Abstract
A previously unidentified gonadotropin-regulated long chain acyl-CoA synthetase (GR-LACS) was cloned and characterized as a 79-kDa cytoplasmic protein expressed in Leydig cells of the rat testis. GR-LACS shares sequence identity with two conserved regions of the LACS and luciferase families, including the ATP/AMP binding domain and the 25-aa fatty acyl-CoA synthetase signature motif, but displays low overall amino acid similarities (23–28%). GR-LACS mRNA is expressed abundantly in Leydig cells of the adult testis and to a lesser degree in the seminiferous tubules in spermatogonia and Sertoli cells. It is also observed in ovary and brain. Immunoreactive protein expression was observed mainly in Leydig cells and minimally in the tubules but was not detected in other tissues. In vivo, treatment with a desensitizing dose of human chorionic gonadotropin caused transcriptional down-regulation of GR-LACS expression in Leydig cells. The expressed protein present in the cytoplasm of transfected cells displayed acyl-CoA synthetase activity for long chain fatty acid substrates. GR-LACS may contribute to the provision of energy requirements and to the biosynthesis of steroid precursors and could participate through acyl-CoA's multiple functions in the regulation of the male gonad.
Luteinizing hormone (LH) supports steroidogenesis and maintains testicular and ovarian function (1). Mediators of LH action exert homologous regulation of receptors, steroidogenic enzymes, and other genes of the Leydig cell. In contrast to the physiological concentrations of gonadotropins that maintain steroidogenesis and gonadal LH/human chorionic gonadotropin (hCG) and prolactin receptors, high exogenous concentrations of hCG down-regulate receptors and steroidogenic enzymes of Leydig cells at steps beyond pregnenolone formation (2–7). The impairment of steroidogenesis that follows the initial receptor-mediated activation is independent of receptor down-regulation and results from changes at the transcriptional levels that cause a marked reduction of enzyme expression. Recently, we cloned a gonadotropin-regulated testicular RNA helicase that is present in Leydig cells and meiotic germinal cells of the testis (8). This protein was found to be markedly up-regulated by hCG by means of cAMP-induced androgen formation in Leydig cells at hCG doses that cause down-regulation of steroidogenic enzymes and receptors.
In a further search for genes regulated by gonadotropin, we have isolated a previously uncharacterized protein that is down-regulated by hCG and is termed gonadotropin-regulated long chain acyl-CoA synthetase (GR-LACS). This member of the LACS family is highly abundant in the Leydig cell and is distinct from the previously characterized mammalian forms of the enzyme (9–13). Its constitutive expression in Leydig cells is negatively regulated by the receptor-mediated action of LH. In addition to its potential contributions to energy production and testicular steroidogenesis, GR-LACS could provide long chain acyl-CoA esters with regulatory effects on enzyme activity, membrane function, and gene expression.
Materials and Methods
Animal Treatment and Cell Preparation.
The gonadotropin-induced down-regulation of LH receptor and steroidogenic enzymes in Leydig cells was produced as described (3) by administration of an s.c. injection of 2.5 μg of hCG to adult male rats. Animals were killed at various times after treatment, and the testes were removed. Leydig cells were prepared by collagenase dispersion and purified by centrifugal elutriation (14). The seminiferous tubules were minced, and the germ cells were collected by centrifugation. Cells were processed for RNA and protein analysis.
Differential Display Analysis, Isolation, and Cloning of GR-LACS.
Total RNA samples prepared from Leydig cells of control and hCG-treated rats were analyzed by differential display technology (8). PCR products with incorporated [33P]dATP (2,000 Ci/mmol, 1 Ci = 37 GBq; DuPont/NEN) were separated in nondenatured 6% sequencing gels. A 3′-untranslated region fragment derived from differential display analysis was used as a probe for screening a rat Leydig cell cDNA library in ZAP Express vector (Stratagene) to obtain full-length cDNA.
Chromosomal Localization.
Chromosomal mapping (rat, mouse, and human lymphocytes) was performed by fluorescence in situ hybridization by using GR-LACS biotinylated full-length cDNA (2.7 kb; ref. 8).
RNase Protection Assay.
RNase protection assays were performed by established methodology (15). The GR-LACS (nucleotides 1,165–1,510) and a rat β-actin cRNA probe were generated by PCR followed by subcloning and in vitro transcription (8). Total RNA (10 μg) samples were applied for hybridization. The protected fragments resolved on 6% sequencing gels were quantified by PhosphorImager analysis.
Northern Blot, mRNA Stability, and Nuclear Run-Off Assay.
mRNA was extracted from rat Leydig cells and from various male and female rat tissues by using Fast-Track mRNA kits (Invitrogen). The mRNA samples (5 μg) were resolved on 1% agarose gels and hybridized with 32P-labeled rat GR-LACS cDNA probe. To investigate the stability of the GR-LACS mRNA, cells were incubated with 10 μg/ml actinomycin D (0–10 h), and mRNA samples were resolved as described above. Nuclear run-off assays were performed as described (16).
In Situ Hybridization Analysis of GR-LACS mRNA in Rat Testis.
In situ hybridization was carried out by following standard protocols (17). Testes were fixed in 4% (vol/vol) paraformaldehyde for 16 h at 22°C. Slides were hybridized to 35S-labeled GR-LACS cRNA (nucleotides 824-2689) probes (sense and antisense) for 16 h at 45°C and processed as described (8).
Western Blot and Immunocytochemistry Analysis.
Western blot and immunocytochemical analyses were performed by using a rabbit polyclonal antibody raised to a GR-LACS peptide (amino acids 16–28). Immunosignals were detected by using an enhanced Chemiluminescent system (Amersham Pharmacia). GR-LACS values were normalized by the corresponding β-actin signals. For immunolocalization of GR-LACS, testes from adult rats were fixed in 4% (vol/vol) paraformaldehyde and embedded in paraffin. Serial sections were used for immunostaining with a peroxidase-labeled avidin-biotin detection system (18).
Subcellular Localization of Rat GR-LACS in Mouse Leydig Tumor Cells (mLTC) and COS-1 Cells by Using Confocal Microscopy.
The full length of the GR-LACS cDNA-coding region subcloned into the pEGFP-N2 vector [GR-LACS-green fluorescent protein (GFP)] or pGFP vector only (2 μg/ml) was transfected into cells with Lipofectamine Plus (GIBCO/BRL). After a 24-h incubation, the cells were examined in an inverted microscope or harvested for Western blot analyses by using a GFP monoclonal antibody (CLONTECH).
In Vitro Transcription/Translation of GR-LACS cDNA.
The full length of GR-LACS cDNA (1 μg) in pBK CMV vector (pBK-GR-LACS) was transcribed and translated in vitro into protein with a Reticulocyte Lysate System (Promega). The empty vector (pBK) was used as the negative control. The 35S-labeled translated protein samples were resolved on 10% SDS polyacrylamide gels.
Analysis of Fatty Acyl-CoA Synthetase (FACS) Activity.
COS-1 cells were transfected with pBK-GR-LACS or pBK. FACS activities were determined in cell extracts as described (19) by using the following radiolabeled fatty acids: [14C]decanoic, [3H]myristic, [14C]lignoceric, [3H]arachidonic, [3H]eicosapentanoic (American Radiolabeled Chemicals, St. Louis), [3H]palmitic, and [3H]oleic (Amersham Pharmacia). The labeled acyl-CoAs were quantitated and normalized by the protein concentration. All experiments were performed three times in triplicate.
Results
Identification, Isolation, and Cloning of a Gonadotropin DownRegulated Expression Protein.
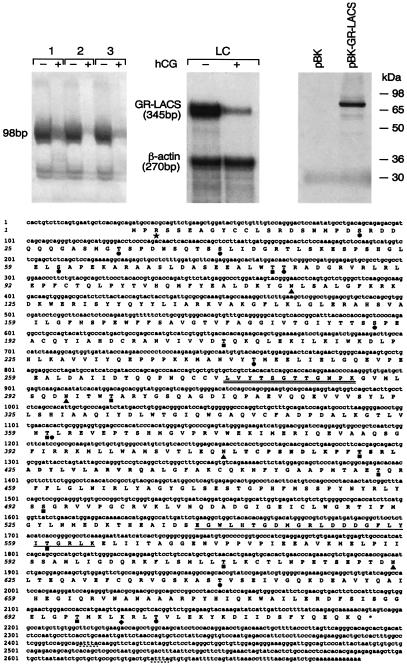
Genes regulated by gonadotropin were obtained by the analysis of differential display patterns of RNA obtained from Leydig cells of rats treated with a single desensitizing dose of hCG (2.5 μg) or of vehicle alone 24 h before killing (8). PCR amplifications using primers 5′-dT11GC-3′ and 5′-CTGCTTGATG-3′ revealed a 98-bp fragment down-regulated by hCG (Fig. 1 Upper Left). RNase protection assays using the 345-bp fragment derived from the coding region of GR-LACS as a probe confirmed that this gene was down-regulated by hCG in testicular Leydig cells (Fig. 1 Upper Center). The protected fragment of β-actin mRNA (270 bp) showed no change.
Figure 1.
(Upper) Down-regulation of GR-LACS by hCG identified by mRNA differential display analysis (Left), confirmed by RNase protection assay in rat testicular Leydig cell mRNA (Center), and in vitro translation of GR-LACS (Right). (Upper left) Differential display analysis of three sets of total RNA samples from Leydig cells of adult rats treated 24 h earlier with a 2.5-μg dose of hCG (+) or vehicle alone (−). (Upper Center) Autoradiography of RNase-protected GR-LACS fragments of 345 bp and a β-actin fragment (270 bp). (Upper Right) The full length of GR-LACS cDNA in pBK-CMV vector (pBK-GR-LACS) or vector only (pBK) was transcribed/translated in vitro. (Lower) GR-LACS cDNA nucleotide, deduced amino acid sequences, and putative functional domains. Consensus sequence domains are indicated below. Double underlined, AMP-binding domain; single underlined, FACS signature motif; ▴, N-glycosylation. Phosphorylation sites: ♦, cAMP/cGMP-dependent protein kinase; ■, protein kinase C; ●, casein kinase II; ★, tyrosine kinase; dash underlined, mRNA destabilizing signals.
To isolate the cDNA sequence of this hCG regulated mRNA, the 32P-labeled 3′-untranslated region PCR fragment was used as a probe to screen a rat Leydig cell cDNA library. The full-length cDNA sequence of 2,689 bp contained an ORF encoding 721 amino acids (Fig. 1 Lower; accession no. AF208125). The size of the nucleotide sequence is consistent with the transcript size (2.7 kb) revealed by Northern blot analysis (Fig. 2). The in vitro transcribed/translated recombinant full-length cDNA of GR-LACS yielded a protein of 79 kDa (Fig. 1 Upper Right), which was in agreement with the expected size from the deduced amino acid sequence.
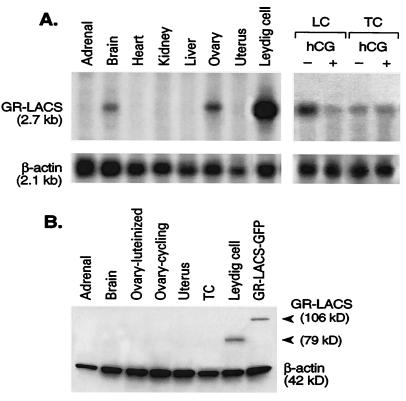
Figure 2.
Northern and Western analyses of GR-LACS gene expression. (A) Northern analysis. mRNA samples isolated from several tissues of adult male rats, the uterus and ovaries of cycling (ovary) female rats, as well as Leydig cells (LC) and testicular tubule cells (TC) obtained from adult rats treated with 2.5 μg of hCG (+) or control (−). Samples were hybridized by using GR-LACS and β-actin cDNA probes. (B) Tissue-specific expression of GR-LACS protein and transient expression of GR-LACS-GFP fusion protein in COS cells were analyzed by Western blot analysis with a specific GR-LACS antibody. kD, kilodalton.
A domain search of GR-LACS protein with the prosite program revealed a putative AMP-binding domain signature (amino acids 276–287). Also, it contained 4 potential N-glycosylation sites, a cAMP/cGMP-dependent protein kinase phosphorylation site, 10 protein kinase C phosphorylation sites, 11 casein kinase II phosphorylation sites, 1 tyrosine kinase phosphorylation site, and 2 potential mRNA destabilizing signals (Fig. 1 Lower). A database search with the fasta software (GCG) revealed an overall amino acid identity of 23–28% with LACSs of several species (9, 10, 12, 13, 20–23). A pileup comparison revealed that GR-LACS contains two regions that are highly conserved in LACSs and luciferases from different species. The most conserved region corresponds to amino acids 276–288 “LxxTSGTTGNPKG” and contains a putative AMP-binding domain. The second conserved region (amino acids 540–564) contains the FACS signature motif (24) of 25 amino acids with the 8 highly conserved amino acid residues (amino acids 540–547). Also, a neighboring sequence with conserved glycine residues in the adjacent region (GExxxxGxxxxxGY, amino acids 513–526) was found in GR-LACS as in other members of this superfamily.
Chromosomal Localization of GR-LACS.
Chromosomal mapping with human, mouse, and rat lymphocytes demonstrated that this gene resides in chromosomes 8q22-24 of rat, 9BD of mouse, and 15q23-24 of human (not shown).
Tissue Distribution of GR-LACS mRNA.
Northern blot analysis detected a single transcript of 2.7 kb in testis, ovary, and brain but not in uterus and the other tissues examined (Fig. 2A Left). The abundance of GR-LACS mRNA was relatively higher in testis than in other tissues. GR-LACS mRNA was predominantly expressed in Leydig cells and, to a lesser extent, in germ cells of adult testis. hCG-induced down-regulation of GR-LACS mRNA was observed only in Leydig cells where LH/hCG receptors are present but not in germ cells (Fig. 2A Right). Western blot analysis with a specific polyclonal antibody revealed the presence of GR-LACS protein in Leydig cells, but not in testicular tubule cells or in other tissues. The antibody also detected the expressed GR-LACS-GFP fusion protein in transiently transfected COS-1 cells (Fig. 2B).
Cellular Localization of GR-LACS mRNA and Protein Expression in Testis.
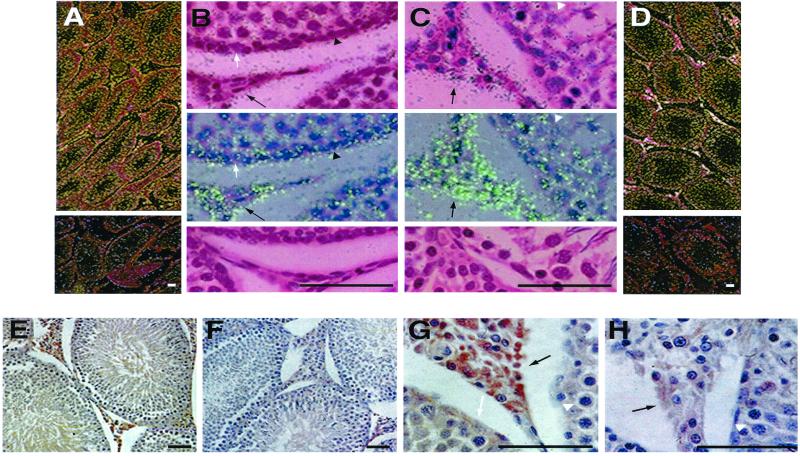
Cellular localization of GR-LACS mRNA within the rat testicular compartments (Fig. 3 A–D) revealed that GR-LACS transcripts were present predominantly in Leydig cells and more sparsely in the tubular compartment. Most of the interstitial cells showed strong signals in 46-day-old animals (Fig. 3 C and D) and less intensity in 30-day-old animals (Fig. 3 A and B). Within the seminiferous tubules, labeling was observed primarily in spermatogonia but also in spermatocytes and Sertoli cells. The immunohistochemistry study revealed that GR-LACS protein was present in interstitial cells of the testis, whereas it was minimally detected in the tubules (Fig. 3 E–H).
Figure 3.
Cellular localization of GR-LACS gene expression in the rat testis as revealed by in situ hybridization and immunostaining. (A and D) In situ hybridization of GR-LACS mRNA in tissue sections of rat testes photographed by dark field microscopy. Testis sections derived from pubertal, day 30 (A) and adult, day 46 (D) were incubated with antisense probe and showed strong positive signals (bright white granules) in interstitial cells and less intense signals in tubule cells. Sense controls (Lower A and D) were negative. (B and C) Higher magnification: day 30 (B) and day 46 (C). (B and C Top) Bright field (dark granules); (B and C Middle) epi-illumination/bright field microscopy (green granules); and (B and C Bottom) the negative sense controls. Dark arrow-head, spermatogonia; white arrow, Sertoli cell; and black arrow, Leydig cell. (Bar = 50 μm.) (E–H) Immunohistochemistry studies of GR-LACS protein in adult rat testis. (E and G) Positive immunostaining (brown signal) was noted almost exclusively in Leydig cells (black arrow) and only traces were seen in the tubules. (F and H) Negative controls. (Bar = 50 μm.)
Gonadotropin Induced Down-Regulation of GR-LACS Gene Expression.
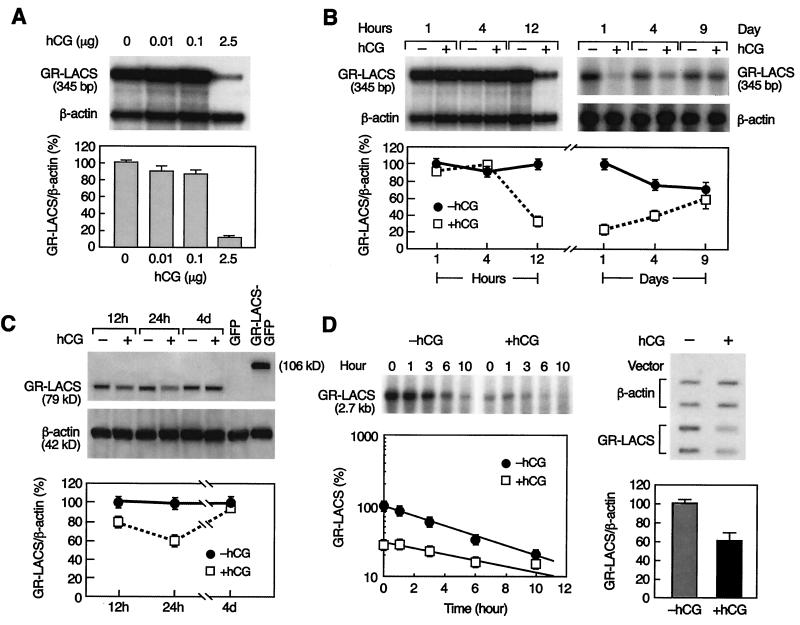
Further studies were conducted to gain insights into the characteristics of the regulatory mechanism(s) of hCG-mediated GR-LACS mRNA down-regulation. Only the high dose of hCG (2.5 μg) induced a marked down-regulation of GR-LACS mRNA (Fig. 4A). This desensitizing dose was used in the time course shown in Fig. 4B. No significant changes in GR-LACS mRNA levels were observed at 1 and 4 h, whereas these levels were significantly reduced at 12 h, reached maximum depression at 24 h, and returned to control levels 9 days after hCG treatment. Western blot analyses showed initial down-regulation of rat Leydig cell GR-LACS protein at 12 h (82% ± 4% of control) with a further decrease at 24 h (63% ± 3% of control) after treatment. Complete recovery was observed at 4 days after hCG treatment at the time when mRNA expression began to recover (Fig. 4C).
Figure 4.
Dose-dependent (A) and time-course studies of GR-LACS mRNA (B), protein (C) regulated by hCG in rat Leydig cells, and stability of GR-LACS mRNA (D Left) and nuclear run-off assay (D Right). RNase protection assays after 24 h in vivo treatment with increasing concentrations of hCG (A) or at various times after a 2.5-μg injection of hCG (B). (C) Western analysis. (D) Northern blot analysis of mRNA (Left) from Leydig cells obtained 12 h after in vivo treatment with 2.5 μg of hCG and incubated with actinomycin D for 0–10 h. Nuclei from control and hCG-desensitized Leydig cells were analyzed by nuclear run-off assay (Right). Autoradiographs (Upper) quantified by PhosphorImager (A–D Lower) were normalized by β-actin (A–C). Mean ± SEM, relative to control. kD, kilodalton.
To explore whether the decrease of GR-LACS mRNA induced by hCG was related to changes in mRNA stability, we assessed GR-LACS mRNA degradation rates. The half-life of GR-LACS mRNA in the hCG-treated group was 6.5 h; in the nontreated group, it was 5.5 h (Fig. 4D Left). This result indicated that down-regulation of GR-LACS mRNA in Leydig cells was not caused by the increase of the mRNA degradation by hCG but rather by a change in transcriptional activity. Nuclear run-off assays demonstrated that newly synthesized GR-LACS 32P-labeled Leydig cell mRNA from hCG-treated animals was significantly decreased when compared with nontreated controls, whereas β-actin remained unchanged (Fig. 4D Right).
Localization and Functional Analyses of GR-LACS Protein Expressed in Transfected Cells.
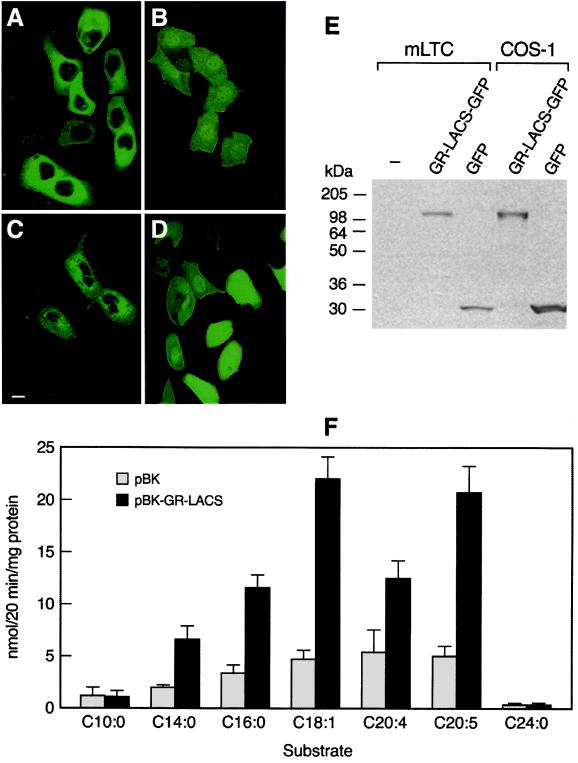
Subcellular localization of transfected GR-LACS tagged with GFP (GR-LACS-GFP) in mLTC and COS-1 cells by confocal microscopy demonstrated that it was localized exclusively in the cytoplasm of both cell lines (Fig. 5 A and C), whereas GFP (control) was present throughout the nuclei, membranes, and cytoplasm (Fig. 5 B and D). The specificity and expression of GR-LACS-GFP were assessed by Western blotting (Fig. 5E). A single 106-kDa band corresponding to the GR-LACS-GFP fusion protein was detected in both mLTC and COS-1 cells transfected with GR-LACS-GFP, whereas a 27-kDa GFP band was detected in GFP-transfected cells.
Figure 5.
(A–D) Subcellular localization of GR-LACS by confocal microscope. (E) Western analysis. (F) Acyl-CoA synthetase activities. Expression of GR-LACS-GFP (A and C) or GFP control (B and D) in mLTC (A and B) and COS-1 (C and D) was examined by confocal microscope. (Bar = 5 μm.) (E) Western blot analysis. Cells transfected with GR-LACS-GFP and GFP or untransfected (−) were analyzed by Western blotting with GFP monoclonal antibody. (F) Acyl-CoA synthetase activity of transfected COS cells. Activity of extracts from cells transfected with pBK (basal activity) or pBK-GR-LACS (conferred activity) assessed by using various fatty acids as substrates. Mean ± SEM, n = 4.
GR-LACS transfection increased myristoyl-CoA (14:0), palmitoyl-CoA (16:0), oleoyl-CoA (18:1), arachidonyl-CoA (20:4), and eicosapentanoyl-CoA (20:5) synthetase activities up to 2-fold over basal activity (P < 0.01). A gradual increase in basal activity was observed with the increasing chain length from C:14 to C:18 (Fig. 5F). No differences in decanoyl-CoA (10:0) synthetase activities were observed between extracts from basal (pBK) and pBK-GR-LACS-transfected cells. Minimal endogenous lignoceroyl-CoA (24:0) synthetase activity was observed in pBK-transfected COS-1 cells, and GR-LACS-pBK transfection failed to confer additional activity. These data demonstrate that GR-LACS has acyl-CoA synthetase activity for saturated and unsaturated long chain fatty acids but displays no activity for the very long chain fatty acid (lignoceric acid, C24:0) and a medium chain fatty acid (decanoic acid, C10:0).
Discussion
This study has identified and characterized a previously uncharacterized gene termed GR-LACS that possesses structural and functional characteristics of FACS and is transcriptionally down-regulated by gonadotropin in Leydig cells of the rat testis. Although GR-LACS shares low overall amino acid sequence similarity (23–28%) with other members of the LACS (9–11) and luciferase families (22, 23), it contains the two conserved functional motifs that are common to these and related enzymes. One of the regions conserved in GR-LACS, LACSs (9–11), and luciferases (22, 23) is the putative domain for the ATP-dependent covalent binding of AMP during the first reaction. The other domain is the 25-aa consensus sequence present in all FACSs; it is recognized as the FACS signature motif [1-DGWLHTGDIGXWXPXGXLKIIDRKK25] (24). This region of GR-LACS contains 13 identical and 2 highly conserved amino acids. A single conserved substitution (E for D) is present at amino acid 1 of the invariant amino acid region (amino acids 1–8) of this motif. This conservative substitution is also present in firefly luciferase (22, 23). Furthermore, amino acids 16, 20, and 23 (G, I, and R), which are important for enzymatic activity, are invariant in GR-LACS. The presence in this protein of a putative AMP/ATP-binding domain of the family of adenylate-forming enzymes, the FACS signature motif and the functional expression of activities for long chain fatty acids demonstrate that GR-LACS is a distinct member of the FACS family.
GR-LACS mRNA is highly expressed in Leydig cells of the pubertal and adult testis and to a lesser extent in the germinal and Sertoli cells. GR-LACS mRNA was also present in the ovary and brain. Immunoreactive GR-LACS protein was expressed abundantly in the Leydig cell and minimally in the seminiferous tubule and was not detected in other tissues. The lack of expression of GR-LACS protein at sites where its mRNA is present could result from either the presence of very low levels of this protein or lack of translation of this mRNA form. Alternatively, the protein may be an alternatively spliced variant form or the product of the use of an alternate initiation–translation site. The fact that a single mRNA species was detected in this study does not preclude the presence of additional mRNA species within the 2.7-kb range of the isolated GR-LACS that could result from alternative splicing. There is a methionine residue (amino acid 31) within the N terminus that could provide an alternative initiation site of translation. In both cases, the protein product may not be recognizable by the specific peptide antibody to the N-terminal region of GR-LACS (amino acids 16–28).
The decrease in Leydig cell GR-LACS mRNA at 12 h after hCG treatment followed the down-regulation of the steroidogenic enzymes, 3β-hydroxysteroid dehydrogenase I (1 h) and II (4 h; ref. 2), and 17α-hydroxylase (3 h; ref. 6), and was concurrent with that of 17β-hydroxysteroid dehydrogenase (7), LH, and prolactin receptors. The down-regulation of GR-LACS mRNA was concurrent with the up-regulation of gonadotropin-regulated testicular RNA helicase (GRTH), a recently isolated testicular helicase that is increased by gonadotropin treatment (8). It is of interest that the significant decrease in mRNA levels at 12 h was not paralleled by changes in protein expression, which was reduced at 24 h of hCG treatment at the time when the mRNA was maximally reduced and recovered (i.e., at 4 days, when GR-LACS mRNA began to increase). This observation is in contrast to previous findings with steroidogenic enzymes, in which, for the most part, changes in mRNA were comparable with those of protein expression (2, 6, 7). These results indicate that the GR-LACS protein is efficiently translated. Whether this discrepancy relates to the increased levels of the GRTH (8) remains to be determined.
The temporal profile and mode of GR-LACS gene regulation by gonadotropin (after or concurrent rather than preceding reduction) indicated that the protein product is unlikely to be a transcriptional regulator of steroidogenic enzymes and LH receptors. Confocal microscopic analysis revealed an exclusively cytoplasmic localization of the expressed protein in transfected cells. Furthermore, transfection of GR-LACS cDNA into cells did not affect the activity of cotransfected receptor and steroidogenic enzyme promoters (not shown). Thus, the available evidence indicates that the observed changes on GR-LACS do not contribute to the down-regulation of steroidogenic enzymes and receptors (2, 4, 6, 7). The transcriptional down-regulation of GR-LACS induced by hCG may result from the direct or indirect nuclear actions of mediators of the trophic hormone stimulus that precede the phase of down-regulation. In contrast to recent findings demonstrating the regulation of the gonadotropin-regulated testicular RNA helicase by androgen, our studies have excluded its participation in the regulation of GR-LACS (not shown).
FACSs play a major role in intermediary metabolism of fatty acid pathways by catalyzing the activation of fatty acyl-CoA thioesters. These enzymes have a central role in cellular homeostasis through their formation of fatty acyl-CoA products that are substrates for β-oxidation and phospholipid biosynthesis. They are also involved in protein transport, enzyme activation, protein acylation, cell signaling, and transcriptional regulation (25, 26). Long chain fatty acids (C12–C20) are activated by LACSs present in the mitochondria and peroxisomes where they undergo β-oxidation and in the microsomes where they are used in the synthesis of complex lipids. GR-LACS displayed enzymatic activity for long chain fatty acids and was shown not to possess very long chain fatty acid activity (>C20) in transfected COS cells.
The mRNA of the GR-LACS gene and its protein, which are down-regulated by gonadotropin, are highly expressed in Leydig cells of pubertal and adult animals. In these cells, androgen biosynthesis is active, depends highly upon the availability of steroid precursors, and has a high energy requirement. Both of these requirements may be contributed by the acyl-CoA synthetase activity of GR-LACS. In the Leydig cell, cholesterol is generated from endogenous precursors rather than derived from circulating lipoproteins, which is the case in other steroidogenic tissues (27). For this reason, GR-LACS potentially could be of importance in the maintenance of adequate cholesterol pools required for steroid biosynthesis in the testis. It is clear that the reduction of GR-LACS gene expression induced by hCG (2.5 μg) does not affect the endogenous accumulation of pregnenolone (3), indicating the adequate availability of endogenous cholesterol for pregnenolone formation in the Leydig cell during down-regulation. The previously observed reduction of testosterone production by this hCG-treatment dose seems to result from the reduced transcription of enzymes distal to pregnenolone formation (2, 3, 6, 7).
These studies have identified and characterized a form of acyl-CoA synthetase that is gonadotropin-regulated in the Leydig cell and displays preferential activity for long chain fatty acids. This form of LACS is distinct from other reported LACS enzymes expressed in the rat brain, liver, adrenal glands, and gonads (20–30% aa identity; refs. 9, 10, 12 and 13). This previously uncharacterized LACS protein may exert multiple actions at specific cellular sites through its acyl-CoA intermediates to fulfill steroidogenic and other functional requirements of testicular Leydig cells.
Abbreviations
- LH
luteinizing hormone
- FACS
fatty acyl-CoA synthetase
- hCG
human chorionic gonadotropin
- GR-LACS
gonadotropin-regulated long chain acyl-CoA synthetase
- GFP
green fluorescent protein
- mLTC
mouse Leydig tumor cells
Footnotes
This paper was submitted directly (Track II) to the PNAS office.
Data deposition: The sequence reported in this paper has been deposited in the GenBank database (accession no. AF208125).
References
- 1.Dufau M L. Annu Rev Physiol. 1988;50:483–508. doi: 10.1146/annurev.ph.50.030188.002411. [DOI] [PubMed] [Google Scholar]
- 2.Tang P Z, Tsai-Morris C H, Dufau M L. Endocrinology. 1998;139:4496–4505. doi: 10.1210/endo.139.11.6316. [DOI] [PubMed] [Google Scholar]
- 3.Cigorraga S B, Dufau M L, Catt K J. J Biol Chem. 1978;253:4297–4304. [PubMed] [Google Scholar]
- 4.Dufau M L, Cigorraga S B, Baukal A J, Bator J M, Sorrell S H, Neubauer J F, Catt K J. J Steroid Biochem. 1979;11:193–199. doi: 10.1016/0022-4731(79)90296-6. [DOI] [PubMed] [Google Scholar]
- 5.Nozu K, Dehejia A, Zawistowich L, Catt K J, Dufau M L. J Biol Chem. 1981;256:12875–12882. [PubMed] [Google Scholar]
- 6.Nishihara M, Winters C A, Buzko E, Waterman M R, Dufau M L. Biochem Biophys Res Commun. 1988;154:151–158. doi: 10.1016/0006-291x(88)90663-8. [DOI] [PubMed] [Google Scholar]
- 7.Tsai-Morris C H, Khanum A, Tang P-Z, Dufau M L. Endocrinology. 1999;140:3534–3542. doi: 10.1210/endo.140.8.6944. [DOI] [PubMed] [Google Scholar]
- 8.Tang P Z, Tsai-Morris C H, Dufau M L. J Biol Chem. 1999;274:37932–37940. doi: 10.1074/jbc.274.53.37932. [DOI] [PubMed] [Google Scholar]
- 9.Suzuki H, Kawarabayasi Y, Kondo J, Abe T, Nishikawa K, Kimura S, Hashimoto T, Yamamoto T. J Biol Chem. 1990;265:8681–8685. [PubMed] [Google Scholar]
- 10.Fujino T, Yamamoto T. J Biochem (Tokyo) 1992;111:197–203. doi: 10.1093/oxfordjournals.jbchem.a123737. [DOI] [PubMed] [Google Scholar]
- 11.Abe T, Fujino T, Fukuyama R, Minoshima S, Shimizu N, Toh H, Suzuki H, Yamamoto T. J Biochem (Tokyo) 1992;111:123–128. doi: 10.1093/oxfordjournals.jbchem.a123707. [DOI] [PubMed] [Google Scholar]
- 12.Kang M J, Fujino T, Sasano H, Minekura H, Yabuki N, Nagura H, Iijima H, Yamamoto T T. Proc Natl Acad Sci USA. 1997;94:2880–2884. doi: 10.1073/pnas.94.7.2880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fujino T, Kang M J, Suzuki H, Iijima H, Yamamoto T. J Biol Chem. 1996;271:16748–16752. doi: 10.1074/jbc.271.28.16748. [DOI] [PubMed] [Google Scholar]
- 14.Aquilano D R, Dufau M L. Endocrinology. 1984;114:499–510. doi: 10.1210/endo-114-2-499. [DOI] [PubMed] [Google Scholar]
- 15.Gilman M. In: Current Protocols in Molecular Biology. Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. New York: Greene & Wiley; 1987. pp. 4.7.1–4.7.6. [Google Scholar]
- 16.Greenberg M E, Bender T P. In: Current Protocols in Molecular Biology. Ausubel F M, Brent R, Kingston R E, Moore D D, Seidman J G, Smith J A, Struhl K, editors. New York: Greene & Wiley; 1987. pp. 4.10.1–4.10.7. [Google Scholar]
- 17.Fox C H, Cottler-Fox M. In: Current Protocols in Immunology. Coligan J, Kruisbeek A, editors. New York: Wiley; 1993. [Google Scholar]
- 18.Fabbri A, Ciocca D R, Ciampani T, Wang J, Dufau M L. Endocrinology. 1995;136:2303–2308. doi: 10.1210/endo.136.5.7720679. [DOI] [PubMed] [Google Scholar]
- 19.Watkins P A, Lu J F, Steinberg S J, Gould S J, Smith K D, Braiterman L T. J Biol Chem. 1998;273:18210–18219. doi: 10.1074/jbc.273.29.18210. [DOI] [PubMed] [Google Scholar]
- 20.Klenk H P, Clayton R A, Tomb J F, White O, Nelson K E, Ketchum K A, Dodson R J, Gwinn M, Hickey E K, Peterson J D, et al. Nature (London) 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- 21.Cole S T, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon S V, Eiglmeier K, Gas S, Barry C E, III, et al. Nature (London) 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 22.Devine J H, Kutuzova G D, Green V A, Ugarova N N, Baldwin T O. Biochim Biophys Acta. 1993;1173:121–132. doi: 10.1016/0167-4781(93)90172-a. [DOI] [PubMed] [Google Scholar]
- 23.Wood K V, Lam Y A, Seliger H H, McElroy W D. Science. 1989;244:700–702. doi: 10.1126/science.2655091. [DOI] [PubMed] [Google Scholar]
- 24.Black P N, Zhang Q, Weimar J D, DiRusso C C. J Biol Chem. 1997;272:4896–4903. doi: 10.1074/jbc.272.8.4896. [DOI] [PubMed] [Google Scholar]
- 25.Shrago E. J Nutr. 2000;130:290S–293S. doi: 10.1093/jn/130.2.290S. [DOI] [PubMed] [Google Scholar]
- 26.Black P N, Faergeman N J, DiRusso C C. J Nutr. 2000;130:305S–309S. doi: 10.1093/jn/130.2.305S. [DOI] [PubMed] [Google Scholar]
- 27.Charreau E H, Calvo J C, Nozu K, Pignataro O, Catt K J, Dufau M L. J Biol Chem. 1981;256:12719–12724. [PubMed] [Google Scholar]