Abstract
The innate immune response is critical for the epithelial antimicrobial barrier. The human β-defensins are small, cationic antimicrobial peptides that are made by epithelial cells and that play a role in mucosal and skin defenses. Human β-defensin 1 (hBD-1) is expressed constitutively in epithelial tissues, whereas hBD-2 and hBD-3 are expressed in response to bacterial stimuli or inflammation. Previous studies showed that hBD-2 was induced by Fusobacterium nucleatum cell wall extract without the involvement of the NF-κB transcription factors, which typically are associated with innate immunity and inflammation. The goal of this study was to characterize signaling pathways involved in hBD-2 induction in response to commensal and pathogenic bacteria. Cultured human oral and foreskin keratinocytes were treated separately with inhibitors of NF-κB, c-Jun N-terminal kinase (JNK), and p38 and then stimulated with oral commensal Streptococcus gordonii, oral pathogens Porphyromonas gingivalis and Actinobacillus actinomycetemcomitans, skin commensal Staphylococcus epidermidis, or skin pathogen Streptococcus pyogenes. Different bacteria induced different levels of hBD-2 and in response to the various inhibitors tested, although certain common patterns were observed for commensal- and pathogen-stimulated cells. hBD-2 induction by all bacteria tested was partially or completely blocked by inhibitors of the JNK and p38 pathways. However, in addition, hBD-2 induction by pathogenic bacteria in both oral and foreskin keratinocytes was blocked by inhibitors of NF-κB. The results indicate that commensal and pathogenic bacteria utilize different pathways in hBD-2 induction and suggest that epithelial cells from different body sites have common signaling mechanisms to distinguish between commensal and pathogenic bacteria.
Epithelial tissues function as the first line of defense between the host and the outside environment, which includes commensal and pathogenic bacteria. Various epithelial tissues of the body are populated with distinct populations of commensal bacteria, and recent conceptual advances suggest that host tissues may promote associations with these commensal bacteria which may be beneficial to the host and may choose not to activate an immune response to eliminate them. However, this molecular dialogue between commensal organisms and the host may be disrupted by certain pathogens (34). Another advance is the concept of pattern recognition receptors continually monitoring microbial colonization and detecting conserved microbial structures, such as lipopolysaccharide (LPS), leading to host innate immune responses (30). Nevertheless, it is not clear how these concepts explain differences in the responses of epithelial cells to commensal and pathogenic organisms.
Gingival epithelium is a stratified squamous epithelium that functions as a barrier against pathogenic oral bacteria. In the oral cavity, epithelial tissues are constantly exposed to a variety of bacteria, but most individuals maintain a healthy homeostasis. Recent studies demonstrated that these tissues protect the host not only by providing a physical barrier but also through innate immune responses in the form of antimicrobial peptides (13, 21, 36). These antimicrobial peptides, defined as containing fewer than 100 amino acids, have a broad spectrum of activity against both gram-negative and gram-positive bacteria as well as against yeasts and viruses (15, 26). In humans, these antimicrobial peptides include defensins and a cathelicidin family member, LL-37 (2, 14). The human defensins include the α-defensins of intestinal and neutrophil origins and the β-defensins of skin and oral mucosa and other epithelia. The defensins function against bacteria by forming pores and disrupting bacterial membrane integrity, although new studies also suggest additional, cytoplasmic targets (12, 18).
The β-defensins are small, cationic antimicrobial peptides made by epithelial cells and expressed in all human epithelial cells tested to date (5). They were first identified in tracheal epithelial cells and were subsequently found in many types of human epithelial cells, including kidney, urinary tract, oral mucosa, and skin cells (10, 16, 25, 41, 44). β-Defensins are up-regulated by bacterial LPS as well as by tumor necrosis factor alpha (TNF-α) and interleukin 1 (IL-1) via the CD-14-mediated signal transduction pathway (10, 35). Human β-defensin 1 (hBD-1) is expressed constitutively in epithelial tissues, whereas hBD-2 and hBD-3 are expressed when epithelial tissues are stimulated with bacteria, Candida albicans, IL-1, or TNF-α (17, 22, 24, 29, 33). Although hBD-2 is induced and expressed only in inflamed sites in most tissues, in the oral epithelium it is expressed in normal, uninflamed gingival tissue as well, presumably because of the continual exposure of this mucosa to bacteria (6). The regulation of expression of multiple additional defensins is not yet known, although genes for 28 human β-defensins have been identified (38). Human β-defensins are suspected to play a role in mucosal and skin defenses through their direct antimicrobial action as well as through potential signaling between innate and acquired immune responses (39, 43). Defensins may act as adjuvants in antibody production (40). Additionally, hBD-2 attracts and stimulates immature dendritic cells via the CCR6 receptor, eventually leading to maturation, activation, and up-regulation of IL-8 (42). Thus, hBD-2 not only has direct antimicrobial properties but also indirectly stimulates a long-term immune response.
Previous studies showed that hBD-2 was induced in cultured human oral keratinocytes (HOK) and human foreskin keratinocytes (HFK) by proinflammatory cytokines, the epithelial cell activator phorbol myristate acetate (PMA), and bacteria, including the skin commensal Staphylococcus epidermidis and a cell wall extract of the oral commensal Fusobacterium nucleatum (11, 17, 23, 24, 29). Up-regulation of hBD-2 in HOK by the F. nucleatum cell wall involved mitogen-activated protein (MAP) kinase signaling pathways but not NF-κB transcription factors (23). The NF-κB transcription factor pathway is important in the cellular response to inflammatory stimuli and to the overall response to pathogens in many cell types, mediating signaling from Toll-like receptors (TLRs) in response to microbial ligands. Our goal was to characterize further the signaling pathways involved in hBD-2 induction in response to commensal and pathogenic bacteria by using both oral and skin keratinocytes. We hypothesized that epithelial cells respond differently to commensal and pathogenic bacteria and that different signaling pathways are involved in hBD-2 up-regulation by commensal and pathogenic bacteria. In this study, we distinguished the utilization of these pathways by using specific inhibitors of each pathway and verified by quantitative methods which pathways are essential for hBD-2 induction. We provide evidence that different bacteria utilize different pathways for hBD-2 induction, and a common pattern that was observed suggests that commensals and pathogens may utilize different pathways for inducing hBD-2.
MATERIALS AND METHODS
Human epithelial cells and bacterial culture conditions.
Healthy gingival samples were obtained from patients undergoing third-molar extraction at the Department of Oral Surgery, School of Dentistry, University of Washington. Fresh human neonatal foreskin samples were collected from the Dermatology Clinic at the University of Washington Medical Center. Tissue was cut into small pieces (2 by 2 mm) and treated with 6 mg of Dispase (Becton Dickinson, Franklin Lakes, N.J.)/ml overnight at 4°C to separate the epithelium from the underlying fibrous connective tissue. The epithelium readily dissociated and was incubated at 37°C in 5 ml of trypsin-EDTA (0.05% trypsin, 0.53 mM EDTA) for 10 min. Subsequently isolated HOK and HFK were grown to 80% confluence in KBM supplemented with KGM (Cambrex, Walkersville, Md.). Porphyromonas gingivalis (ATCC 33277) cells were cultured under anaerobic conditions (85% N2, 10% H2, 5% CO2) at 37°C in Trypticase soy broth (BBL, Sparks, Md.) supplemented with 1 g of yeast extract, 5 mg of hemin, and 1 mg of menadione per liter. S. epidermidis and Streptococcus pyogenes were grown in Trypticase soy broth at 37°C under static conditions. Actinobacillus actinomycetemcomitans was grown in Todd-Hewitt broth supplemented with 1 g of yeast extract per 100 ml at 37°C under anaerobic conditions. Bacterial numbers were determined by measuring density with a Klett-Summerson photometer.
Inhibitors.
Table 1 lists the inhibitors used to block specific signaling pathways. The concentrations used for 1-pyrrolidinecarbodithioic acid (PDTC), MG132, and SB203580 were determined based on an earlier study (23). For c-Jun N-terminal kinase (JNK) I (JNKI) and JNKII, doses of 50 nM, 100 nM, 1 μM, and 10 μM were tested for their ability to block hBD-2 induction in HOK by the F. nucleatum cell wall; 1 μM was determined to be the concentration that efficiently blocked hBD-2 induction without cytotoxicity.
TABLE 1.
Specific inhibitors used in this study
| Inhibitor | Target | Function | Concn (μM) |
|---|---|---|---|
| PDTC | NF-κB | Blocks dissociation of IκB | 50 |
| MG132 | NF-κB | Blocks dissociation of IκB; also activates JNK and p38 | 10 |
| JNKI | JNK | Inhibits phosphorylation of JNK | 1 |
| JNKII | JNK | Inhibits phosphorylation of JNK; also partially inhibits TNF-α and IL-2 | 1 |
| SB203580 | p38 | Inhibits phosphorylation of p38 | 50 |
Conditions for RT-PCR.
Total RNA was extracted from keratinocytes by using an RNAqueous kit (Ambion, Austin, Tex.) according to the manufacturer's suggestions. Reverse transcription was performed with 1 μg of total RNA, 1× reverse transcriptase (RT) buffer, 250 nM oligo(dT) primer, 10 mM deoxynucleoside triphosphate (dNTP) mix, 50 U of RT, and 13 U of RNase inhibitor. Initial denaturation of RNA secondary structure was carried out at 72°C for 2 min, followed by annealing of the primer and template at 42°C for 1 h. The temperature was subsequently raised to 95°C for 10 min in order to inactivate RT. Alternatively, cells were lysed by using a Cells-to-cDNA II kit (Ambion) according to the manufacturer's suggestions, and 5-μl samples of the lysates were used as templates for reverse transcription. Controls without RT were included in each experiment. Amplification of the resulting cDNA was carried out with a 50-μl PCR mixture containing 3 μl of cDNA, 1× PCR buffer, 1.5 mM MgCl2, 10 mM dNTP mix, 250 nM each sense and antisense primers, and 2.5 U of Taq DNA polymerase. The PCR conditions were denaturation at 94°C for 30 s, annealing at 57°C for 30 s, and elongation at 72°C for 2 min for 35 cycles. A housekeeping gene, that for ribosomal protein (RPO), was used as a control to determine the total RNA level. The oligonucleotides for hBD-2 and RPO were previously described (24).
Conditions for real-time PCR.
The resulting cDNA was analyzed by using an iCycler (Bio-Rad, Hercules, Calif.) and a QuantiTech SYBR green PCR kit (Qiagen, Valencia, Calif.) according to the manufacturers' suggestions. The SYBR green mixture contained 100 mM KCl, 40 mM Tris-HCl (pH 8.4), 0.4 mM each dNTP, 50 U of Taq DNA polymerase/ml, 6 mM MgCl2, SYBR green I, and 20 nM fluorescein. The reaction was set up in a 96-well plate, with each well containing 25 μl of the SYBR green mixture, 5 μl of cDNA, and 250 nM each primer. The amplification conditions were initial denaturation at 95°C for 15 min followed by 40 cycles of denaturation at 95°C for 15 s, annealing at 57°C for 15 s, and elongation at 72°C for 30 s. Melting curve analysis was performed in order to confirm that the detected signal was that of SYBR green binding to the expected amplification product and not to the possible primer-dimers. The amplified product was run on an agarose gel to confirm that no spurious products were amplified during the cycles. Real-time PCR amplification was performed in duplicate, and an average was calculated. An amplification plot from a series of dilutions of hBD-2 or RPO plasmids was used to obtain a linear correlation between the threshold cycle, at which fluorescence is determined to be statistically significant above the background, and the log amount of template present. The relative ratio of hBD-2 to RPO for each sample blocked with an inhibitor was calculated and compared with the relative ratio for a bacterium-stimulated sample, set at 100% (23).
RESULTS
hBD-2 is induced by various bacteria.
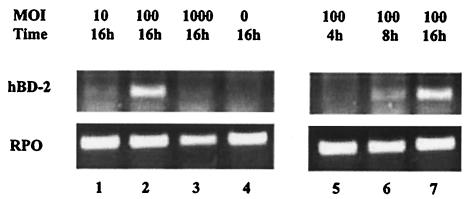
Several species of bacteria were tested for their ability to induce hBD-2 in cultured oral or skin keratinocytes. Commensal bacteria included Streptococcus gordonii and F. nucleatum (oral) and S. epidermidis (skin). For pathogenic bacteria, we tested various strains of the skin pathogen S. pyogenes and the oral pathogens P. gingivalis and A. actinomycetemcomitans. For P. gingivalis, multiplicities of infection (MOIs) of 1, 10, 100, and 1,000 were tested, and hBD-2 was induced at MOIs of between 10 and 100 (Fig. 1). At an MOI of 1,000, extensive lysis of keratinocytes was evident. At an MOI of 100, the highest level of hBD-2 induction was observed when cells were stimulated overnight for 16 h (Fig. 1). Likewise, the other bacterial species were tested for their ability to induce hBD-2 at MOIs of 1, 10, and 100, and the highest level of hBD-2 induction was observed at an MOI of 100. Thus, an MOI of 100 was used in subsequent experiments.
FIG. 1.
Dose and time responses of HOK stimulated with P. gingivalis. Lanes: 1, MOI of 10 at 16 h of stimulation; 2, MOI of 100 at 16 h; 3, MOI of 1,000 at 16 h; 4, unstimulated; 5, MOI of 100 at 4 h; 6, MOI of 100 at 8 h; 7, MOI of 100 at 16 h. An MOI of 100 at 16 h is shown twice, since it was part of each condition. RPO was used as a control for the total RNA level.
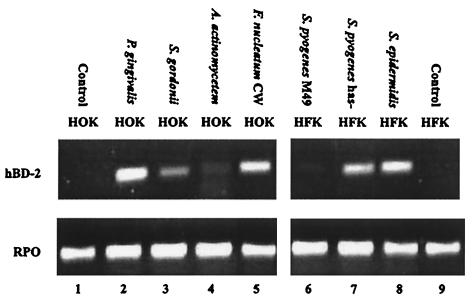
After 16 h of stimulation with bacteria, total RNA was isolated from keratinocytes and subjected to RT-PCR to detect hBD-2 mRNA expression. Each species of bacteria was tested at least three times, each with keratinocytes from different subjects, to confirm its ability to induce hBD-2. All of the species tested induced hBD-2, but at various levels (Fig. 2). For HFK, the highest level of hBD-2 induction was seen with S. epidermidis, whereas for HOK, the highest level of induction was seen with the F. nucleatum cell wall and P. gingivalis (Fig. 2). Four strains of S. pyogenes were initially tested for their ability to induce hBD-2 in cultured HFK. The highest level of induction was seen with S. pyogenes strain CS101 has mutant. Thus, this strain was used in subsequent experiments. The CS101 has mutant is a capsule-deficient strain and thus is expected to be less pathogenic than the other S. pyogenes strains tested. For both HFK and HOK, commensals were more effective in up-regulating hBD-2 than were pathogens, with the exception of P. gingivalis.
FIG. 2.
hBD-2 induction by various bacterial species at an MOI of 100. A. actinomycetem, A. actinomycetemcomitans; CW, cell wall extract.
Activation of NF-κB transcription factors and MAP kinases JNK and p38 during hBD-2 induction.
Earlier reports suggested that multiple pathways are involved in hBD-2 induction. hBD-2 induction by TNF-α and IL-1 presumably involves NF-κB downstream from specific proinflammatory cytokine receptors (9, 33). In contrast, hBD-2 induction by F. nucleatum involves MAP kinases JNK and p38, and MAP kinase ERK is involved in hBD-2 induction by PMA (23). In the present study, an evaluation of whole-cell protein extracts from oral keratinocytes stimulated for 4 h with S. gordonii, P. gingivalis, and A. actinomycetemcomitans confirmed that both JNK and p38 were activated during hBD-2 induction (data not shown). The same results were obtained for skin keratinocytes stimulated with S. epidermidis and S. pyogenes. Analysis of nuclear extracts from both oral and skin keratinocytes stimulated for 4 h with the bacteria used in this study also confirmed nuclear translocation of the p65 subunit of NF-κB during hBD-2 induction (data not shown), suggesting possible involvement of all of these pathways.
MAP kinases JNK and p38 are involved in hBD-2 induction by commensal bacteria.
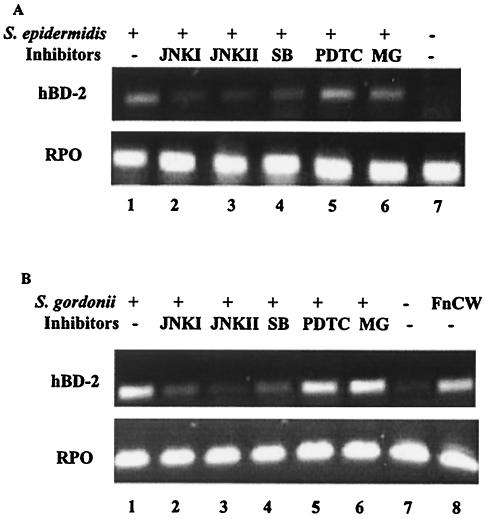
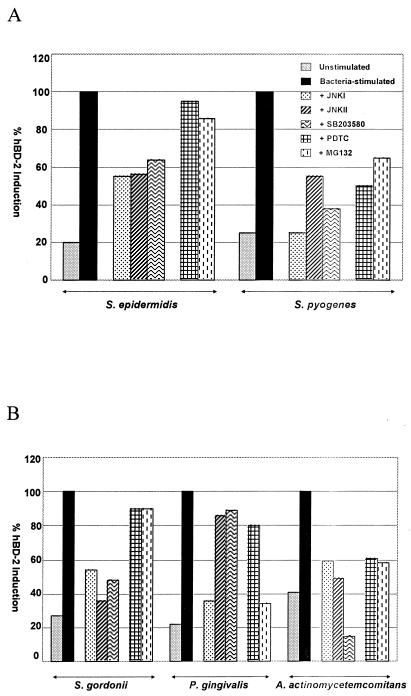
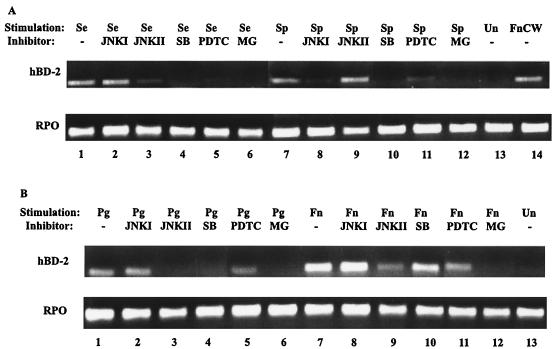
The use of different pathways in the induction of hBD-2 can be distinguished by specific inhibitors. Cultured keratinocytes were incubated with specific inhibitors for 1 h and then stimulated for 16 h with selected bacteria. Strong induction of hBD-2 by S. gordonii and S. epidermidis was observed (Fig. 3). Induction of hBD-2 by these commensal bacteria was inhibited by the MAP kinase inhibitors JNKI, JNKII, and SB203580 (Fig. 3). However, the NF-κB inhibitors PDTC and MG132 had little effect on the induction of hBD-2 by these commensal bacteria (Fig. 3). Thus, we observed for both oral and skin keratinocytes distinct differences in the inhibition by MAP kinase inhibitors and NF-κB inhibitors of hBD-2 induction by the commensal bacteria S. gordonii and S. epidermidis. The inhibition by MAP kinase inhibitors but not by NF-κB inhibitors of hBD-2 induction by commensal bacteria is in agreement with the results of earlier studies with an F. nucleatum cell wall extract (23) and confirms and extends the previous findings.
FIG. 3.
Effects of inhibitors on epithelial cells stimulated with commensal bacteria. (A) HFK stimulated with S. epidermidis. (B) HOK stimulated with S. gordonii. Lanes: 1, stimulated with bacteria only; 2 through 6, treated for 1 h prior to bacterial stimulation with JNKI, JNKII, SB203580 (SB), PDTC, and MG132 (MG), respectively; 7, unstimulated; 8, stimulated with F. nucleatum cell wall extract (FnCW) only.
MAP kinases and NF-κB transcription factors are both involved in hBD-2 induction by pathogenic bacteria.
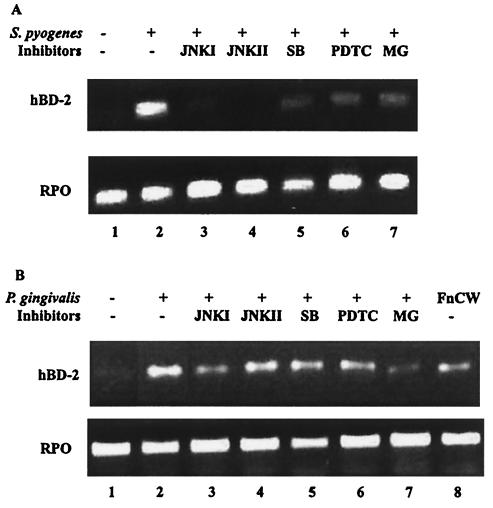
The involvement of MAP kinase pathways and the NF-κB pathway in hBD-2 induction by the oral pathogenic bacteria P. gingivalis and A. actinomycetemcomitans and by the skin pathogenic bacterium S. pyogenes was examined. P. gingivalis is a gram-negative coccobacillus and an etiologic agent of severe adult periodontitis. A. actinomycetemcomitans is a gram-negative bacterium implicated in early juvenile periodontitis. S. pyogenes, a gram-positive coccus, causes impetigo and necrotizing faciitis. As was seen with commensal bacteria, hBD-2 induction by pathogenic bacteria also was inhibited by the MAP kinase inhibitors JNKI, JNKII, and SB203580 in both oral and skin keratinocytes (Fig. 4). The decreased level of hBD-2 induction in response to the MAP kinase inhibitors was more prominent in HFK stimulated with S. pyogenes than in HOK stimulated with P. gingivalis (Fig. 4). When JNKII and SB203580 were used together in HOK prior to stimulation by P. gingivalis, a synergistic effect was seen (data not shown). hBD-2 induction by these bacteria also was inhibited by the NF-κB inhibitors PDTC and MG132 (Fig. 4), in contrast to their effects on hBD-2 induction by commensal bacteria (Fig. 3). PDTC and MG132 reduced the level of hBD-2 induction in HFK stimulated with S. pyogenes (Fig. 4A). Likewise, in HOK stimulated with A. actinomycetemcomitans, both PDTC and MG132 reduced the level of hBD-2 induction (Fig. 5B). The same results were observed for P. gingivalis-stimulated HOK, although more inhibition was observed when MG132 was used (Fig. 4B). These results suggest that the pathogens use the NF-κB pathway as an additional pathway for hBD-2 induction in both oral and skin keratinocytes.
FIG. 4.
Effects of inhibitors on epithelial cells stimulated with pathogenic bacteria. (A) HFK stimulated with S. pyogenes CS101 has mutant. (B) HOK stimulated with P. gingivalis. Lanes: 1, unstimulated; 2, stimulated with bacteria only; 3 through 7, treated for 1 h prior to bacterial stimulation with JNKI, JNKII, SB203580 (SB), PDTC, and MG132 (MG), respectively; 8, stimulated with F. nucleatum cell wall extract (FnCW) only.
FIG. 5.
Real-time PCR analysis of the effects of inhibitors on commensals and pathogens. (A) HFK stimulated with S. epidermidis or S. pyogenes CS101 has mutant. (B) HOK stimulated with S. gordonii, P. gingivalis, or A. actinomycetemcomitans. Cells were treated with inhibitors for 1 h prior to bacterial stimulation. The data represent the means of three experiments.
In order to determine whether keratinocytes from one body site would recognize commensal bacteria from another body site as pathogenic and induce hBD-2 by utilizing NF-κB transcription factors, we stimulated HOK with skin commensal S. epidermidis or skin pathogen S. pyogenes. hBD-2 induction by both bacteria in HOK was inhibited by inhibitors of JNK, p38, and NF-κB (Fig. 6A). Similar results were observed when HFK were stimulated with the oral commensal F. nucleatum cell wall or oral pathogen P. gingivalis (Fig. 6B). These results indicate that keratinocytes recognize any bacteria from other body sites as pathogens and utilize NF-κB transcription factors as well as MAP kinases in hBD-2 induction. NF-κB is involved in the inflammatory response, and our results are consistent with an inflammatory response to pathogens independent of classification as gram positive or gram negative.
FIG. 6.
Epithelial cells respond to commensal bacteria from another body site as if they were pathogenic bacteria in hBD-2 induction. (A) HOK stimulated with skin commensal S. epidermidis (Se) or skin pathogen S. pyogenes CS101 has mutant (Sp). (B) HFK stimulated with oral pathogen P. gingivalis (Pg) or oral commensal F. nucleatum cell wall (Fn). Lanes: 1 and 7, stimulated with bacteria only; 2 and 8, 3 and 9, 4 and 10, 5 and 11, and 6 and 12, treated for 1 h prior to bacterial stimulation with JNKI, JNKII, SB203580 (SB), PDTC, and MG132 (MG), respectively; 13, unstimulated (Un); 14, stimulated with F. nucleatum cell wall extract (FnCW) only.
Quantitative real-time PCR.
In order to confirm the RT-PCR data with a quantitative analysis, we performed real-time PCR with SYBR green I dye. A quantitative assessment of end-point values obtained by RT-PCR can be only semiquantitative, because the results can be influenced by limiting reagents, small differences in reaction components, or cycling parameters. Real-time PCR data are obtained during the exponential phase of PCR amplification, thus providing a more reliable quantitative analysis. The initial copy number of the gene of interest is based on the threshold cycle, which is inversely proportional to the log of the initial copy number in the template. Because SYBR green I fluoresces when it binds nonspecifically to double-stranded DNA, melting curve analysis was performed to confirm that the detection of fluorescence occurred above the melting temperature of possible primer-dimers (data not shown). Visualization on an agarose gel confirmed the purity of the amplified product and confirmed that no spurious products were present (data not shown). Figure 5 shows the various levels of hBD-2 induction by the strains of bacteria used in this study. Various responses were obtained when the effects of MAP kinase inhibitors on the induction of hBD-2 by commensal and pathogenic bacteria were tested (Fig. 5). For S. gordonii-stimulated HOK, all three inhibitors of MAP kinases gave similar inhibition levels. For cells stimulated with A. actinomycetemcomitans, the inhibitor of MAP kinase p38 (SB203580) reduced the level of hBD-2 induction the most, whereas with P. gingivalis, JNKI blocked hBD-2 induction the most. Consistent with the results obtained by RT-PCR, hBD-2 induction was decreased by NF-κB inhibitors in both oral and skin keratinocytes stimulated with pathogens (Fig. 5). For S. pyogenes- and A. actinomycetemcomitans-stimulated cells, both PDTC and MG132 reduced hBD-2 induction, while for P. gingivalis-stimulated cells, MG132 was more effective in inhibiting hBD-2 induction, consistent with the RT-PCR findings (Fig. 5). These real-time PCR results illustrate the diversity of pathways used in hBD-2 induction and the utilization of the NF-κB pathway in hBD-2 induction by pathogenic bacteria in the oral epithelium and the epidermis.
DISCUSSION
In this study, we found that hBD-2 is induced by both commensal and pathogenic bacterial species in cultured keratinocytes derived from the gingiva of the oral cavity and from the epidermis. In the oral epithelium, a stratified epithelium, hBD-2 also is induced by TNF-α, IL-1, and PMA, but bacterial LPS is a poor stimulant (24, 29). In contrast, in the tracheal epithelium, a pseudostratified epithelium, bacterial LPS up-regulates hBD-2 mRNA transcription, and antibodies against CD14, a cell surface receptor for LPS, inhibit this transcription (9, 10). CD14 also interacts with TLR4 to activate NF-κB (37). These findings suggest that TLRs may be involved in the epithelial innate immune response and hBD-2 induction. Nevertheless, LPSs from F. nucleatum and Escherichia coli were poor stimulants of hBD-2 in cultured HOK even when human serum was added as a source of LPS binding protein or soluble CD14 (24). In human skin keratinocytes, E. coli LPS weakly induced hBD-2 signaling, but the induction was greatly increased when monocyte-derived cells were used as intermediaries between LPS and the epidermal keratinocytes (27) because of the amplified epidermal response to LPS through IL-1-mediated signaling and the action of IL-1 as the dominant inducer of hBD-2 (27). Further investigations are needed to test more extensively the possible roles of TLRs in hBD-2 induction, which may differ in epithelial cells derived from different sources. Regardless of the possible involvement of LPS in hBD-2 induction, both gram-negative and gram-positive bacteria induced hBD-2 in cultured HOK (Fig. 2). Furthermore, the effects of MAP kinase and NF-κB inhibitors on hBD-2 induction were the same in cultured keratinocytes stimulated with gram-negative (F. nucleatum) and gram-positive (S. epidermidis and S. gordonii) commensals (Fig. 3) (23).
The expression of hBD-2 is associated with the differentiation of epithelial cells (6). The promoter region of hBD-2 contains multiple NF-κB and AP-1 binding sites (28). Because AP-1 is a transcription factor involved in cell proliferation and differentiation and because MAP kinases activate AP-1, it is reasonable to expect that MAP kinases will regulate hBD-2 gene transcription. MAP kinase signaling activity also is known to be associated with the invasion process of pathogenic bacteria, such as Salmonella, Neisseria gornorrheae, and enteropathogenic E. coli (8, 31, 32). Although MAP kinase ERK is involved in hBD-2 induction by PMA, we did not observe the activation of ERK during hBD-2 induction by the bacteria tested here. In hBD-2 induction by pathogens, various inhibitory levels were observed with MAP kinase inhibitors, but decreased hBD-2 expression was observed consistently with NF-κB transcription factor inhibitors (Fig. 5). Other studies reported the involvement of NF-κB in hBD-2 up-regulation in intestinal and tracheal epithelial cells stimulated with Salmonella and E. coli LPSs, respectively (3, 33). However, different cell types were used in those studies. Other studies were done with nonstratified or immortalized cell lines, which may differ from the primary cell lines derived from the stratified epithelia used here.
Our data illustrate the diversity of pathways used in hBD-2 induction in both oral and skin keratinocytes, and the consistent pattern observed indicates that commensal and pathogenic bacteria may use different pathways for the induction of hBD-2. The variety of responses that we observed also may indicate that different components of bacteria act with different receptors, such as TLRs. TLR2 is involved in the inflammatory response to lipoteichoic acid and lipopeptides, and TLR4 is involved in the response to LPS (4, 19). Studies with inhibitors distinguish only downstream events and not initial receptor-bacterium interactions. Therefore, it is possible that both commensal and pathogenic bacteria use the same TLRs initially but that different components of bacteria have different affinities for certain phosphorylation cascades or require different cofactors or adaptor proteins, thus taking different routes later in the pathway. Further investigations are needed to understand whether commensals and pathogens interact with different TLRs or other cell surface receptors or with a combination of several types of receptors.
Compared to the other pathogenic bacteria tested here, A. actinomycetemcomitans and S. pyogenes, P. gingivalis showed only a slight decrease in hBD-2 induction in response to PDTC. This response seems to be unique to P. gingivalis. This organism has mechanisms to down-regulate the host immune response, such as suppression of IL-8 expression induced by commensal bacteria (7, 20). In contrast to E. coli LPS, P. gingivalis LPS does not induce gamma interferon or IL-12 (34). Therefore, compared to other pathogenic gram-negative bacteria, P. gingivalis may have a different way of interacting with the host innate immune response. Additionally, P. gingivalis produces numerous proteases, including arginine-specific (RgpA and RgpB) and lysine-specific (Kgp) gingipain proteases. Therefore, we cannot rule out the possibility that P. gingivalis does not utilize a pathogen-associated molecular pattern but rather uses proteases for the induction of hBD-2. This enzymatic activity may confer on P. gingivalis its unique additional mechanisms of interaction with HOK. Krisanaprakornkit et al. reported that a P. gingivalis crude cell wall extract treated with protease inhibitors did not induce hBD-2 in cultured HOK (24). Although we report in this study that a viable intact P. gingivalis culture induced hBD-2, P. gingivalis cells washed in phosphate-buffered saline failed to induce hBD-2 in HOK at any MOI (data not shown). All of these findings suggest a possible role of P. gingivalis proteases in hBD-2 induction, and we are currently investigating whether a specific P. gingivalis protease is involved in hBD-2 induction in HOK.
The oral mucosa is constantly exposed to a variety of bacteria but still generally maintains a healthy balance. Both constitutive and inducible β-defensins are expressed in HOK, suggesting the existence of general and specific innate host defense systems in response to infection. Dale et al. suggested that oral mucosal cells are in an activated state with respect to hBD-2 expression and that this state is part of the normal barrier function of the oral epithelium (6). In contrast, in the epidermis, hBD-2 expression is associated primarily with inflammation or a diseased state (1). Our data suggest that hBD-2 is induced by both commensal and pathogenic bacteria and that NF-κB transcription factors are not involved in hBD-2 induction by commensals. It is possible that commensals use more discrete pathways, not involving genes associated with innate immunity or inflammation, for the induction of hBD-2. This study provides evidence that the oral epithelium and the epidermis use the same signaling pathways for hBD-2 induction and that there are individualized epithelial cell responses to different bacterial species.
Acknowledgments
We thank Carol Belton and Choe Sangchi Tang, Comprehensive Center for Oral Health Research, University of Washington, for providing HOK and Phil Fleckman and Anna Pirrone, Department of Dermatology, University of Washington, for providing HFK. We thank Gary Darmstadt for providing the S. pyogenes strains. We also thank Richard Darveau for helpful discussions.
This work was supported by NIDCR grants R01 DE 013573 and P60 DE 013061.
Editor: W. A. Petri, Jr.
REFERENCES
- 1.Ali, R. S., A. Falconer, M. Ikram, C. E. Bissett, R. Cerio, and A. G. Quinn. 2001. Expression of the peptide antibiotics human beta defensin-1 and human beta defensin-2 in normal human skin. J. Investig. Dermatol. 117:106-111. [DOI] [PubMed] [Google Scholar]
- 2.Bals, R., X. Wang, M. Zasloff, and J. M. Wilson. 1998. The peptide antibiotic LL-37/hCAP-18 is expressed in epithelia of the human lung where it has broad antimicrobial activity at the airway surface. Proc. Natl. Acad. Sci. USA 95:9541-9546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Becker, M. N., G. Diamond, M. W. Verghese, and S. H. Randell. 2000. CD14-dependent lipopolysaccharide-induced beta-defensin-2 expression in human tracheobronchial epithelium. J. Biol. Chem. 275:29731-29736. [DOI] [PubMed] [Google Scholar]
- 4.Brightbill, H. D., D. H. Libraty, S. R. Krutzik, R. B. Yang, J. T. Belisle, J. R. Bleharski, M. Maitland, M. V. Norgard, S. E. Plevy, S. T. Smale, P. J. Brennan, B. R. Bloom, P. J. Godowski, and R. L. Modlin. 1999. Host defense mechanisms triggered by microbial lipoproteins through Toll-like receptors. Science 285:732-736. [DOI] [PubMed] [Google Scholar]
- 5.Dale, B. A. 2002. Periodontal epithelium: a newly recognized role in health and disease. Periodontology 2000 30:70-78. [DOI] [PubMed] [Google Scholar]
- 6.Dale, B. A., J. R. Kimball, S. Krisanaprakornkit, F. Roberts, M. Robinovitch, R. O'Neal, E. V. Valore, T. Ganz, G. M. Anderson, and A. Weinberg. 2001. Localized antimicrobial peptide expression in human gingiva. J. Periodontal Res. 36:285-294. [DOI] [PubMed] [Google Scholar]
- 7.Darveau, R. P., C. M. Belton, R. A. Reife, and R. J. Lamont. 1998. Local chemokine paralysis, a novel pathogenic mechanism for Porphyromonas gingivalis. Infect. Immun. 66:1660-1665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Grado, M., C. M. Rosenberger, A. Gauthier, B. A. Vallance, and B. B. Finlay. 2001. Enteropathogenic Escherichia coli infection induces expression of the early growth response factor by activating mitogen-activated protein kinase cascades in epithelial cells. Infect. Immun. 69:6217-6224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Diamond, G., V. Kaiser, J. Rhodes, J. P. Russell, and C. L. Bevins. 2000. Transcriptional regulation of beta-defensin gene expression in tracheal epithelial cells. Infect. Immun. 68:113-119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Diamond, G., J. P. Russell, and C. L. Bevins. 1996. Inducible expression of an antibiotic peptide gene in lipopolysaccharide-challenged tracheal epithelial cells. Proc. Natl. Acad. Sci. USA 93:5156-5160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dinulos, J. G., L. Mentele, L. P. Fredericks, B. A. Dale, and G. L. Darmstadt. 2003. Keratinocyte expression of human beta defensin 2 following bacterial infection: role in cutaneous host defense. Clin. Diagn. Lab. Immunol. 10:161-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Friedrich, C. L., A. Rozek, A. Patrzykat, and R. E. Hancock. 2001. Structure and mechanism of action of an indolicidin peptide derivative with improved activity against gram-positive bacteria. J. Biol. Chem. 276:24015-24022. [DOI] [PubMed] [Google Scholar]
- 13.Ganz, T. 1999. Defensins and host defense. Science 286:420-421. [DOI] [PubMed] [Google Scholar]
- 14.Hancock, R. E., and M. G. Scott. 2000. The role of antimicrobial peptides in animal defenses. Proc. Natl. Acad. Sci. USA 97:8856-8861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hancock, R. E. W. 1997. Peptide antibiotics. Lancet 349:418-422. [DOI] [PubMed] [Google Scholar]
- 16.Harder, J., J. Bartels, E. Christophers, and J. M. Schroder. 1997. A peptide antibiotic from human skin. Nature 387:861. [DOI] [PubMed] [Google Scholar]
- 17.Harder, J., U. Meyer-Hoffert, L. M. Teran, L. Schwichtenberg, J. Bartels, S. Maune, and J. M. Schroder. 2000. Mucoid Pseudomonas aeruginosa, TNF-alpha, and IL-1beta, but not IL-6, induce human beta-defensin-2 in respiratory epithelia. Am. J. Respir. Cell Mol. Biol. 22:714-721. [DOI] [PubMed] [Google Scholar]
- 18.Hill, C. P., J. Yee, M. E. Selsted, and D. Eisenberg. 1991. Crystal structure of defensin HNP-3, an amphiphilic dimer: mechanisms of membrane permeabilization. Science 251:1481-1485. [DOI] [PubMed] [Google Scholar]
- 19.Hoshino, K., O. Takeuchi, T. Kawai, H. Sanjo, T. Ogawa, Y. Takeda, K. Takeda, and S. Akira. 1999. Cutting edge: Toll-like receptor 4 (TLR4)-deficient mice are hyporesponsive to lipopolysaccharide: evidence for TLR4 as the Lps gene product. J. Immunol. 162:3749-3752. [PubMed] [Google Scholar]
- 20.Huang, G. T., D. Kim, J. K. Lee, H. K. Kuramitsu, and S. K. Haake. 2001. Interleukin-8 and intercellular adhesion molecule 1 regulation in oral epithelial cells by selected periodontal bacteria: multiple effects of Porphyromonas gingivalis via antagonistic mechanisms. Infect. Immun. 69:1364-1372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Huttner, K. M., and C. L. Bevins. 1999. Antimicrobial peptides as mediators of epithelial host defense. Pediatr. Res. 45:785-794. [DOI] [PubMed] [Google Scholar]
- 22.Jurevic, R. J., M. Bai, R. B. Chadwick, T. C. White, and B. A. Dale. 2003. Single-nucleotide polymorphisms (SNPs) in human beta-defensin 1: high-throughput SNP assays and association with Candida carriage in type I diabetics and nondiabetic controls. J. Clin. Microbiol. 41:90-96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Krisanaprakornkit, S., J. R. Kimball, and B. A. Dale. 2002. Regulation of human beta-defensin-2 in gingival epithelial cells: the involvement of mitogen-activated protein kinase pathways, but not the NF-kappaB transcription factor family. J. Immunol. 168:316-324. [DOI] [PubMed] [Google Scholar]
- 24.Krisanaprakornkit, S., J. R. Kimball, A. Weinberg, R. P. Darveau, B. W. Bainbridge, and B. A. Dale. 2000. Inducible expression of human beta-defensin 2 by Fusobacterium nucleatum in oral epithelial cells: multiple signaling pathways and role of commensal bacteria in innate immunity and the epithelial barrier. Infect. Immun. 68:2907-2915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krisanaprakornkit, S., A. Weinberg, C. N. Perez, and B. A. Dale. 1998. Expression of the peptide antibiotic human beta-defensin 1 in cultured gingival epithelial cells and gingival tissue. Infect. Immun. 66:4222-4228. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lehrer, R. I., and T. Ganz. 2002. Defensins of vertebrate animals. Curr. Opin. Immunol. 14:96-102. [DOI] [PubMed] [Google Scholar]
- 27.Liu, L., A. A. Roberts, and T. Ganz. 2003. By IL-1 signaling, monocyte-derived cells dramatically enhance the epidermal antimicrobial response to lipopolysaccharide. J. Immunol. 170:575-580. [DOI] [PubMed] [Google Scholar]
- 28.Liu, L., C. Zhao, H. H. Heng, and T. Ganz. 1997. The human beta-defensin-1 and alpha-defensins are encoded by adjacent genes: two peptide families with differing disulfide topology share a common ancestry. Genomics 43:316-320. [DOI] [PubMed] [Google Scholar]
- 29.Mathews, M., H. P. Jia, J. M. Guthmiller, G. Losh, S. Graham, G. K. Johnson, B. F. Tack, and P. B. McCray, Jr. 1999. Production of beta-defensin antimicrobial peptides by the oral mucosa and salivary glands. Infect. Immun. 67:2740-2745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Medzhitov, R., C. Janeway. 2000. The Toll receptor family and microbial recognition. Trends Microbiol. 8:452-456. [DOI] [PubMed] [Google Scholar]
- 31.Mynott, T. L., B. Crossett, and S. R. Prathalingam. 2002. Proteolytic inhibition of Salmonella enterica serovar Typhimurium-induced activation of the mitogen-activated protein kinases ERK and JNK in cultured human intestinal cells. Infect. Immun. 70:86-95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Naumann, M., T. Rudel, B. Wieland, C. Bartsch, and T. F. Meyer. 1998. Coordinate activation of activator protein 1 and inflammatory cytokines in response to Neisseria gonorrhoeae epithelial cell contact involves stress response kinases. J. Exp. Med. 188:1277-1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.O'Neil, D. A., E. M. Porter, D. Elewaut, G. M. Anderson, L. Eckmann, T. Ganz, and M. F. Kagnoff. 1999. Expression and regulation of the human beta-defensins hBD-1 and hBD-2 in intestinal epithelium. J. Immunol. 163:6718-6724. [PubMed] [Google Scholar]
- 34.Roberts, F. A., and R. P. Darveau. 2002. Beneficial bacteria of the periodontium. Periodontology 2000 30:40-50. [DOI] [PubMed] [Google Scholar]
- 35.Russell, J. P., G. Diamond, A. P. Tarver, T. F. Scanlin, and C. L. Bevins. 1996. Coordinate induction of two antibiotic genes in tracheal epithelial cells exposed to the inflammatory mediators lipopolysaccharide and tumor necrosis factor alpha. Infect. Immun. 64:1565-1568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Schonwetter, B. S., E. D. Stolzenberg, and M. A. Zasloff. 1995. Epithelial antibiotics induced at sites of inflammation. Science 267:1645-1648. [DOI] [PubMed] [Google Scholar]
- 37.Schroder, N. W., B. Opitz, N. Lamping, K. S. Michelsen, U. Zahringer, U. B. Gobel, and R. R. Schumann. 2000. Involvement of lipopolysaccharide binding protein, CD14, and Toll-like receptors in the initiation of innate immune responses by Treponema glycolipids. J. Immunol. 165:2683-2693. [DOI] [PubMed] [Google Scholar]
- 38.Schutte, B. C., J. P. Mitros, J. A. Bartlett, J. D. Walters, H. P. Jia, M. J. Welsh, T. L. Casavant, and P. B. McCray, Jr. 2002. Discovery of five conserved beta-defensin gene clusters using a computational search strategy. Proc. Natl. Acad. Sci. USA 99:2129-2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Scott, M. G., C. M. Rosenberger, M. R. Gold, B. B. Finlay, and R. E. Hancock. 2000. An alpha-helical cationic antimicrobial peptide selectively modulates macrophage responses to lipopolysaccharide and directly alters macrophage gene expression. J. Immunol. 165:3358-3365. [DOI] [PubMed] [Google Scholar]
- 40.Tani, K., W. J. Murphy, O. Chertov, R. Salcedo, C. Y. Koh, I. Utsunomiya, S. Funakoshi, O. Asai, S. H. Herrmann, J. M. Wang, L. W. Kwak, and J. J. Oppenheim. 2000. Defensins act as potent adjuvants that promote cellular and humoral immune responses in mice to a lymphoma idiotype and carrier antigens. Int. Immunol. 12:691-700. [DOI] [PubMed] [Google Scholar]
- 41.Valore, E. V., C. H. Park, A. J. Quayle, K. R. Wiles, P. B. McCray, Jr., and T. Ganz. 1998. Human beta-defensin-1: an antimicrobial peptide of urogenital tissues. J. Clin. Investig. 101:1633-1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yang, D., O. Chertov, S. N. Bykovskaia, Q. Chen, M. J. Buffo, J. Shogan, M. Anderson, J. M. Schroder, J. M. Wang, O. M. Howard, and J. J. Oppenheim. 1999. Beta-defensins: linking innate and adaptive immunity through dendritic and T cell CCR6. Science 286:525-528. [DOI] [PubMed] [Google Scholar]
- 43.Yang, D., O. Chertov, and J. J. Oppenheim. 2001. The role of mammalian antimicrobial peptides and proteins in awakening of innate host defenses and adaptive immunity. Cell. Mol. Life Sci. 58:978-989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zhao, C., I. Wang, and R. I. Lehrer. 1996. Widespread expression of beta-defensin hBD-1 in human secretory glands and epithelial cells. FEBS Lett. 396:319-322. [DOI] [PubMed] [Google Scholar]