse health effects associated with chronic iAs exposure. Several previous studies have linked the AS3MT/M287T polymorphism with lower tDMAs/tMAs ratios or higher percentage of MAs in urine (Engström et al., 2007, 2011; Hernández et al., 2008a,b; Lindberg et al., 2007; Valenzuela et al., 2009) and with evidence of increased susceptibility to the toxic effects of iAs (Drobná et al., 2012; Hernández and Marcos, 2008; Sampayo-Reyes et al., 2010; Valenzuela et al., 2009). From these findings, investigators posited that the M287T polymorphism modifies the kinetics of AS3MT-catalyzed methylation so that the concentration of the intermediate tMAs increases relative to the concentration of tDMAs. However, the present study of wtAS3MT and AS3MT/M287T-catalyzed methylation reactions found that the M287T polymorphism had little effect on tDMAs/tMAs ratios. tDMAs/tMAs ratios for wtAS3MT and AS3MT/M287T-catalyzed methylation reactions differed significantly only in reactions containing 2 μM iAsIII and 1 mM GSH (Figure 5). The lack of a consistent effect of genotype on the methylation capacity of AS3MT indicates that differences in the catalytic efficiences of wtAS3MT and AS3MT/M287T do not fully account for differences in tDMAs/tMAs ratios in urine. It is likely that processes affecting the binding and retention of methylated arsenicals in tissues that occur between AS3MT-catalyzed methylation and excretion of methylated metabolites in urine also influence the ratios of these metabolites in urine. A recent genome wide association study supports the concept that multiple genetically determined aspects of iAs metabolism could contribute to the both metabolic and toxicity phenotypes (Pierce et al., 2012). Among these modifiers of arsenic metabolism could be interindividual variation in the control of GSH metabolism. Altered GSH availability could modulate the catalytic activities of both wtAS3MT and AS3MT/M287T.
Although TCEP, a synthetic non-thiol reductant, also supported the AS3MT-catalyzed methylation of iAsIII and MAsIII, lower rates of conversion were found with this reductant than with TR/Trx/NADPH. Notably, addition of GSH to reaction mixtures containing either TR/Trx/NADPH or TCEP as reductant evoked similar changes in the pattern of metabolites and in the kinetic properties of reactions catalyzed by either form of hAS3MT. This suggests that the modulatory action of GSH did not depend upon a specific interaction between reductant and GSH but rather upon its independent effect on catalysis by the protein. Understanding the distinct roles of dithiol reductants and GSH in control of catalysis by hAS3MT will depend on elucidating the structure of the enzyme and the molecular basis of methylation and substrate reduction processes that it catalyses.
In summary, this study found that a common polymorphism in the hAS3MT gene was associated with differences in the pattern of methylated metabolites generated in reaction mixtures that contained a dithiol reductant and variant forms of the enzyme. This provides a plausible biochemical link between hAS3MT genotype and As methylation phenotype as reflected by the pattern of arsenical metabolites in urine (Engstrom et al., 2011). This relation between hAS3MT genotype and phenotype can be extended to a disease susceptibility phenotype. Statistically significant increases in risk of skin lesions or of carotid atherosclerosis have been associated with the AS3MT/M287T polymorphism (Hsieh et al., 2011; Valenzuela et al., 2009) and in children with susceptibility to DNA damage (Sampayo-Reyes et al., 2010). Similarly, individuals with AS3MT/M287T polymorphisms were more likely than wtAS3MT carriers to be diagnosed with diabetes in our recent study in the Zimapan and Lagunera regions (Mexico) where residents are exposed to iAs in drinking water (Del Razo et al., 2011). Consistent with results of the present study, carriers of AS3MT/M287T had higher levels of DMAsIII in urine (Drobná et al., 2012). Taken together, this data suggest that increased production of DMAsIII is at least in part responsible for an increased susceptibility of AS3MT/M287T carriers to chronic iAs toxicity. Thus, understanding the molecular basis of variation in capacity for AS3MT-catalyzed conversion of iAs into its methylated metabolites is a first step in linking genotype to phenotypic variation in disease susceptibility and better evaluation of risk associated with exposure to this metalloid.
Supplementary Material
Highlights.
Human AS3MT and AS3MT(M287T) require a dithiol reductant for optimal activity.
Both enzymes methylate arsenite to tri- and pentavalent methylated metabolites.
Neither enzyme requires glutathione (GSH) to methylate arsenite or methylarsonite.
However, in presence of a dithiol addition of 1mM GSH increases methylation rates.
In presence of 1mM GSH, AS3MT(M287T) produces more dimethylarsinite than AS3MT.
Acknowledgements
This work was supported by NIH grant No. 2 R01 ES010845 to M.S and the UNC Nutrition Obesity Research Center grant No. DK056350. The authors thank Dr. William Cullen (University of British Columbia, Canada) for providing custom synthesized MAsIII and DMAsIII for this study. This manuscript has been reviewed in accordance with the policy of the National Health and Environmental Effects Research Laboratory, U.S. Environmental Protection Agency, and approved for publication. Approval does not signify that the contents necessarily reflect the views and policies of the Agency, nor does mention of trade names or commercial products constitute endorsement or recommendation for use.
Abbreviations
- AAS
atomic absorption spectrometry
- AdoMet
S-adenosylmethionine
- ANOVA
analysis of variance
- As
arsenic
- AsIII
trivalent As
- AsV
pentavalent As
- AS3MT
Arsenic (+3 oxidation state) methyltransferase
- iAs
inorganic arsenic
- iAsIII
arsenite
- iAsV
arsenate
- CT
cryotrapping
- DMAs
dimethylarsenic
- DMAsIII
dimethylarsinous acid
- DMAsV
dimethylarsinic acid
- GSH
glutathione
- hAS3MT
human AS3MT
- HG
hydride generation
- MAs
monomethylarsenic
- MAsIII
methylarsonous acid
- MAsV
methylarsonic acid
- rAs3mt
rat As3mt
- TCEP
tris (2-carboxyethyl) phosphine hydrochloride
- tDMAs
DMAsIII + DMAsV
- tMAs
MAsIII + MAsV
- TMAs
trimethylarsenic
- TMAsIII
trimethylarsine
- TMAsVO
trimethylarsine oxide
- TR
thioredoxin reductase
- Trx
thioredoxin
- Wt
wild-type
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Agusa T, Iwata H, Fujihara J, Kunito T, Takeshita H, Minh TB, Trang PT, Viet PH, Tanabe S. Genetic polymorphisms in AS3MT and arsenic metabolism in residents of the Red River Delta, Vietnam. Toxicol. Appl. Pharmacol. 2009;236:131–141. doi: 10.1016/j.taap.2009.01.015. [DOI] [PubMed] [Google Scholar]
- Challenger F. Biological methylation. Adv. Enzymol. 1951;12:429–491. doi: 10.1002/9780470122570.ch8. [DOI] [PubMed] [Google Scholar]
- Chen B, Arnold LL, Cohen SM, Thomas DJ, Le XC. Mouse arsenic (+3 oxidation state) methyltransferase genotype affects metabolism and tissue dosimetry of arsenicals after arsenite administration in drinking water. Toxicol. Sci. 2011;124:320–326. doi: 10.1093/toxsci/kfr246. [DOI] [PubMed] [Google Scholar]
- Chung CJ, Hsueh YM, Bai CH, Huang YK, Huang YL, Yang MH, Chen CJ. Polymorphisms in arsenic metabolism genes, urinary arsenic methylation profile and cancer. Cancer Causes Control. 2009;20:1653–1661. doi: 10.1007/s10552-009-9413-0. [DOI] [PubMed] [Google Scholar]
- Cullen WR, McBride BC, Reglinski J. The reduction of trimethylarsine oxide to trimethylarsine by thiols: a mechanistic model for the biological reduction of arsenicals. J. Inorg. Biochem. 1984;21:45–60. [Google Scholar]
- Del Razo LM, Styblo M, Cullen WR, Thomas DJ. Determination of trivalent methylated arsenicals in biological matrices. Toxicol. Appl. Pharmacol. 2001;174:282–293. doi: 10.1006/taap.2001.9226. [DOI] [PubMed] [Google Scholar]
- Del Razo LM, García-Vargas GG, Valenzuela OL, Hernandez-Castellanos E, Sánchez-Peña LC, Drobná Z, Loomis, D., Stýblo M. Exposure to arsenic in drinking water is associated with increased prevalence of diabetes: a cross-sectional study in the Zimapán and Lagunera Regions in Mexico. Environ. Health. 2011;10:73. doi: 10.1186/1476-069X-10-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Devesa V, Del Razo LM, Adair B, Drobná Z, Waters SB, Hughes MF, Styblo M, Thomas DJ. Comprehensive analysis of arsenic metabolites by pH-specific hydride generation atomic absorption spectrometry. J. Anal. At. Spectrom. 2004;19:1460–1467. [Google Scholar]
- Drobná Z, Del Razo LM, García-Vargas GG, Valenzuela OL, Hernandez-Castellanos E, Sánchez-Peña LC, Stýblo M, Loomis D. AS3MT polymorphism and prevalence of diabetes among residents of the Zimapan and Lagunera regions in Mexico. J. Expo. Sci. Environ. Epidemiol. 2012 doi: 10.1038/jes.2012.103. (revised ms submitted) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engström KS, Broberg K, Concha G, Nermell B, Warholm M, Vahter M. Genetic polymorphisms influencing arsenic metabolism: Evidence from Argentina. Environ. Health Perpect. 2007;115:599–605. doi: 10.1289/ehp.9734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engström K, Vahter M, Mlakar SJ, Concha G, Nermell B, Raqib R, Cardozo A, Broberg K. Polymorphisms in arsenic(+III oxidation state) methyltransferase (AS3MT) predict gene expression of AS3MT as well as arsenic metabolism. Environ. Health Perspect. 2011;119:182–188. doi: 10.1289/ehp.1002471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujihara J, Fujii Y, Agusa T, Kunito T, Yasuda T, Moritani T, Takeshita H. Ethnic differences in five intronic polymorphisms associated with arsenic metabolism within human arsenic (+3 oxidation state) methyltransferase (AS3MT) gene. Toxicol. Appl. Pharmacol. 2009;234:41–46. doi: 10.1016/j.taap.2008.09.026. [DOI] [PubMed] [Google Scholar]
- Gallogly MM, Mieyal JJ. Mechanisms of reversible protein glutathionylation in redox signaling and oxidative stress. Curr. Opin. Pharmacol. 2007;7:381–391. doi: 10.1016/j.coph.2007.06.003. [DOI] [PubMed] [Google Scholar]
- Hayakawa T, Kobayashi Y, Cui X, Hirano S. A new metabolic pathway of arsenite: arsenite-glutathione complexes are substrates for human arsenic methyltransferase Cyt19. Arch Toxicol. 2005;79:183–191. doi: 10.1007/s00204-004-0620-x. [DOI] [PubMed] [Google Scholar]
- Hernández A, Marcos R. Genetic variations associated with interindividual sensitivity in the response to arsenic exposure. Pharmacogenomics. 2008;9:1113–1132. doi: 10.2217/14622416.9.8.1113. [DOI] [PubMed] [Google Scholar]
- Hernández A, Xamena N, Sekaran C, Tokunaga H, Sampayo-Reyes A, Quinteros D, Creus A, Marcos R. High arsenic metabolic efficiency in AS3MT287Thr allele carriers. Pharmacogenet. Genomics. 2008a;18:349–355. doi: 10.1097/FPC.0b013e3282f7f46b. [DOI] [PubMed] [Google Scholar]
- Hernández A, Xamena N, Surrallés J, Sekaran C, Tokunaga H, Quinteros D, Creus A, Marcos R. Role of the Met(287)Thr polymorphism in the AS3MT gene on the metabolic arsenic profile. Mutat. Res. 2008b;637:80–92. doi: 10.1016/j.mrfmmm.2007.07.004. [DOI] [PubMed] [Google Scholar]
- Hernández-Zavala A, Matoušek T, Drobná Z, Adair BM, Dĕdina J, Thomas DJ, Stýblo M. Speciation of arsenic in biological matrices by automated hydride generation-cryotrapping-atomic absorption spectrometry with multiple microflame quartz tube atomizer (multiatomizer) J. Anal. At. Spectrom. 2008;23:342–351. doi: 10.1039/b706144g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh YC, Lien LM, Chung WT, Hsieh FI, Hsieh PF, Wu MM, Tseng HP, Chiou HY, Chen CJ. Significantly increased risk of carotid atherosclerosis with arsenic exposure and polymorphisms in arsenic metabolism genes. Environ Res. 2011;111:804–810. doi: 10.1016/j.envres.2011.05.003. [DOI] [PubMed] [Google Scholar]
- Hughes MF, Edwards BC, Herbin-Davis KM, Saunders J, Styblo M, Thomas DJ. Arsenic (+3 oxidation state) methyltransferase genotype affects steady-state distribution and clearance of arsenic in arsenate-treated mice. Toxicol. Appl. Pharmacol. 2010;249:217–223. doi: 10.1016/j.taap.2010.09.017. [DOI] [PubMed] [Google Scholar]
- Hwang YH, Chen YH, Su YN, Hsu CC, Chen YH, Yuan TH. Genetic polymorphism of As3MT and delayed urinary DMA excretion after organic arsenic intake from oyster ingestion. J. Environ. Monit. 2010;12:1247–1254. doi: 10.1039/c000844c. [DOI] [PubMed] [Google Scholar]
- Lin S, Cullen WR, Thomas DJ. Methylarsenicals and arsinothiols are potent inhibitors of mouse liver thioredoxin reductase. Chem Res Toxicol. 1999;12:924–930. doi: 10.1021/tx9900775. [DOI] [PubMed] [Google Scholar]
- Lin S, Del Razo LM, Styblo M, Wang C, Cullen WR, Thomas DJ. Arsenicals inhibit thioredoxin reductase in cultured rat hepatocytes. Chem Res Toxicol. 2001;14:305–311. doi: 10.1021/tx0001878. [DOI] [PubMed] [Google Scholar]
- Lin S, Shi Q, Nix FB, Styblo M, Beck MA, Herbin-Davis KM, Hall LL, Simeonsson JB, Thomas DJ. A novel S-Adenosyl-L-methionine: arsenic(III) methyltransferase from rat liver cytosol. J. Biol. Chem. 2002;277:10795–10803. doi: 10.1074/jbc.M110246200. [DOI] [PubMed] [Google Scholar]
- Lindberg AL, Kumar R, Goessler W, Thirumaran R, Gurzau E, Koppova K, Rudnai P, Leonardi G, Fletcher T, Vahter M. Metabolism of low-dose inorganic arsenic in a central European population: influence of sex and genetic polymorphisms. Environ. Health Perspect. 2007;115:1081–1086. doi: 10.1289/ehp.10026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marapakala K, Qin J, Rosen BP. Identification of catalytic residues in the As(III) S-adenosylmethionine methyltransferase. Biochemistry. 2012;51:944–951. doi: 10.1021/bi201500c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matoušek T, Hernández-Zavala A, Svoboda M, Langerová L, Adair BM, Drobná Z, Thomas DJ, Stýblo M, Dĕdina J. Oxidation state specific generation of arsines from methylated arsenicals based on L-cysteine treatment in buffered media for speciation analysis by hydride generation - automated cryotrapping - gas chromatography-atomic absorption spectrometry with the multiatomizer. Spetrochim. Acta Part B. 2008;63:396–406. doi: 10.1016/j.sab.2007.11.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Messens J, Silver S. Arsenate reduction: thiol cascade chemistry with convergent evolution. J Mol Biol. 2006;362:1–17. doi: 10.1016/j.jmb.2006.07.002. [DOI] [PubMed] [Google Scholar]
- Pierce BL, Kibriya MG, Tong L, Jasmine F, Argos M, Roy S, Paul-Brutus R, Rahaman R, Rakibuz-Zaman M, Parvez F, Ahmed A, Quasem I, Hore SK, Alam S, Islam T, Slavkovich V, Gamble MV, Yunus M, Rahman M, Baron JA, Graziano JH, Ahsan H. Genome-wide association study identifies chromosome 10q24.32 variants associated with arsenic metabolism and toxicity phenotypes in Bangladesh. PLoS Genet. 2012;82:e1002522. doi: 10.1371/journal.pgen.1002522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roos G, Foloppe N, Van Laer K, Wyns L, Nilsson L, Geerlings P, Messens J. How thioredoxin dissociates its mixed disulfide. PLoS Comput Biol. 2009;5(8):e1000461. doi: 10.1371/journal.pcbi.1000461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampayo-Reyes A, Hernández A, El-Yamani N, López-Campos C, Mayet-Machado E, Rincón-Castañeda CB, Limones-Aguilar, Mde L, López-Campos JE, de León MB, González-Hernández S, Hinojosa-Garza D, Marcos R. Arsenic induces DNA damage in environmentally exposed Mexican children and adults. Influence of GSTO1 and AS3MT polymorphisms. Toxicol Sci. 2010;117:63–71. doi: 10.1093/toxsci/kfq173. [DOI] [PubMed] [Google Scholar]
- Song X, Geng Z, Zhu J, Li C, Hu X, Bian N, Zhang X, Wang Z. Structure-function roles of four cysteine residues in the human arsenic (+3 oxidation state) methyltransferase (hAS3MT) by site-directed mutagenesis. Chem Biol Interact. 2009;179:321–328. doi: 10.1016/j.cbi.2008.12.018. [DOI] [PubMed] [Google Scholar]
- Song X, Geng Z, Li X, Hu X, Bian N, Zhang X, Wang Z. New insights into the mechanism of arsenite methylation with the recombinant human arsenic (+3) methyltransferase (hAS3MT) Biochimie. 2010;92:1397–1406. doi: 10.1016/j.biochi.2010.07.002. [DOI] [PubMed] [Google Scholar]
- Styblo M, Delnomdedieu M, Thomas DJ. Biological mechanisms and toxicological consequences of the methylation of arsenic. In: Cherian MG, Goyer RA, editors. Toxicology of Metals-Biochemical Aspects, Handbook of Experimental Pharmacology. vol. 115. Springer Verlag; Berlin: 1995. pp. 407–433. [Google Scholar]
- Sun QA, Wu Y, Zappacosta F, Jeang KT, Lee BJ, Hatfield DL, Gladyshev VN. Redox regulation of cell signaling by selenocysteine in mammalian thioredoxin reductases. J. Biol. Chem. 1999;274:24522–24530. doi: 10.1074/jbc.274.35.24522. [DOI] [PubMed] [Google Scholar]
- Thomas DJ, Styblo M, Lin S. The cellular metabolism and systemic toxicity of arsenic. Toxicol. Appl. Pharmacol. 2001;176:127–144. doi: 10.1006/taap.2001.9258. [DOI] [PubMed] [Google Scholar]
- Thomas DJ, Li J, Waters SB, Xing W, Adair BM, Drobna Z, Devesa V, Styblo M. Arsenic (+3 oxidation state) methyltransferase and methylation of arsenicals. Exp. Biol. Med. 2007;232:3–13. [PMC free article] [PubMed] [Google Scholar]
- Vahter M. Methylation of inorganic arsenic in different mammalian species and population groups. Sci. Prog. 1999;82:69–88. doi: 10.1177/003685049908200104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela OL, Borja-Aburto VH, Garcia-Vargas GG, Cruz MB, Calderon-Aranda EM, Del Razo LM. Urinary trivalent methylated arsenic species in population chronically exposed to inorganic arsenic. Environ. Heath Perspect. 2005;113:250–254. doi: 10.1289/ehp.7519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valenzuela OL, Drobná Z, Hernández-Castellanos E, Sánchez-Peña LC, García-Vargas GG, Borja-Aburto VH, Stýblo M, Del Razo LM. Association of AS3MT polymorphisms and the risk of premalignant arsenic skin lesions. Toxicol. Appl. Pharmacol. 2009;239:200–207. doi: 10.1016/j.taap.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters SB, Devesa-Perez V, Del Razo LM, Styblo M, Thomas DJ. Endogenous reductants support catalytic function of recombinant rat cyt19, an arsenic methyltransferase. Chem. Res. Toxicol. 2004a;17:404–409. doi: 10.1021/tx0342161. [DOI] [PubMed] [Google Scholar]
- Waters SB, Devesa V, Fricke MW, Creed JT, Styblo M, Thomas DJ. Glutathione modulates recombinant rat arsenic (+3 oxidation state) methyltransferase-catalyzed formation of trimethylarsine oxide and trimethylarsine. Chem. Res. Toxicol. 2004b;17:1621–1629. doi: 10.1021/tx0497853. [DOI] [PubMed] [Google Scholar]
- Wood TC, Salavagionne OE, Mukherjee B, Wang L, Klumpp AF, Thomae BA, Eckloff BW, Schaid DJ, Wieben ED, Weinshilboum RM. Human arsenic methyltransferase (AS3MT) pharmacogenetics. Gene resequencing and functional genomics studies. J. Biol. Chem. 2006;281:7364–7373. doi: 10.1074/jbc.M512227200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.