Abstract
Mycobacterium ulcerans is an environmental organism which is responsible for the disease Buruli ulcer, a necrotizing skin disease emerging in west Africa. M. ulcerans produces the polyketide-derived macrolide mycolactone, which is required for the immunosuppression and tissue damage which characterizes Buruli ulcer. We have extracted lipids from the cell envelope and culture filtrate from 52 isolates of Mycobacterium species, analyzed them with thin-layer chromatography, and tested them in a murine fibroblast cell line (L929) cytotoxicity assay to investigate whether these mycobacterial species produce mycolactone. For these studies chloroform-methanol (2:1, vol/vol) extracts were prepared from representative fast- and slow-growing mycobacterial species. Isolates tested included 16 uncharacterized, slow-growing, environmental mycobacterial species isolated from areas in which M. ulcerans infection is endemic. Although several strains of mycobacteria studied produced cytopathic lipids, none of these produced a phenotype on cultured cells consistent with that produced by mycolactone. Two mycobacterial species, M. scrofulaceum and M. kansasii, and eight of the environmental mycobacterial isolates contained cell-associated lipids cytopathic to fibroblasts at concentrations of 33 to 1,000 μg/ml. In contrast, mycolactone produces cytotoxicity at less than 2 ng/ml. Analysis of 16S rRNA sequences from the eight environmental isolates suggests that these are novel mycobacterial species. Results from these studies suggest that, although production of cytopathic lipids is relatively common among mycobacterial species, the production of mycolactone as a cell-associated or secreted molecule appears so far to be restricted to M. ulcerans.
Mycobacterium ulcerans is the causative agent of Buruli ulcer, an emerging infection in west Africa. Buruli ulcer is the third most common mycobacterial disease in immunocompetent people. The disease is characterized by progressive, severe necrotizing skin lesions that do not respond to antimicrobial therapy and require surgical excision as treatment. The first stage of disease is indicated by a papule or nodule. As the disease progresses, ulceration occurs, accompanied by necrosis of the subcutaneous fat and vascular damage. Extensive sloughing and in some cases autoamputation of an infected limb may occur (18). M. ulcerans is an acid-fast, slow-growing, environmental organism. Infection by M. ulcerans is associated with residence in swampy areas. The mode of transmission has not been identified, but evidence suggests that bacteria may be introduced to the host through minor wounds which come in contact with infected water (17). Recent evidence has also implicated the involvement of several species of aquatic insects as potential vectors for M. ulcerans (10; F. Portaels, P. Elsen, A. Guimaraes-Peres, P. A. Fonteyne, and W. M. Meyers, Letter, Lancet 353:986, 1999).
M. ulcerans produces the polyketide-derived macrolide mycolactone, which is required for the organism's virulence. Mycolactone exhibits a specific cytopathic effect on murine L929 fibroblasts characterized by cell rounding within 24 h and cell cycle arrest in the G0/G1 phase of the cell cycle, followed by apoptosis in 72 h (4). Mycolactone is a relatively polar lipid, obtained by extracting bacteria or culture filtrate with chloroform-methanol (2:1, vol/vol) and enriched for by adding ice-cold acetone, a process which removes the phospholipids and leaves the acetone-soluble lipids (ASLs) (3).
Lipids are important virulence determinants for many microorganisms. For example, lipopolysaccharide (LPS) is a major virulence factor of gram-negative bacteria (6). Mycobacterium species lack LPS, but their cell envelope is composed of several biologically active lipids, many of which are polyketide derived. Phthiocerol dimycocerosate in M. tuberculosis and phenolic glycolipid PGL-1 in M. leprae are two examples of cell wall-associated polyketide-derived virulence determinants which may play a role in virulence (1, 14).
Macrolides are a specific class of polyketides which contain a lactone ring. This class of molecules includes many useful antibiotics (e.g., erythromycin), antifungals (e.g., amphotericin B), and immunosuppressants (e.g., rapamycin). Bacterially encoded macrolides are made by nonpathogenic soil bacteria, such as Streptomyces and Saccharopolyspora spp., as secondary metabolites. Mycolactone is the only macrolide produced by a human pathogen as well as the only known macrolide produced by a mycobacterial species (5, 8).
Although clinical data suggest that M. ulcerans is the only mycobacterial species which produces a secreted toxin, studies have never been conducted on a large number of mycobacterial species to determine whether mycolactone or related molecules might be present. Two recent findings led us to investigate the distribution of mycolactone among mycobacterial species. Mycolactone is a hybrid polyketide comprising two polyketides, one of which cyclizes to form the core lactone (5, 11). We have shown that the core lactone precursor to mycolactone can be detected on the cell surface of M. ulcerans and that this molecule is cytopathic at concentrations above 10 μg/ml (11). Second, the presence of a large numbers of insertion sequence (IS) elements in M. ulcerans raises the possibility that some or all of the genes encoding mycolactone may have been acquired by horizontal transfer from other bacteria (15). If these bacteria lacked factors required for colonization of the human host, mycolactone production would probably not have been detected. We were particularly interested in investigating the mycolactone phenotype of environmental mycobacteria obtained from aquatic sources in areas of endemicity for Buruli ulcer.
In this study we have extracted lipids from a collection of 61 mycobacterial isolates representing 30 species of mycobacteria and used cytopathic activity (CPA) and thin-layer chromatography (TLC) along with mass spectroscopy (MS) to determine if any of these mycobacteria make mycolactone or mycolactone-related molecules. Included in this collection of strains are 16 slow-growing, uncharacterized, environmental isolates of mycobacteria, which were isolated from water in regions of endemicity for M. ulcerans infection within the Democratic Republic of Congo. Although we found that several mycobacterial species, including eight of the Congo environmental isolates (CEI), produce cytopathic lipids, the specific cytopathicity phenotype produced by these lipids was markedly different from that produced by mycolactone. Finally, phylogenetic analysis of the environmental isolates based on 16S rRNA sequence shows that these uncharacterized CEI are new species of Mycobacterium, none of which are closely related to M. ulcerans.
MATERIALS AND METHODS
Bacterial cultures.
M. paratuberculosis K10 was obtained from John Bannantine, National Animal Disease Center, Ames, Iowa. M. abscessus isolates 353, 706, and 44196 were obtained from Barry Kreiswirth, Public Health Research Institute, Newark, N.J. All other known Mycobacterium species that were tested were obtained from the Trudeau Mycobacterial Culture Collection (TMCC) and stored at −80°C in our laboratory. Mycobacteria were grown without shaking in Middlebrook 7H9 medium (Difco, Detroit, Mich.) supplemented with 10% Middlebrook oleic acid-bovine serum albumin-dextrose-catalase enrichment (OADC supplement) at 37°C in 175-cm2 tissue culture flasks (Falcon) and incubated at the optimal temperature for the individual species. Environmental mycobacteria were isolated from water environments in Congo and were received on Lowenstein-Jensen slants. These strains were grown at 32°C in Middlebrook 7H9 with OADC supplement in 25-cm2 tissue culture flasks and transferred at a 1:100 dilution to 175-cm2 flasks.
Preparation of ASLs.
ASLs were harvested from late-log-phase cultures (approximately 3 weeks for TMCC species and 12 to 24 weeks for CEI) to obtain the highest yield of secondary metabolites. This corresponds to the growth phase when macrolide production is at a peak in M. ulcerans (11). The bacterial cell wall was stripped of lipids by extraction with chloroform-methanol (2:1, vol/vol). Polyketides are relatively polar lipids within this extract. To further enrich for polyketides, ASLs were prepared from lipids by adding ice-cold acetone to remove phospholipids (2). Briefly, aliquots of 200 ml from late-log-phase cultures of the mycobacteria tested were pelleted, dried, and weighed. The dried pellet was extracted with chloroform-methanol (2:1, vol/vol) for 2 h at room temperature. Cultures were once again spun to pellet the bacterial debris and separate the organic matter. The organic layer was dried down with a Rotoevaporator (Buchi) at 56°C and resuspended in ice-cold acetone. ASLs were analyzed by silica TLC plates (Alltech) with a solvent system of chloroform-methanol-water (90:10:1, vol/vol/vol). Lipids were visualized with ceric sulfate-ammonium molybdate in 2 M sulfuric acid stain as previously described (11).
Purification of cytopathic lipids.
ASLs which demonstrated any cytopathic effect were run on silica TLC plates in three different solvent systems, 90:10, 93:7, and 95:5 (vol/vol) chloroform-methanol, to separate individual lipids. Preparative separation of individual lipid species was achieved with a Chromatotron (Harrison Research, Palo Alto, Calif.). Briefly, the appropriate solvent was added to a separatory funnel and run through the Chromatotron until 25 ml of solvent was collected. ASLs were resuspended in 2 ml of solvent and loaded into the Chromatotron, and 5-ml fractions of solvent eluates (14) were collected. The samples were analyzed by TLC and visualized with ceric sulfate-ammonium molybdate in 2 M sulfuric acid stain. Lipid fractions obtained by this method were tested in duplicate on L929 murine fibroblasts for CPA.
Cytopathicity assays.
L929 murine fibroblast cells were maintained in Dulbecco modified Eagle medium supplemented with 5% heat-inactivated calf serum in 25-cm2 tissue culture flasks at 37°C with 5% CO2 as previously described (2). ASL samples in 100% ethanol were diluted with tissue culture medium, and serial dilutions (1:2) were added to L929 cells at 1/20 of the volume. Samples were tested at concentrations from 20,000 μg/ml to less than 1 ng/ml. Cytopathicity was defined broadly. A sample was scored positive for cytopathicity if it caused altered cell morphology of greater than 90% of the cells in the monolayer. Partial phenotypes (i.e., 25 and 50% cell alteration) were not observed, and CPA was determined as the lowest concentration of lipid required for altered cell morphology. At 24 and 48 h posttreatment, cells were examined microscopically to determine the presence of specific mycolactone-mediated cytopathicity. This phenotype has been previously established as the minimal concentration of ASLs per milliliter which caused inhibition of cell growth by 24 h and 90% cell rounding with loss of the monolayer by 48 h (2-4). Some non-M. ulcerans mycobacterial ASLs repeatedly produced altered cell morphology of greater than 90% of the monolayer although the kinetics and morphology were not similar to those produced by mycolactones. These lipids were categorized as cytopathic. All cytopathic ASLs were tested at least three times in multiple dilutions. Culture filtrates (CF) were also assayed for cytopathicity. For these assays a 50% dilution of CF in tissue culture medium was added to cultured cells, from which further twofold dilutions were made. Cytopathicity was assessed as described above. All samples were tested in multiple dilutions at least twice.
Identification of environmental mycobacterial species.
Environmental isolates were tested for the presence of the ISs IS2404 and IS2606 to determine whether they were M. ulcerans. PCR amplification of IS2404 was performed with primers MU5 (5′-AGCGACCCCAGTGGATTGGT-3′) and MU6 (5′-CGGTGATCAAGCGTTCACGA-3′), which amplified a region of approximately 492 bp, and amplification of IS2606 was performed with primers MU7 (5′-GGCCTGGCGGATTGCTCAAGG-3′) and MU8 (5′-CGTAGATGTGGGCGAAATGG-3′), which amplified a region of approximately 332 bp, as previously described (16).
To obtain taxonomic information for the uncharacterized mycobacterial species, PCR amplifications of a 577-bp region of the 16S rRNA gene were performed with primers pA (5′-AGAGTTTGATCCTGGCTCAG-3′) and MSHE (5′-GCGACAAACCACCTACGAG-3′) as previously described (7).
Cloning of 16S rRNA.
Genomic DNA was extracted from eight of the CEI (01-627, 01-628, 01-629, 01-632, 01-633, 01-636, 01-664, and 01-665) by a DNA extraction method for mycobacteria previously described (12). PCR products from the 16S rRNA primers were transformed into Escherichia coli with the TA TOPO cloning kit (Invitrogen, Carlsbad, Calif.), and transformants were selected by 5-bromo-4-chloro-3-indolyl-β-d-galactopyranoside (X-Gal)-mediated blue-white screening and ampicillin resistance. Positive clones were grown in 25 ml of Luria-Bertani broth-ampicillin (50 μg/ml) at 37°C with shaking at 250 rpm. The Wizard Plus Minipreps DNA purification system (Promega, Madison, Wis.) was used to isolate plasmid DNA. Purified plasmid DNA was bidirectionally sequenced at the University of Tennessee Molecular Biology Research Facility (Knoxville).
Phylogenetic analysis.
The 16S rRNA sequences of the eight CEI with cytopathic lipids were compared with those of closely related Mycobacterium species and Nocardia species, whose sequences were taken from the GenBank database.
Sequence alignments were performed with Clustal W (Megalign, version 5; DNAStar, Madison, Wis.). A phylogenetic tree was constructed and bootstrap values, with 500 permutations, were determined with the computer program Molecular Evolutionary Genetics Analysis (MEGA), version 2.1 (9). The neighbor-joining (NJ) method with Jukes-Cantor distances was used to construct the tree. Nocardia asteroides and Nocardia farcinica were used as outgroups to root the tree.
Nucleotide sequence accession numbers.
The nucleotide sequences for 01-627, 01-628, 01-629, 01-632, 01-633, 01-636, 01-664, and 01-665 have been assigned the GenBank accession no. AY312270, AY312271, AY312272, AY312273, AY312274, AY312275, AY312276, and AY312277, respectively.
RESULTS
TLC analysis of mycobacteria for detection of mycolactone.
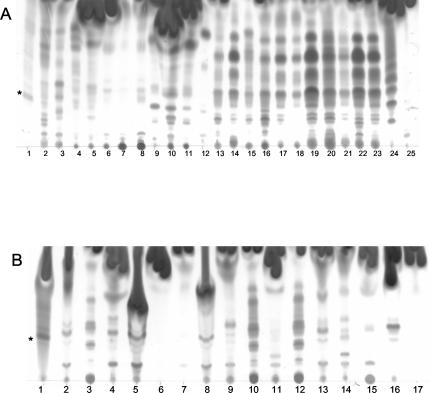
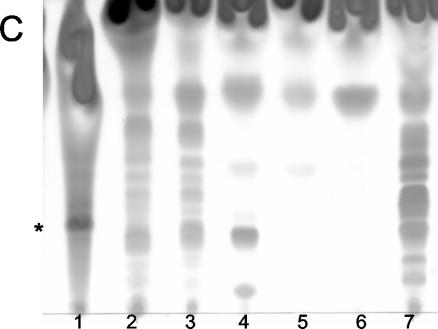
Mycolactone is a light yellow UV-active lipid with a refractive index (Rf) of 0.23 in chloroform-methanol-water (90:10:1, vol/vol/vol) (4). TLC analysis of unstained lipids showed that, although the strains tested contained many lipids which were UV active, none contained a light yellow UV-active component with an Rf similar to that of mycolactone (data not shown). Since the core lactone lacks conjugated double bonds and is therefore not pigmented, the core could not be detected by this method (11). TLC analysis of stained lipids from slow- and fast-growing mycobacterial species showed that most of the mycobacteria tested contained many polar lipids with Rf values less than 0.5 (Fig. 1). In fact, most of these mycobacteria contained a greater quantity and variety of polar lipids than M. ulcerans. However, only thin-layer chromatograms of ASLs from M. ulcerans contained a lipid species indicating the presence of mycolactone (Fig. 1).
FIG. 1.
Silica TLC of ASLs from all species of Mycobacterium tested, run in chloroform-methanol-water (90:10:1, vol/vol/vol) and visualized by oxidative charring in a ceric molybdate-10% sulfuric acid stain. (A) ASLs extracted from the slow-growing TMCC strains. Strains are represented in lanes as follows: 1, 1615; 2, 706; 3, 1464; 4, 1467; 5, 1456; 6, 1318; 7, 1319; 8, 1324; 9, 1203; 10, 1204; 11, 1214; 12, 1601; 13, 1302; 14, 1305; 15, 1306; 16, 1307; 17, 1312; 18, 1314; 19, 1315; 20, 1316; 21, 1320; 22, 1321; 23, 1323; 24, 1481; 25, 1541. *, mycolactone. (B) ASLs extracted from the slow-growing CEI. Strains are represented in lanes as follows: 1, 1615; 2, 01-626; 3, 01-627; 4, 01-628; 5, 01-629; 6, 01-630; 7, 01-631; 8, 01-632; 9, 01-633; 10, 01-634; 11, 01-635; 12, 01-636; 13, 01-664; 14, 01-665; 15, 01-666; 16, 01-667; 17, 01-668. (C) ASLs extracted from fast-growing TMCC strains. Strains are represented in lanes as follows: 1, 1615; 2, 1524; 3, 1543; 4, 1529; 5, 1530; 6, 1516; 7, 1515.
Analysis of ASLs for mycolactone-mediated cytopathicity.
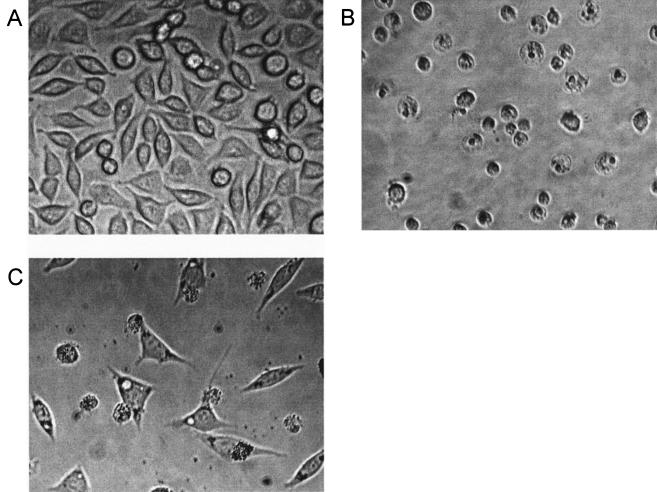
ASLs from 63% of the mycobacterial isolates tested (32 of 52) were not cytopathic for L929 cells, even at concentrations greater than 100 μg/ml. Cytopathicity, when it occurred, was much more likely to be associated with slow-growing rather than fast-growing mycobacterial species. None of the nine isolates representing six species of fast-growing Mycobacterium species contained cytopathic lipids despite the fact that several species tested can cause skin infections in humans. Fast-growing mycobacterial species tested were M. abscessus isolates 353, 706, 44196, and TMC 1424; M. chelonae TMC 1543; M. fortuitum TMC 1529 and TMC 1530; M. phlei TMC 1516; and M. smegmatis TMC 1515. Among the 11 known slow-growing mycobacterial species tested, only two contained cytopathic isolates. ASLs from 3 out of 3 M. kansasii isolates and from 7 out of 11 M. scrofulaceum isolates produced cytopathic effects on L929 cells (Table 1). Both of these species are associated with human disease. In addition it was surprising to find that ASLs from 8 out of the 16 CEI isolates tested representing at least eight different species (01-627, 01-628, 01-629, 01-632, 01-633, 01-636, 01-664, and 01-665) were cytopathic (Table 1). However the CPA of these ASLs differed greatly from that produced by mycolactone in potency, kinetics, and cytopathicity phenotype. None of the lipids tested approached the potency of mycolactone. Whereas M. ulcerans 1615 ASLs were cytopathic at 2.12 ng/ml, the cytopathic ASLs from other mycobacterial isolates exhibited CPA at concentrations ranging from 20,000 to 330 μg/ml (Table 1). Thus M. ulcerans 1615 ASLs are more than 100,000-fold more active than ASLs from any other isolate tested (Table 1). More importantly, the cytopathic effect produced by non-M. ulcerans ASLs differed both in terms of kinetics and morphological phenotype observed from that of mycolactone. Mycolactone-mediated cytopathicity develops slowly, and cell rounding is not prominent for 18 to 24 h (3). In contrast cytopathicity observed for ASLs from non-M. ulcerans bacteria was obvious by 4 to 6 h. Differences in cell morphology also clearly distinguished the effects of ASLs from M. ulcerans from those of other cytopathic lipids. Whereas mycolactone-treated cells were characterized by eccentric nuclei and clear cytoplasm (Fig. 2B), vacuolization was the prominent feature produced by ASLs from non-M. ulcerans isolates (Fig. 2C). Control L929 fibroblasts appeared as rhomboid cells which formed a confluent monolayer by 48 h (Fig. 2A). Mycolactone had a readily identifiable and specific effect on L929 cells, characterized by cell rounding within 24 h, inhibition of cell growth, and detachment of the monolayer within 48 h (2) (Fig. 2B). None of the other lipids tested exhibited this phenotype. In contrast, cytopathic ASLs from species other than M. ulcerans produced extensive cytoplasmic vacuolization. Vacuoles could be detected as early as 4 h although maximum vacuolization did not occur until 24 h. Despite extensive vacuolization, cells remained attached to the monolayer and retained their typical rhomboid morphology (Fig. 2). Vacuolization does not occur in cells treated with M. ulcerans ASLs, purified mycolactone, or mycolactone core (2, 3, 11). These results show that neither mycolactone nor the core mycolactone lipid was present as cell-associated lipids in any of the mycobacterial isolates tested other than M. ulcerans.
TABLE 1.
Mycobacterium species used in this study and the results of the ASL cytopathicity assays for the slow-growing mycobacteriaa
| Isolate, species | Source | CPAc of ASLs (μg/ml) |
|---|---|---|
| 1615, M. ulcerans (ATCC 35840) | TMCC | 0.0021 |
| 706, M. avium (ATCC 35714) | TMCC | —d |
| K10, M. paratuberculosis | NADCb | — |
| 1464, M. intracellulare (ATCC 35768) | TMCC | — |
| 1467, M. intracellulare (ATCC 35770) | TMCC | — |
| 1456, M. gastri (ATCC 15754) | TMCC | — |
| 1318, M. gordonae (ATCC 35756) | TMCC | — |
| 1319, M. gordonae (ATCC 35757) | TMCC | — |
| 1324, M. gordonae (ATCC 14470) | TMCC | 10,000 |
| 1203, M. kansasii (ATCC 35776) | TMCC | 1,250 |
| 1204, M. kansasii (ATCC 12478) | TMCC | 5,000 |
| 1214, M. kansasii (ATCC 35777) | TMCC | — |
| 1601, M. microti (ATCC 35781) | TMCC | 10,000 |
| 1302, M. scrofulaceum (ATCC 35785) | TMCC | — |
| 1305, M. scrofulaceum (ATCC 35786) | TMCC | 10,000 |
| 1306, M. scrofulaceum (ATCC 35787) | TMCC | 10,000 |
| 1307, M. scrofulaceum (ATCC 35788) | TMCC | 20,000 |
| 1312, M. scrofulaceum (ATCC 35790) | TMCC | 10,000 |
| 1314, M. scrofulaceum (ATCC 35791) | TMCC | 20,000 |
| 1315, M. scrofulaceum (ATCC 35792) | TMCC | 20,000 |
| 1316, M. scrofulaceum (ATCC 35793) | TMCC | — |
| 1320, M. scrofulaceum (ATCC 35794) | TMCC | — |
| 1321, M. scrofulaceum (ATCC 35795) | TMCC | — |
| 1323, M. scrofulaceum (ATCC 19981) | TMCC | — |
| 1481, M. nonchromogenicium (ATCC 19530) | TMCC | — |
| 1541, M. flavescens (ATCC 14474) | CEI | — |
| 01-626 | CEI | 2,500 |
| 01-627 | CEI | 2,500 |
| 01-628 | CEI | 10,000 |
| 01-629 | CEI | — |
| 01-630 | CEI | — |
| 01-631 | CEI | 1,600 |
| 01-632 | CEI | 10,000 |
| 01-633 | CEI | — |
| 01-634 | CEI | — |
| 01-635 | CEI | 2,500 |
| 01-636 | CEI | 2,500 |
| 01-664 | CEI | 330 |
| 01-665 | CEI | — |
| 01-666 | CEI | — |
| 01-667 | CEI | — |
| 01-668 |
Cytopathicity of ASLs of a chloroform-methanol (2:1, vol/vol) extract from Mycobacterium spp. on L929 fibroblasts after 48 h.
NADC, National Animal Disease Center, Ames, Iowa.
CPA is defined as the minimal amount of ASLs added to cause altered morphology of 90% of the cells and detachment within 48 h.
—, no cytopathic effect seen at <20 μg/μl.
FIG. 2.
Effect of ASLs on L929 fibroblasts at 48 h. Original magnification, ×200. (A) Control monolayer. (B) Effect of 1615 ASLs at 2.12 ng/ml. (C) CEI 01-628 ASLs (10,000 μg).
Analysis of mycobacterial CF for the presence of mycolactone.
George et al. have previously shown that mycolactone is present both on the cell envelope and in the CF of M. ulcerans (2). Despite the fact that several isolates of the mycobacterial species tested above contained cytopathic cell-associated lipids, cytopathicity could not be detected in CF from any isolate aside from M. ulcerans. Since ASLs were not prepared from CF, any cytotoxic lipid present would have had to be very potent or present in large amounts to produce detectable activity in the CF. These results suggest, however, that non-M. ulcerans strains which produce cell-associated cytopathic lipids do not secrete or slough off large amounts of these lipids into the CF. In summary, data from cytopathicity assays using partially purified mycolactones from the cell pellet (ASLs) and CF suggest that only M. ulcerans produces mycolactone.
Characterization of cytopathic lipids in non-M. ulcerans species.
Cultures of Mycobacterium species which contained cytopathic ASLs were expanded to 1 liter to obtain sufficient lipids for analysis of individual lipid species. Unfortunately, we were unable to assign CPA to specific lipids in most cases due to the poor growth of many isolates and the small amounts of lipids produced. Most of the environmental mycobacteria grew very slowly and only at low temperatures. In some cases 6 months of growth was required before cultures reached late log phase.
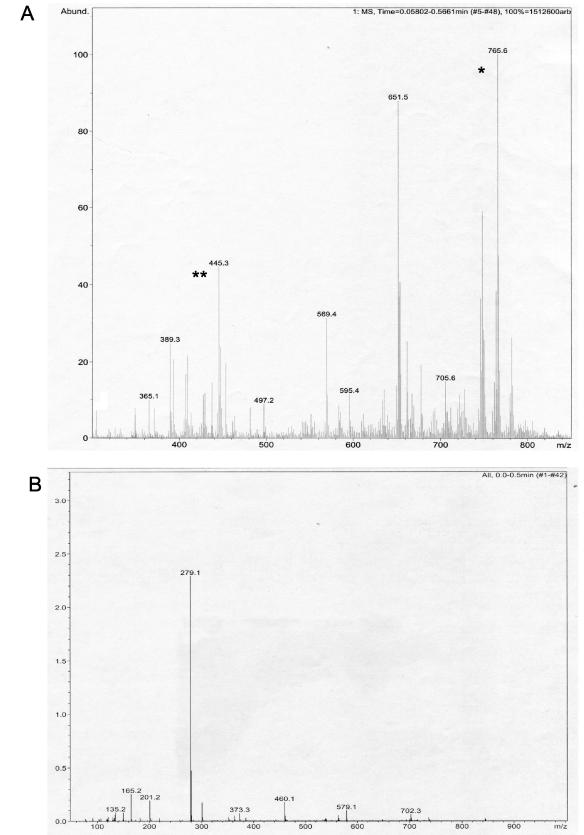
A specific cytopathic lipid was partially purified from CEI 01-628 ASLs by Chromatotron separation (Fig. 3). In this isolate cytopathicity was assigned to a lipid with an Rf of 0.14 in a solvent system of chloroform-methanol (93:7, vol/vol). This lipid was cytopathic at 2.5 × 103 μg/ml. The cytopathicity phenotype of the purified lipid species from CEI 01-628 was identical to the phenotype produced by the ASLs (data not shown). The cytopathic fraction of CEI 01-628 was less polar than mycolactone, which has an Rf of 0.08 when run in chloroform-methanol (93:7, vol/vol). A sufficient quantity of lipid could not be obtained for structural analysis. However, MS analysis was performed on this lipid component of CEI 01-628 to further distinguish it from mycolactone and the core lactone (Fig. 4A). Mycolactone produces a molecular ion at m/z 765.5 [M+Na]+, whereas the core lactone is detected as a peak at m/z 447 [M+Na]+ (3, 11) (Fig. 4A). The cytopathic fraction of 01-628 was devoid of mycolactone ions. However, a major peak for this lipid from CEI 01-628 was seen at m/z 279.1 [M+Na]+ (Fig. 4B). Thus data from MS, TLC, and cell assays provide conclusive evidence that the cytopathic lipid isolated from CEI 01-628 is neither mycolactone nor core lactone.
FIG. 3.
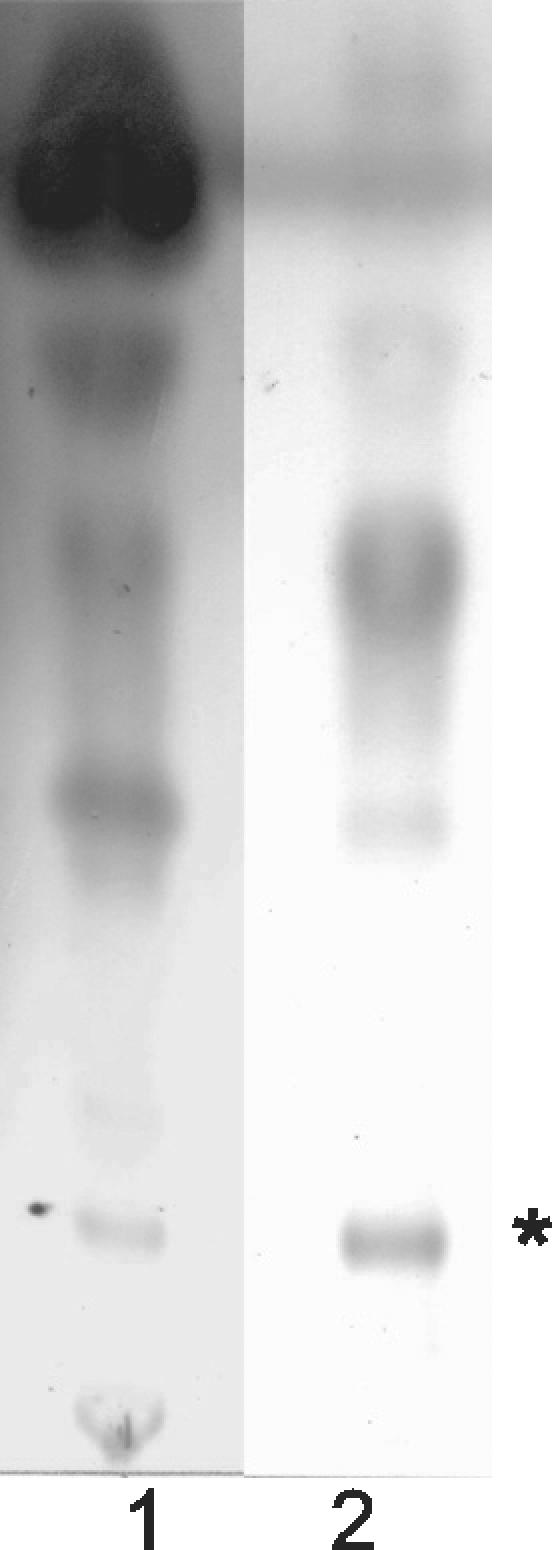
TLC analysis of CEI 01-628 ASLs. Lane 1, 01-628 whole ASLs; lane 2, 01-628 ASL cytopathic fraction. *, cytopathic lipid.
FIG. 4.
MS analysis of ASLs. (A) M. ulcerans 1615. (B) 01-628. *, mycolactone; **, core lactone.
CEI with cytopathic lipids are identified as new Mycobacterium species.
All 16 CEI were isolated from aquatic sources in the Democratic Republic of Congo during the late 1970s. These strains were isolated from the lower Congo region, between Kinshasa and Moanda, and were characterized as slow-growing mycobacteria with an optimal growth temperature of 33°C and sensitivity to dapsone. At the time of isolation, they were tested against all known species of Mycobacterium by nonmolecular methods and did not match any known mycobacterial species (13). Most of these isolates grow extremely slowly in our laboratory. The growth and colonial morphology of the CEI varied considerably, and most isolates do not look similar to M. ulcerans in pigment or colonial morphology. Further, whereas M. ulcerans cannot grow at 37°C, some of the CEI grew very slowly at 37°C (Table 2) (13).
TABLE 2.
Growth and morphology of uncharacterized slow-growing mycobacterial strains from aquatic sources from the Democratic Republic of Congo
| Strain | Growth morphologya/pigmentation†c | Growthd at:
|
Cytopathicityb | |
|---|---|---|---|---|
| 32°C | 37°C | |||
| 1615 (M. ulcerans) | Rough/nonchromogenic | + | −c | Yes |
| 01-626 | Rough/nonchromogenic | + | +/− | No |
| 01-627 | Smooth/scotochromogenic | + | + | Yes |
| 01-628 | Rough/nonchromogenic | + | +/− | Yes |
| 01-629 | Rough/photochromogenic | + | +/− | Yes |
| 01-630 | Smooth and rough/scotochromogenic | + | +/− | No |
| 01-631 | Rough/scotochromogenic | + | +/− | No |
| 01-632 | Rough/nonchromogenic | + | +/− | Yes |
| 01-633 | Smooth and rough/nonchromogenic | + | +/− | Yes |
| 01-634 | Smooth/scotochromogenic | + | +/− | No |
| 01-635 | Smooth/scotochromogenic | + | +/− | No |
| 01-636 | Smooth/scotochromogenic | + | + | Yes |
| 01-664 | Rough/nonchromogenic | + | +/− | Yes |
| 01-665 | Rough/scotochromogenic | + | +/− | Yes |
| 01-666 | Smooth/nonchromogenic | + | +/− | No |
| 01-667 | Smooth/scotochromogenic | + | +/− | No |
| 01-668 | Smooth and rough/scotochromogenic | + | +/− | No |
Growth morphology as seen on Middlebrook 7H10 medium.
Cytopathicity on L929 fibroblasts.
Nonchromogenic, lacks production of pigment; scotochromogenic, produces pigment with or without light present; photochromogenic, produces pigment only after exposure to light.
+, growth; −, no growth; +/−, weak growth.
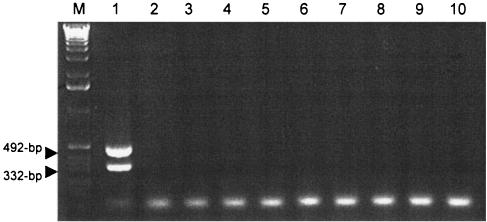
Modern taxonomic methods rely on molecular tools which were not available in the 1970s, and these tools were used to reexamine the taxonomic status of eight CEI isolates which produced cytopathic ASLs. M. ulcerans contains two ISs (IS2404 and IS2606) that are almost entirely unique to M. ulcerans (16). Although PCR detection of these two IS elements has become the “gold standard” used for diagnosis and identification of M. ulcerans, limited effort has been made to investigate whether these IS elements might be present in environmental mycobacterial strains. With one exception, M. ulcerans has been isolated only from patient lesions although it is presumed to have an environmental reservoir. The possibility exists that there may be M. ulcerans strains in the environment that have not been detected because they do not have genes for mycolactone. Therefore, PCR was used to determine whether IS2404 or IS2606 was present in the eight CEI which produced cytopathic lipids. Because CEI species were isolated from aquatic habitats in areas of endemicity for M. ulcerans, it was important to determine whether these isolates contained IS2404 or IS2606 elements, as a negative finding would strengthen the validity of the IS-based PCR test for M. ulcerans (Portaels et al., letter). As expected, PCR results show that genomic DNA from M. ulcerans 1615 contains both the 492-bp fragment of IS2404 and the 332-bp fragment of IS2606. However, none of the CEI with cytopathic lipids contained either of these IS elements (Fig. 5). The remaining CEI (01-626, 01-630, 01-631, 01-634, 01-635, 01-666, 01-667, and 01-668) were also tested for IS2404 and IS2606 and were found negative for both IS elements (data not shown).
FIG. 5.
PCR analysis of eight CEI with cytopathic lipids for IS2404 and IS2606. Lane M, 1-kb ladder (Invitrogen); lane 1, M. ulcerans; lane 2, 01-627; lane 3, 01-628; lane 4, 01-629; lane 5, 01-632; lane 6, 01-633; lane 7, 01-636; lane 8, 01-664; lane 9, 01-665; lane 10, 492-bp fragment, IS2404; 332-bp fragment, IS2606.
Taxonomic relationships of environmental isolates.
To further characterize the environmental isolates which produced cytopathic lipids, partial DNA sequences for the 16S rRNA genes in these isolates were obtained and used for taxonomic studies. PCR was performed using genomic DNA of these eight CEI with primers based on bp 1 to 577 of the M. tuberculosis 16S rRNA gene (16S ribosomal DNA [rDNA]) as previously described (7). The primers used to amplify 16S rDNA in this study, pA and MSHE, were chosen because of the length of the fragment amplified and because they include sequences from two hypervariable regions in Mycobacterium species16S rRNA genes, A and B. Hypervariable region A is between bp 129 and 276, and hypervariable region B in between bp 430 and 495, with E. coli 16S rDNA as a reference point for nucleotides. BLAST analysis of 16S rRNA obtained from the CEI showed that these sequences were unique mycobacterial sequences and that each of these eight CEI were new species of Mycobacterium.
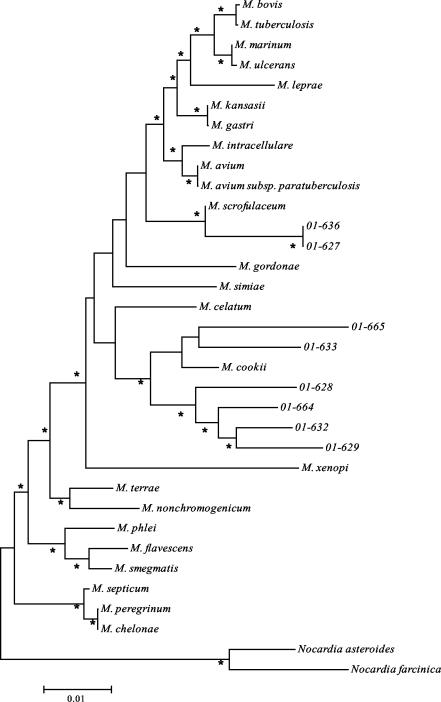
Alignments of the partial sequences of the eight CEI were performed using 24 other Mycobacterium species in order to place the new isolates in a taxonomic tree. These alignments were performed using ClustalW in the program Megalign (DNAStar). A phylogenetic tree was constructed from these species with the MEGA program using the NJ method with Jukes-Cantor distances, and N. asteroides and N. farcinica were used as outgroups to root the tree. Bootstrap values were calculated with 500 permutations (Fig. 6).
FIG. 6.
NJ tree based on partial 16S rRNA sequences from CEI. Closely related species were detected by BLAST analysis. The tree was rooted by using N. asteroides and N. farcinica as outgroups. *, bootstrap value >50 (out of 100).
It was apparent from this phylogenetic analysis that none of the eight cytopathic CEI were closely related to M. ulcerans. However, three distinct groups of strains could be distinguished among the CEI. Two of the CEI, 01-636 and 01-627, were closely related to M. scrofulaceum. The other six CEI (01-628, 01-629, 01-632, 01-633, and 01-664) showed relationship to M. cookii and M. celatum, with 01-665 and 01-633 being more closely related to M. cookii.
DISCUSSION
Previous work has identified mycolactone, a polyketide-derived macrolide, as a virulence determinant in M. ulcerans (2, 3). Until the discovery of mycolactone in M. ulcerans, macrolides had been isolated only from soil bacteria, such as the Streptomyces species, or from fungi (8). The pathology of Buruli ulcer is unique in many respects, and the fact that it is an extracellular infection is in stark contrast to the intracellular lifestyle of other mycobacterial pathogens. This unique pathology, which is associated with mycolactone production, might suggest that the molecule would not be found in other mycobacterial pathogens. However, if the molecule were present in a mycobacterial species with little ability to colonize a human host, it might not have come to attention. For this reason we investigated whether similar molecules might be made by other mycobacteria and focused attention particularly on aquatic mycobacteria. A further rationale for these studies is provided by the fact that M. ulcerans contains an unusually large number of IS elements (15). The presence of such a large number of IS elements raises the possibility that the mycolactone gene cluster may have been acquired by horizontal transfer from another environmental organism. The presence of such a molecule in an environmental mycobacterial species could easily remain undetected. To prove the nonexistence of a molecule in a group of bacteria is considerably more difficult than to prove its existence. Thus our failure to find mycolactone in any of the mycobacterial strains tested must be evaluated in light of the limitations of the study.
The list of purified mycobacterial polyketides is rapidly expanding (1, 14). All of the mycobacterial polyketides so far characterized have been found on the cell envelope, and many can also be detected in culture filtrate. For this reason we limited our search for mycolactones to the cell surface and culture filtrate. If such molecules were produced as cytoplasmic lipids, we would not have detected them. We think a cytoplasmic location for such hydrophobic molecules would be unusual. Second, we required that a molecule either be present in sufficient amount or have high enough potency to be detected by bioassays. A molecule which required greater than milligram amounts for activity would not have been identified in these studies. We have also biased our studies by looking for a particular cytopathicity phenotype. We have previously shown that there is a family of mycolactone congeners produced by different isolates of M. ulcerans which differ in potency and have shown the core lactone is active only at microgram amounts. All of these molecules, however, have the same kinetics and cytopathicity phenotype originally described for mycolactone (11). The structure of mycolactone shows that this molecule is a hybrid polyketide which is likely to be encoded by at least two separate polyketide synthase genes. All samples were assayed at an initial concentration far in excess of that required to detect core activity. An additional limitation to these studies is that, if mycolactone were produced in very small amounts under the growth conditions used, it would not have been detected. We chose to assay late-log cultures because evidence from both Streptomyces literature and M. ulcerans shows that macrolides are secondary metabolites usually produced in greatest amounts during this growth stage (1, 8). Taking the above considerations into account, our results provide strong evidence that mycolactone production is not common among mycobacteria and suggest that mycolactone may be unique to M. ulcerans.
A surprising finding from this study was that many of the environmental mycobacteria tested produced lipids which were cytopathic for cells. Although none of these lipids produced mycolactone-mediated cytopathicity characterized by growth arrest and apoptotic cell death, many CEI produced lipids which were clearly cytopathic for L929 cells. Further, all cytopathic lipids from CEI produced similar effects on cells, characterized by rapid onset and extensive vacuolization, and these effects were associated with lipid extracts from multiple isolates of M. kansasii and M. scrofulaceum. The role these cytopathic lipids play in the lives of CEI is unknown. However, the designation environmental isolate often simply means that the isolate is rarely isolated from humans and is somewhere else in the environment. Whether such bacteria exist as commensals or pathogens of aquatic animals or plants is unknown.
Acknowledgments
This work was supported by the National Institute of Allergy and Infectious Diseases, National Institutes of Health, grant number AI49418.
We acknowledge Armand Mve-Obiang and Brian Ranger for their outstanding technical assistance. CEI were provided by Francoise Portaels.
Editor: J. T. Barbieri
REFERENCES
- 1.Cox, J. S., B. Chen, M. McNeil, and W. R. Jacobs, Jr. 1999. Complex lipid determines tissue-specific replication of Mycobacterium tuberculosis in mice. Nature 402:79-83. [DOI] [PubMed] [Google Scholar]
- 2.George, K. M., L. P. Barker, D. M. Welty, and P. L. Small. 1998. Partial purification and characterization of biological effects of a lipid toxin produced by Mycobacterium ulcerans. Infect. Immun. 66:587-593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.George, K. M., D. Chatterjee, G. Gunawardana, D. Welty, J. Hayman, R. Lee, and P. L. Small. 1999. Mycolactone: a polyketide toxin from Mycobacterium ulcerans required for virulence. Science. 283:854-857. [DOI] [PubMed] [Google Scholar]
- 4.George, K. M., L. Pascopella, D. M. Welty, and P. L. Small. 2000. A Mycobacterium ulcerans toxin, mycolactone, causes apoptosis in guinea pig ulcers and tissue culture cells. Infect. Immun. 68:877-883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gunawardana, G., D. Chatterjee, K. M. George, P. Brennan, D. Whittern, and P. L. Small. 1999. Characterization of novel macrolide toxins, mycolactones A and B, from a human pathogen, Mycobacterium ulcerans. J. Am. Chem. Soc. 121:6092-6093. [Google Scholar]
- 6.Heumann, D., and T. Roger. 2002. Initial responses to endotoxins and gram-negative bacteria. Clin. Chim. Acta 323:59-72. [DOI] [PubMed] [Google Scholar]
- 7.Hughes, M. S., N. W. Ball, L. A. Beck, G. W. de Lisle, R. A. Skuce, and S. D. Neill. 1997. Determination of the etiology of presumptive feline leprosy by 16S rRNA gene analysis. J. Clin. Microbiol. 35:2464-2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Katz, L., and S. Donadio. 1993. Polyketide synthesis: prospects for hybrid antibiotics. Annu. Rev. Microbiol. 47:875-912. [DOI] [PubMed] [Google Scholar]
- 9.Kumar, S., K. Tamura, I. B. Jakobsen, and M. Nei. 2001. MEGA2: molecular evolutionary genetics analysis software. Bioinformatics 17:1244-1245. [DOI] [PubMed] [Google Scholar]
- 10.Marsollier, L., R. Robert, J. Aubry, J. P. Saint Andre, H. Kouakou, P. Legras, A. L. Manceau, C. Mahaza, and B. Carbonnelle. 2002. Aquatic insects as a vector for Mycobacterium ulcerans. Appl. Environ. Microbiol. 68:4623-4628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mve-Obiang, A., R. E. Lee, F. Portaels, and P. L. Small. 2003. Heterogeneity of mycolactones produced by clinical isolates of Mycobacterium ulcerans: implications for virulence. Infect. Immun. 71:774-783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mve-Obiang, A., M. Mestdagh, and F. Portaels. 2001. DNA isolation from chloroform/methanol-treated mycobacterial cells without lysozyme and proteinase K. BioTechniques 30:272-274, 276. [DOI] [PubMed] [Google Scholar]
- 13.Portaels, F. 1980. Study of unclassified dapsone sensitive mycobacteria isolated from the environment in Zaire. Ann. Soc. Belg. Med. Trop. 60:381-386. [PubMed] [Google Scholar]
- 14.Rambukkana, A., G. Zanazzi, N. Tapinos, and J. L. Salzer. 2002. Contact-dependent demyelination by Mycobacterium leprae in the absence of immune cells. Science 296:927-931. [DOI] [PubMed] [Google Scholar]
- 15.Stinear, T., J. K. Davies, G. A. Jenkin, F. Portaels, B. C. Ross, F. Oppedisano, M. Purcell, J. A. Hayman, and P. D. Johnson. 2000. A simple PCR method for rapid genotype analysis of Mycobacterium ulcerans. J. Clin. Microbiol. 38:1482-1487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stinear, T., B. C. Ross, J. K. Davies, L. Marino, R. M. Robins-Browne, F. Oppedisano, A. Sievers, and P. D. Johnson. 1999. Identification and characterization of IS2404 and IS2606: two distinct repeated sequences for detection of Mycobacterium ulcerans by PCR. J. Clin. Microbiol. 37:1018-1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Thangaraj, H. S., M. R. Evans, and M. H. Wansbrough-Jones. 1999. Mycobacterium ulcerans disease; Buruli ulcer. Trans. R. Soc. Trop. Med. Hyg. 93:337-340. [DOI] [PubMed] [Google Scholar]
- 18.van der Werf, T. S., W. T. van der Graaf, J. W. Tappero, and K. Asiedu. 1999. Mycobacterium ulcerans infection. Lancet 354:1013-1018. [DOI] [PubMed] [Google Scholar]