Abstract
Protein kinase C δ (PKC δ) is normally activated by diacylglycerol produced from receptor-mediated hydrolysis of inositol phospholipids. On stimulation of cells with H2O2, the enzyme is tyrosine phosphorylated, with a concomitant increase in enzymatic activity. This activation does not appear to accompany its translocation to membranes. In the present study, the tyrosine phosphorylation sites of PKC δ in the H2O2-treated cells were identified as Tyr-311, Tyr-332, and Tyr-512 by mass spectrometric analysis with the use of the precursor-scan method and by immunoblot analysis with the use of phosphorylation site-specific antibodies. Tyr-311 was the predominant modification site among them. In an in vitro study, phosphorylation at this site by Lck, a non-receptor-type tyrosine kinase, enhanced the basal enzymatic activity and elevated its maximal velocity in the presence of diacylglycerol. The mutation of Tyr-311 to phenylalanine prevented the increase in this maximal activity, but replacement of the other two tyrosine residues did not block such an effect. The results indicate that phosphorylation at Tyr-311 between the regulatory and catalytic domains is a critical step for generation of the active PKC δ in response to H2O2.
Protein kinase C (PKC) comprises a family of more than ten serine/threonine protein kinases that are involved in a variety of signal transduction pathways (1). Each isoform has the regulatory and catalytic domains in the amino- and carboxyl-terminal halves, respectively. The isoforms are divided into three groups, cPKC, nPKC, and aPKC, because of the structural differences in their regulatory domains. The cPKC and nPKC isoforms are activated by diacylglycerol produced from receptor-mediated hydrolysis of inositol phospholipids and are the prime targets of tumor-promoting phorbol esters that bind to the cysteine-rich sequence, named the C1 region, in the regulatory domain. In general, a number of protein kinases are known to be controlled by phosphorylation (2), and the PKC family members have three phosphorylation motif sites mostly conserved among the family (3). One is a threonine residue in the activation loop of the catalytic domain, and the others are serine and threonine residues located in the carboxyl-terminal end region, named the turn and hydrophobic motifs, respectively.
The PKC isoforms are further phosphorylated on tyrosine upon stimulation of the cells (4), and the role of tyrosine phosphorylation has been investigated for PKC δ, a member of the nPKC group (5). That is, PKC δ is phosphorylated on tyrosine in v-ras-transformed keratinocytes (6) and in various cells stimulated with phorbol ester, growth factors, and hormones (7–14). However, controversial results are reported on the functional consequence of the tyrosine phosphorylation reaction induced by these membrane-coupled signaling processes. In keratinocytes, tyrosine phosphorylation reduces its catalytic activity (6, 12), whereas in other cases the modification reaction enhances the enzymatic activity (4, 7, 8) or even alters its enzymatic specificity (9, 15, 16). We have reported that PKC δ is tyrosine phosphorylated in the H2O2-treated cells and that the enzyme recovered is constitutively active and is independent of diacylglycerol (4). Consistently, PKC δ does not translocate to membranes, but apparently stays in the cytosol after H2O2 stimulation (17). Furthermore, H2O2-induced apoptosis is enhanced in the cell line stably overproducing the PKC isoform (18). Therefore, it is plausible that PKC δ is activated by tyrosine phosphorylation in the H2O2-treated cells in a mechanism unrelated to receptor-coupled hydrolysis of inositol phospholipids, and that the enzyme thus activated has some role in the regulation of cellular functions.
Thus far, Tyr-52 and Tyr-187 of PKC δ have been identified as the phosphorylation sites after stimulation by IgE antigen (10) and platelet-derived growth factor (19), respectively. PKC δ is in fact phosphorylated by various tyrosine kinases, such as Fyn (1, 12, 20), Src (12, 13, 20–23), Lyn (10, 22), Abl (24, 25), and growth factor receptors (7, 12). Tyr-155 (10), Tyr-311 (23), and Tyr-565 (10) are proposed to be the phosphorylation sites for the Src family protein kinases. We have found that PKC δ is phosphorylated on tyrosine in the carboxyl-terminal half of the molecule (4). It may be possible that phosphorylation on different tyrosine residues produces distinct results with regard to the kinetic properties of the enzyme. In this study, the tyrosine as well as the serine and threonine phosphorylation sites of PKC δ in the H2O2-treated cells were identified, and the kinetics of the tyrosine-phosphorylated enzyme were investigated.
Materials and Methods
Expression Vectors.
FLAG-epitope-tagged expression plasmid of rat PKC δ was constructed as described (4), and the protein product was designated as FLAG-PKC δ. Tyr-311, Tyr-332, Tyr-451, Tyr-512, and Tyr-523 of PKC δ were replaced with phenylalanine by site-directed mutagenesis to make FLAG-epitope-tagged mutant molecules. The mutants replacing Tyr-311, Tyr-332, Tyr-512, Tyr-311/Tyr-332, Tyr-311/Tyr-332/Tyr-451, Tyr-311/Tyr-332/Tyr-512, and Tyr-311/Tyr-332/Tyr-523 were designated as Y311F, Y332F, Y512F, Y311/332F, Y311/332/451F, Y311/332/512F, and Y311/332/523F, respectively. The cDNA insert of murine Lck (26), kindly donated by R. M. Perlmutter (University of Washington, Seattle) was cloned into the FLAG-epitope-tagged expression plasmid. The DNA sequences of these constructs were confirmed by the dideoxy chain-termination method with a DNA Sequencing System Model 373A (Applied Biosystems).
Cell Culture and Transfection.
COS-7 cells were maintained in DMEM supplemented with 10% FCS at 37°C in a 5% CO2 incubator. The cells were transfected with the expression plasmids by electroporation and cultured for 48 h before experiments. Where indicated, the cells were treated with 5 mM H2O2 for 10 min.
Generation of Antibodies.
Phosphorylation site-specific antisera were raised against the phosphopeptides having an additional cysteine residue at the amino-terminal end of the amino acid sequence of rat PKC δ essentially as described (27, 28): CPETVGIpYQGFEK (amino acids 305–316), CIPDNNGTpYGKIWEG (amino acids 325–338), CGTPDpYIAPEILQG (amino acids 508–520), CNEKQPLpSFSDKNL (amino acids 637–649), and CTAFKGFpSFVNPKY (amino acids 656–668), where pS and pY indicate phosphoserine and phosphotyrosine, respectively. The phosphopeptides were coupled to keyhole limpet hemocyanin and used to immunize rabbits. The phosphorylation site-specific antibodies were purified from antisera by successive column chromatographies with the use of affinity resins coupled with each phosphopeptide and nonphosphopeptide. The resulting antibodies were designated as anti-pY311, anti-pY332, anti-pY512, anti-pS643, and anti-pS662, respectively.
Mass Spectrometric Analysis.
COS-7 cells expressing FLAG-PKC δ were treated with H2O2 and were homogenized by sonication in 20 mM Tris⋅HCl (pH 7.5) containing 1 mM EDTA, 1 mM EGTA, 1 mM DTT, 150 mM NaCl, 10 mM NaF, 1 mM Na3VO4, 20 mM β-glycerophosphate, and 50 μg/ml phenylmethylsulfonyl fluoride. The supernatant was subjected to FLAG M2 affinity column chromatography (Sigma), and FLAG-PKC δ was eluted by 100 mM glycine⋅HCl at pH 3.0. The eluate was neutralized with 1 M Tris⋅HCl at pH 8.0, and NaF, Na3VO4, and β-glycerophosphate were added to the solution at the final concentrations in the homogenization buffer. The purification procedures above were carried out at 0–4°C. PKC δ isolated (6 μg) was digested with 1 μg of lysyl endoprotease (Wako Pure Chemical Industries, Osaka) in 100 mM Tris⋅HCl (pH 9.0) containing 2 M urea at 37°C for 16 h. The reaction was stopped by the addition of formic acid at a final concentration of 0.1%. The resulting peptide mixture was subjected to liquid chromatography/mass spectrometry (LC/MS) and MS/MS analysis. LC/MS analysis was carried out on an electrospray triple quadrupole mass spectrometer (PE Sciex API-III) equipped with a nanospray ion source (Protana, Odense, Denmark), and MS/MS analysis was performed on a hybrid-type mass spectrometer (Micromass Q-TOF) with a nanospray ion source supplied by the manufacturer. The LC/MS system was constructed as described (29), with minor modifications. A capillary column packed with C18 material (PepMap C18, 75 μm × 150 mm, LCPackings) was eluted with a linear gradient of H2O-acetonitrile in the presence of 0.1% formic acid at a flow rate of 0.2 μl/min. The column eluate was injected directly into the electrospray ion source with a pulled fused silica capillary source (New Objectives, Cambridge, MA). The spectrometer was operated in a negative-ion mode to detect phosphopeptides with precursor scanning; the details of the procedures will be described elsewhere. To obtain sequence data from the phosphopeptides thus identified, the peptide mixtures were desalted on 1 μl of Poros R3 column (Applied Biosystems) self-packed in pulled glass capillaries (30). The peptides were eluted directly into spraying capillaries and were subjected to nanospray MS/MS analysis in the positive-ion mode.
Immunoprecipitation and Immunoblot Analysis.
The following procedures were carried out at 0–4°C, as described (31). The transfected cells were washed with PBS and lyzed in 20 mM Tris⋅HCl (pH 7.5) containing 1 mM EDTA, 1 mM EGTA, 10 mM 2-mercaptoethanol, 1% Triton X-100, 150 mM NaCl, 10 mM NaF, 1 mM Na3VO4, and 50 μg/ml phenylmethylsulfonyl fluoride. The lysate was centrifuged, and the supernatant was incubated for 1 h with an anti-FLAG monoclonal antibody (Sigma). Then, protein A-Sepharose (Amersham Pharmacia) was added to the mixture and incubated for 30 min. The immunoprecipitates were collected by centrifugation and were washed four times each time with 20 mM Tris⋅HCl (pH 7.5) containing 150 mM NaCl and 1% Triton X-100. The samples were boiled in SDS sample buffer, and proteins were separated by SDS/PAGE and transferred to an Immobilon P membrane (Millipore). Immunoblot analysis was carried out with the use of the anti-FLAG antibody, a monoclonal anti-phosphotyrosine antibody (anti-PY) (4G10; Upstate Biotechnology, Lake Placid, NY), the antibody directed against phosphorylated Thr-505 of PKC δ (anti-pT505) (New England Biolabs), or the phosphorylation site-specific antibodies described above as the primary antibody. The alkaline phosphatase-conjugated anti-rabbit or anti-mouse antibody (Promega) was used as the secondary antibody. The color reaction was carried out with the use of 5-bromo-4-chloro-3-indolyl-phosphate and nitro blue tetrazolium as substrates, as described (31).
Protein Kinase Assay.
FLAG-PKC δ, the phosphorylation site mutants of PKC δ, and Lck were immunoprecipitated from transfected COS-7 cells. Each PKC δ molecule and Lck immobilized on the resin were mixed and incubated for 20 min at 30°C in the reaction mixture containing 50 mM Tris⋅HCl (pH 7.5), 10 mM MnCl2, and 1 mM ATP. After washing, aliquots of the proteins immobilized on the resin were subjected to either immunoblot analysis or the protein kinase assay. The PKC activity was assayed by measuring the incorporation of 32Pi into calf thymus H1 histone from [γ-32P]ATP in the reaction mixture containing 20 mM Tris⋅HCl (pH 7.5), 10 mM MgCl2, 20 μM ATP, 15–50 kBq of [γ-32P]ATP, and 200 μg/ml H1 histone (4). The incubation was carried out for 5 min at 30°C. The phosphorylated proteins were separated by SDS/PAGE and were visualized and quantitated by measurement of the intensity of photostimulated luminescence with a Bio-imaging Analyzer (Fuji). Where indicated, the PKC activity was measured in the presence of phosphatidylserine and diacylglycerol.
Results
Phosphorylation Sites of PKC δ.
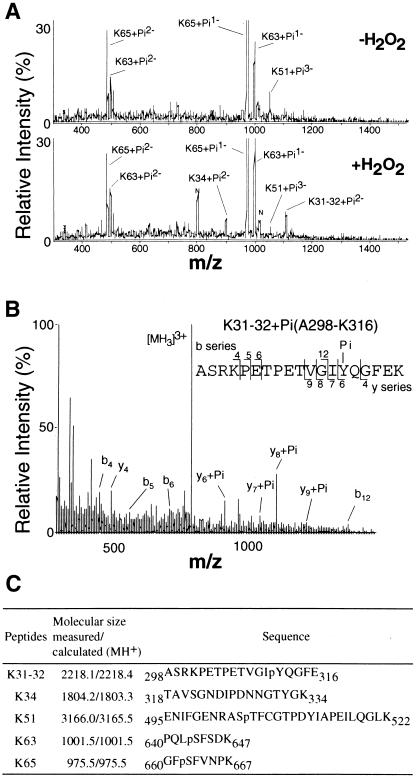
PKC δ was phosphorylated on serine and threonine in the unstimulated cells (3), and the H2O2 treatment enhanced phosphorylation on serine and threonine residues and induced additional phosphorylation on tyrosine, with a concomitant increase in enzymatic activity (4). To analyze the activation mechanism of PKC δ, the phosphorylation sites of the PKC isoform were identified by the precursor-scan nano LC/MS method. FLAG-PKC δ was recovered from COS-7 cells that were treated with H2O2 or vehicle and was digested with lysyl endoprotease. The resulting phosphopeptides were then subjected to identification of the phosphorylation sites (Fig. 1). In the precursor scan mode, in which precursor ions producing fragment ion of m/z 79 (PO3−) are selectively detected, only phosphorylated peptides were recorded. The peaks were identified by comparing their masses with those of the known peptide sequences and by determining their partial sequences by MS/MS analysis. Three phosphopeptides, K51 (Glu-495–Lys-522, [M-3H]3− ion at m/z 1054.0), K63 (Pro-640–Lys-647, [M-2H]2− ion at m/z 499.0, and [M-H]1− ion at m/z 999.0), and K65 (Gly-660–Lys-667, [M-2H]2− ion at 486.5 and [M-H]1− ion at m/z 973.5), were detected in the PKC isoform obtained from the untreated control cells (Fig. 1A). These peptide fragments each contained a phosphorylation motif site of Thr-505, Ser-643, and Ser-662 in the activation loop, the turn motif, and the hydrophobic motif, respectively.
Figure 1.
Mass spectrometric analysis of the phosphorylation sites of PKC δ recovered from H2O2-treated COS-7 cells. (A) Precursor-ion-scan spectra for m/z −79 of the lysyl endoprotease digests of PKC δ. The phosphopeptides isolated from the control (Upper) and H2O2-treated (Lower) cells were analyzed. All scans were combined where significant peaks were detected. (B) Identification of the phosphorylation sites in phosphopeptide K31–32. Peptide mixtures were desalted and subjected to nanospray MS/MS analysis. The product ion spectrum of the precursor ion at m/z 740 produced by the collision-induced dissociation was obtained with the Q-TOF mass spectrometer. (C) The phosphorylation sites of PKC δ. The phosphopeptides identified in PKC δ obtained from the H2O2-treated cells are summarized.
When the cells were treated with H2O2, two additional phosphopeptides, K31–32 (Ala-298–Lys-316, [M-2H]2− ion at m/z 1107.5) and K34 (Thr-318–Lys-334, [M-2H]2− ion at m/z 900.0), were detected. Both peptides were derived from the hinge region of PKC δ between the regulatory and catalytic domains. In the peptide K31–32, Tyr-311 was identified as the major phosphorylation site by MS/MS analysis (Fig. 1B). The presence of y4 and y6 + Pi ions narrows down the phosphorylation site to Tyr-Gln; only the former is the possible phosphorylation site. On the other hand, the phosphorylation site of K34 could not be identified because of the low abundance of the phosphopeptide. However, this phosphopeptide has only one tyrosine residue, Tyr-332, and was not recovered from the untreated cells. Thus, this tyrosine residue could be the plausible phosphorylation site in the H2O2-treated cells. The phosphorylation sites in K51, K63, and K65 were identified as Thr-505, Ser-643, and Ser-662, respectively, by MS/MS analysis (data not shown). K51 from the activation loop also contains a tyrosine residue, Tyr-512. Most of the phosphoryl group in K51 was found on Thr-505, but phosphorylation at Tyr-512 could not be excluded. The phosphopeptides as well as the phosphorylation sites identified as described above by mass spectrometric analysis are summarized in Fig. 1C.
Mutation Analysis.
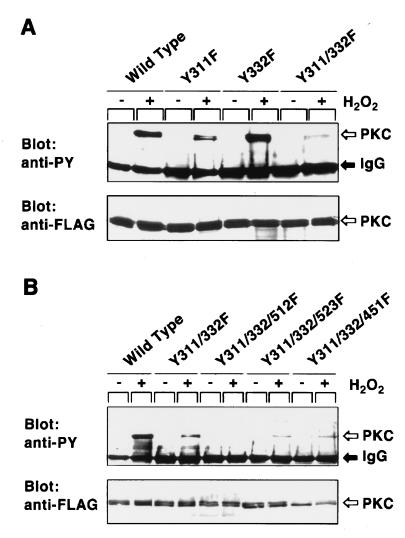
The mutant molecules in which Tyr-311 and Tyr-332 are replaced by phenylalanine were expressed in COS-7 cells (Fig. 2A). Consistent with the results of mass spectrometric analysis, H2O2-induced tyrosine phosphorylation was markedly reduced for the mutant Y311F, whereas the mutation of Tyr-332 showed a subtle effect. Tyrosine phosphorylation of the double mutant Y311/322F was lower than that of the single mutant Y311F, but the double-mutant molecule was still tyrosine phosphorylated. Our previous study (4) predicted that Tyr-512 and Tyr-532 in the catalytic domain are phosphorylated, because these tyrosine residues are conserved among the PKC isoforms, all of which were more or less tyrosine phosphorylated on the H2O2 treatment, and because tyrosine phosphorylation of PKC δ was reduced when these residues were replaced by phenylalanine. Consistent with this prediction, the mutant molecule Y311/332/512F was totally inert for tyrosine phosphorylation, whereas the mutant molecule Y311/332/532F was still phosphorylated on tyrosine when treated with H2O2 (Fig. 2B). Replacement of other tyrosine residues conserved in the catalytic domain of the all PKC isoforms, such as Tyr-451 and Tyr-469, did not affect tyrosine phosphorylation. The results of mutation analysis described above indicate that three tyrosine residues, Tyr-311 and Tyr-332 in the hinge region and Tyr-512 in the catalytic domain, are the tyrosine phosphorylation sites of PKC δ in the H2O2-treated cells.
Figure 2.
Tyrosine phosphorylation of the PKC δ mutants in H2O2-treated COS-7 cells. (A) Tyrosine phosphorylation of the single- and double-point mutants. (B) Tyrosine phosphorylation of the triple-point mutants. Cells transfected with each expression plasmid were incubated with H2O2, and the epitope-tagged proteins were immunoprecipitated by the anti-FLAG antibody. Immunoblot analysis was carried out with the use of the anti-PY (Upper) or anti-FLAG (Lower) antibody.
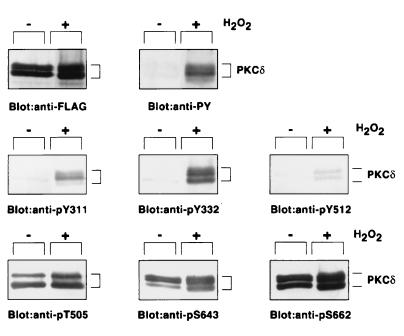
Immunoblot Analysis.
The sites of phosphorylation of PKC δ were investigated further with the use of antibodies raised against phosphopeptides corresponding to each phosphorylation site (Fig. 3). FLAG-PKC δ recovered from serum-starved COS-7 cells appeared to be a doublet, as reported (32). Tyrosine phosphorylation of these protein bands was confirmed by immunoblot analysis with the anti-phosphotyrosine antibody. Namely, FLAG-PKC δ obtained from the H2O2-treated cells, but not the enzyme from the untreated cells, was recognized by the anti-pY311, anti-pY332, and anti-pY512 antibodies. The immunoreaction of these phosphorylation site-specific antibodies disappeared after absorption of the antibodies with each phosphopeptide used as an antigen (data not shown).
Figure 3.
Immunoblot analysis of PKC δ recovered from H2O2-treated COS-7 cells. Cells expressing FLAG-PKC δ were cultured in DMEM containing 0.1% BSA for 18 h before the H2O2 treatment, and FLAG-PKC δ was immunoprecipitated by the anti-FLAG antibody. Immunoblot analysis was carried out with the use of the anti-FLAG, anti-PY, or each phosphorylation site-specific antibody.
In contrast, PKC δ immunoprecipitated from both the H2O2-treated and control cells was recognized by the anti-pT505, anti-pS643, and anti-pS662 antibodies. It was noted that phosphorylation on serine and threonine residues was enhanced by the H2O2 treatment. These results are compatible with those of phosphoamino acid analysis in the previous study (4).
Tyrosine Phosphorylation in Vitro.
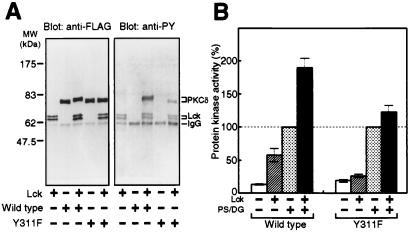
PKC δ was phosphorylated in vitro by a nonreceptor-type tyrosine kinase Lck, which is known to be activated in the H2O2-treated cells (33) (Fig. 4A). The wild-type PKC δ was phosphorylated efficiently by Lck, as judged by immunoblot analysis and its band shift. Y311F, the mutant molecule replacing the major phosphorylation site, was phosphorylated by Lck only slightly. Tyrosine phosphorylation elevated the basal activity of the wild-type enzyme in the absence of diacylglycerol and phosphatidylserine (Fig. 4B). With the lipid activators, the tyrosine-phosphorylated enzyme showed a much higher activity than the nonphosphorylated enzyme. Y311F depended on the lipid activators, but the activity was not effected by the incubation with Lck.
Figure 4.
Tyrosine phosphorylation and activation of PKC δ by Lck in vitro. FLAG-PKC δ and Y311F immunoprecipitated from transfected COS-7 cells were incubated with or without Lck in the presence of ATP and MnCl2. (A) Tyrosine phosphorylation. Immunoblot analysis was carried out with the use of the anti-FLAG or anti-PY antibody. (B) Protein kinase activity. PKC activity was measured with the use of H1 histone as substrate in the presence or absence of 8 μg/ml phosphatidylserine and 0.8 μg/ml diacylglycerol. The activity of the enzyme without incubation with Lck in the presence of phosphatidylserine and diacylglycerol was used as 100%. The results shown are the means ± SE of five independent experiments.
Kinetic Analysis.
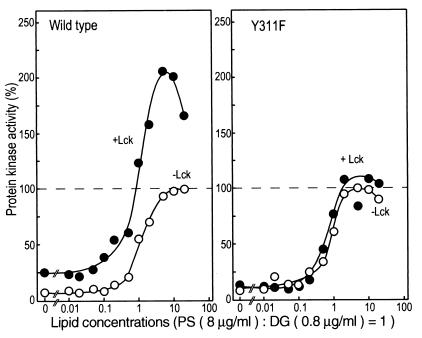
The tyrosine-phosphorylated PKC δ was activated further by the addition of diacylglycerol and phosphatidylserine, and hence the kinase activity was assayed in the presence of various concentrations of the lipid activators (Fig. 5). Diacylglycerol and phosphatidylserine at a ratio of 1:10 (0.8 and 8 μg/ml, respectively, in Fig. 4B) have normally been used for the standard assay (32). Both the wild-type and Y311F enzymes expressed in COS-7 cells required higher concentrations of the lipids to obtain maximal activity, presumably because the immunoprecipitated enzymes need more lipid activators than do the soluble enzymes. For the Y311F enzyme, incubation with Lck did not affect its enzymatic activity significantly, whereas the wild-type enzyme showed an elevated maximal velocity when tyrosine was phosphorylated. It was thus concluded that tyrosine phosphorylation not only enhances the basal activity in the absence of diacylglycerol, but also elevates its maximal velocity in the presence of the lipid activators.
Figure 5.
Properties of PKC δ tyrosine phosphorylated by Lck. FLAG-PKC δ and Y311F immunoprecipitated from transfected COS-7 cells were incubated with or without Lck, and PKC activity was measured with the use of H1 histone as substrate in the presence of various concentrations of phosphatidylserine (PS) and diacylglycerol (DG). The maximal activity of the enzyme without incubation with Lck was used as 100%.
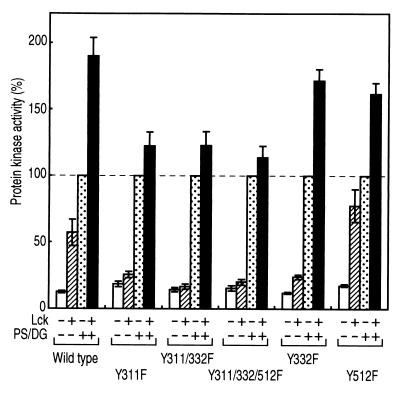
The mutation of the other two tyrosine residues, Tyr-332 and Tyr-512, did not prevent the increase in the maximal velocity in the presence of the lipid activators after tyrosine phosphorylation by Lck (Fig. 6). Y512F showed properties similar to those of the wild-type enzyme. Y332F did not show an increase in the basal activity, implying that Tyr-332 may play some role in the activation of PKC δ. An additional mutation of Tyr-332 or the changes in both Tyr-332 and Tyr-512 did not cause a significant difference from the single mutant Y332F, indicating again that Tyr-311 is the major phosphorylation site in vitro as well.
Figure 6.
Protein kinase activity of the PKC mutants. FLAG-PKC δ and each mutant immunoprecipitated from transfected COS-7 cells were incubated with or without Lck, and PKC activity was measured with the use of H1 histone as substrate in the presence or absence of 8 μg/ml phosphatidylserine and 0.8 μg/ml diacylglycerol. The activity of the enzyme without incubation with Lck in the presence of phosphatidylserine and diacylglycerol was used as 100%. The results shown are the means ± SE of five independent experiments.
Discussion
In this study, the sites of tyrosine phosphorylation of PKC δ in the H2O2-treated cells were identified as Tyr-311, Tyr-332, and Tyr-512. It was also shown that Thr-505, Ser-643, and Ser-662 of PKC δ in the activation loop, the turn motif, and the hydrophobic motif, respectively (3), are phosphorylated in the quiescent and stimulated cells. Although a quantitative estimation is not possible in this kind of analysis, it may be concluded that Ser-643 and Ser-662 are constitutively phosphorylated to a significant extent—up to 30%, judging from the peak intensities in the positive mode spectra. The stoichiometry was much less for the other sites, but a significant increase in phosphorylation was detected in the two peptides, K31–32 and K34, in the H2O2-treated cells. With regard to tyrosine phosphorylation, Tyr-311 and Tyr-332 are located in the hinge region between the regulatory and catalytic domains of the enzyme. Tyr-311 was the predominant phosphorylation site, and the modification of Tyr-332 was less prominent than that of Tyr-311. The fragment K51 including Tyr-512 and Thr-505 was recovered as a phosphopeptide by the precursor scan method, but most of the phosphoryl group was found on Thr-505. Presumably, the stoichiometry of phosphorylation at Tyr-512 was lower than that of the other two tyrosine phosphorylation sites in the cells. In the previous study (4), two fragments with approximate molecular masses of 45 and 25 kDa were detected by the antibody against phosphotyrosine as well as the antibody recognizing the carboxyl-terminal end of PKC δ, which were generated by partial tryptic digestion of PKC δ recovered from the H2O2-treated cells. It is plausible that the 25-kDa fragment is phosphorylated at Tyr-512, because this fragment was too small to contain Tyr-311 and Tyr-332. Immunoblot analysis using the phosphorylation site-specific antibodies clearly indicated that these three tyrosine residues are phosphorylated on H2O2 stimulation. Furthermore, analysis of the mutated molecules revealed that there is no other detectable tyrosine phosphorylation site in the PKC isoform in the H2O2-treated cells.
Tyr-52 and Tyr-187 in the regulatory domain are shown to be phosphorylated in vivo by stimulation of the cells with IgE antigen (10) and platelet-derived growth factor (19), respectively, and Tyr-155 and Tyr-565 are the in vitro phosphorylation sites for the Src family protein kinase Lyn (10). Thus, PKC δ may be phosphorylated by more than one tyrosine kinase in the different signaling pathways. In the cells expressing the active mutant of Src, it has been shown that PKC δ is tyrosine phosphorylated and degraded, and that the replacement of Tyr-311 reduces the modification reaction and its subsequent degradation (23). Tyrosine phosphorylation may facilitate the proteolysis of PKC δ, because the sequence Asp-Ile-Pro-Asp-Asn (amino acids 324–328) matching the caspase-3 recognition motif is located between the two tyrosine phosphorylation sites in the hinge region (34). The cleavage product of PKC δ was not detected, however, in the Chinese hamster ovary cell line treated with H2O2 that overproduced PKC δ (18).
PKC δ recovered from the H2O2-treated cells is constitutively active and independent of the lipid activator (4). Consistently, PKC δ phosphorylated at Tyr-311 in the hinge region in vitro showed an enhanced enzymatic activity. In PKC βII, Thr-314 and Thr-324 in the hinge region are autophosphorylated, and this reaction may influence the protein kinase activity (35). The modification in the region between the regulatory and catalytic domains may be a common mechanism for the regulation of this protein kinase family. PKC δ phosphorylated at Tyr-311 in vitro was further activated, however, by diacylglycerol. It is possible that phosphorylation at other sites, such as Thr-505, Ser-643, and Ser-662, is required for the conversion of PKC δ to the lipid activator-independent form, because phosphorylation on serine and threonine of the enzyme significantly increased in the H2O2-treated cells (4). Although phosphorylation at Thr-505 in the activation loop is not a prerequisite for its basal and activator-dependent enzymatic activity (36), the modification of this site, together with the adjacent residue of Tyr-512, may contribute to activation of the enzyme. If serine phosphorylation on the carboxyl-terminal end region is required for full enzyme activation, the modification in both the hinge and carboxyl-terminal end regions on tyrosine and serine, respectively, may be integral to the conformational change of the enzyme that induces its activation. Alternatively, redox modification of the enzyme by reactive oxygen species has been suggested to have an important role in the regulation of PKC δ activity (37). Further structural and biochemical studies are required to clarify the multiple activation mechanisms of PKC δ in response to various signals.
Acknowledgments
We thank R. M. Perlmutter for providing the Lck cDNA, Y. Ono for discussions, and J. Jidai for secretarial assistance. This study was supported in part by research grants from the Scientific Research Funds of the Ministry of Education, Science, Sports and Culture of Japan; the Special Coordination Funds for Promoting Science and Technology of the Science and Technology Agency of Japan; New Lead Research Laboratories, Sankyo Company, Ltd.; and the Japan Foundation for Applied Enzymology.
Abbreviations
- PKC
protein kinase C
- LC
liquid chromatography
- MS
mass spectrometry
References
- 1.Nishizuka Y. FASEB J. 1995;9:486–496. [PubMed] [Google Scholar]
- 2.Johnson L N, Noble M E, Owen D J. Cell. 1996;85:149–158. doi: 10.1016/s0092-8674(00)81092-2. [DOI] [PubMed] [Google Scholar]
- 3.Parekh D B, Ziegler W, Parker P J. EMBO J. 2000;19:496–503. doi: 10.1093/emboj/19.4.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Konishi H, Tanaka M, Takemura Y, Matsuzaki H, Ono Y, Kikkawa U, Nishizuka Y. Proc Natl Acad Sci USA. 1997;94:11233–11237. doi: 10.1073/pnas.94.21.11233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gschwendt M. Eur J Biochem. 1999;259:555–564. doi: 10.1046/j.1432-1327.1999.00120.x. [DOI] [PubMed] [Google Scholar]
- 6.Denning M F, Dlugosz A A, Howett M K, Yuspa S H. J Biol Chem. 1993;268:26079–26081. [PubMed] [Google Scholar]
- 7.Li W, Mischak H, Yu J-C, Wang L-M, Mushinski J F, Heidaran M A, Pierce J H. J Biol Chem. 1994;269:2349–2352. [PubMed] [Google Scholar]
- 8.Li W, Yu J-C, Michieli P, Beeler J F, Ellmore N, Heidaran M A, Pierce J H. Mol Cell Biol. 1994;14:6727–6735. doi: 10.1128/mcb.14.10.6727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haleen-Smith H, Chang E-Y, Szallasi Z, Blumberg P M, Rivera J. Proc Natl Acad Sci USA. 1995;92:9112–9116. doi: 10.1073/pnas.92.20.9112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Szallasi Z, Denning M F, Chang E-Y, Rivera J, Yuspa S H, Lehel C, Olah Z, Anderson W B, Blumberg P M. Biochem Biophys Res Commun. 1995;214:888–894. doi: 10.1006/bbrc.1995.2370. [DOI] [PubMed] [Google Scholar]
- 11.Soltoff S P, Toker A. J Biol Chem. 1995;270:13490–13495. doi: 10.1074/jbc.270.22.13490. [DOI] [PubMed] [Google Scholar]
- 12.Denning M F, Dlugosz A A, Threadgill D W, Magnuson T, Yuspa S H. J Biol Chem. 1996;271:5325–5331. doi: 10.1074/jbc.271.10.5325. [DOI] [PubMed] [Google Scholar]
- 13.Shanmugam M, Krett N L, Peters C A, Maizels E T, Murad F M, Kawakatsu H, Rosen S T, Hunzicker-Dunn M. Oncogene. 1998;16:649–654. doi: 10.1038/sj.onc.1201684. [DOI] [PubMed] [Google Scholar]
- 14.Brodie C, Bogi K, Acs P, Lorenzo P S, Baskin L, Blumberg P M. J Biol Chem. 1998;273:30713–30718. doi: 10.1074/jbc.273.46.30713. [DOI] [PubMed] [Google Scholar]
- 15.Ács P, Beheshti M, Szallasi Z, Li L, Yuspa S H, Blumberg P M. Carcinogenesis. 2000;21:887–891. doi: 10.1093/carcin/21.5.887. [DOI] [PubMed] [Google Scholar]
- 16.Kronfeld I, Kazimirsky G, Lorenzo P S, Garfield S H, Blumberg P M, Brodie C. J Biol Chem. 2000;275:35491–35498. doi: 10.1074/jbc.M005991200. [DOI] [PubMed] [Google Scholar]
- 17.Ohmori S, Shirai Y, Sakai N, Fujii M, Konishi H, Kikkawa U, Saito N. Mol Cell Biol. 1998;18:5263–5271. doi: 10.1128/mcb.18.9.5263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Konishi H, Matsuzaki H, Takaishi H, Yamamoto T, Fukunaga M, Ono Y, Kikkawa U. Biochem Biophys Res Commun. 1999;264:840–846. doi: 10.1006/bbrc.1999.1579. [DOI] [PubMed] [Google Scholar]
- 19.Li W, Chen X-H, Kelley C A, Alimandi M, Zhang J, Chen Q, Bottaro D P, Pierce J H. J Biol Chem. 1996;271:26404–26409. doi: 10.1074/jbc.271.42.26404. [DOI] [PubMed] [Google Scholar]
- 20.Gschwendt M, Kielbassa K, Kittstein W, Marks F. FEBS Lett. 1994;347:85–89. doi: 10.1016/0014-5793(94)00514-1. [DOI] [PubMed] [Google Scholar]
- 21.Zang Q, Lu Z M, Curto M, Barile N, Shalloway D, Foster D A. J Biol Chem. 1997;272:13275–13280. doi: 10.1074/jbc.272.20.13275. [DOI] [PubMed] [Google Scholar]
- 22.Song J S, Swann P G, Szallasi Z, Blank U, Blumberg P M, Rivera J. Oncogene. 1998;16:3357–3368. doi: 10.1038/sj.onc.1201886. [DOI] [PubMed] [Google Scholar]
- 23.Blake R A, Garcia-Paramio P, Parker P J, Courtneidge S A. Cell Growth Differ. 1999;10:231–241. [PubMed] [Google Scholar]
- 24.Yuan Z M, Utsugisawa T, Ishiko T, Nakada S, Huang Y, Kharbanda S, Weichselbaum R, Kufe D. Oncogene. 1998;16:1643–1648. doi: 10.1038/sj.onc.1201698. [DOI] [PubMed] [Google Scholar]
- 25.Sun X, Wu F, Datta R, Kharbanda S, Kufe D. J Biol Chem. 2000;275:7470–7473. doi: 10.1074/jbc.275.11.7470. [DOI] [PubMed] [Google Scholar]
- 26.Marth J D, Peet R, Krebs E G, Perlmutter R M. Cell. 1985;43:393–404. doi: 10.1016/0092-8674(85)90169-2. [DOI] [PubMed] [Google Scholar]
- 27.Nishizawa K, Yano T, Shibata M, Ando S, Saga S, Takahashi T, Inagaki M. J Biol Chem. 1991;266:3074–3079. [PubMed] [Google Scholar]
- 28.Czernik A J, Girault J A, Nairn A C, Chen J, Snyder G, Kebabian J, Greengard P. Methods Enzymol. 1991;201:264–283. doi: 10.1016/0076-6879(91)01025-w. [DOI] [PubMed] [Google Scholar]
- 29.Taniguchi H, Manenti S, Suzuki M, Titani K. J Biol Chem. 1994;269:18299–18302. [PubMed] [Google Scholar]
- 30.Wilm M, Neubauer G, Mann M. Anal Chem. 1966;68:527–533. doi: 10.1021/ac950875+. [DOI] [PubMed] [Google Scholar]
- 31.Konishi H, Matsuzaki H, Tanaka M, Ono Y, Tokunaga C, Kuroda S, Kikkawa U. Proc Natl Acad Sci USA. 1996;93:7639–7643. doi: 10.1073/pnas.93.15.7639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ogita K, Miyamoto S, Yamaguchi K, Koide H, Fujisawa N, Kikkawa U, Sahara S, Fukami Y, Nishizuka Y. Proc Natl Acad Sci USA. 1992;89:1592–1596. doi: 10.1073/pnas.89.5.1592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hardwick J S, Sefton B M. Proc Natl Acad Sci USA. 1995;92:4527–4531. doi: 10.1073/pnas.92.10.4527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Emoto Y, Manome Y, Meinhardt G, Kisaki H, Kharbanda S, Robertson M, Ghayur T, Wong W W, Kamen R, Weichselbaum R, et al. EMBO J. 1995;14:6148–6156. doi: 10.1002/j.1460-2075.1995.tb00305.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Flint A J, Paladini R D, Koshland D E., Jr Science. 1990;249:408–411. doi: 10.1126/science.2377895. [DOI] [PubMed] [Google Scholar]
- 36.Stempka L, Girod A, Muller H J, Rincke G, Marks F, Gschwendt M, Bossemeyer D. J Biol Chem. 1997;272:6805–6811. doi: 10.1074/jbc.272.10.6805. [DOI] [PubMed] [Google Scholar]
- 37.Gopalakrishna R, Jaken S. Free Radical Biol Med. 2000;28:1349–1361. doi: 10.1016/s0891-5849(00)00221-5. [DOI] [PubMed] [Google Scholar]