Abstract
Signaling crosstalk between complement and Toll-like receptors (TLRs) normally serves to coordinate host immunity. However, the periodontal bacterium Porphyromonas gingivalis expresses C5 convertase-like enzymatic activity and adeptly exploits complement-TLR crosstalk to subvert host defenses and escape elimination. Intriguingly, this defective immune surveillance leads to the remodeling of the periodontal microbiota to a dysbiotic state that causes inflammatory periodontitis. Understanding the mechanisms by which P. gingivalis modulates complement function to cause dysbiosis offers new targets for complement therapeutics.
Introduction
Microbial pathogens which successfully disarm, suppress, or delay host defenses target preferentially innate immunity (Finlay and McFadden, 2006) and particularly key systems such as complement and the Toll-like receptor (TLR) family of pattern-recognition receptors (Hajishengallis and Lambris, 2011). Collectively, complement and TLRs sense pathogens through “pattern recognition” and “missing-self recognition” strategies and trigger the activation of antimicrobial and inflammatory responses, as well as the initiation of the adaptive immune response (Kawai and Akira, 2010; Ricklin et al., 2010). Until relatively recently, both systems were primarily investigated as separate entities. However, a substantial body of literature has demonstrated extensive crosstalk between complement and TLR signaling pathways (Hajishengallis and Lambris, 2010; Hawlisch et al., 2005; la Sala et al., 2005; Zhang et al., 2007). Both synergistic and antagonistic interactions between complement and TLRs have been described which apparently serve to invigorate the host response or regulate it to prevent unwarranted inflammatory responses (Ricklin, et al., 2010). Therefore, pathogens targeting complement and/or TLRs have the opportunity to infiltrate an extensive network of signaling pathways in ways that could impair innate host defense and, moreover, deregulate the induction of adaptive immunity or divert it in ways that favor their survival.
In this review we focus on Porphyromonas gingivalis, a gram-negative oral anaerobic bacterium recently shown to manipulate complement-TLR crosstalk to enhance its survival and virulence (Hajishengallis et al., 2007; Hajishengallis et al., 2011; Liang et al., 2011; Wang et al., 2007; Wang et al., 2010) (Figure 1). Importantly, these subversion mechanisms are highly relevant in the context of the collective virulence of the microbial community in which P. gingivalis resides. Specifically, P. gingivalis can act as a “keystone pathogen” which remodels the commensal microbiota into a dysbiotic partner that disrupts the homeostatic balance with the host tissue leading to destructive inflammation in periodontitis (Hajishengallis, et al., 2011) (Figure 2).
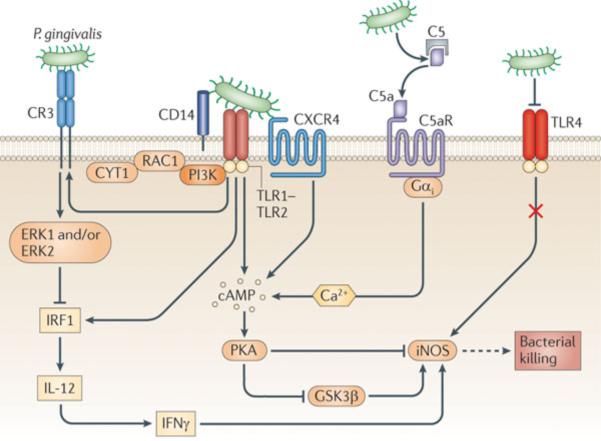
Figure 1. Exploitation of signaling crosstalk by P. gingivalis.
P. gingivalis interacts with Toll-like receptor (TLR)-2 (as part of a CD14–TLR2–TLR1 signaling complex) and with TLR4. The activation of TLR4 is suppressed by the bacterium's atypical lipopolysaccharide which acts as an antagonist. The TLR2 response is manipulated through crosstalk with other innate receptors. By means of Arg-specific cysteine proteinases that release biologically active C5a from C5, P. gingivalis activates the C5a receptor (C5aR) and induces intracellular Ca2+ signaling which synergistically enhances the otherwise weak cAMP responses induced by TLR2 activation alone. Maximal cAMP induction requires the participation of CXC-chemokine receptor 4 (CXCR4), which is activated directly by the bacterium's fimbriae. The resulting activation of the cAMP-dependent protein kinase A (PKA) inactivates glycogen synthase kinase-3β (GSK3β) and inhibits the inducible nitric oxide synthase (iNOS)-dependent killing of the pathogen in macrophages. An additional pathway induced downstream of TLR2 is an inside-out signaling pathway, mediated by RAC1, phosphatidylinositol-3 kinase (PI3K) and cytohesin 1 (CYT1), which transactivates complement receptor-3 (CR3). Activated CR3 binds P. gingivalis and induces extracellular signal-regulated kinase-1/ERK2 signaling, which in turn selectively downregulates IL-12 p35 and p40 mRNA expression through suppression of interferon regulatory factor 1 (IRF1). Inhibition of bioactive IL-12, and secondarily IFNγ, leads to impaired immune clearance of P. gingivalis.
From Hajishengallis and Lambris, 2011, Nature Reviews Immunology, 11, 187–200 (used by permission).
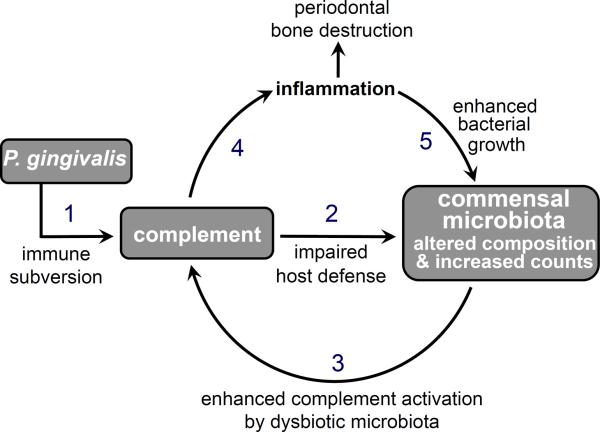
Figure 2. P. gingivalis exploits complement and causes dysbiotic inflammation.
P. gingivalis subverts complement and impairs host defense leading to altered composition and increased numbers of periodontal commensal bacteria which, in turn, cause complement-dependent periodontal inflammation and bone loss. The inflammatory environment is favorable to further bacterial growth as it provides the dysbiotic microbiota with a nutrient-rich gingival inflammatory exudate. Numbers indicate a possible sequence of events, although the overall process represents a self-feeding cycle that drives and sustains persistent inflammation.
Periodontitis is a biofilm-driven chronic inflammatory disease that affects the tooth-supporting tissues (periodontium) leading, in some cases, to tooth loss (Pihlstrom et al., 2005). This oral disease affects the majority of adults, whereas an estimated 10–15% develops severe periodontitis, which increases the patients' risk for atherosclerosis, aspiration pneumonia, diabetes, adverse pregnancy outcomes, and perhaps rheumatoid arthritis (Genco and Van Dyke, 2010; Lalla and Papapanou, 2011; Lundberg et al., 2010; Pihlstrom, et al., 2005; Tonetti et al., 2007; Xiong et al., 2006).
Periodontitis is characterized by dramatic changes in the numbers and composition of the periodontal bacterial community relative to health (Moore et al., 1982; Socransky, 1977; Socransky and Haffajee, 2005). It has recently been proposed that periodontitis fundamentally represents disruption of host-microbial homeostasis caused by dysbiosis of the periodontal microbiota (Darveau, 2010; Hajishengallis, et al., 2011). According to this notion, changes in the relative abundance of individual components of the microbiota compared to their abundancies in health can lead to alterations in the host-microbial crosstalk sufficient to initiate inflammatory disease.
Although P. gingivalis is a quantitatively minor component of periodontal pathogenic biofilms, its presence has been associated with progressive bone loss in periodontitis patients (Chaves et al., 2000; Doungudomdacha et al., 2000; Kumar et al., 2006; Moore, et al., 1982; Moore et al., 1991; Socransky et al., 1998). However, the fundamental mechanism by which P. gingivalis may contribute to a polymicrobial disease such as periodontitis has remained elusive until recently. As alluded to above, useful insights have been provided by a study in the mouse model of periodontitis which demonstrated that P. gingivalis can act as a kesystone pathogen which reshapes an otherwise harmless periodontal microbiota into a disease-provoking microbiota (Hajishengallis, et al., 2011).
The capacity of P. gingivalis to act as a keystone member of the periodontal microbiota is dependent, in great part, upon its ability to exploit complement and TLRs (Hajishengallis, et al., 2011) (Table 1). Subversion of complement and TLRs might also account for the ability of P. gingivalis to relocate to systemic tissues and contribute to disease. In this regard, P. gingivalis is a common isolate from aspiration pneumonia and lung abscesses (Finegold, 1991; Okuda et al., 2005) and has been detected in a viable state in atherosclerotic plaques (Kozarov et al., 2005). Below we summarize and discuss recently established mechanisms whereby P. gingivalis can manipulate complement to sabotage its functions and its productive interactions with TLRs. In this way, the host response is diverted to favor the pathogen and its microbial community leading to dysbiosis and destructive inflammation in the periodontal tissue.
Table 1.
Subversion of complement and TLRs by P. gingivalis
| Mechanism | Effector molecules | Refs. | |
|---|---|---|---|
| 1 | Inhibition of complement activation by degrading the central complement component (C3) | Gingipains, especially HRgpA & RgpB | (Popadiak, et al., 2007; Potempa, et al., 2009) |
| 2 | Hijacking negative regulators of complement (C4b-binding protein) | HRgpA | (Potempa, et al., 2008) |
| 3 | Proactive generation of C5a for instigating subversive C5aR-TLR2 crosstalk | HRgpA & RgpB | (Liang, et al., 2011; Wang, et al., 2010) |
| 4 | Transactivation of CR3 and exploitation of CR3-mediated phagocytosis | Fimbriae | (Liang, et al., 2011; Wang, et al., 2007) |
| 5 | Suppression of TLR2-induced IL-12 via CR3 binding or C5aR activation | Fimbriae or HRgpA & RgpB | (Hajishengallis, et al., 2007; Liang, et al., 2011) |
| 6 | TLR4 evasion by expressing dephosphorylated tetra-acylated lipid A | Lipid A 1- and 4-phosphatases & deacylase | (Coats, et al., 2009) |
| 7 | TLR4 antagonism by expressing monophosphorylated tetra-acylated lipid A | Lipid A 4-phosphatase & deacylase (lipid A 1-phosphatase suppressed by hemin) | (Coats, et al., 2005; Coats, et al., 2009) |
Complement inhibition by P. gingivalis
The interactions of P. gingivalis with complement are complex and include both inhibitory and stimulatory effects (Krauss et al., 2010) (Table 1). This is not surprising given that complement mediates diverse functions (Ricklin, et al., 2010), some of which could involve effective antimicrobial action (targets of inactivation), while certain complement-dependent inflammatory pathways could benefit the periodontal bacteria (targets of activation). P. gingivalis can suppress complement activation, regardless of the initiation pathway involved (classical, lectin, or alternative) through gingipain-dependent degradation of C3 (Popadiak et al., 2007). All three gingipain enzymes can cause complement inactivation, although the Arg-specific enzymes (HRgpA and RgpB) are more potent than the Lys-specific gingipain (Kgp) (Popadiak, et al., 2007). As a consequence of complement inhibition, the deposition of opsonins or the membrane attack complex on the surface of P. gingivalis is suppressed, whereas ablation of its gingipain activity by chemical or genetic means reverses these effects (Potempa and Pike, 2009). For increased safety against complement, this bacterium seems to hijack physiological inhibitors of the complement cascade. Indeed, P. gingivalis uses its HRgpA to capture the circulating C4b-binding protein on the bacterial cell surface, and in this way to negatively regulate the classical pathway C3 convertase (Potempa et al., 2008).
It should be noted that other periodontal bacteria associated with periodontitis, such as Prevotella intermedia, Tannerella forsythia, and Treponema denticola, also possess mechanisms for escaping complement. Interpain A (InpA), a streptopain-like cysteine protease secreted by some Prevotella intermedia strains, degrades C3 and contributes to resistance against the antibacterial activity of complement (Potempa et al., 2009). Karilysin, a metalloproteinase expessed by T. forsythia confers resistance to killing by human complement by acting at different stages of complement activation: Specifically, karilysin inhibits the classical and lectin pathways by degrading MBL, ficolin-2, ficolin-3, and C4, whereas it blocks the terminal pathway by degrading C5 (Jusko et al., 2012). Importantly, both InpA and karilysin synergize with gingipains in these functions suggesting that P. gingivalis, P. intermedia, and T. forsythia together can better promote the survival in the dental biofilm of bystander bacterial species, which could otherwise be readily eliminated by the bactericidal activity of complement. T. denticola expresses a 11.4-kDa cell surface lipoprotein which can bind factor H (McDowell et al., 2009). This function suggests that T. denticola may escape complement-dependent killing by means of surface-bound, biologically active factor H. Interestingly, however, after full-length factor H becomes associated with T. denticola, the bacterium uses its serine protease dentilisin to cleave factor H and generate a 50-kDa factor H fragment that remains attached to the bacterial surface (McDowell, et al., 2009). An interesting question is whether the attached fragment retains useful complement inhibitory activity to protect T. denticola against complement. Alternatively, through this mechanism, dentilisin may in fact inactivate factor H and deregulate complement, thereby promoting local inflammation which could also be beneficial for periodontal bacteria (see next section).
Intriguingly, the HRgpA and RgpB gingipains of P. gingivalis display C5 convertase-like activity and generate biologically active C5a, whereas C5b is destroyed (Wingrove et al., 1992). In heat-inactivated human serum, P. gingivalis can rapidly generate >30 nM of C5a (Wang, et al., 2010). This activity seems counterproductive for the adaptive fitness of this pathogen, since C5a is probably the most potent effector of the complement cascade and generally promotes host defense, e.g., through chemotactic recruitment and activation of leukocytes (Guo and Ward, 2005; Hopken et al., 1996). Strikingly, however, P. gingivalis exploits C5a to manipulate TLR signaling (below).
Hijacking of C5aR-TLR2 crosstalk
The TLR system senses P. gingivalis primarily through TLR2, as shown by in vitro and in vivo observations, which additionally have excluded a significant role for TLR4 (Burns et al., 2006; Hajishengallis et al., 2006). The lack of significant TLR4 involvement in the host response to P. gingivalis is counterintuitive given that this is a gram-negative bacterium that expresses a lipopolysaccharide. However, P. gingivalis utilizes specific lipid A 1- and 4-phosphatases and a deacylase which in concert generate a tetra-acylated and dephosphorylated lipid A structure that is biologically inert (Coats et al., 2009). Moreover, high concentrations of hemin (as can be found in inflamed periodontal sites) suppress the lipid A 1-phosphatase activity and lead to the generation of a mono-phosphorylated lipid A, which acts as a TLR4 antagonist (Coats et al., 2005; Coats, et al., 2009) (Table 1). These unique LPS features allow P. gingivalis to evade or antagonize TLR4 activation at the receptor level. Moreover, this pathogen has evolved sophisticated complement-dependent strategies to selectively intercept only a subset of TLR2 signaling events (Kagan, 2010; Wang, et al., 2010). In this way, P. gingivalis can filter out pathways that could result in its elimination, whereas it can selectively maintain or enhance host responses that favor its survival.
Specifically, P. gingivalis instigates a C5aR-TLR2 subversive crosstalk that undermines the killing function of macrophages. Indeed, P. gingivalis-generated C5a locally activates C5aR and stimulates Gαi-dependent intracellular Ca2+ signaling which synergistically enhances an otherwise weak cAMP response by P. gingivalis-induced TLR2 activation alone. In this crosstalk pathway, which additionally involves the participation of the CXC chemokine receptor-4, elevated and sustained production of cAMP leads to the activation of the cAMP-dependent protein kinase A which inactivates the glycogen synthase kinase-3β and impairs nitric oxide-dependent killing of P. gingivalis (Hajishengallis et al., 2008; Wang, et al., 2010).
The P. gingivalis-induced C5aR-TLR2 crosstalk also regulates cytokine production in favor of the pathogen (Liang, et al., 2011). Specifically, P. gingivalis proactively and selectively inhibits TLR2-induced IL-12p70, whereas the same C5aR-TLR2 crosstalk upregulates inflammatory and bone-resorptive cytokines (IL-1β, IL-6, and TNF). In vivo, the ability of P. gingivalis to manipulate TLR2 activation via the C5a-C5aR axis allows it to escape IL-12p70-dependent immune clearance and to cause inflammatory bone loss in a murine model of experimental periodontitis (Liang, et al., 2011). Therefore, by targeting C5aR, P. gingivalis not only can promote its survival but also cause destructive inflammation.
Because the C5aR-TLR2 crosstalk inhibits only a subset of TLR2 signaling events, C5aR was characterized as a “TLR modulatory receptor” as opposed to “TLR inhibitory receptors”, such as IL-10 receptor or TGF-β receptor, which block most, if not all, inflammatory responses (Kagan, 2010). In fact, it would not be in the “best interest” of P. gingivalis to induce a generalized immunosuppression, since such action would spare the bacterium from leukocytes but, on the other hand, would probably starve it to elimination. In this regard, P. gingivalis is a proteolytic and asaccharolytic organism the survival and growth of which depend crucially on inflammatory tissue break down products (degraded proteins) and gingival inflammatory fluid-derived hemin, a source of essential iron (Krauss, et al., 2010). Therefore, inflammation is exploited by P. gingivalis as a means for survival, although in a chronic setting this causes collateral damage to the periodontium.
Exploitation of the TLR2-CR3 inside-out signaling pathway
P. gingivalis appears to use a number of distinct but reinforcing mechanisms to escape leukocyte killing. Thus, in addition to exploiting the C5aR-TLR2 crosstalk, it can also take advantage of the TLR2 inside-out signaling pathway for complement receptor 3 (CR3) transactivation. CR3 is a β2 integrin (CD11b–CD18) the binding activity of which is tightly regulated: In resting cells, CR3 displays a low-affinity conformation, whereas a rapid and transient shift to a high-affinity state can be triggered through inside-out signaling by chemokine or anaphylatoxin receptors (Abram and Lowell, 2009). More recently, TLRs were also shown to mediate inside-out signaling for CR3 activation (Han et al., 2010; Harokopakis and Hajishengallis, 2005; Sendide et al., 2005). Tight regulation of the CR3 binding activity is important since this is a functionally versatile molecule involved in the phagocytosis of apoptotic cells, leukocyte trafficking, and regulation of cytokine production (Ricklin, et al., 2010).
However, CR3 activation may come under the control of P. gingivalis: The pathogen induces TLR2-mediated transactivation of CR3 through an inside-out pathway that involves Rac1, PI3K and cytohesin-1 (Harokopakis and Hajishengallis, 2005; Harokopakis et al., 2006). This mechanism allows P. gingivalis to bind activated CR3 for a relatively safe “outside-in” entry into macrophages. Indeed, CR3-deficient macrophages are superior to wild-type macrophages in the intracellular killing of this oral pathogen (Hajishengallis et al., 2006; Wang, et al., 2007). Moreover, pharmacological inhibition of CR3 greatly facilitates the intracellular killing of P. gingivalis (Hajishengallis, et al., 2007). Antagonistic blockade of CR3 also suppresses P. gingivalis-induced periodontal bone loss in the mouse periodontitis model (Hajishengallis, et al., 2007).
Although CR3 is a phagocytic receptor, it is not linked to strong microbicidal mechanisms such as those activated by FcγR-mediated phagocytosis (Caron and Hall, 1998; Lowell, 2006). This is possibly because CR3 is heavily committed with the phagocytosis of iC3b-coated apoptotic cells, which are not normally recognized as danger (Kim et al., 2004; Mevorach et al., 1998). Furthermore, at least under certain conditions, CR3-derived phagosomes do not fuse with lysosomes (Gatfield and Pieters, 2000). These observations may explain why CR3 is a target of exploitation by P. gingivalis. Moreover, CR3 binding by the fimbriae of P. gingivalis inhibits IL-12p70 production by macrophages (Hajishengallis, et al., 2007) (Figure 1). Not surprisingly, therefore, the TLR2-CR3 crosstalk pathway is exploited by several pathogens in addition to P. gingivalis (Harokopakis and Hajishengallis, 2005; Oliva et al., 2009; Sendide, et al., 2005).
In the context of the periodontitis-atherosclerosis connection (Seymour et al., 2007) and the observation of viable P. gingivalis in atherosclerotic plaques (Kozarov, et al., 2005), it is intriguing to hypothesize that the intracellular persistence of P. gingivalis in macrophages (Huang et al., 2009; Wang, et al., 2007) might allow this organism to exploit these cells as “Trojan horses” to relocate to systemic tissues and subsequently infect permissive cells (e.g., endothelial cells). Although this hypothesis remains to be tested, the capacity of P. gingivalis for cell exit and infection of new host cells has already been demonstrated (Li et al., 2008; Yilmaz, 2008).
P. gingivalis as an orchestrator of complement-dependent dysbiotic inflammation
As pointed out above, there are stark differences in the composition of the periodontal microbiota in health and disease (Darveau, 2010; Moore, et al., 1982; Socransky, 1977). The assumption that these differences signified bacterial specificity in the etiology of periodontitis led to significant progress, including the identification of a number of bacterial species as putative periodontal pathogens. In addition to P. gingivalis, Treponema denticola and Tannerella forsythia were also shown to be strongly associated with periodontitis. These three species together comprise the so-called “red complex” and are frequently isolated together from diseased sites (Holt and Ebersole, 2005; Socransky, et al., 1998). Over the years much of periodontal research focused on identifying virulence factors of these three bacteria in the context of a conventional infectious disease (Holt and Ebersole, 2005).
However, the dramatic differences in the composition of the microbiota in periodontal disease versus health could alternatively be interpreted by a non-classical view of microbial etiology of disease. According to this viewpoint, periodontal pathogens may not directly cause periodontitis but rather reshape the normally symbiotic microbiota into a dysbiotic one that disrupts the normal homeostatic relationship with the host tissue. In this context, P. gingivalis was suspected to be an orchestrator of periodontal disease by virtue of its sophisticated strategies to subvert host immunity. Specifically, the capacity of P. gingivalis to impair innate immunity could alter the growth and development of the entire periodontal biofilm. The overgrowth of a dysbiotic biofilm, stabilized by the presence of P. gingivalis, could in turn break down the host-microbial homeostasis in the periodontium and trigger destructive inflammation. This hypothesis led to the studies showing that P. gingivalis can act as a keystone member of the disease-provoking periodontal microbial community (Hajishengallis, et al., 2011). The “keystone species” concept was originally proposed by the ecologist R.T. Paine in 1969 (Paine, 1969). In the ecological literature, keystones are species whose effects on their communities are inordinately large relative to their abundance. Such species are thought to constitute the “keystone” of the community's structure in a way reminiscent of a keystone in an arch (reviewed by (Hajishengallis et al., 2012).
Consistent with the above discussed hypothesis, the colonization of the murine periodontium by P. gingivalis was shown to be accompanied by significant alterations in the numbers and community organization of the commensal bacteria, followed by inflammation and destruction of the tooth-supporting bone (Hajishengallis, et al., 2011). Intriguingly, P. gingivalis caused these dysbiotic alterations while present only at very low numbers, specifically < 0.01% of the total bacterial counts. Moreover, P. gingivalis failed to cause periodontitis by itself in germ-free mice despite its ability to colonize their periodontium (Hajishengallis, et al., 2011). Therefore, the dysbiotic microbiota that develops in the presence of P. gingivalis is most likely the eventual cause of the disease.
A major question addressed in the same study was whether complement was involved in P. gingivalis-instigated dysbiosis. Since C5aR was shown earlier to be critical for the ability of P. gingivalis to subvert leukocytes (Liang, et al., 2011; Wang, et al., 2010) (Figure 1), it was hypothesized that C5aR-deficient mice would be resistant to P. gingivalis-induced dysbiosis. Indeed, this bacterium failed to alter the commensal microbiota in the absence of C5aR. Moreover, an isogenic mutant of P. gingivalis lacking gingipain activity failed to alter the commensal microbiota in normal mice (Hajishengallis, et al., 2011). In both cases, the failure to transform the normal commensal microbiota into a dysbiotic state prevented the development of periodontal inflammation and bone loss. Therefore, mice are protected from P. gingivalis-induced dysbiosis when the pathogen does not have access to C5aR or lacks the enzyme to exploit this receptor. In contrast, under conditions that allow C5aR exploitation by P. gingivalis (i.e., wild-type mice orally inoculated with the wild-type organism), there is uncontrolled growth of commensal bacteria which leads to complement-dependent inflammation that destroys the periodontal bone (Hajishengallis, et al., 2011). Periodontal inflammation may also generate tissue breakdown products (e.g., degraded proteins and hemin), which serve bacterial nutritional needs and can stabilize the transition to a disease-provoking microbiota (Figure 2). In this regard, the inflammatory changes in the periodontium can be better exploited by proteolytic and asaccharolytic bacteria, that is, species associated with periodontal disease rather than health (Darveau, 2010). Those bacteria that cannot utilize these nutrients, or for which host inflammation is detrimental, are likely to be outcompeted or eliminated. As outlined above, complement is centrally involved in this self-feeding vicious cycle of periodontal dysbiosis and destructive inflammation (Figure 2).
Therapeutic implications in human periodontitis
Complement components can be found in active form in the gingival crevicular fluid at up to 70% of their concentration in serum, although certain components can be found at much higher levels reflecting local production in the periodontium (Potempa, et al., 2009). Clinical and histological observations suggest that complement may be involved in the pathogenesis of human periodontitis (Hajishengallis, 2010). Indeed, inflamed gingiva or samples of crevicular fluid from periodontitis patients have increased levels of activated complement fragments relative to control samples from healthy individuals (Beikler et al., 2008; Courts et al., 1977; Niekrash and Patters, 1986; Nikolopoulou-Papaconstantinou et al., 1987; Patters et al., 1989; Rautemaa and Meri, 1996; Schenkein and Genco, 1977). Moreover, experimental induction of gingival inflammation in human volunteers causes progressive elevation of complement cleavage products correlating with increased clinical indices of inflammation (Patters, et al., 1989).
Although the data from the human studies are correlative, the findings that C5aR-deficient mice are resistant to experimental periodontitis establish an intimate link between complement and periodontal pathogenesis (Hajishengallis, et al., 2011; Liang, et al., 2011). Importantly, treatment of P. gingivalis-colonized mice with a C5aR antagonist leads to the elimination of P. gingivalis from the periodontium and reverses the dysbiotic changes that are required for the development of periodontitis (Hajishengallis, et al., 2011). This observation provides a causal link between complement and periodontitis and offers proof-of-concept that dysbiotic diseases could be treated by specific targeting of keystone pathogens. From a periodontal point of view, the same observation suggests that complement therapeutics may find application in the treatment of human periodontitis.
Conclusions
The subversive action of P. gingivalis on complement may explain, at least in part, its ability to persist in the periodontium and establish dysbiotic inflammation. P. gingivalis is perhaps the first documented case of a keystone pathogen in microbial ecology as it fulfills the criteria of low abundance and major influence on the microbial community, which is remodeled in a way that promotes periodontal pathogenesis (Hajishengallis, et al., 2011). Moreover, available evidence suggests that keystone pathogens may underlie the etiology of also other polymicrobial inflammatory diseases (Hajishengallis et al., 2012). Although established in the mouse model, the keystone pathogen concept is consistent with observations in non-human primates and in humans (reviewed by Darveau et al., 2012). Briefly, immunization of Macaca fascicularis with a gingipain-based vaccine resulted in suppression of the counts of indigenous P. gingivalis accompanied by a reduction in the total subgingival bacterial load and protection against bone loss (Page et al., 2007). Moreover, P. gingivalis is a quantitatively minor component of human periodontitis-associated biofilms (Doungudomdacha, et al., 2000; Kumar, et al., 2006; Moore, et al., 1982), despite its association with progressive bone loss in periodontitis patients (Chaves, et al., 2000; Moore, et al., 1991). Nevertheless, even at this low abundance, P. gingivalis exploits complement and triggers dysbiotic changes that set the stage for inflammatory periodontitis. The ongoing elucidation of the mechanisms underlying these interactions may facilitate the development of novel therapeutic interventions in human periodontitis.
Acknowledgements
The authors are supported by grants from the U.S. National Institutes of Health, DE015254, DE018292, DE021580, and DE021685 (GH) and GM062134, AI030040, AI072106, AI068730 (JDL).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abram CL, Lowell CA. The ins and outs of leukocyte integrin signaling. Annu Rev Immunol. 2009;27:339. doi: 10.1146/annurev.immunol.021908.132554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beikler T, Peters U, Prior K, Eisenacher M, Flemmig TF. Gene expression in periodontal tissues following treatment. BMC Med Genomics. 2008;1:30. doi: 10.1186/1755-8794-1-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burns E, Bachrach G, Shapira L, Nussbaum G. Cutting Edge: TLR2 is required for the innate response to Porphyromonas gingivalis: Activation leads to bacterial persistence and TLR2 deficiency attenuates induced alveolar bone resorption. J. Immunol. 2006;177:8296. doi: 10.4049/jimmunol.177.12.8296. [DOI] [PubMed] [Google Scholar]
- Caron E, Hall A. Identification of two distinct mechanisms of phagocytosis controlled by different Rho GTPases. Science. 1998;282:1717. doi: 10.1126/science.282.5394.1717. [DOI] [PubMed] [Google Scholar]
- Chaves ES, Jeffcoat MK, Ryerson CC, Snyder B. Persistent bacterial colonization of Porphyromonas gingivalis, Prevotella intermedia, and Actinobacillus actinomycetemcomitans in periodontitis and its association with alveolar bone loss after 6 months of therapy. J Clin Periodontol. 2000;27:897. doi: 10.1034/j.1600-051x.2000.027012897.x. [DOI] [PubMed] [Google Scholar]
- Coats SR, Pham TT, Bainbridge BW, Reife RA, Darveau RP. MD-2 mediates the ability of tetra-acylated and penta-acylated lipopolysaccharides to antagonize Escherichia coli lipopolysaccharide at the TLR4 signaling complex. J Immunol. 2005;175:4490. doi: 10.4049/jimmunol.175.7.4490. [DOI] [PubMed] [Google Scholar]
- Coats SR, Jones JW, Do CT, Braham PH, Bainbridge BW, To TT, Goodlett DR, Ernst RK, Darveau RP. Human Toll-like receptor 4 responses to P. gingivalis are regulated by lipid A 1- and 4'- phosphatase activities. Cell Microbiol. 2009;11:1587. doi: 10.1111/j.1462-5822.2009.01349.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courts FJ, Boackle RJ, Fudenberg HH, Silverman MS. Detection of functional complement components in gingival crevicular fluid from humans with periodontal diseases. J Dent Res. 1977;56:327. doi: 10.1177/00220345770560032001. [DOI] [PubMed] [Google Scholar]
- Darveau RP. Periodontitis: a polymicrobial disruption of host homeostasis. Nat Rev Microbiol. 2010;8:481. doi: 10.1038/nrmicro2337. [DOI] [PubMed] [Google Scholar]
- Darveau RP, Hajishengallis G, Curtis MA. Porphyromonas gingivalis as a potential community activist for disease. J Dent Res. 2012;91 doi: 10.1177/0022034512453589. Epub ahead of print: doi: 10.1177/0022034512453589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doungudomdacha S, Rawlinson A, Douglas CW. Enumeration of Porphyromonas gingivalis, Prevotella intermedia and Actinobacillus actinomycetemcomitans in subgingival plaque samples by a quantitative-competitive PCR method. J Med Microbiol. 2000;49:861. doi: 10.1099/0022-1317-49-10-861. [DOI] [PubMed] [Google Scholar]
- Finegold SM. Aspiration pneumonia. Rev Infect Dis. 1991;13(Suppl 9):S737. doi: 10.1093/clinids/13.supplement_9.s737. [DOI] [PubMed] [Google Scholar]
- Finlay BB, McFadden G. Anti-immunology: evasion of the host immune system by bacterial and viral pathogens. Cell. 2006;124:767. doi: 10.1016/j.cell.2006.01.034. [DOI] [PubMed] [Google Scholar]
- Gatfield J, Pieters J. Essential role for cholesterol in entry of mycobacteria into macrophages. Science. 2000;288:1647. doi: 10.1126/science.288.5471.1647. [DOI] [PubMed] [Google Scholar]
- Genco RJ, Van Dyke TE. Prevention: Reducing the risk of CVD in patients with periodontitis. Nat Rev Cardiol. 2010;7:479. doi: 10.1038/nrcardio.2010.120. [DOI] [PubMed] [Google Scholar]
- Guo RF, Ward PA. Role of C5a in inflammatory responses. Annu Rev Immunol. 2005;23:821. doi: 10.1146/annurev.immunol.23.021704.115835. [DOI] [PubMed] [Google Scholar]
- Hajishengallis G. Complement and periodontitis. Biochem Pharmacol. 2010;80:1992. doi: 10.1016/j.bcp.2010.06.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Lambris JD. Crosstalk pathways between Toll-like receptors and the complement system. Trends Immunol. 2010;31:154. doi: 10.1016/j.it.2010.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Lambris JD. Microbial manipulation of receptor crosstalk in innate immunity. Nat. Rev. Immunol. 2011;11:187. doi: 10.1038/nri2918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Darveau RP, Curtis MA. The keystone pathogen hypothesis. Nat Rev Microbiol. 2012;10 doi: 10.1038/nrmicro2873. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Shakhatreh MA, Wang M, Liang S. Complement receptor 3 blockade promotes IL-12-mediated clearance of Porphyromonas gingivalis and negates its virulence in vivo. J Immunol. 2007;179:2359. doi: 10.4049/jimmunol.179.4.2359. [DOI] [PubMed] [Google Scholar]
- Hajishengallis G, Wang M, Harokopakis E, Triantafilou M, Triantafilou K. Porphyromonas gingivalis fimbriae proactively modulate β2 integrin adhesive activity and promote binding to and internalization by macrophages. Infect Immun. 2006;74:5658. doi: 10.1128/IAI.00784-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Wang M, Liang S, Triantafilou M, Triantafilou K. Pathogen induction of CXCR4/TLR2 cross-talk impairs host defense function. Proc Natl Acad Sci U S A. 2008;105:13532. doi: 10.1073/pnas.0803852105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Krauss JL, Liang S, McIntosh ML, Lambris JD. Pathogenic microbes and community service through manipulation of innate immunity. Adv Exp Med Biol. 2012;946:69. doi: 10.1007/978-1-4614-0106-3_5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hajishengallis G, Tapping RI, Harokopakis E, Nishiyama S-I, Ratti P, Schifferle RE, Lyle EA, Triantafilou M, Triantafilou K, Yoshimura F. Differential interactions of fimbriae and lipopolysaccharide from Porphyromonas gingivalis with the Toll-like receptor 2-centred pattern recognition apparatus. Cell. Microbiol. 2006;8:1557. doi: 10.1111/j.1462-5822.2006.00730.x. [DOI] [PubMed] [Google Scholar]
- Hajishengallis G, Liang S, Payne MA, Hashim A, Jotwani R, Eskan MA, McIntosh ML, Alsam A, Kirkwood KL, Lambris JD, Darveau RP, Curtis MA. Low-abundance biofilm species orchestrates inflammatory periodontal disease through the commensal microbiota and complement. Cell Host Microbe. 2011;10:497. doi: 10.1016/j.chom.2011.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Han C, Jin J, Xu S, Liu H, Li N, Cao X. Integrin CD11b negatively regulates TLR-triggered inflammatory responses by activating Syk and promoting degradation of MyD88 and TRIF via Cbl-b. Nat Immunol. 2010;11:734. doi: 10.1038/ni.1908. [DOI] [PubMed] [Google Scholar]
- Harokopakis E, Hajishengallis G. Integrin activation by bacterial fimbriae through a pathway involving CD14, Toll-like receptor 2, and phosphatidylinositol-3-kinase. Eur J Immunol. 2005;35:1201. doi: 10.1002/eji.200425883. [DOI] [PubMed] [Google Scholar]
- Harokopakis E, Albzreh MH, Martin MH, Hajishengallis G. TLR2 transmodulates monocyte adhesion and transmigration via Rac1- and PI3K-mediated inside-out signaling in response to Porphyromonas gingivalis fimbriae. J Immunol. 2006;176:7645. doi: 10.4049/jimmunol.176.12.7645. [DOI] [PubMed] [Google Scholar]
- Hawlisch H, Belkaid Y, Baelder R, Hildeman D, Gerard C, Kohl J. C5a negatively regulates toll-like receptor 4-induced immune responses. Immunity. 2005;22:415. doi: 10.1016/j.immuni.2005.02.006. [DOI] [PubMed] [Google Scholar]
- Holt SC, Ebersole JL. Porphyromonas gingivalis, Treponema denticola, and Tannerella forsythia: the “red complex”, a prototype polybacterial pathogenic consortium in periodontitis. Periodontol 2000. 2005;38:72. doi: 10.1111/j.1600-0757.2005.00113.x. [DOI] [PubMed] [Google Scholar]
- Hopken UE, Lu B, Gerard NP, Gerard C. The C5a chemoattractant receptor mediates mucosal defence to infection. Nature. 1996;383:86. doi: 10.1038/383086a0. [DOI] [PubMed] [Google Scholar]
- Huang MT, Taxman DJ, Holley-Guthrie EA, Moore CB, Willingham SB, Madden V, Parsons RK, Featherstone GL, Arnold RR, O'Connor BP, Ting JP. Critical role of apoptotic speck protein containing a caspase recruitment domain (ASC) and NLRP3 in causing necrosis and ASC speck formation induced by Porphyromonas gingivalis in human cells. J Immunol. 2009;182:2395. doi: 10.4049/jimmunol.0800909. [DOI] [PubMed] [Google Scholar]
- Jusko M, Potempa J, Karim AY, Ksiazek M, Riesbeck K, Garred P, Eick S, Blom AM. 2012 A metalloproteinase karilysin present in the majority of Tannerella forsythia isolates inhibits all pathways of the complement system. J Immunol. 188:2338. doi: 10.4049/jimmunol.1101240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kagan JC. “Complementing” toll signaling. Sci Signal. 2010;3:e15. doi: 10.1126/scisignal.3120pe15. [DOI] [PubMed] [Google Scholar]
- Kawai T, Akira S. The role of pattern-recognition receptors in innate immunity: update on Toll-like receptors. Nat Immunol. 2010;11:373. doi: 10.1038/ni.1863. [DOI] [PubMed] [Google Scholar]
- Kim S, Elkon KB, Ma X. Transcriptional suppression of interleukin-12 gene expression following phagocytosis of apoptotic cells. Immunity. 2004;21:643. doi: 10.1016/j.immuni.2004.09.009. [DOI] [PubMed] [Google Scholar]
- Kozarov EV, Dorn BR, Shelburne CE, Dunn WA, Jr., Progulske-Fox A. Human atherosclerotic plaque contains viable invasive Actinobacillus actinomycetemcomitans and Porphyromonas gingivalis. Arterioscler. Thromb. Vasc. Biol. 2005;25:e17. doi: 10.1161/01.ATV.0000155018.67835.1a. [DOI] [PubMed] [Google Scholar]
- Krauss JL, Potempa J, Lambris JD, Hajishengallis G. Complementary Tolls in the periodontium: how periodontal bacteria modify complement and Toll-like receptor responses to prevail in the host. Periodontol 2000. 2010;52:141. doi: 10.1111/j.1600-0757.2009.00324.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar PS, Leys EJ, Bryk JM, Martinez FJ, Moeschberger ML, Griffen AL. Changes in periodontal health status are associated with bacterial community shifts as assessed by quantitative 16S cloning and sequencing. J Clin Microbiol. 2006;44:3665. doi: 10.1128/JCM.00317-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- la Sala A, Gadina M, Kelsall BL. G(i)-protein-dependent inhibition of IL-12 production is mediated by activation of the phosphatidylinositol 3-kinase-protein 3 kinase B/Akt pathway and JNK. J Immunol. 2005;175:2994. doi: 10.4049/jimmunol.175.5.2994. [DOI] [PubMed] [Google Scholar]
- Lalla E, Papapanou PN. Diabetes mellitus and periodontitis: a tale of two common interrelated diseases. Nat. Rev. Endocrinol. 2011;7:738. doi: 10.1038/nrendo.2011.106. [DOI] [PubMed] [Google Scholar]
- Li L, Michel R, Cohen J, Decarlo A, Kozarov E. Intracellular survival and vascular cell-to-cell transmission of Porphyromonas gingivalis. BMC Microbiol. 2008;8:26. doi: 10.1186/1471-2180-8-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang S, Krauss JL, Domon H, McIntosh ML, Hosur KB, Qu H, Li F, Tzekou A, Lambris JD, Hajishengallis G. The C5a receptor impairs IL-12-dependent clearance of Porphyromonas gingivalis and is required for induction of periodontal bone loss. J Immunol. 2011;186:869. doi: 10.4049/jimmunol.1003252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowell CA. Rewiring phagocytic signal transduction. Immunity. 2006;24:243. doi: 10.1016/j.immuni.2006.03.002. [DOI] [PubMed] [Google Scholar]
- Lundberg K, Wegner N, Yucel-Lindberg T, Venables PJ. Periodontitis in RA-the citrullinated enolase connection. Nat Rev Rheumatol. 2010;6:727. doi: 10.1038/nrrheum.2010.139. [DOI] [PubMed] [Google Scholar]
- McDowell JV, Huang B, Fenno JC, Marconi RT. Analysis of a unique interaction between the complement regulatory protein factor H and the periodontal pathogen Treponema denticola. Infect Immun. 2009;77:1417. doi: 10.1128/IAI.01544-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mevorach D, Mascarenhas JO, Gershov D, Elkon KB. Complement-dependent clearance of apoptotic cells by human macrophages. J. Exp. Med. 1998;188:2313. doi: 10.1084/jem.188.12.2313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore WE, Holdeman LV, Smibert RM, Hash DE, Burmeister JA, Ranney RR. Bacteriology of severe periodontitis in young adult humans. Infect Immun. 1982;38:1137. doi: 10.1128/iai.38.3.1137-1148.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moore WE, Moore LH, Ranney RR, Smibert RM, Burmeister JA, Schenkein HA. The microflora of periodontal sites showing active destructive progression. J Clin Periodontol. 1991;18:729. doi: 10.1111/j.1600-051x.1991.tb00064.x. [DOI] [PubMed] [Google Scholar]
- Niekrash CE, Patters MR. Assessment of complement cleavage in gingival fluid in humans with and without periodontal disease. J Periodontal Res. 1986;21:233. doi: 10.1111/j.1600-0765.1986.tb01455.x. [DOI] [PubMed] [Google Scholar]
- Nikolopoulou-Papaconstantinou AA, Johannessen AC, Kristoffersen T. Deposits of immunoglobulins, complement, and immune complexes in inflamed human gingiva. Acta Odontol Scand. 1987;45:187. doi: 10.3109/00016358709098858. [DOI] [PubMed] [Google Scholar]
- Okuda K, Kimizuka R, Abe S, Kato T, Ishihara K. Involvement of periodontopathic anaerobes in aspiration pneumonia. J. Periodontol. 2005;76:2154. doi: 10.1902/jop.2005.76.11-S.2154. [DOI] [PubMed] [Google Scholar]
- Oliva C, Turnbough CL, Jr., Kearney JF. CD14-Mac-1 interactions in Bacillus anthracis spore internalization by macrophages. Proc Natl Acad Sci U S A. 2009;106:13957. doi: 10.1073/pnas.0902392106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Page RC, Lantz MS, Darveau R, Jeffcoat M, Mancl L, Houston L, Braham P, Persson GR. Immunization of Macaca fascicularis against experimental periodontitis using a vaccine containing cysteine proteases purified from Porphyromonas gingivalis. Oral Microbiol Immunol. 2007;22:162. doi: 10.1111/j.1399-302X.2007.00337.x. [DOI] [PubMed] [Google Scholar]
- Paine RT. A note on trophic complexity and community stability. Am Nat. 1969;103:91. [Google Scholar]
- Patters MR, Niekrash CE, Lang NP. Assessment of complement cleavage in gingival fluid during experimental gingivitis in man. J Clin Periodontol. 1989;16:33. doi: 10.1111/j.1600-051x.1989.tb01609.x. [DOI] [PubMed] [Google Scholar]
- Pihlstrom BL, Michalowicz BS, Johnson NW. Periodontal diseases. Lancet. 2005;366:1809. doi: 10.1016/S0140-6736(05)67728-8. [DOI] [PubMed] [Google Scholar]
- Popadiak K, Potempa J, Riesbeck K, Blom AM. Biphasic effect of gingipains from Porphyromonas gingivalis on the human complement system. J. Immunol. 2007;178:7242. doi: 10.4049/jimmunol.178.11.7242. [DOI] [PubMed] [Google Scholar]
- Potempa J, Pike RN. Corruption of innate immunity by bacterial proteases. J. Innate Immun. 2009;1:70. doi: 10.1159/000181144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potempa M, Potempa J, Okroj M, Popadiak K, Eick S, Nguyen KA, Riesbeck K, Blom AM. Binding of complement inhibitor C4b-binding protein contributes to serum resistance of Porphyromonas gingivalis. J Immunol. 2008;181:5537. doi: 10.4049/jimmunol.181.8.5537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potempa M, Potempa J, Kantyka T, Nguyen KA, Wawrzonek K, Manandhar SP, Popadiak K, Riesbeck K, Eick S, Blom AM. Interpain A, a cysteine proteinase from Prevotella intermedia, inhibits complement by degrading complement factor C3. PLoS Pathog. 2009;5:e1000316. doi: 10.1371/journal.ppat.1000316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rautemaa R, Meri S. Protection of gingival epithelium against complement-mediated damage by strong expression of the membrane attack complex inhibitor protectin (CD59) J Dent Res. 1996;75:568. doi: 10.1177/00220345960750010901. [DOI] [PubMed] [Google Scholar]
- Ricklin D, Hajishengallis G, Yang K, Lambris JD. Complement: a key system for immune surveillance and homeostasis. Nat Immunol. 2010;11:785. doi: 10.1038/ni.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schenkein HA, Genco RJ. Gingival fluid and serum in periodontal diseases. II. Evidence for cleavage of complement components C3, C3 proactivator (factor B) and C4 in gingival fluid. J Periodontol. 1977;48:778. doi: 10.1902/jop.1977.48.12.778. [DOI] [PubMed] [Google Scholar]
- Sendide K, Reiner NE, Lee JS, Bourgoin S, Talal A, Hmama Z. Cross-talk between CD14 and complement receptor 3 promotes phagocytosis of mycobacteria: regulation by phosphatidylinositol 3-kinase and cytohesin-1. J. Immunol. 2005;174:4210. doi: 10.4049/jimmunol.174.7.4210. [DOI] [PubMed] [Google Scholar]
- Seymour GJ, Ford PJ, Cullinan MP, Leishman S, Yamazaki K. Relationship between periodontal infections and systemic disease. Clin Microbiol Infect. 2007;13(Suppl 4):3. doi: 10.1111/j.1469-0691.2007.01798.x. [DOI] [PubMed] [Google Scholar]
- Socransky SS. Microbiology of periodontal disease -- present status and future considerations. J Periodontol. 1977;48:497. doi: 10.1902/jop.1977.48.9.497. [DOI] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD. Periodontal microbial ecology. Periodontol 2000. 2005;38:135. doi: 10.1111/j.1600-0757.2005.00107.x. [DOI] [PubMed] [Google Scholar]
- Socransky SS, Haffajee AD, Cugini MA, Smith C, Kent RL., Jr. Microbial complexes in subgingival plaque. J. Clin. Periodontol. 1998;25:134. doi: 10.1111/j.1600-051x.1998.tb02419.x. [DOI] [PubMed] [Google Scholar]
- Tonetti MS, D'Aiuto F, Nibali L, Donald A, Storry C, Parkar M, Suvan J, Hingorani AD, Vallance P, Deanfield J. Treatment of periodontitis and endothelial function. N Engl J Med. 2007;356:911. doi: 10.1056/NEJMoa063186. [DOI] [PubMed] [Google Scholar]
- Wang M, Shakhatreh MA, James D, Liang S, Nishiyama S, Yoshimura F, Demuth DR, Hajishengallis G. Fimbrial proteins of Porphyromonas gingivalis mediate in vivo virulence and exploit TLR2 and complement receptor 3 to persist in macrophages. J Immunol. 2007;179:2349. doi: 10.4049/jimmunol.179.4.2349. [DOI] [PubMed] [Google Scholar]
- Wang M, Krauss JL, Domon H, Hosur KB, Liang S, Magotti P, Triantafilou M, Triantafilou K, Lambris JD, Hajishengallis G. Microbial hijacking of complement-toll-like receptor crosstalk. Sci Signal. 2010;3:ra11. doi: 10.1126/scisignal.2000697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wingrove JA, DiScipio RG, Chen Z, Potempa J, Travis J, Hugli TE. Activation of complement components C3 and C5 by a cysteine proteinase (gingipain-1) from Porphyromonas (Bacteroides) gingivalis. J. Biol. Chem. 1992;267:18902. [PubMed] [Google Scholar]
- Xiong X, Buekens P, Fraser WD, Beck J, Offenbacher S. Periodontal disease and adverse pregnancy outcomes: a systematic review. Bjog. 2006;113:135. doi: 10.1111/j.1471-0528.2005.00827.x. [DOI] [PubMed] [Google Scholar]
- Yilmaz O. The chronicles of Porphyromonas gingivalis: the microbium, the human oral epithelium and their interplay. Microbiology. 2008;154:2897. doi: 10.1099/mic.0.2008/021220-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X, Kimura Y, Fang C, Zhou L, Sfyroera G, Lambris JD, Wetsel RA, Miwa T, Song WC. Regulation of Toll-like receptor-mediated inflammatory response by complement in vivo. Blood. 2007;110:228. doi: 10.1182/blood-2006-12-063636. [DOI] [PMC free article] [PubMed] [Google Scholar]