Abstract
Mycobacterium tuberculosis possesses five genes with significant homology to the resuscitation-promoting factor (Rpf) of Micrococcus luteus. The M. luteus Rpf is a secreted ∼16-kDa protein which restores active growth to cultures of M. luteus rendered dormant by prolonged incubation in stationary phase. More recently, the Rpf-like proteins of M. tuberculosis have been shown to stimulate the growth of extended-stationary-phase cultures of Mycobacterium bovis BCG. These data suggest that the Rpf proteins can influence the growth of mycobacteria; however, the studies do not demonstrate specific functions for the various members of this protein family, nor do they assess the function of M. tuberculosis Rpf homologues in vivo. To address these questions, we have disrupted each of the five rpf-like genes in M. tuberculosis Erdman, and analyzed the mutants for their growth in vitro and in vivo. In contrast to M. luteus, for which rpf is an essential gene, we find that all of the M. tuberculosis rpf deletion mutant strains are viable; in addition, all show growth kinetics similar to Erdman wild type both in vitro and in mouse organs following aerosol infection. Analysis of rpf expression in M. tuberculosis cultures from early log phase through late stationary phase indicates that expression of the rpf-like genes is growth phase-dependent, and that the expression patterns of the five M. tuberculosis rpf genes, while overlapping to various degrees, are not uniform. We also provide evidence that mycobacterial rpf genes are expressed in vivo in the lungs of mice acutely infected with virulent M. tuberculosis.
Mycobacterium tuberculosis is a globally important human pathogen responsible for a staggering burden of disease. Recent estimates from the World Health Organization (WHO) indicate that there are approximately 8 million new cases of tuberculosis each year, the complications of which result in 2 million deaths annually (WHO fact sheet no. 104 [http://www.who.int/mediacentre/factsheets/who104/en/]). The WHO further estimates that, between 2002 and 2020, on the order of 109 individuals will be newly infected with M. tuberculosis if present control efforts are not improved. The remarkable success of M. tuberculosis as a human pathogen is attributable at least in part to the capacity of this organism to persist for months or years within the host even in the setting of a robust host immune response. It is believed that the majority of persons infected with M. tuberculosis harbor a latent infection. In approximately 2 to 23% of these latently infected but otherwise-healthy individuals, the infection will reactivate and cause clinically apparent disease within their lifetime (7). Given that a third of the world's population is thought to harbor latent infection with M. tuberculosis, and therefore provide a large reservoir for potential disease reactivation (3), an understanding of mycobacterial persistence is critical to the global control of tuberculosis.
Although tuberculous persistence and reactivation play significant roles in the pathogenesis of tuberculosis, relatively little is known regarding the molecular mechanisms which regulate mycobacterial physiology in these disease states. Animal models have revealed specific host effector functions which contribute to controlling latent infection, including those mediated by CD4+ T cells, reactive nitrogen intermediates, and tumor necrosis factor alpha (5), yet much remains to be learned about the mycobacterial physiology of persistence. In particular, it remains to be defined whether the bacteria in the persistent phase of infection are dormant or slowly replicating; equally unclear are the level of metabolic activity of quiescent M. tuberculosis and the genetic mechanisms which maintain latency or trigger reactivation.
M. tuberculosis encodes a family of five proteins which share homology with the resuscitation-promoting factor (Rpf) of the nonsporulating GC-rich gram-positive bacterium Micrococcus luteus. Based on in vitro studies of M. luteus reactivation, the M. tuberculosis Rpfs are potential regulators of persistence and reactivation. M. luteus, when held in spent growth medium for many months, lost viability when plated on agar plates such that only ∼1 in 104 microscopically observable bacteria was able to form a colony on solid medium; however, the bacteria were found to be dormant rather than dead, as adding back sterile supernatant from an actively growing M. luteus culture increased the viable count by several orders of magnitude (13, 14). It was later reported that an ∼16- to 17-kDa secreted protein, isolated from M. luteus culture supernatant, had resuscitating activity at picomolar concentrations (22). Database searches reveal genes similar to the M. luteus rpf in a variety of other GC-rich bacteria, including Streptomyces coelicolor, Corynebacterium glutamicum, Mycobacterium leprae, M. tuberculosis, and Mycobacterium bovis, the last two of which each possess five genes with regions of high homology to the M. luteus rpf. The rpf-like genes of M. tuberculosis H37Rv are distributed throughout the chromosome, with gene designations Rv0867c (rpfA), Rv1009 (rpfB), Rv1884c (rpfC), Rv2389c (rpfD), and Rv2450c (rpfE) (http://genolist.pasteur.fr /TubercuList/). More recently, it has been shown that each of the five Rpf-like proteins of M. tuberculosis, when expressed as a histidine-tagged fusion in and purified from Escherichia coli, was able to stimulate the growth of extended-stationary-phase (5-month-old) cultures of M. bovis BCG at picomolar concentrations (25). In these studies the growth of the M. bovis BCG culture was absolutely dependent upon provision of the recombinant M. tuberculosis Rpf to the medium. The authors also showed that addition of affinity-purified polyclonal anti-Rpf antibodies inhibited the growth of cultures inoculated with 2-month-old M. tuberculosis Academia strain and 6-week-old M. bovis BCG and that this inhibition could be overcome by supplying exogenous Rpf (25).
The data described above strongly suggest that the M. tuberculosis Rpf family may play a role in stimulating growth of stationary-phase bacteria and may therefore function in vivo in reactivation of latent infection. The M. luteus Rpf is known to be secreted; of the five M. tuberculosis proteins, Rv1009 has a prokaryotic lipoprotein lipid attachment site (Prosite PS00013) at its N terminus and may be affixed to the cell membrane. The remaining four Rpf-like proteins each possess, depending upon the prediction scheme used (9, 22, 25), a putative prokaryotic signal sequence and may therefore be secreted or membrane associated. The Rpf-like proteins have been detected by Western blotting in culture supernatants of M. bovis BCG (25), and a Rv2450c::phoA′ fusion was shown to direct the export of the PhoA moiety (9). Given the evidence that the Rpf-like proteins have an extracellular function and that the mature forms of the proteins exhibit growth-stimulatory activity, it is possible that the proteins are being utilized by M. tuberculosis as a means of communication between neighboring bacilli in a paracrine fashion, perhaps to convey information regarding population density or environmental conditions, since intercellular communication among prokaryotes is a well-documented phenomenon (15, 34, 35, 38).
In this study, we have generated individual rpf deletion mutants for each of the five M. tuberculosis rpf-like genes, using the phage-based technology of specialized transduction (1), and have examined the in vitro growth characteristics of the knockout strains as well as their growth kinetics in a murine model of chronic persistent infection. We have also examined the expression of the rpf-like genes in M. tuberculosis grown in vitro, as well as in the lungs of mice with acute tuberculosis.
MATERIALS AND METHODS
Bacterial strains and growth conditions.
M. tuberculosis Erdman was grown at 37°C in liquid Middlebrook 7H9 medium supplemented with 0.2% glycerol, 0.05% Tween 80, and 10% OADC enrichment (Becton Dickinson) or on solid Middlebrook 7H10 medium (Difco) supplemented with 0.5% glycerol and 10% OADC. Where appropriate, hygromycin (Roche) was added at 50 μg/ml and kanamycin (Sigma) was added at 25 μg/ml. Stock strains were grown to an optical density at 600 nm (OD600) of ∼1 in roller bottles, divided into 1-ml aliquots in cryovials, and stored at −70°C until needed. M. tuberculosis CI4, a clinical strain (28), was similarly treated. Recombinant DNA work was carried out using E. coli host strains DH5α (Invitrogen) or HB101 (Gibco BRL). E. coli was grown on solid or in liquid Luria-Bertani medium supplemented with ampicillin (100 μg/ml) or hygromycin (150 μg/ml) where appropriate.
Construction of rpf disruption mutants of M. tuberculosis using specialized transduction.
The rpf disruption mutant strains were constructed using the method of specialized transduction essentially as described elsewhere (1, 2). Briefly, substrates for allelic exchange were generated by amplifying the upstream and downstream flanking regions of each of the five rpf homologues using the primer pairs listed in Table 1, yielding products of the lengths indicated. PCR products were cloned into vector pCR2.1 using the TA cloning kit (Invitrogen) and sequenced at the Albert Einstein College of Medicine sequencing facility. Restriction sites were engineered into the PCR primers to facilitate cloning. PCR products were cloned into pjsc284 to create pjmt17, pjmt18, pjmt19, pjmt20, and pjmt21, such that a hygromycin resistance cassette replaced almost the entirety of the coding region of, respectively, Rv2389c, Rv1009, Rv2450c, Rv1884c, and Rv0867c (Table 2). For Rv1009, an additional knockout construct, pjmt18A, was engineered for the following reason. The original construct, pjmt18, was designed so as to eliminate as much of the Rpf conserved domain as possible; this approach resulted in the removal of the first four amino acids, including the initiating methionine, from the predicted coding sequence of the next downstream gene, ksgA, therefore almost certainly interfering with expression of this gene. To preserve the coding sequence of ksgA and to potentially leave intact the upstream promoter sequence for this gene, another Rv1009 deletion construct (pjmt18A), which retains the final 190 nucleotides prior to the termination codon of Rv1009, was prepared.
TABLE 1.
Primers used in constructing the rpf deletion mutant and rpf-complemented strains
| Primer name | Gene and flank locationa | Primer sequenced | Product size(s): (bp) |
|---|---|---|---|
| 30 | Rv2389c upstream (R) | 5′-CTCGAGCAGGGACGATTGAGG-3′ | 626 |
| 31 | Rv2389c upstream (F) | 5′-ACTAGTTCAGGTCGAGGTTGAC-3′ | |
| 32 | Rv2389c downstream (F) | 5′-TCTAGAAAGCAAACCCGGTGTC-3′ | 644 |
| 33 | Rv2389c downstream (R) | 5′-GGTACCAAATATGACTACTGAGAT-3′ | |
| 34 | Rv1009 upstream (R) | 5′-TCTAGACGCAACATCGGTGATTG-3′ | 641 |
| 35 | Rv1009 upstream (F) | 5′-GGTACCGGTAAACCGCTGATG-3′ | |
| 36 | Rv1009 downstream (F) | 5′-CTCGAGCGGGTGCGCGCTG-3′ | 630, 809b |
| 36A | Rv1009 downstream (F revised) | 5′-CTCGAGCAACACCGGCAACG-3′ | |
| 37 | Rv1009 downstream (R) | 5′-ACTAGTCCGGAATAGACACGC-3′ | |
| 38 | Rv2450c upstream (R) | 5′-CTCGAGCCGCCGCGGCTG-3′ | 622 |
| 39 | Rv2450c upstream (F) | 5′-ACTAGTTGGTGTCGGTCAGCT-3′ | |
| 40 | Rv2450c downstream (F) | 5′-TCTAGAACGTTGTTCCTTTCGC-3′ | 615 |
| 41 | Rv2450c downstream (R) | 5′-GGTACCTATGAATCACAGTGCG-3′ | |
| 42 | Rv1884c upstream (R) | 5′-CTCGAGTATTCCGCGCTGACG-3′ | 644 |
| 43 | Rv1884c upstream (F) | 5′-ACTAGTCAGCCGTTGGCGTG-3′ | |
| 44 | Rv1884c downstream (F) | 5′-TCTAGAACCGGATTGATCAGGC-3′ | 618 |
| 45 | Rv1884c downstream (R) | 5′-GGTACCTTACCCGTCCACGT-3′ | |
| 46 | Rv0867c upstream (R) | 5′-AGATCTGCTGACGGGCTGAGG-3′ | 607 |
| 47 | Rv0867c upstream (F) | 5′-ACTAGTGCTGTCGCGCGGGG-3′ | |
| 48 | Rv0867c downstream (F) | 5′-TCTAGACATACGTTAGGTAATTCC-3′ | 642 |
| 49 | Rv0867c downstream (R) | 5′-GGTACCCTGCGATCTGGCGC-3′ | |
| 52 | Rv1009c (F) | 5′-AATTGGCCATGTTGCGCCTGG-3′ | 1,106 |
| 53 | Rv1009c (R) | 5′-GCCAAGCTTTCAGCGCGCACC-3′ | |
| 60 | Rv1010 (ksgA)c (F) | 5′-CTGGATCCTATGTGCTGCACGAG-3′ | 972 |
| 61 | Rv1010 (ksgA)c (R) | 5′-GGCAAGCTTTCAGCTCGCCGA-3′ |
R, reverse primer; F, forward primer.
The product of the 36-37 primer pair is 630 bp; the product of the 36A-37 primer pair is 809 bp.
Complementation strain.
Restriction sites are underlined.
TABLE 2.
Plasmids and strains used in this study
| Plasmid or strain | Relevant characteristic(s) | Source or reference |
|---|---|---|
| Plasmids | ||
| pCR2.1 | Ampr; cloning vector for PCR products | Invitrogen |
| pjsc284 | Hygr; cosmid vector for cloning allelic exchange substrates; has oriE, single λ-cos site, PacI site for introduction into shuttle phasmid; res-hyg-res cassette flanked by γδ-res sites | Derivative of pYUB854, 1 |
| pMV261 | Kanr; pMV206 shuttle plasmid with hsp60 promoter | 33 |
| pMV306 | Kanr; site-specific integrating shuttle vector | 33 |
| phAE87 | TM4-based phasmid shuttle vector with temperature-sensitive regulon | 2 |
| pjmt17 | Hygr; pjsc284 containing flanking sequences of Rv2389c gene | This study |
| pjmt18 | Hygr; pjsc284 containing flanking sequences of Rv1009 gene | This study |
| pjmt18A | Hygr; pjsc284, but containing revised flanking sequence of Rv1009 gene | This study |
| pjmt19 | Hygr; pjsc284 containing flanking sequences of Rv2450c gene | This study |
| pjmt20 | Hygr; pjsc284 containing flanking sequences of Rv1884c gene | This study |
| pjmt21 | Hygr; pjsc284 containing flanking sequences of Rv0867c gene | This study |
| Strains | ||
| mc2155 | M. smegmatis | 32 |
| Erdman | M. tuberculosis | Trudeau Institute (Saranac Lake, N.Y.) |
| mc23125 | Erdman ΔRv2389c::res-hyg-res | This study |
| mc23126 | Erdman ΔRv1009::res-hyg-res | This study |
| mc23126A | Erdman ΔRv1009::res-hyg-res; revised construct | This study |
| mc23127 | Erdman ΔRv2450c::res-hyg-res | This study |
| mc23128 | Erdman ΔRv1884c::res-hyg-res | This study |
| mc23129 | Erdman ΔRv0867c::res-hyg-res | This study and reference 1a |
| mc23301 | mc23126 (attB::Rv1009) | This study |
| mc23302 | mc23126 (attB::ksgA) | This study |
| mc23301A | mc23126A (attB::Rv1009) | This study |
| mc23302A | mc23126A (attB::ksgA) | This study |
In reference 1, this strain is designated mc23510.
The plasmids described above (pjmt17, pjmt18, pjmt18A, pjmt19, pjmt20, and pjmt21) were digested with PacI and ligated into PacI-digested phAE87 DNA. The ligation mixtures were packaged into lambda in vitro (Gigapack III XL; Stratagene) and transduced into E. coli HB101 cells, which were then plated onto Luria-Bertani agar supplemented with hygromycin. Cosmid DNA prepared from the resulting colonies was used to electroporate Mycobacterium smegmatis mc2155, and plaques obtained at 30°C were amplified to make a high-titer lysate as described previously (1). The resulting phage were used to infect log-phase M. tuberculosis Erdman at the nonpermissive temperature of 37°C according to published methods (1, 8). DNA prepared from hygromycin-resistant colonies was digested with the restriction enzymes specified in Fig. 1, and Southern blots were probed with the flanking regions of the genes of interest to confirm the presence of the deletion. [α-32P]dCTP-labeled probes were prepared using Ready-To-Go DNA labeling beads (Pharmacia). The following flank region probes were used: mc23125, 644-bp downstream flank; mc23126, 630-bp downstream flank; mc23126A, 809-bp downstream flank; mc23127, 615-bp downstream flank; mc23128, 644-bp upstream flank; and mc23129, 607-bp upstream flank. The plasmids and strains used in this study are described in Table 2.
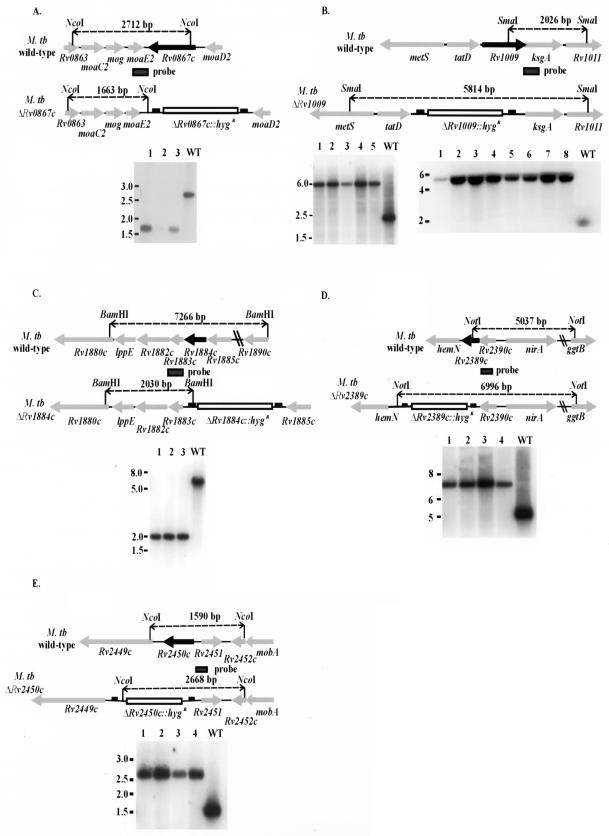
FIG. 1.
Construction of rpf deletion mutants by allelic replacement. Shown in each panel is a schematic illustrating the chromosomal organization of the wild-type (upper) and rpf deletion mutant (lower) strains for Rv0867c (A), Rv1009 (B), Rv1884c (C), Rv2389c (D), and Rv2450c (E), in which each of the individual rpf-like genes has been disrupted by a hygromycin resistance cassette. The schematics also indicatethe restriction enzymes used to digest the genomic DNA for Southern blot analysis and the sizes of expected hybridizing bands for each strain (indicated by the dashed lines). Below each diagram of the chromosome is a Southern blot showing digested genomic DNA for several independent clones of the Δrpf mutant (numbered) and for wild-type Erdman (always the lane on the far right [WT]) probed with the flanking region of the gene. The migration of molecular size markers (in kilobases) is indicated to the left of each blot. The location of each probe is indicated in the schematic above. Below the ΔRv1009 diagram in panel B are two Southern blots: on the left is mc23126 (ΔRv1009) and on the right is mc23126A (ΔRv1009 rev).
Complementation of Rv1009 knockouts.
The two Rv1009 knockouts (mc23126 and mc23126A) were complemented with either (i) the Rv1009 coding sequence, amplified with primer pair 52-53 (Table 1) or (ii) the Rv1010 (ksgA) coding sequence, amplified with primer pair 60-61 (Table 1) using Erdman genomic DNA as template. After sequencing, the PCR product was cloned downstream of the mycobacterial hsp60 promoter into the vector pMV261 using the restriction sites indicated (Table 1). This generated a construct harboring the cloned genes (Rv1009 or Rv1010 [ksgA]) driven by the hsp60 promoter. The hsp60-Rv1009 or hsp60-ksgA construct was then excised from pMV261 using the HindIII and XbaI sites and subcloned into the integrative vector pMV306. The resulting plasmids were used to transform the corresponding knockout strains; kanamycin-resistant (Kanr) colonies were selected, and the presence of the complementing gene, integrated at the attB site, was confirmed by Southern blot analysis. The strain designations for the various complemented strains are listed in Table 2.
In vitro growth studies.
Starter cultures of Erdman wild type and of each of the rpf deletion mutant strains (mc23125, mc23126, mc23127, mc23128, and mc23129) were grown in 10 ml of Middlebrook 7H9 at 37°C with agitation to an OD600 of ∼1.0 (∼3 × 108 to 5 × 108 CFU/ml). A 1-ml aliquot of each culture was pelleted by centrifugation and resuspended in 1 ml of fresh 7H9 medium. The aliquot was sonicated two times for 10 s each to disrupt clumps and added to 40 ml of Middlebrook 7H9 (complete medium containing Tween 80, OADC, and glycerol as described above) in an amount estimated to give a final concentration of 1 × 106 to 2 × 106 CFU/ml. The inoculum was also serially diluted in 7H9-Tween 80 and plated onto 7H10 agar so that the precise input CFU could be calculated. The cultures were grown in 250-ml sterile disposable Erlenmeyer flasks (catalog no. 430183; Corning) with gentle shaking at 37°C. Aliquots were removed at various time periods and sonicated; a portion was used to measure the OD600, and the remainder was serially diluted and plated to determine the number of CFU per milliliter. To assess colony size of the various rpf deletion mutants, mc23126 and mc23126A were grown in 7H9 supplemented with hygromycin and mc23301, mc23301A, mc23302, and mc23302A were grown in 7H9 supplemented with both hygromycin and kanamycin. M. tuberculosis Erdman, the wild-type control, was cultured in 7H9 medium without antibiotics. At an appropriate time postinoculation, cultures were serially diluted in 7H9 containing 0.05% Tween 80 and plated onto 7H10 agar in the absence of antibiotics. Colony size was assessed 3 to 4 weeks after plating.
Mouse infections.
7H9 broth (containing hygromycin for the knockout strains or hygromycin plus kanamycin for the complemented strain) was inoculated with frozen stocks of each strain and grown to an OD600 of ∼1.0. The bacteria were pelleted by centrifugation, washed three times in phosphate-buffered saline containing 0.05% Tween 80, and sonicated briefly using a cup horn sonicator. Female C57BL/6 mice (10 to 12 weeks old; Jackson Laboratories) were infected with ∼500 to 1,000 CFU of the various strains of M. tuberculosis by aerosol using a closed-air aerosolization system (In-Tox Products, Albuquerque, N.Mex.) as previously described (28). At various times after infection, mice were sacrificed and portions (one quarter to one half) of the lungs, spleens, and livers were homogenized in phosphate-buffered saline containing 0.05% Tween 80. Tissue bacterial load was assessed by plating serial dilutions of the homogenates onto 7H10 agar, containing hygromycin as appropriate for the various knockout strains. Portions of the lungs, liver, and spleen were also fixed overnight in 10% phosphate-buffered formalin, embedded in paraffin, sectioned, and stained with hematoxylin and eosin. Four mice per group were sacrificed at each time point except at 24 h postinfection, when three mice were sacrificed per group.
Extraction of RNA and gene expression analysis.
A log-phase starter culture of M. tuberculosis Erdman strain was used to inoculate the working culture at an approximate density of 106 CFU/ml (based on the estimate that an OD600 of 1 ≈ 3 × 108 to 5 × 108 CFU/ml) in complete 7H9 medium. At various times postinoculation, bacteria were harvested by centrifugation, the supernatant decanted and the pellet resuspended in TRIzol reagent (Invitrogen Life Technologies). The M. tuberculosis resuspended in TRIzol reagent was then transferred to 2-ml screw-cap tubes (Sarstedt) containing 0.1-mm-diameter zirconia-silica beads (Biospec Products, Inc.) and subjected to bead beating using a mini Bead Beater (Biospec Products, Inc.) at maximum speed for 5 min. After bead beating, the material was transferred to 15-ml RNase-free tubes (Sarstedt) and chloroform was added at a ratio of 1 ml of chloroform per 5 ml of TRIzol reagent. The tubes were centrifuged at 2,400 × g for 15 min using a tabletop centrifuge (Forma Scientific, Inc.), after which the aqueous phase was transferred to 1.5-ml microcentrifuge tubes and the nucleic acids were precipitated with isopropanol. The pellets were washed with ethanol, dried briefly, and resuspended in 180 μl of nuclease-free water (Ambion). The RNA preparation was treated with 50 U of DNase (10 U/μl; Roche) for 60 min at 37°C, reextracted with TRIzol reagent and chloroform, again precipitated with isopropanol, and redissolved in 180 μl of nuclease-free water. The RNA was subjected to a second DNase treatment and extraction protocol exactly as above. The RNA preparations were then treated with DNase a third time using the DNA-free kit (Ambion) according to the manufacturer's instructions. An ∼0.5- to 0.7-μg portion of each non-reverse-transcribed sample was subjected to PCR analysis to assure that no product was evident on an ethidium bromide-stained agarose gel after 30 cycles of amplification before proceeding with reverse transcription (RT) of the RNAs. Mycobacterial RNAs from infected mouse lungs were similarly prepared, using tissues which had been homogenized in TRIzol reagent (26).
One microgram of total RNA was reverse-transcribed using SuperScript II RNaseH− reverse transcriptase (Invitrogen Life Technologies) in a total reaction volume of 20 μl as directed by the manufacturer (for the 4-month-old Erdman culture and for the in vivo expression analysis, 5 μg of total RNA was used in the 20-μl of RT reaction mixture). The reaction mixture contained reverse transcriptase buffer, 10 mM dithiothreitol, 1 μl of RNase inhibitor (40 U/μl; Promega), deoxynucleoside triphosphate mix (0.5 mM concentration [each] of dATP, dCTP, dTTP, and dGTP), 0.08 A260 unit of random primers (AMV First Strand cDNA Synthesis kit for RT-PCR; Roche), and 200 U of SuperScript II RT. Fivefold serial dilutions of the RT reaction were prepared and used as the template in a PCR totaling 50 μl. The reaction mixture was prepared using the Qiagen Taq PCR Core kit and consisted of PCR buffer containing 1.5 mM MgCl2, Q solution, a 200 μM concentration of each deoxynucleoside triphosphate, 1.5 U of Taq DNA polymerase, and a 0.4 μM concentration of each primer. Primers used were as follows: 66 forward (5′-CCGGGCCTTGGGTTTTC-3′) and 67 reverse (5′-CAGGCGACGTGGGATTC-3′) for Rv2389c; 68 forward (5′-CGGCCAACGGGTATTACGGTGGTGT-3′) and 69 reverse (5′-GCGGGTGGCGAGGTCAGC-3′) for Rv1009; 70 forward (5′-CGGAGCCGGCGGAGTATCG-3′) and 71 reverse (5′-CCAGCCGGTATCGCCAATG-3′) for Rv2450c; 72 forward (5′-AGGTGGCCGGCTTGAACTG-3′) and 73 reverse (5′CACGGCATCCATGTCGCTCTCCAC-3′) for Rv1884c; and 74 forward (5′-GGCCACCTGATCCCATTCC-3′) and 75 reverse (5′-CGCCACCGTAAGCCCACCAC-3′) for Rv0867c. The amplification protocol involved denaturation at 95°C for 5 min, followed by 30 cycles of denaturation at 95°C for 20 s, annealing of primers at 55°C for 30 s and primer extension at 72°C for 30 s; after completion of the 30th cycle, the extension reaction was continued for another 3 min at 72°C. For the in vivo expression analysis, the PCR was continued for 40 cycles. A positive control consisting of M. tuberculosis genomic DNA was included in each set of reactions, as well as a negative control with no template. Also included for each set of primers was a no-RT control, consisting of 33 ng of total RNA, to verify again the absence of DNA contamination. We chose 16S rRNA and 23S rRNA as internal controls for the time course experiment, to demonstrate that a band of similar intensity was obtained from a similar amount of starting material (33 ng of total RNA) at each time point. The primers used and their sequences were as follows: 16S forward, 5′-GAGATACTCGAGTGGCGAAC-3′; 16S reverse, 5′-GGCCGGCTACCCGTCGTC-3′; 23S forward, 5′-GAAGAATGAGCCTGCGAGTC-3′; and 23S reverse, 5′-GGTCCAGAACACGCCACTAT-3′.
RESULTS
Organization of the rpf gene loci and construction of the rpf deletion mutant strains and complemented strains.
A schematic illustrating the chromosomal organizations of the rpf-like genes in the parental and rpf deletion mutant strains is shown in Fig. 1A to E. The M. tuberculosis rpf homologues are distributed throughout the chromosome and are surrounded by genes of widely differing or unknown function, such that it is difficult to discern any functional role for the Rpfs based on the context of the neighboring genes. Two of the genes, Rv0867c (Fig. 1A) and Rv2450c (Fig. 1E), are likely expressed as monocistronic transcripts. For Rv2450c the next downstream gene is transcribed in the opposite orientation, and Rv2450c is annotated as being preceded by a ribosome binding site and followed by a stem-loop structure which may be a transcriptional terminator. Rv0867c is expressed convergently with its upstream neighbor and, although it is expressed in the same direction as its downstream neighbor, the open reading frames are separated by nearly 450 nucleotides, and the intergenic region contains a possible ribosome binding site for Rv0867c. That Rv2450c and Rv0867c are likely to be expressed independently of their neighbors is important, as it signifies that any phenotype observed for the deletion mutants is more likely to be a direct result of the loss of the rpf-like gene than of polar effects on surrounding genes. The immediate neighbors of Rv2450c are of unknown function, while Rv0867c is embedded within a cluster of genes involved with molybdopterin metabolism. For the remaining three rpf homologues the situation is more complex; based on gene context all are likely to exist in multigene operons. The coding sequence of Rv2390c overlaps Rv2389c by 3 nucleotides, while 106 nucleotides separates Rv2389c (Fig. 1D) from hemN (Rv2388c), a putative coproporphyrinogen oxidase, an enzyme which plays a role in protoheme biosynthesis. Rv1884c (Fig. 1C) appears to be part of a six- or seven-gene operon, including the Ag85B precursor (Rv1886c), chorismate mutase (Rv1885c), and a possible hydroxysteroid dehydrogenase (Rv1882c) among other genes.
The Rv1009 gene (Fig. 1B) possesses a 28-nucleotide overlap between the carboxy-terminal coding sequence of Rv1009 (including the termination codon) and the amino-terminal coding sequence of ksgA (Rv1010), an arrangement preserved in M. bovis and M. leprae. Such a close proximity is often found in coordinately regulated genes. The ksgA gene is annotated for homology with its E. coli counterpart (37) which encodes a methyltransferase that methylates two adenosine residues near the 3′ end of 16S rRNA, a modification which confers sensitivity to the antibiotic kasugamycin (11, 12) and is the only rRNA modification which is shared by prokaryotes and eukaryotes (16).
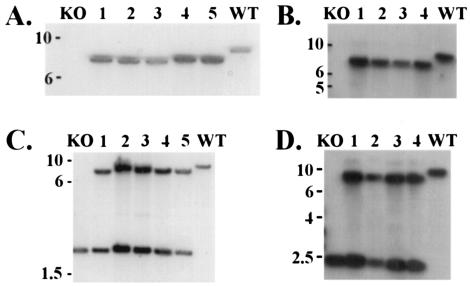
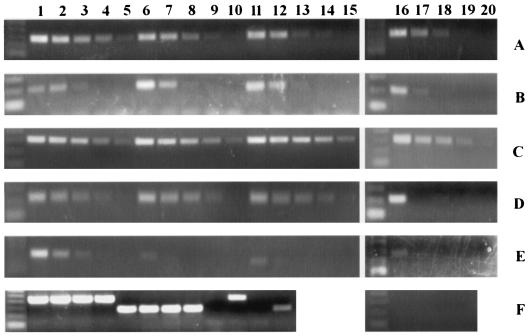
We used the method of specialized transduction (1) to generate null mutations of each of the five rpf-like genes in M. tuberculosis Erdman, removing >90% of each coding sequence and replacing this region with a hygromycin resistance cassette flanked by res sites. The disruption of each of the five rpf-like genes was confirmed by Southern blot analysis (Fig. 1, lower panels) using the upstream- or downstream-flanking sequences of the genes as probes and selecting restriction enzymes which yielded hybridization products of easily distinguishable sizes. As shown in Fig. 1, three to eight hygromycin-resistant colonies were selected for each deletion mutant, and all showed hybridizing bands of the predicted sizes, indicating loss of the rpf-like genes. Each of the rpf disruption mutant strains was also complemented with the entire coding sequence placed under control of the hsp60 promoter integrated as a single copy at the attB site. Shown in Fig. 2 are Southern blots documenting the integration of the appropriate rpf-complementing plasmid at the attB site for the two ΔRv1009 deletion mutants. There are four panels shown because each of the two ΔRv1009 strains (mc23126 and mc23126A) was complemented, separately, with Rv1009 and ksgA coding sequences. The ΔRv1009 strains have two hybridizing bands when probed with the ksgA gene, because the great majority (mc23126) or the entirety (mc23126A) of the ksgA coding sequence was left intact in the knockout strains; the complementation was performed out of concern that polar effects of the Rv1009 knockout on ksgA might interfere with expression of this downstream, overlapping gene. The Southern blots documenting complementation of the remaining rpf deletion mutant strains are not shown.
FIG. 2.
Southern blot analysis of complementation of ΔRv1009. Shown are Southern blots of the complemented strains for mc23126 (ΔRv1009) and mc23126A (ΔRv1009 rev), each probed with the coding sequence of the complementing gene, and each panel showing (i) the original knockout strain (KO) in the far left lane, (ii) several independent complemented clones (numbered) in the middle lanes, and (iii) Erdman wild type (WT) in the far right lane. In all panels the migration of molecular size markers (in kilobases) is indicated at left. (A) For analysis of mc23301(ΔRv1009 complemented with Rv1009), chromosomal DNA was digested with EcoRI and probed with the Rv1009 coding sequence (primer pair 52/53 PCR product), yielding no hybridizing band in the original knockout strain, a 7,340-bp band in the complemented strains (lanes 1 to 5) and an 8,730-bp band for Erdman WT. (B) For analysis of mc23301A (ΔRv1009 rev complemented with Rv1009), chromosomal DNA was digested as in panel A above, the probe was again the Rv1009 coding sequence, and the expected band sizes are as noted in panel A above. In this panel the complemented strains are in lanes 1 to 4. (C) For analysis of mc23302 (ΔRv1009 complemented with ksgA), genomic DNA was digested with EcoRI and probed with the ksgA coding sequence (primer pair 60-61 PCR product), yielding a single band of 8,730 bp for Erdman WT, a single band of 1,960 bp for the original knockout strain (ΔRv1009), and two bands of 7,223 and 1,960 bp for the complemented strains (lanes 1 to 5). The complemented strains have two hybridizing bands because nearly the entire ksgA coding sequence was left intact in this knockout; the complementation was performed due to concerns about polar effects on ksgA expression. Therefore, each complemented strain has one band which comigrates with the band present in the original knockout strain and one additional band corresponding to insertion of a second copy of the ksgA gene at the attB site. (D) For analysis of mc23302A (ΔRv1009 rev complemented with ksgA), genomic DNA was digested and probed as above in panel C and the expected band sizes are as noted above; here the complemented strains are in lanes 1 to 4.
Individual rpf-like genes are nonessential for growth in vitro, but the ΔRv1009 mutant gives a small colony phenotype.
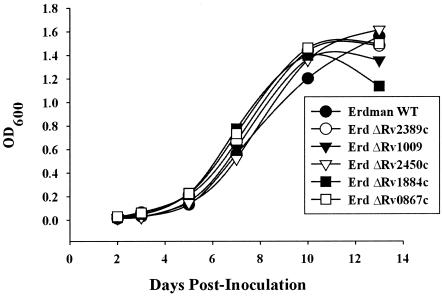
Growth characteristics of the five rpf deletion mutants were compared to the wild-type strain in vitro. When grown in liquid Middlebrook 7H9 medium with shaking at 37°C and inoculated at an initial density of ∼106 CFU/ml, each of the mutant strains grew in a manner indistinguishable from wild type through the first 14 days postinoculation, as shown in Fig. 3. This experiment was performed twice with similar results. In one of the two experiments, analysis of day 45 cultures, which were in extended stationary phase, revealed similar numbers of CFU for wild-type Erdman and each of the five rpf deletion mutants (data not shown). With respect to the assessment of growth by CFU, it is worth noting that the accuracy of this approach is compromised because of the high propensity of the M. tuberculosis to clump in old cultures.
FIG. 3.
In vitro growth kinetics of M. tuberculosis Erdman wild type and the five rpf deletion mutant strains in Middlebrook 7H9 medium at 37°C with agitation. The ΔRv1009 strain used in this experiment was mc23126. We also found that strain mc23126A (ΔRv1009 rev) grew in a manner indistinguishable from wild type in 7H9 medium and that the number of CFU at each time point was similar for each of the strains tested (not shown).
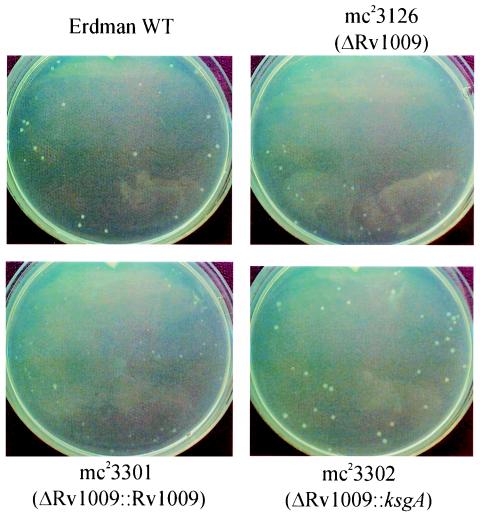
When plated onto Middlebrook 7H10 solid medium, each of the rpf deletion mutant strains displayed a colony morphology similar to wild type, with the exception of the Rv1009 disruption mutant strains, mc23126 and mc23126A. The strain mc23126, from which the Rv1009 coding sequence was deleted but which had also lost the extreme N-terminal coding sequence of Rv1010 (ksgA), including the initiating methionine, was noted to yield a small colony phenotype on 7H10 medium (Fig. 4, upper right) when compared with Erdman wild type (Fig. 4, upper left). The colony size phenotype was maximal at about the time the wild-type colonies became easily visible (about 3 weeks of incubation); subsequently, at 4 to 5 weeks and beyond, the colonies of the mc23126 strain achieved a size similar to that of the wild type. The colony size was restored by complementation with the ksgA coding sequence (Fig. 4, lower right), whereas complementation with the Rv1009 coding sequence (Fig. 4, lower left) actually resulted in slightly smaller colonies. The strain mc23126A, also having the majority of the Rv1009 coding region deleted but retaining the final 190 nucleotides and leaving the ksgA reading frame completely intact yielded colonies only slightly smaller than wild-type, and again, complementation with ksgA, but not Rv1009, restored the original colony size (data not shown). Taken together, the results suggest that, although the deletion of the Rv1009 gene from M. tuberculosis results in smaller colony size, this phenotype is likely to be due to polar effects on the expression of the immediately downstream and overlapping ksgA gene, since the colony size is returned essentially to normal by integration of an additional copy of ksgA. It is noteworthy that this apparent defect in growth of mc23126 on agar is not observed when bacilli are cultured in liquid medium (Fig. 3). The reason behind this discrepancy is unclear, but could be due to the differences in growth conditions encountered in liquid broth within a rolling bottle versus those on the stationary surface of solid agar.
FIG. 4.
Colony size of the original ΔRv1009 strain (mc23126) compared with Erdman wild type and the effect on colony size of complementation with Rv1009 and with ksgA. Cultures were grown and transferred onto plates with Middlebrook 7H10 agar, and colonies were photographed at 23 days postinoculation as described in Materials and Methods. The strains in each panel are indicated. There were similar numbers of colonies on each plate.
Growth and survival of rpf deletion mutant strains in mouse tissues.
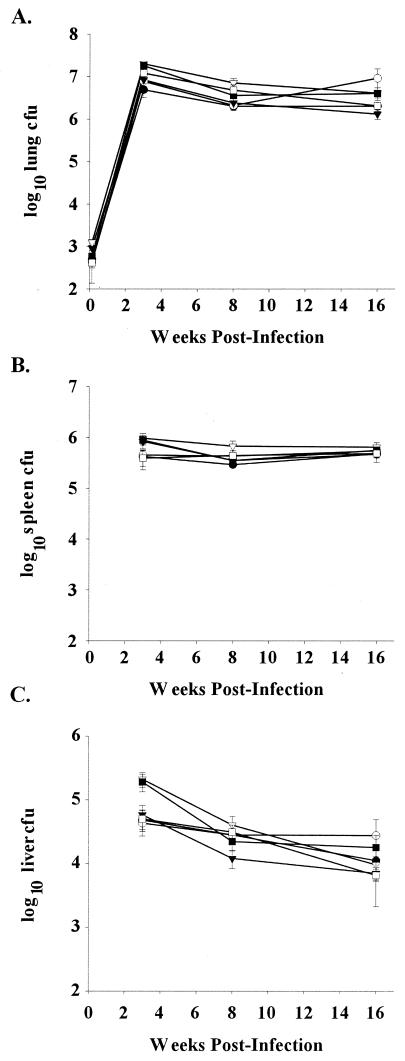
We used a low-dose murine aerosol infection model of chronic persistent infection (28) to examine the in vivo phenotypes of the M. tuberculosis rpf deletion mutant strains. This model permits assessment of the ability of the mutant strains to persist in mouse tissues. Enumeration of bacteria within the lung, spleen, and liver (Fig. 5A to C) revealed that the mutant strains proliferated similarly to the wild type within these organs, reaching titers of approximately 106 to 107 in the lung and between 105 and 106 in the spleen by 3 weeks postinfection and persisting at similar levels until 16 weeks. This experiment was repeated a total of three times for strains mc23125, mc23127, mc23128, and mc23129 and two times for mc23126A. We also examined the histopathology of lung, spleen, and liver for each of the rpf deletion mutant strains at 3, 8, and 16 weeks postinfection and observed no apparent difference when compared with Erdman wild type in terms of the granulomatous inflammation, the cell types observed and the extent of tissue damage (data not shown). Overall this aerosol infection of C57BL/6 mice shows that deletion of any single rpf gene in M. tuberculosis results in no observable defects in growth or persistence up to 16 weeks postinfection.
FIG. 5.
Disruption of individual rpf-like genes does not alter bacterial burden in organs of C57BL/6 mice through 16 weeks postinfection. Bacterial numbers in the lung (A), spleen (B), and liver (C) of C57BL/6 mice infected by aerosol with either Erdman wild type (WT) (•), mc23125 (ΔRv2389c) (○), mc23126A (ΔRv1009rev) (▾), mc23127 (ΔRv2450c) (▿), mc23128 (ΔRv1884c) (▪), or mc23129 (ΔRv0867c) (□) were determined. Data (means ± standard error [error bars] of three to four mice per group) are representative of three independent experiments.
In vitro expression of the rpf-like genes.
Given our findings that the M. tuberculosis Δrpf mutants showed growth kinetics indistinguishable from Erdman wild type up to 16 weeks post-aerosol infection, we considered that the lack of a growth phenotype might be due to functional redundancy within this gene family; that is, perhaps loss of one Rpf homologue is compensated by the continued or enhanced expression of one or more of the remaining homologues. In the in vitro studies of Mukamolova et al. (25) all five Rpf-like proteins of M. tuberculosis share the growth-promoting activity. The notion of functional redundancy is further suggested by the fact that, although several of the M. tuberculosis Rpfs have unique features, such as a proline-alanine-rich carboxy terminus for Rv0867c or a lipoprotein lipid attachment site for Rv1009, all share an ∼70-amino-acid conserved region of high homology (∼70% identity). In addition, while the single M. luteus rpf gene is essential (24), the individual rpf-like genes of M. tuberculosis are dispensable for in vitro or in vivo growth (results of this study).
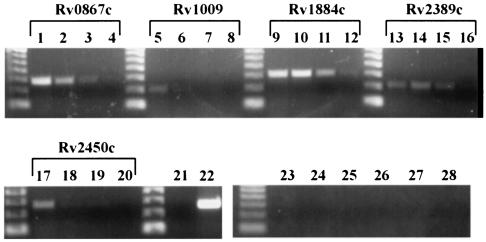
To further explore the issue of functional redundancy, we investigated expression of the rpf-like genes in axenic culture over time, from early exponential phase into stationary phase. We inoculated a culture with 1.75 × 106 CFU/ml of M. tuberculosis Erdman and obtained samples at day 4 (OD600 = 0.125, 3.1 × 107 CFU/ml), day 6 (OD600 = 0.574; 5.55 × 108 CFU/ml), day 8 (OD600 = 1.408; 1.05 × 109 CFU/ml), and day 12 (OD600 = 2.512; 2.75 × 109 CFU/ml), and prepared RNA from these samples for quantitative dilutional PCR. As shown in Fig. 6, transcripts for all of the rpf-like genes are detectable at the earliest time point (columns 1 to 5), but relative expression levels for the genes differ at later time points (columns 6 to 10, 11 to 15, and 16 to 20). Notably, Rv2450c (row E) is expressed maximally in early log phase, at day 4, with expression falling markedly by day 6 and remaining low through day 12. Overall, beyond the early log phase of growth, Rv2450c exhibits the weakest level of expression among the five M. tuberculosis rpf homologues. The relatively weak expression of Rv2450c has been reported by a recent study that examined rpf expression by actively growing M. tuberculosis H37Rv at a single unspecified time point (25). Three of the other rpf-like genes, Rv0867c (row A), Rv1009 (row B), and Rv2389c (row D), have higher expression at days 4 through 6 and reduced, though still detectable, expression at day 12. Only Rv1884c (row C) appears to be expressed at undiminished levels at day 12 when compared with day 4. Therefore the M. tuberculosis rpf-like genes, with the exception of Rv1884c, follow the expression pattern of the M. luteus rpf, which is characterized by diminishing expression as the culture progresses from exponential growth into stationary phase (24). However, whereas the relatively fast-growing M. luteus did not express detectable levels of rpf in stationary-phase (24 h) cultures (24), expression of all five of the M. tuberculosis rpf-like genes could be detected in extended-stationary-phase (4-month-old) cultures (Fig. 7). Although the time course represents exclusively in vitro data and does not necessarily reflect the expression during growth in mouse tissues in vivo, the overlapping expression patterns observed for several of the rpf-like genes may explain, at least in part, the lack of an in vivo growth phenotype for each of the five M. tuberculosis mutant strains with deletion of a single rpf homologue.
FIG. 6.
Expression of the M. tuberculosis rpf-like genes in vitro as assessed by quantitative dilutional PCR. RNA was prepared from Erdman grown in 7H9 medium, and 1 μg of total RNA was reverse transcribed and serially diluted fivefold for use as template in PCRs to amplify each of the rpf-like genes. Primers, as described in Materials and Methods, were used to amplify portions of Rv0867c (A), Rv1009 (B), Rv1884c (C), Rv2389c (D), and Rv2450c (E). (A to E) Lanes 1 to 5 contain PCR products of serial dilutions of cDNA from day 4 (OD600 = 0.125), lanes 6 to 10 are from day 6 (OD600 = 0.574), lanes 11 to 15 are from day 8 (OD600 = 1.408), and lanes 16 to 20 are from day 12 (OD600 = 2.512). (F) PCR products of cDNAs from day 4 (lanes 1 and 5), day 6 (lanes 2 and 6), day 8 (lanes 3 and 7), and day 12 (lanes 4 and 8) using 16S rRNA primers (lanes 1 to 4, 9, and 10) and 23S rRNA primers (lanes 5 to 8, 11, and 12). Lanes 9 and 11 contain no template (negative controls), while lanes 10 and 12 contain Erdman genomic DNA as the template. Lanes 16 to 19 show that no PCR product was obtained from non-reverse-transcribed RNA from day 4 (lane16), day 6 (lane 17), day 8 (lane 18), or day 12 (lane 19) cultures, using primers to amplify Rv1884c. A 50-bp ladder (Invitrogen) is shown in the far left lane of each gel.
FIG. 7.
Expression of M. tuberculosis rpf homologues in a late-stationary-phase (4-month-old) Erdman culture. RNA was prepared from Erdman grown in 7H9 medium, and 5 μg of total RNA was reverse transcribed and serially diluted fivefold for use as template in PCRs to amplify each of the rpf-like genes. Primers, as described in Materials and Methods, were used to amplify portions of Rv0867c (lanes 1 to 4), Rv1009 (lanes 5 to 8), Rv1884c (lanes 9 to 12), Rv2389c (lanes 13 to 16), and Rv2450c (lanes 17 to 20). Lane 21 is a negative control containing no template, and lane 22 shows amplification of 23S rRNA from the same cDNA template used to amplify the rpf genes. Lanes 23 to 28 contain non-reverse-transcribed RNA as the template, using primers to amplify Rv2389c (lane 23), Rv1009 (lane 24), Rv2450c (lane 25), Rv1884c (lane 26), Rv0867c (lane 27), and 23S rRNA (lane 28). A 50-bp ladder (Invitrogen) is shown to the left of each series of samples.
In vivo expression of the M. tuberculosis rpf genes.
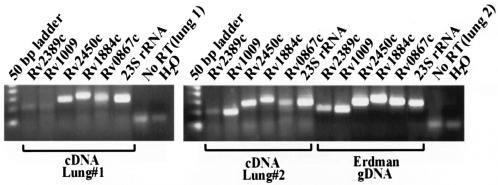
To further evaluate the biological relevance of the mycobacterial Rpf protein family, studies have been initiated to examine the expression of all five rpf genes in the lungs of C57BL/6 mice infected with a clinical strain of M. tuberculosis (CI4) (28). For these studies, total RNA extracted from the lungs of CI4-infected mice at 2 weeks postinfection was analyzed by RT-PCR. As shown in Fig. 8, expression of each of the five M. tuberculosis rpf-like genes is detected in the lungs of infected mice during this early exponential growth phase of infection. RNA samples which were not subjected to reverse transcription gave no detectable PCR product. Although not quantitative, the results indicate that the various members of the rpf gene family, in a clinical strain of M. tuberculosis, are expressed during an acute infection in mice, at a level such that they are clearly detectable using 1.25 μg of total RNA (host and bacterial) as template for the RT-PCR.
FIG. 8.
In vivo expression of M. tuberculosis rpf-like genes. RNA was prepared from lungs of mice infected with a clinical strain of M. tuberculosis, and total RNA (equivalent of 1.25 μg per PCR mixture) was reverse transcribed and used as the template in PCRs to amplify each of the rpf-like genes or to amplify 23S rRNA, as indicated by the gene designation above each lane. Also shown are the results of PCR amplification using RNA which had not been reverse transcribed (No RT), employing 23S rRNA primers, for both lung 1 and lung 2. The far right lane of each gel shows the results of PCR amplification using 23S rRNA primers with water (H2O) as template.
DISCUSSION
This study reports the construction, by phage-mediated allelic exchange technology (1), of isogenic M. tuberculosis mutants with interruptions in each of the five rpf-like genes, and presents the results of a phenotypic analysis of the resulting strains. There exists strong evidence that the M. luteus Rpf functions as a regulator of growth (22, 23) and the gene is essential for Micrococcus viability (24). Further, the M. tuberculosis Rpf-like proteins have recently been demonstrated to exert a similar growth-stimulatory activity on aged cultures of M. bovis BCG (25). Our results indicate that individual rpf-like genes can be deleted from the chromosome of M. tuberculosis with no significant alteration of growth in liquid culture (Fig. 3). Disruption of Rv1009, however, results in a small colony phenotype, although this is likely due to polar effects on ksgA (Fig. 4). The individual rpf deletion mutants had no attenuation of growth, compared to wild-type Erdman, in the organs of C57BL/6 mice through 16 weeks postinfection (Fig. 5). The immunopathology elicited by the M. tuberculosis rpf deletion mutants is also indistinguishable from that observed in tissues of wild-type-infected mice (data not shown). Results of in vitro studies showed that the expression kinetics of M. tuberculosis rpf genes fall into distinct patterns that overlap to various degrees and that these genes are expressed by bacilli in extended-stationary-phase cultures. In vivo studies revealed that all five rpf genes are expressed in the lungs of mice acutely infected with M. tuberculosis.
The lack of a phenotype regarding in vitro growth or in vivo growth in acute and chronic persistent pulmonary infections of mice suggests that the function of the various M. tuberculosis Rpfs may be entirely or partially redundant. If this were the case, loss of a specific function as a result of disruption of a specific rpf-like gene may be compensated for by continued expression of one or more of the remaining homologues. Our analysis of the in vitro expression of these genes from early exponential phase through stationary phase does reveal that the expression patterns of the five homologues are overlapping. Although suggestive of potentially overlapping functions, clearly the results are open to additional interpretations, and as yet we have not directly addressed the effect that deletion of a specific rpf-like gene has on the expression of the remaining homologues. Interestingly, while overlapping, the in vitro expression kinetics of the five M. tuberculosis rpf genes from early log phase to late stationary phase fall into three readily discernible patterns (Fig. 6): (i) of the five homologues, the expression of Rv2450c, while peaking early (maximal at the first time point obtained, 4 days after inoculation of the culture), consistently exhibited the lowest levels of expression thereafter; in contrast, (ii) the expression of Rv1884c is consistently high throughout the 8-day time frame of the study (from day 4 to day 12 postinoculation); while (iii) the expression of Rv0867c, Rv1009, and Rv2389c peaks at 4 to 6 days after initiation of the culture and then diminishes as the bacilli transit into stationary phase (12-day-old cultures). The distinct in vitro expression profiles exhibited by the five rpf genes indicate that these M. tuberculosis genes may be differentially regulated. The differential expression patterns, which vary depending upon the growth phase of the cultures (Fig. 6), suggest that individual members of the M. tuberculosis Rpf family may serve distinct functions at specific phases of infection, although at least for individual rpf knockouts the functions appear to be nonessential.
In addition to the data obtained from gene expression studies presented above (Fig. 6), evidence exists, albeit indirect, to support the notion that M. tuberculosis rpf genes are indeed differentially regulated. This evidence derives primarily from whole-genome expression profiling studies. For instance, Rv2450c is the only rpf induced in M. tuberculosis by 15 min of acid shock (pH 5.5) (4), while Rv1884c shows increased expression in H37Rv compared with a mutant disrupted in the sigE gene (20), the latter encoding an extracytoplasmic function sigma factor induced by heat shock, sodium dodecyl sulfate treatment (19), and the intracellular milieu of cultured human primary macrophages (10). Additionally, both Rv1884c and Rv2450c may exhibit modestly up-regulated expression upon shift from 20 to 0.2% oxygen (30; supplementary material at http://schoolniklab.stanford.edu/projects/tb.html). Together, these data strongly suggest that the expression of M. tuberculosis rpf genes is differentially regulated.
As noted above, the diverse gene contexts of the M. tuberculosis rpf genes render predictions of function based upon activities of the neighboring gene products untenable. The individual, unique features of the Rpfs, however, suggest possible functional differences among the various members. For example, the Rv2450c coding sequence contains a proline-rich region at its amino terminus (21 of the first 100 amino acids), while Rv0867c contains a highly proline- and alanine-rich carboxy terminus (a proline content of 25.3% in the C-terminal 292 amino acids), a feature shared with a number of other mycobacterial proteins, included among them the secreted 45/47-kDa antigen complex of M. tuberculosis (17, 18, 27, 36, 39). The functions of these mycobacterial proline-rich proteins are unknown, though a number have been shown to be immunologically important secreted antigens, stimulating strong antibody and delayed-type hypersensitivity responses (18, 27). Rv1009 is unique among the rpf-like genes in possessing a lipoprotein lipid attachment site, while Rv1884c and Rv2389c have no recognizable conserved motifs beyond the shared Rpf domain and putative signal sequences.
Biochemical characterization of the various Rpfs is essential to elucidate the precise function of each individual member of this family of putative growth-regulatory proteins. Based on the apparent lability of the in vitro resuscitation activity of M. tuberculosis Rpfs (25), this task is unlikely to be straightforward. The reported functional instability of the Micrococcus Rpf was significant enough that, for the relatively lengthy M. tuberculosis studies, prescreening for activity using the faster-growing M. luteus was necessary (31). The apparent lability of the resuscitation activity of the M. tuberculosis Rpfs may have contributed to our own failure in consistently demonstrating growth-promoting activity in experimental systems designed to resuscitate stationary-phase cultures of M. smegmatis or M. tuberculosis using E. coli-expressed recombinant Rpfs (J. M. Tufariello and J. Chan, unpublished results). To further complicate data interpretation, in some of these resuscitation studies an apparent resuscitating activity was observed with addition of bovine serum albumin or Rpf-unrelated purified mycobacterial proteins to negative control cultures (J. M. Tufariello and J. Chan, unpublished results), emphasizing the importance of appropriate controls in the interpretation of these very difficult assays.
Much remains to be learned regarding the functions and regulatory mechanisms of the M. tuberculosis Rpfs and the biological relevance of these proteins in M. tuberculosis growth and pathogenesis. Our finding that all five Rpf homologues are expressed in the lungs of mice infected with CI4, a clinical strain of M. tuberculosis, suggests a biological role for these putative growth-regulatory proteins. Formal proof of biological relevance would require the demonstration that rpf deletion mutants display certain in vivo phenotypes. As discussed, the lack of a growth phenotype in M. tuberculosis strains with disruption of a single rpf suggests functional redundancy. The testing of this hypothesis requires evaluation of the in vitro and in vivo growth characteristics of M. tuberculosis strains harboring disruptions of multiple rpf genes. We are in the process of constructing such multi-rpf deletion mutants by unmarking the existing single knockout strains using the γδ-resolvase expression plasmid (1) and building in successive deletions using the phages we have already generated. Finally, it is noteworthy that all five rpf homologues are expressed in M. tuberculosis obtained from extended-stationary-phase cultures (Fig. 7). Though speculative, this observation suggests the possibility that Rpfs may play a role in the reactivation of quiescent bacilli. Characterization of the in vivo expression of the M. tuberculosis rpf genes and of the capacity of M. tuberculosis Δrpf mutants to reactivate using established murine models (6, 21, 29) will help evaluate this intriguing possibility.
Acknowledgments
We thank Jiayong Xu and Biao Wu for expert technical assistance with the aerosol infections, Hideaki Ohno for generously providing RNA from M. tuberculosis-infected mouse lungs, Fred Tsen for valuable assistance with preparation of the manuscript and figures, and Christopher Basler for critical reading of the manuscript and for many helpful discussions. We also thank Jordan Kriakov for providing the temperature-sensitive phage phAE87.
This work was funded by NIH grants AI49375 (J.M.T.), AI26170 (W.R.J.), and AI36990 (J.C.).
Editor: W. A. Petri, Jr.
REFERENCES
- 1.Bardarov, S., S. Bardarov, Jr., M. S. Pavelka, Jr., V. Sambandamurthy, M. Larsen, J. Tufariello, J. Chan, G. Hatfull, and W. R. Jacobs, Jr. 2002. Specialized transduction: an efficient method for generating marked and unmarked targeted gene disruptions in Mycobacterium tuberculosis, M. bovis BCG and M. smegmatis. Microbiology 148:3007-3017. [DOI] [PubMed] [Google Scholar]
- 2.Bardarov, S., J. Kriakov, C. Carriere, S. Yu, C. Vaamonde, R. A. McAdam, B. R. Bloom, G. F. Hatfull, and W. R. Jacobs, Jr. 1997. Conditionally replicating mycobacteriophages: a system for transposon delivery to Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 94:10961-10966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Dye, C., S. Scheele, P. Dolin, V. Pathania, and M. C. Raviglione. 1999. Consensus statement. Global burden of tuberculosis: estimated incidence, prevalence, and mortality by country. W.H.O. Global Surveillance and Monitoring Project. JAMA 282:677-686. [DOI] [PubMed] [Google Scholar]
- 4.Fisher, M. A., B. B. Plikaytis, and T. M. Shinnick. 2002. Microarray analysis of the Mycobacterium tuberculosis transcriptional response to the acidic conditions found in phagosomes. J. Bacteriol. 184:4025-4032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Flynn, J. L., and J. Chan. 2001. Immunology of tuberculosis. Annu. Rev. Immunol. 19:93-129. [DOI] [PubMed] [Google Scholar]
- 6.Flynn, J. L., C. A. Scanga, K. E. Tanaka, and J. Chan. 1998. Effects of aminoguanidine on latent murine tuberculosis. J. Immunol. 160:1796-1803. [PubMed] [Google Scholar]
- 7.Gedde-Dahl, T. 1952. Tuberculous infection in the light of tuberculin matriculation. Am. J. Hyg. 56:139-214. [DOI] [PubMed] [Google Scholar]
- 8.Glickman, M. S., J. S. Cox, and W. R. Jacobs, Jr. 2000. A novel mycolic acid cyclopropane synthetase is required for cording, persistence, and virulence of Mycobacterium tuberculosis. Mol. Cell 5:717-727. [DOI] [PubMed] [Google Scholar]
- 9.Gomez, M., S. Johnson, and M. L. Gennaro. 2000. Identification of secreted proteins of Mycobacterium tuberculosis by a bioinformatic approach. Infect. Immun. 68:2323-2327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Graham, J. E., and J. E. Clark-Curtiss. 1999. Identification of Mycobacterium tuberculosis RNAs synthesized in response to phagocytosis by human macrophages by selective capture of transcribed sequences (SCOTS). Proc. Natl. Acad. Sci. USA 96:11554-11559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Helser, T. L., J. E. Davies, and J. E. Dahlberg. 1971. Change in methylation of 16S ribosomal RNA associated with mutation to kasugamycin resistance in Escherichia coli. Nat. New Biol. 233:12-14. [DOI] [PubMed] [Google Scholar]
- 12.Helser, T. L., J. E. Davies, and J. E. Dahlberg. 1972. Mechanism of kasugamycin resistance in Escherichia coli. Nat. New Biol. 235:6-9. [DOI] [PubMed] [Google Scholar]
- 13.Kaprelyants, A. S., and D. B. Kell. 1993. Dormancy in stationary-phase cultures of Micrococcus luteus: flow cytometric analysis of starvation and resuscitation. Appl. Environ. Microbiol. 59:3187-3196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kaprelyants, A. S., G. V. Mukamolova, and D. B. Kell. 1994. Estimation of dormant Micrococcus luteus cells by penicillin lysis and by resuscitation in cell-free spent medium at high dilution. FEMS Microbiol. Lett. 115:347-352. [Google Scholar]
- 15.Kleerebezem, M., L. E. N. Quadri, O. P. Kulpers, and W. M. de Vos. 1997. Quorum sensing by peptide pheromones and two-component signal-transduction systems in gram-positive bacteria. Mol. Microbiol. 24:895-904. [DOI] [PubMed] [Google Scholar]
- 16.Lafontaine, D., J. Delcour, A. L. Glasser, J. Desgres, and J. Vandenhaute. 1994. The DIM1 gene responsible for the conserved m6(2)Am6(2)A dimethylation in the 3′-terminal loop of 18 S rRNA is essential in yeast. J. Mol. Biol. 241:492-497. [DOI] [PubMed] [Google Scholar]
- 17.Laqueyrerie, A., P. Militzer, F. Romain, K. Eiglmeier, S. Cole, and G. Marchal. 1995. Cloning, sequencing, and expression of the apa gene coding for the Mycobacterium tuberculosis 45/47-kilodalton secreted antigen complex. Infect. Immun. 63:4003-4010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Manca, C., K. Lyashchenko, R. Colangeli, and M. L. Gennaro. 1997. MTC28, a novel 28-kilodalton proline-rich secreted antigen specific for the Mycobacterium tuberculosis complex. Infect. Immun. 65:4951-4957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manganelli, R., E. Dubnau, S. Tyagi, F. R. Kramer, and I. Smith. 1999. Differential expression of 10 sigma factor genes in Mycobacterium tuberculosis. Mol. Microbiol. 31:715-724. [DOI] [PubMed] [Google Scholar]
- 20.Manganelli, R., M. I. Voskuil, G. K. Schoolnik, and I. Smith. 2001. The Mycobacterium tuberculosis ECF sigma factor sigmaE: role in global gene expression and survival in macrophages. Mol. Microbiol. 41:423-437. [DOI] [PubMed] [Google Scholar]
- 21.Mohan, V. P., C. A. Scanga, K. Yu, H. M. Scott, K. E. Tanaka, E. Tsang, M. C. Tsai, J. L. Flynn, and J. Chan. 2001. Effects of tumor necrosis factor alpha on host immune response in chronic persistent tuberculosis: possible role for limiting pathology. Infect. Immun. 69:1847-1855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mukamolova, G. V., A. S. Kaprelyants, D. I. Young, M. Young, and D. B. Kell. 1998. A bacterial cytokine. Proc. Natl. Acad. Sci. USA 95:8916-8921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Mukamolova, G. V., S. S. Kormer, D. B. Kell, and A. S. Kaprelyants. 1999. Stimulation of the multiplication of Micrococcus luteus by an autocrine growth factor. Arch. Microbiol. 172:9-14. [DOI] [PubMed] [Google Scholar]
- 24.Mukamolova, G. V., O. A. Turapov, K. Kazarian, M. Telkov, A. S. Kaprelyants, D. B. Kell, and M. Young. 2002. The rpf gene of Micrococcus luteus encodes an essential secreted growth factor. Mol. Microbiol. 46:611-621. [DOI] [PubMed] [Google Scholar]
- 25.Mukamolova, G. V., O. A. Turapov, D. I. Young, A. S. Kaprelyants, D. B. Kell, M. Young, K. Kazarian, and M. Telkov. 2002. A family of autocrine growth factors in Mycobacterium tuberculosis. Mol. Microbiol. 46:623-635. [DOI] [PubMed] [Google Scholar]
- 26.Ohno, H., G. F. Zhu, V. P. Mohan, D. Chu, S. Kohno, W. R. Jacobs, Jr., and J. Chan. 2003. The effects of reactive nitrogen intermediates on gene expression in Mycobacterium tuberculosis. Cell. Microbiol. 5:637-648. [DOI] [PubMed] [Google Scholar]
- 27.Romain, F., A. Laqueyrerie, P. Militzer, P. Pescher, P. Chavarot, M. Lagranderie, G. Auregan, M. Gheorghiu, and G. Marchal. 1993. Identification of a Mycobacterium bovis BCG 45/47-kilodalton antigen complex, an immunodominant target for antibody response after immunization with living bacteria. Infect. Immun. 61:742-750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scanga, C. A., V. P. Mohan, K. Tanaka, D. Alland, J. L. Flynn, and J. Chan. 2001. The inducible nitric oxide synthase locus confers protection against aerogenic challenge of both clinical and laboratory strains of Mycobacterium tuberculosis in mice. Infect. Immun. 69:7711-7717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Scanga, C. A., V. P. Mohan, K. Yu, H. Joseph, K. Tanaka, J. Chan, and J. L. Flynn. 2000. Depletion of CD4+ T cells causes reactivation of murine persistent tuberculosis despite continued expression of IFN-gamma and NOS2. J. Exp. Med. 192:347-358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sherman, D. R., M. Voskuil, D. Schnappinger, R. Liao, M. I. Harrell, and G. K. Schoolnik. 2001. Regulation of the Mycobacterium tuberculosis hypoxic response gene encoding alpha-crystallin. Proc. Natl. Acad. Sci. USA 98:7534-7539. (Erratum, 98:15393.) [DOI] [PMC free article] [PubMed]
- 31.Shleeva, M. O., K. Bagramyan, M. V. Telkov, G. V. Mukamolova, M. Young, D. B. Kell, and A. S. Kaprelyants. 2002. Formation and resuscitation of “non-culturable” cells of Rhodococcus rhodochrous and Mycobacterium tuberculosis in prolonged stationary phase. Microbiology 148:1581-1591. [DOI] [PubMed] [Google Scholar]
- 32.Snapper, S. B., R. E. Melton, S. Mustafa, T. Kieser, and W. R. Jacobs, Jr. 1990. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 4:1911-1919. [DOI] [PubMed] [Google Scholar]
- 33.Stover, C. K., V. F. de la Cruz, T. R. Fuerst, J. E. Burlein, L. A. Benson, L. T. Bennett, G. P. Bansal, J. F. Young, M. H. Lee, G. F. Hatfull, et al. 1991. New use of BCG for recombinant vaccines. Nature 351:456-460. [DOI] [PubMed] [Google Scholar]
- 34.Sturme, M. H. J., M. Kleerebezem, J. Nakayama, A. D. L. Akkermans, E. E. Vaughan, and W. M. de Vos. 2002. Cell to cell communication by autoinducing peptides in gram-positive bacteria. Antonie Leeuwenhoek 81:233-243. [DOI] [PubMed] [Google Scholar]
- 35.Swift, S., J. P. Throup, P. Williams, G. P. C. Salmond, and G. S. A. B. Stewart. 1996. Quorum sensing: a population-density component in the determination of bacterial phenotype. Trends Biochem. Sci. 21:214-219. [PubMed] [Google Scholar]
- 36.Thole, J. E., L. F. Stabel, M. E. Suykerbuyk, M. Y. De Wit, P. R. Klatser, A. H. Kolk, and R. A. Hartskeerl. 1990. A major immunogenic 36,000-molecular-weight antigen from Mycobacterium leprae contains an immunoreactive region of proline-rich repeats. Infect. Immun. 58:80-87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Buul, C. P. J. J., and P. H. van Knippenberg. 1985. Nucleotide sequence of the ksgA gene of Escherichia coli: comparison of methyltransferases affecting dimethylation of adenosine in ribosomal RNA. Gene 38:65-72. [DOI] [PubMed]
- 38.Whitehead, N. A., A. M. L. Barnard, H. Slater, N. J. L. Simpson, and G. P. C. Salmond. 2001. Quorum-sensing in gram-negative bacteria. FEMS Microbiol. Rev. 25:365-404. [DOI] [PubMed] [Google Scholar]
- 39.Yamaguchi, R., K. Matsuo, A. Yamazaki, C. Abe, S. Nagai, K. Terasaka, and T. Yamada. 1989. Cloning and characterization of the gene for immunogenic protein MPB64 of Mycobacterium bovis BCG. Infect. Immun. 57:283-288. [DOI] [PMC free article] [PubMed] [Google Scholar]