Abstract
The relative abilities of pneumococcal serotypes and strains (clones) to cause acute otitis media (AOM) were investigated by comparing the serotypes and genotypes of pneumococci recovered from cases of AOM (n = 149) in children <2 years of age with those from nasopharyngeal carriage (n = 288) in age-matched controls from the same region. The odds ratio (OR) for association of pooled vaccine serotypes with AOM was found to be slightly elevated over unity, although this was not significantly different from that of pooled nonvaccine or vaccine-related serotypes. Comparing individual serotypes, 19F and 23F had 2- to 2.5-fold higher ORs, although these were not markedly different from the ORs of nonvaccine serotypes. None of the major clones had an OR that was significantly greater than the average, and the differences in ORs among serotypes and clones were much less than those for invasive disease, suggesting little variation in their ability to cause AOM. We conclude that serotype replacement may reduce the long-term efficacy of these vaccines against AOM.
Streptococcus pneumoniae (the pneumococcus) is one of the major bacterial causes of acute otitis media (AOM) in children, being responsible for between 30 and 50% of all cases (3, 13). The economic cost of AOM is high and is estimated to be $5 billion per annum in the United States (4). One of the contributing factors to this high cost is the high number of antibiotic prescriptions for AOM, which is reflected in an increasing prevalence of antibiotic resistance in pneumococci, which in turn may complicate the treatment of AOM. Multivalent conjugate vaccines have recently been developed which cover the pneumococcal serotypes most commonly associated with invasive disease in developed countries. Clinical trials of these vaccines have demonstrated 97% efficacy against invasive pneumococcal disease caused by isolates of vaccine serotypes (1). A separate study conducted in Finland found 57% efficacy against AOM caused by pneumococci of vaccine serotypes (7). Serotypes other than those covered by the vaccines cause a significant proportion of AOM, and the overall reduction in pneumococcal AOM in vaccinated children was 34%, with a modest reduction in AOM due to all causes of about 6% (7).
An important finding of almost all trials of the conjugate pneumococcal vaccines is not only a significant reduction in nasopharyngeal (NP) carriage of pneumococci of vaccine serotypes but also a concomitant increase in carriage of nonvaccine serotypes (12; S. K. Obaro, R. A. Adegbola, W. A. Banya, and B. M. Greenwood, Letter, Lancet 348:271-272, 1996). The mechanism, extent, duration, and consequences of this phenomenon (serotype replacement) are subjects of current debate (17; Obaro et al., letter).
S. pneumoniae is an antigenically diverse species in which more than 90 serotypes have been identified. However, the prevalence with which the serotypes are recovered from patients with invasive disease varies greatly (10), presumably because some serotypes have a much greater propensity to cause invasive disease than others. If conjugate vaccines provide protection against those serotypes that are most invasive and reduce carriage and transmission of these serotypes, their implementation should lead to a sustained reduction in invasive disease, as serotype replacement would increase exposure to serotypes that are considered much less invasive than the vaccine serotypes. However, for mucosal infections like AOM, it is much less clear whether there is a wide variation in the abilities of different serotypes to cause disease. The association of the common childhood serotypes (e.g., types 6B, 9V, 14, 19F, and 23F) with AOM is not necessarily evidence for any special propensity of these to cause AOM, as these serotypes are the ones most commonly carried in the nasopharynx of children (8, 19) and their association with AOM could merely reflect the fact that they are the most likely to gain access to the middle ear from the nasopharynx. Thus, even if all serotypes are equally able to cause AOM, the majority of episodes of this disease would be caused by the most frequently carried childhood serotypes.
It is important to know whether isolates of the vaccine serotypes have a special ability to cause AOM, or whether almost any pneumococcus that colonizes the nasopharynx has a similar ability to cause AOM, as it impacts the long-term efficacy of conjugate vaccines against this common childhood disease. If isolates of vaccine serotypes cause most AOM because they have a special ability to do so, compared to nonvaccine serotypes, there should be a sustained reduction in AOM even if serotype replacement occurs. However, if isolates of nonvaccine serotypes were just as capable of causing AOM as those of vaccine serotypes, serotype replacement following mass vaccination would simply lead to AOM eventually being caused predominantly by nonvaccine serotypes rather than by vaccine serotypes, without substantially reducing the overall prevalence of disease.
We have asked whether pneumococcal serotypes, and individual pneumococcal strains (clones), have an equal ability to cause AOM, or whether some have a much greater propensity to cause this disease, by comparing the frequency of serotypes and clones of vaccine type (VT) and non-VT (NVT) pneumococci in the middle ear fluid (MEF) of children suffering from AOM with the frequency of these serotypes and clones among samples from NP carriage in the same group of Finnish children. Our results show relatively small differences in the ability of pneumococcal serotypes and clones to cause AOM, indicating that most serotypes, including nonvaccine serotypes, cause AOM at a frequency that is proportional to their prevalence in NP carriage. The implications for the long-term efficacy of conjugate vaccines against AOM are discussed.
MATERIALS AND METHODS
Strains.
Strains of S. pneumoniae were isolated from samples taken from Finnish children as part of the FinOM Cohort Study between 1994 and 1997, in which 329 unselected children from the Tampere region of Finland were monitored from 2 to 24 months of age as part of a study of the natural history of AOM. Ten NP swabs were obtained, with one being obtained at each of the following ages: 2, 3, 4, 5, 6, 9, 12, 15, 18, and 24 months. Whenever symptoms of acute infection suggesting AOM occurred, the parents were encouraged to bring the child to the study clinic, in which one or two specially trained study physicians diagnosed AOM by pneumatic otoscopy using fixed clinical criteria and tympanometry as an aid (11). According to data collected at scheduled visits, 86% of all cases of AOM occurring in these children during the follow-up were captured at the study clinic. Whenever AOM with effusion occurred, a myringotomy was performed and an MEF sample was aspirated for etiological diagnosis. Pneumococci were cultured immediately from both NP and MEF samples on selective plates and identified by standard methods in the bacteriology laboratory of the National Public Health Institute in Oulu, Finland, as described in detail previously (11, 19). The strains were serotyped using antisera from the Statens Seruminstitut, Copenhagen, Denmark, and were stored as glycerol stocks at −80°C.
The strain from the MEF of each AOM event (n = 201) was characterized by multilocus sequence typing (MLST). Multiple strains from the same child were included only if they were isolated at least 30 days apart or if they were different strains (defined by MLST), in order to avoid repeated samples from the same episode of AOM. Samples from AOM events in children >19.5 months old were also excluded due to a lack of NP samples from the same age group for comparison (see below). With these definitions, there were 151 episodes of AOM in the study cohort. For comparison with this data set, 297 S. pneumoniae controls were randomly selected from the 397 NP isolates obtained from the 329 children in the FinOM Cohort Study as described above, matched by age group (<4.5, 4.5 to 7.5, 7.5 to 10.5, 10.5 to 13.5, 13.5 to 16.5, and 16.5 to 19.5 months). Two further strains were removed from the MEF data set and nine strains were removed from the NP data set on the grounds that sequence data obtained during MLST showed them to be members of a species related to but distinct from true pneumococci (see below). The final data included 149 isolates from episodes of AOM and 288 control isolates from NP carriage.
MLST.
Genomic DNA was isolated using DNeasy tissue kits (Qiagen Inc., Valencia, Calif.) and stored at −20°C. The allelic profile was determined by amplifying internal fragments of the seven housekeeping loci used in the pneumococcal MLST scheme (Taq polymerase and 10× buffer from Qiagen, 50 nM concentrations of the deoxynucleoside triphosphates [Geneamp; Applied Biosystems, Foster City, Calif.], and the PCR conditions described previously [6]). The PCR products were precipitated with 20% polyethylene glycol 8000-2.5 M NaCl (Sigma) and the fragments were sequenced on both strands using the same primers and BigDye II terminators (Applied Biosystems). The products of the sequencing reactions were precipitated with 185 mM sodium acetate in 70% ethanol and were resuspended in 10 μl of HiDi Formamide (Applied Biosystems) and loaded onto an ABI Prism 3700 sequencer. Sequences were analyzed using STARS (obtainable from www.mlst.net), a modified Staden interface developed by Man-Suen Chan for use with MLST projects. Alleles at each locus were assigned, and sequence types (STs) were determined, using the pneumococcal MLST database (www.mlst.net).
Nontypeable presumptive pneumococcal isolates were separated into those that were authentic pneumococci and those that were similar to, but genetically distinct from, authentic pneumococci, by examination of the alleles at the seven MLST loci (14). Authentic pneumococci had alleles at five or more loci that were found in serotypeable pneumococci in the MLST database (or if novel were similar to pneumococcal alleles), whereas those that were not authentic pneumococci had alleles at two or more loci with <97% identity with the most similar pneumococcal allele in the MLST database.
Statistical analysis.
Diversity of STs was assessed using Simpson's index of diversity D (16) as follows:
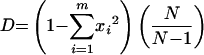 |
where xi is the frequency of the ith ST, m is the number of STs, and N is the total number of isolates; 95% confidence intervals (CI) were estimated by the method of Grundmann et al. (9) as follows: CI = (D − 2 · σ2, D + 2 · σ2), where
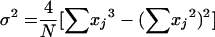 |
and where xj is the frequency of the jth ST and N is the total number of isolates in the sample population.
Diversity of alleles at each locus was studied in an analogous way using the above equations, where xi is the frequency of the ith allele and so on. This measure for allelic diversity is equivalent to Nei's h measure of heterozygosity per locus (15) and was calculated for each MLST locus (h values). The mean diversity over the seven MLST loci (H) was also calculated.
To adjust for potential confounding by age, controls were matched by age group (see above). Separate analyses were performed for pooled vaccine, vaccine-related serotypes, and nonvaccine serotypes, as well as individual serotypes and STs found at least 10 times in the combined sample of 149 cases (AOM) and 288 controls (NP); less-numerous types were pooled in a single class. For each analysis (pooled VTs, serotypes, or STs), the distributions of types among cases and controls were compared by determining the ratio of odds for exposure in cases to that in controls. Specifically. to obtain a common reference for all comparisons, the odds for any type were calculated as the geometric mean of pairwise odds of the type with respect to all types in the analysis (this was realized by using the deviation contrast in the SPSS software [18]). Without adjustment for age, the odds ratio (OR) for type i may thus be calculated:
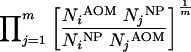 |
where NjAOM and NjNP are the observed numbers of type j from cases (AOM) and controls (NP), respectively, and m is the total number of types in the analysis (10 for serotypes and 12 for STs). Logistic regression was used in the actual analysis to adjust for age matching.
Although the two samples are not wholly independent (26 of 149 [17%] strains from episodes of AOM were recovered during the same 3-month period as strains from NP carriage from the same child), this did not markedly affect the estimates of ORs reported here. ORs of >1 (or <1) indicate an increased (or decreased) likelihood of a pneumococcal type being retrieved from patients with AOM in comparison to the mean odds of AOM due to pneumococci of all types. No corrections were made for multiple comparisons.
RESULTS
Comparison of vaccine and nonvaccine serotypes.
The study was designed to compare the distribution of serotypes and individual clones (STs) of pneumococci causing AOM in children with the distribution of those being carried by children of the same age, in the same locality, during the same time period. All 437 isolates, 149 AOM case isolates and 288 NP control isolates, were characterized by MLST and by serotyping. Nine serotypes were represented by at least 10 isolates in the combined data set. Seven of these were vaccine serotypes or vaccine-related serotypes, and two were nonvaccine serotypes (11A and 35F). Comparison of the presence of VT and NVT strains in cases and controls allows us to estimate the OR for association of types with AOM. These are shown in Table 1. Pooled vaccine serotypes show an OR significantly greater than unity, indicating an association with AOM, but when compared with pneumococci of nonvaccine serotype, the 95% CI overlap (although this is likely to be due to the sample size in this study). The lack of a significant difference between vaccine and nonvaccine serotypes suggests that the difference in OR is small. Vaccine-related types were intermediate between vaccine and nonvaccine serotypes (Table 1).
TABLE 1.
ORs for AOM of pooled vaccine and nonvaccine serotypesa
| Serotype | No. of isolates from:
|
Total no. of isolates | OR (95% CI) | |
|---|---|---|---|---|
| Carriage | AOM | |||
| VT | 156 | 97 | 253 | 1.35 (1.03-1.77) |
| R | 55 | 24 | 79 | 0.95 (0.66-1.37) |
| NVT | 77 | 28 | 105 | 0.78 (0.55-1.10) |
Numbers of isolates of VTs, vaccine-related (R) serotypes, and NVTs in the AOM and carriage samples are shown with the associated ORs for AOM and the 95% CI, based on the logistic regression model. Total numbers of isolates recovered from NP carriage and AOM were 288 and 149, respectively. ORs of >1 indicate a greater propensity to cause AOM; ORs of <1 signify a reduced propensity to cause AOM. ORs significantly different from unity are shown in boldface type.
Diversity of isolates from patients with AOM compared to that of NP carriage isolates.
If the isolates from patients with AOM are a reflection of those that are being carried, the diversities of the isolates in the AOM and NP data sets should be similar. Diversity was assessed by calculating Simpson's index of diversity, D, for genotypes (STs) and individual MLST loci (equivalent to Nei's h). The 95% CI for D were estimated (9), and for the diversity of STs in the NP data set (DNP = 0.98 ± 0.002) and in the AOM data set (DAOM = 0.98 ± 0.003) the CI overlapped, indicating that there was no significant difference in the genotypic diversities of these two populations. The diversity at each of the seven individual MLST loci (h) and the mean of these values (H) were also similar for the AOM and NP data sets, and in all cases the CI overlapped (data not shown).
While the overall genetic diversities of the two samples were similar, their compositions in terms of the STs of the isolates recovered from patients with AOM (cases) and carriage (controls) could still be very different. Therefore, the distribution of the STs of isolates from patients with AOM was compared to that from carriage. Table 2 shows that all of the major STs were represented in both the AOM and NP data sets. This was also the case for most of the less abundant STs; none of the 26 STs that included at least five isolates were recovered only from patients with AOM. Three of these 26 STs were recovered exclusively from carriage isolates; two were vaccine or vaccine-related serotypes (ST 481, serotype 6A, seven isolates; ST 273, serotype 6B, five isolates), and the other was an NVT (ST 487, serotype 35F, five isolates). Thus, only 3 of the 26 most-abundant STs were found in only one of the data sets.
TABLE 2.
Properties and ORs of STs recovered from carriage and AOM isolatesa
| ST | Allele no. at MLST loci
|
Serotype(s) | No. of isolates from:
|
Total no. of isolates | OR (95% CI)b | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| aroE | gdh | gki | recP | spi | xpt | ddl | Carriage | AOM | ||||
| 62 | 2 | 5 | 29 | 12 | 16 | 3 | 14 | 11A | 17 | 5 | 22 | 0.50 (0.19-1.32) |
| 36 | 1 | 8 | 4 | 1 | 1 | 4 | 6 | 23F | 14 | 6 | 20 | 0.73 (0.29-1.86) |
| 37 | 1 | 8 | 6 | 2 | 6 | 4 | 6 | 23F | 8 | 11 | 19 | 2.42 (0.99-5.90) |
| 199 | 8 | 13 | 14 | 4 | 17 | 4 | 14 | 19A, 19F, 15 | 7, 2, 4 | 1, 3, 2 | 19 | 0.79 (0.31-2.02) |
| 124 | 7 | 5 | 1 | 8 | 14 | 11 | 14 | 14 | 11 | 6 | 17 | 0.96 (0.37-2.50) |
| 138 | 7 | 5 | 8 | 5 | 10 | 6 | 14 | 6B | 14 | 3 | 17 | 0.36 (0.11-1.19) |
| 485 | 1 | 5 | 1 | 1 | 1 | 1 | 8 | 19F | 11 | 5 | 16 | 0.78 (0.28-2.17) |
| 507 | 1 | 8 | 6 | 2 | 6 | 20 | 6 | 23F | 9 | 4 | 13 | 0.78 (0.25-2.41) |
| 309 | 8 | 10 | 2 | 5 | 9 | 48 | 6 | 19F | 4 | 6 | 10 | 2.61 (0.78-8.78) |
| 460 | 5 | 7 | 4 | 10 | 10 | 1 | 27 | 6A | 6 | 4 | 10 | 1.15 (0.34-3.83) |
| 488 | 2 | 13 | 9 | 1 | 6 | 28 | 14 | 6A | 4 | 6 | 10 | 2.70 (0.81-8.99) |
| 66 | 2 | 8 | 2 | 4 | 6 | 1 | 1 | 9N, 23F, 19F | 5, 0, 0 | 1, 1, 2 | 9 | |
| 162 | 7 | 11 | 10 | 1 | 6 | 8 | 14 | 9V | 5 | 4 | 9 | |
| 504 | 1 | 8 | 1 | 2 | 6 | 56 | 6 | 23F | 4 | 5 | 9 | |
| 423 | 1 | 5 | 4 | 12 | 5 | 3 | 8 | 19F | 5 | 3 | 8 | |
| 205 | 10 | 5 | 4 | 5 | 13 | 10 | 18 | 4 | 6 | 2 | 8 | |
| 481 | 5 | 1 | 4 | 2 | 10 | 1 | 27 | 6A | 7 | 0 | 7 | |
| 15 | 1 | 5 | 4 | 5 | 5 | 3 | 8 | 15 | 4 | 3 | 7 | |
| 497 | 7 | 25 | 4 | 2 | 48 | 20 | 28 | 6B | 3 | 3 | 6 | |
| 520 | 1 | 1 | 4 | 1 | 18 | 16 | 17 | 22, R | 4, 0 | 0, 2 | 6 | |
| 490 | 2 | 13 | 9 | 1 | 6 | 19 | 14 | 6A, 6B | 3, 2 | 0, 1 | 6 | |
| 176 | 7 | 13 | 8 | 6 | 10 | 6 | 14 | 6B | 4 | 1 | 5 | |
| 492 | 7 | 5 | 1 | 1 | 6 | 31 | 9 | 6B | 4 | 1 | 5 | |
| 191 | 8 | 9 | 2 | 1 | 6 | 1 | 17 | 7F | 4 | 1 | 5 | |
| 487 | 2 | 1 | 1 | 1 | 10 | 1 | 14 | 35F | 5 | 0 | 5 | |
| 273 | 5 | 6 | 2 | 2 | 6 | 1 | 14 | 6B | 5 | 0 | 5 | |
| Other | 107 | 57 | 164 | |||||||||
| Total | 288 | 149 | 437 | |||||||||
STs recovered five or more times from the combined carriage and AOM data sets are shown together with their-allelic profiles and serotypes.
ORs and 95% CI based on the logistic regression model are shown for STs recovered ≥10 times from the combined data set.
There was no significant difference in distribution of AOM and NP isolates within the 11 STs represented by at least 10 isolates (Fisher's exact test; P = 0.18). The isolates from patients with AOM and the carriage isolates were therefore not significantly different in their overall diversity or in their genotypes.
Serotypes and clones present in the NP and AOM data sets.
The major STs and serotypes identified by MLST in the NP and AOM data sets are shown in Tables 2 and 3, respectively. Full versions of these tables are available from the authors on request. The combined data set contained 104 different STs. There were 11 STs that included 10 or more isolates and 31 STs for which only a single isolate was sampled. In the majority of cases, all isolates with the same ST had the same serotype. However, there were some exceptions, where isolates of differing serotypes were found within a single ST (STs 66, 199, 490, and 520). In addition to those strains shown in Table 2, three isolates of ST 71 that expressed the 15A capsule were found, and one expressed the 11A capsule. Only one of these, a 15A isolate, was recovered from MEF. All potential cases of serotype switching were confirmed by both reserotyping and resequencing the MLST loci.
TABLE 3.
ORs for AOM of major pneumococcal serotypesa
| Serotype | Isolates (n)
|
Total | OR (95% CI) | No. of STs | Most common STs (n) | |
|---|---|---|---|---|---|---|
| Carriage | AOM | |||||
| 19Fb | 38 | 34 | 72 | 2.46 (1.43-4.21) | 16 | 485 (16), 309 (9) |
| 23Fb | 41 | 29 | 70 | 1.96 (1.13-3.40) | 8 | 36 (20), 37 (19) |
| 6Bb | 48 | 12 | 60 | 0.67 (0.35-1.30) | 16 | 138 (17), 497 (6) |
| 6A | 35 | 19 | 54 | 1.51 (0.82-2.78) | 17 | 460 (10), 488 (10) |
| 11A | 24 | 8 | 32 | 0.89 (0.40-1.98) | 6 | 62 (22) |
| 14b | 14 | 11 | 25 | 2.18 (0.99-4.81) | 4 | 124 (17) |
| 35F | 15 | 1 | 16 | 0.18 (0.03-1.13) | 4 | 487 (5) |
| 19A | 9 | 2 | 11 | 0.60 (0.15-2.48) | 2 | 199 (8) |
| 9N | 8 | 2 | 10 | 0.67 (0.16-2.79) | 3 | 66 (6) |
| Other | 56 | 31 | 87 | 1.47 (0.88-2.48) | 31 | 162 (9) |
| Total | 288 | 149 | 437 | |||
Numbers of isolates of the most prevalent serotypes in the AOM and carriage isolates are shown with the associated ORs for progression to AOM and the 95% CI, based on the logistic regression model. Only serotypes recovered at least 10 times are shown. “Other” refers to the isolates of all other serotypes. ORs significantly different from unity are shown in boldface type.
Serotype present in the licensed 7-valent conjugate vaccine.
Association of serotypes and clones with AOM and carriage.
While the ORs reported above indicate a slightly elevated association of AOM with carriage of VT pneumococci, this does not exclude the possibility that an individual serotype(s) could be strongly associated with either AOM or carriage but that this is obscured when numerous different capsular types (which may have quite different immunological properties) are pooled.
To investigate any possible differences in the propensity of individual serotypes or STs to cause disease, ORs were estimated for the most common serotypes and STs (Tables 2 and 3). Serotypes 19F and 23F (Table 3) showed significantly elevated ORs. The lowest OR was associated with serotype 35F. For the individual clones (STs), ORs were calculated for those STs present 10 times or more in the combined data set. Even for these clones, wide CI make it hard to draw definitive conclusions. However, the serotype 23F clone, ST 37, may be associated with AOM, and STs 309 and 488, which are, respectively, 19F and 6A clones, may be also, although in both these cases our interpretation must be limited by the small numbers sampled which limit statistical power. In contrast, the 6B clone, ST 138, may be associated with carriage. Note, however, that 95% confidence limits for the ORs of all STs studied included 1, and while this may be a consequence of sample size, it suggests that any differences in ability to cause AOM are slight.
DISCUSSION
In this work we have compared the abilities of vaccine and nonvaccine serotypes, as well as individual serotypes and clones (STs), to cause AOM by comparing the pneumococci causing AOM in young children with those that are carried by children of the same age, in the same region, during the same time period. This approach provides much more meaningful information than assessing the disease potential of serotypes by looking at their rank prevalence in causing disease, as the latter approach fails to take into account any differences in exposure to each serotype. Thus, a serotype that has a high disease potential will cause little disease if exposure to it is rare, and conversely, serotypes with relatively low disease potential may be the most frequently recovered from disease if exposure to them is very common (5).
We wished to assess whether there were serotypes or clones of S. pneumoniae with a special propensity to cause AOM or whether different serotypes and clones had an approximately equal ability to cause AOM, to enable us to assess the likely long-term impact of conjugate vaccines on AOM due to S. pneumoniae. Pneumococcal serotypes in most cases include a number of genetically diverse clones (6), and precise molecular characterization of the strains from disease and carriage can be used to look at the ability of individual clones to cause disease and can address any variation in disease potential within a serotype. Furthermore, where a clone includes isolates of two different serotypes, any difference in disease potential can be assessed and, as the isolates are indistinguishable by MLST, can be postulated to be due to the difference in capsular type (although the possibility of variation at other, unsequenced virulence genes cannot be excluded).
There were several indications from this study that there are relatively small differences in the ability of different serotypes or clones to cause AOM. While pooled VTs were found to be more likely to cause AOM, the difference was small, and indeed the CI for the ORs of VTs and NVTs were found to overlap, although this is likely to be a consequence of the small sample size. Differences in the abilities of individual serotypes to cause AOM were suggested by comparison of their ORs (for example, serotypes 14 and 35F), but again the differences were small, and only serotypes 19F and 23F showed a small but significant association with AOM (Table 3). Because relatively few isolates of nonvaccine serotypes were found in this study, it is hard to directly compare the type-specific ORs of these with those of individual vaccine serotypes. However, the OR of serotype 11A, a common cause of AOM among Finnish children, was similar to that of many of the vaccine serotypes, demonstrating that nonvaccine serotypes may not be less able to cause AOM than those included within the 7-valent conjugate vaccine.
This study has revealed considerable diversity in both serotypes and clones within those pneumococci retrieved from cases of AOM and NP carriage. As a consequence, many serotypes and STs were not present in sufficient numbers to furnish statistical power to detect an association with carriage or AOM, and it is likely that a larger sample size would have detected significant differences between the ORs of these. However, we would not expect it to uncover major clones strongly associated with either of these states, but rather to improve our estimates of the small differences we may infer. In contrast, large differences were found in a recent study of invasive pneumococcal disease in the Oxford region, which used a sample size similar to that in this study. A large range of ORs was found for both serotypes (between 0.1 and 12.1) and individual STs (between 0.1 and 8.3). In the present study, ORs for serotypes ranged between 0.18 and 2.46 and for STs between 0.36 and 2.7 (Tables 2 and 3). This suggests that the sample size here should be able to detect large differences, if they exist, and the failure to find such differences indicates that there is less variation in the ability of STs and serotypes to cause AOM than to cause invasive disease.
Similarly, isolates from invasive disease were significantly less diverse than those from carriage, and there was a highly nonrandom distribution of isolates of the major clones among the samples from carriage and invasive disease. In contrast, no differences were found in our study, suggesting that the isolates that cause AOM are not clearly different from those that are carried in the nasopharynx. There was no evidence of major clones which are carried but do not cause AOM.
Serotypes 19F and 23F appeared to be slightly more able to cause AOM than isolates of the other serotypes that were prevalent in our data set (Table 3). There is some evidence that possession of the serotype 19F capsule may predispose slightly towards the development of AOM. ST 66 included isolates of serotypes 23F, 19F, and 9N, indicating a history of serotype switching within this clone. If we consider the distribution of serotypes between carriage and AOM isolates in this serotypically mixed clone (Table 2), all but one of the six isolates of serotype 9N are from carriage, whereas all of the serotype 19F and 23F isolates are from patients with AOM. This suggests that serotype 19F and 23F isolates may be more commonly recovered from patients with AOM than isolates with the same genotype that express the serotype 9N capsule. Any greater propensity of serotype 19F to cause AOM is unfortunate, as in clinical trials the present formulations of the conjugate vaccines were least effective against this serotype (point estimate for efficacy against AOM due to serotype 19F = 25%) (7).
There is some evidence for a difference in the ability of clones of the same serotype to cause AOM, which would suggest a contribution of genotype as well as capsular type to the ability to cause AOM. STs 37, 36, and 507 all express 23F capsules, but the ORs for these STs may be different (Table 2), although a larger sample is needed to confirm this. For invasive disease, there were no apparent differences among clones of the same serotype, and serotype may be an important, and perhaps the predominant, marker of invasive disease potential (5). In mucosal infections, such as AOM, the situation appears to be different, and serotype may play a lesser role in determining the ability of a strain to cause disease. A difference in the importance of capsular serotype is supported by the fact that nontypeable or unencapsulated pneumococci are very rarely associated with invasive disease, whereas these strains are relatively frequently recovered from patients with AOM (four unencapsulated pneumococci were recovered in this study).
If the capsule plays a less important role in disease, serotype replacement may be expected to reduce the effectiveness of the conjugate vaccines against AOM. Some support for this possibility comes from the Finnish trial, in which an increase in carriage of nonvaccine serotypes was mirrored by a significant increase in AOM due to these serotypes (7). However, over the course of the trial this was not sufficient to eliminate the reduction in AOM, and comparison of tympanostomy tube placements in vaccinated and unvaccinated children from 2 to 4 or 5 years of age showed a significant reduction in the vaccinated children (A. Palmu, J. Verho, P. H. Makela, and T. Kilpi, Abstr. 3rd Int. Symp. Pneumococci Pneumococcal Dis., 2002, p. 72). Also note that a completely vaccinated population would be expected to result in greater selective pressure in favor of nonvaccine serotypes than has been the case in any of the trials reported to date, in which the vaccinated population mixed freely with the nonvaccinated majority. This may result in a more extensive replacement phenomenon than we have seen in the efficacy studies.
The efficacy of conjugate vaccines against pneumonia has not been thoroughly assessed, and much less is known about the strains and serotypes from pneumonia than about those from invasive disease. Protection against bacteremic pneumonia caused by vaccine serotypes appears to be as effective as that against invasive disease, which is consistent with protection against bacteremia (2). However, nonbacteremic pneumonia may more closely resemble a mucosal infection than an invasive infection. Nonbacteremic pneumonia is one of the most significant contributors to the morbidity and mortality of pneumococcal disease (although less serious than meningitis or bacteremic pneumonia, it is far more common). It will be important to assess whether nonbacteremic pneumonia is more similar to invasive disease or a mucosal infection such as AOM, where there appears to be much less variation in the ability of serotypes and clones to cause disease. If the latter is true, we might expect serotype replacement to reduce the effectiveness of conjugate vaccines against this form of pneumonia, since the replacing nonvaccine serotypes may have considerable pathogenic potential.
In conclusion, we have demonstrated that there appear to be smaller differences in the abilities of different pneumococcal serotypes and clones to cause AOM than is found with invasive disease, and following serotype replacement, this has the potential to reduce the efficacy of the conjugate vaccines against AOM. Surveillance for changes in the serotypes causing AOM, and for loss of vaccine efficacy, is required following mass vaccination, but tympanocentesis is now rarely undertaken, and the numbers of isolates obtained from patients with AOM may not be sufficient to observe any changes that occur.
Acknowledgments
This work was supported by a grant from the Wellcome Trust (grant 030662). The FinOM Studies were supported by Aventis Pasteur, Merck & Co., Inc., and Wyeth-Lederle Vaccines.
B.G.S. is a Wellcome Trust Principal Research Fellow.
We thank the FinOM Study staff for data collection, laboratory work, and data management and all families who participated.
Editor: J. N. Weiser
REFERENCES
- 1.Black, S., H. Shinefield, B. Fireman, E. Lewis, P. Ray, J. R. Hansen, L. Elvin, K. M. Ensor, J. Hackell, G. Siber, F. Malinoski, D. Madore, I. Chang, R. Kohberger, W. Watson, R. Austrian, K. Edwards, et al. 2000. Efficacy, safety and immunogenicity of heptavalent pneumococcal conjugate vaccine in children. Pediatr. Infect. Dis. J. 19:187-195. [DOI] [PubMed] [Google Scholar]
- 2.Black, S. B., H. R. Shinefield, S. Ling, J. Hansen, B. Fireman, D. Spring, J. Noyes, E. Lewis, P. Ray, J. Lee, and J. Hackell. 2002. Effectiveness of heptavalent pneumococcal conjugate vaccine in children younger than five years of age for prevention of pneumonia. Pediatr. Infect. Dis. J. 21:810-815. [DOI] [PubMed] [Google Scholar]
- 3.Block, S. L. 1997. Causative pathogens, antibiotic resistance and therapeutic considerations in acute otitis media. Pediatr. Infect. Dis. J. 16:449-456. [DOI] [PubMed] [Google Scholar]
- 4.Bondy, J., S. Berman, J. Glazner, and D. Lezotte. 2000. Direct expenditures related to otitis media diagnoses: extrapolations from a pediatric medicaid cohort. Pediatrics 105:E72. [DOI] [PubMed] [Google Scholar]
- 5.Brueggemann, A. B., D. T. Griffiths, E. Meats, T. Peto, D. W. Crook, and B. G. Spratt. 2003. Clonal relationships between invasive and carriage Streptococcus pneumoniae, and serotype and clone-specific differences in invasive disease potential. J. Infect. Dis. 187:1424-1432. [DOI] [PubMed] [Google Scholar]
- 6.Enright, M. C., and B. G. Spratt. 1998. A multilocus sequence typing scheme for Streptococcus pneumoniae: identification of clones associated with serious invasive disease. Microbiology 144:3049-3060. [DOI] [PubMed] [Google Scholar]
- 7.Eskola, J., T. Kilpi, A. Palmu, J. Jokinen, J. Haapakoski, E. Herva, A. Takala, H. Kayhty, P. Karma, R. Kohberger, G. Siber, and P. H. Makela. 2001. Efficacy of a pneumococcal conjugate vaccine against acute otitis media. N. Engl. J. Med. 344:403-409. [DOI] [PubMed] [Google Scholar]
- 8.Gray, B. M., G. M. Converse III, and H. C. Dillon, Jr. 1980. Epidemiologic studies of Streptococcus pneumoniae in infants: acquisition, carriage, and infection during the first 23 months of life. J. Infect. Dis. 142:923-933. [DOI] [PubMed] [Google Scholar]
- 9.Grundmann, H., S. Hori, and G. Tanner. 2001. Determining confidence intervals when measuring genetic diversity and the discriminatory abilities of typing methods for microorganisms. J. Clin. Microbiol. 39:4190-4192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hausdorff, W. P., J. Bryant, P. R. Paradiso, and G. R. Siber. 2000. Which pneumococcal serogroups cause the most invasive disease: implications for conjugate vaccine formulation and use, part I. Clin. Infect. Dis. 30:100-121. [DOI] [PubMed] [Google Scholar]
- 11.Kilpi, T., E. Herva, T. Kaijalainen, R. Syrjanen, and A. K. Takala. 2001. Bacteriology of acute otitis media in a cohort of Finnish children followed for the first two years of life. Pediatr. Infect. Dis. J. 20:654-662. [DOI] [PubMed] [Google Scholar]
- 12.Klugman, K. P. 2001. Efficacy of pneumococcal conjugate vaccines and their effect on carriage and antimicrobial resistance. Lancet Infect. Dis. 1:85-91. [DOI] [PubMed] [Google Scholar]
- 13.Luotonen, J., E. Herva, P. Karma, M. Timonen, M. Leinonen, and P. H. Makela. 1981. The bacteriology of acute otitis media in children with special reference to Streptococcus pneumoniae as studied by bacteriological and antigen detection methods. Scand. J. Infect. Dis. 13:177-183. [DOI] [PubMed] [Google Scholar]
- 14.Meats, E., A. B. Brueggemann, M. C. Enright, K. Sleeman, D. T. Griffiths, D. W. Crook, and B. G. Spratt. 2003. Stability of serotypes during nasopharyngeal carriage of Streptococcus pneumoniae. J. Clin. Microbiol. 41:386-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nei, M. 1987. Molecular evolutionary genetics. Columbia University Press, New York, N.Y.
- 16.Simpson, E. H. 1949. Measurement of diversity. Nature 163:688. [Google Scholar]
- 17.Spratt, B. G., and B. M. Greenwood. 2000. Prevention of pneumococcal disease by vaccination: does serotype replacement matter? Lancet 356:1210-1211. [DOI] [PubMed] [Google Scholar]
- 18.SPSS Inc. 1999. SPSS regression models 9.0. SPSS Inc., Chicago, Ill.
- 19.Syrjanen, R. K., T. M. Kilpi, T. H. Kaijalainen, E. E. Herva, and A. K. Takala. 2001. Nasopharyngeal carriage of Streptococcus pneumoniae in Finnish children younger than 2 years old. J. Infect. Dis. 184:451-459. [DOI] [PubMed] [Google Scholar]


