Abstract
Fluorescence resonance energy transfer (FRET) under in vivo conditions is a well-established technique for the evaluation of populations of protein bound/unbound nucleic acid (NA) molecules or NA hybridization kinetics. However, in vivo FRET has not been applied to in vivo quantitative conformational analysis of NA thus far. Here we explored parameters critical for characterization of NA structure using single-pair (sp)FRET in the complex cellular environment of a living Escherichia coli cell. Our measurements showed that the fluorophore properties in the cellular environment differed from those acquired under in vitro conditions. The precision for the interprobe distance determination from FRET efficiency values acquired in vivo was found lower (∼31%) compared to that acquired in diluted buffers (13%). Our numerical simulations suggest that despite its low precision, the in-cell FRET measurements can be successfully applied to discriminate among various structural models. The main advantage of the in-cell spFRET setup presented here over other established techniques allowing conformational analysis in vivo is that it allows investigation of NA structure in various cell types and in a native cellular environment, which is not disturbed by either introduced bulk NA or by the use of chemical transfectants.
INTRODUCTION
Inside a living cell, nucleic acids (NA) are exposed to a very complex environment and their structures are, depending on their sequence, modulated by non-specific factors such as viscosity, molecular crowding or by specific interactions with ions and small molecular weight compounds (1). While the complexity of the intracellular environment as a general property of every living organism is generally appreciated, there is a lack of appropriate tools to analyze NA structures in a cellular context.
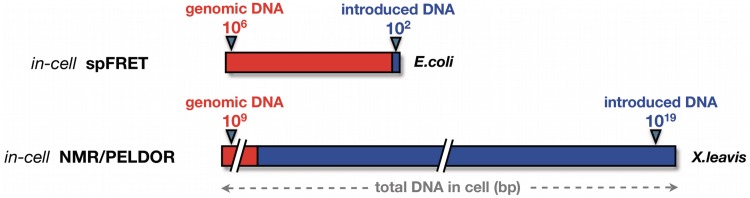
The first attempt to characterize nucleic acid structures at high resolution under native conditions was recently performed by Hansel et al. using nuclear magnetic resonance (NMR) spectroscopy inside living Xenopus laevis oocytes (2). Recently, Krstic et al. extended the technique of the in-cell NMR to pulsed electron–electron double-resonance (PELDOR) spectroscopy (3). In both techniques, exogenous DNA/RNA constructs were mechanically introduced into the X. laevis oocytes/eggs, followed by NMR or PELDOR investigations. Although both methods provide unique information on NA structure ‘in living cells’, this phrase does not necessarily mean ‘in a native cellular environment’. Due to the low inherent sensitivity of NMR and PELDOR detection and high cellular background, the methods require unnaturally large quantities (∼250–500 µM) of exogenous NA to be deposited into the cells (2,3). As illustrated in Figure 1, the number of NA base-pairs deposited into the cell for the in-cell NMR/PELDOR experiments was more than 15 000 times higher than the number of base-pairs in the genomic DNA (2–4). Although the injected cells for in-cell NMR/PELDOR experiments are viable, the addition of such a high concentration of NA might modulate the intracellular environment and bias the structural readout. In practical terms, applications of in-cell NMR and PELDOR spectroscopy are presently limited to X. laevis oocytes and eggs, which are large enough to allow for the introduction of high concentrations of exogenous NA via injection (2–4).
Figure 1.
Schematic representation of requirements of in-cell NMR/PELDOR and in-cell spFRET on concentration of the exogenous NA introduced into cellular environment. The concentrations are displayed relative to average size of genomic DNA of E. coli and X. laevis.
Undoubtedly, the development of new techniques that would allow characterization of NA structure under physiological conditions in vivo is of general interest. Ideally, such techniques would avoid the deposition of large quantities of NA in the cellular environment, allow measurements in a variety of cell types and provide complementary information to the existing techniques of in-cell NMR and PELDOR.
One of the most obvious candidates among the available methods that in principle allow quantitative characterization of NA structure under native conditions is fluorescence resonance energy transfer (FRET) (5–7). FRET has been successfully used to obtain quantitative long-range information on NA structure under in vitro conditions (8–11). A number of studies also demonstrated that FRET measurements could be performed inside living cells (12–14). Although these state-of-the-art in vivo FRET applications are currently limited to the evaluation of populations of protein bound/unbound NA molecules or NA–NA hybridization kinetics and usually require the introduction of a bulk amount of fluorescently labeled NA into cells with the use of chemical transfectants, they clearly reveal the potential for quantitative characterization of nucleic acid structure using in vivo FRET (12–16). Rapid advances in live-cell imaging, demonstrated e.g. by direct observation of the kinetics of transcription of single nascent mRNA molecules (17), have now enabled acquiring quantitative FRET data in living cells at single molecule level.
Here, we explored the possibility of quantitative characterization of nucleic acid structure inside living cells at the single-molecule level using single-pair FRET (spFRET). The proposed approach avoids the need to introduce bulk amounts of DNA into the cells and the use of chemical transfectants, which both disturb composition of the intracellular environment. The parameters critical for characterization of NA structure using spFRET were evaluated for the ATTO-680/ATTO-740 donor–acceptor system based on the experimentally determined variation of FRET efficiencies for a series of terminally labeled DNA duplexes with a length of 8–18 bp, both in vitro and in vivo, in Escherichia coli cells. The application potential and limitations of in-cell spFRET for structural analysis of NA are discussed.
MATERIALS AND METHODS
DNA constructs
Unmodified DNA oligonucleotides were purchased from Generi-Biotech (Hradec Kralove, Czech Republic). Fluorescently labeled DNA oligonucleotides coupled with fluorescent dyes ATTO680 and ATTO740 (ATTO-TEC GmbH) via flexible C6 linkers at the 5′-termini were purchased from Sigma-Aldrich. Table 1 lists the DNA constructs used in this study. For both in vitro and in vivo spFRET measurements, both single-stranded DNA conjugated either with ATTO 680 or ATTO 740 was dissolved in buffer A (0.01% Tween 20, 200 µm sodium l-ascorbate, 0.3 mM EDTA, 10 mM TRIS, pH 7.4). Subsequently, the ATTO680–DNA was mixed with ATTO740–DNA of corresponding size at a ratio of 1:1.15. To allow annealing of complementary strands, the mixture was heated to 95°C for 5 min and left to slowly cool to room temperature.
Table 1.
DNA constructs employed for spFRET measurements
| DNA length [bp] | ATTO680-5′ -> 3′ | ATTO740-5′ -> 3 |
|---|---|---|
| 8 | CCTGCAGG | CCTGCAGG |
| 10 | CCTGCAGTGG | CCACTGCAGG |
| 12 | CCTGCAGTACGG | CCGTACTGCAGG |
| 14 | CCTGCACGACCTGG | CCAGGTCGTGCAGG |
| 16 | CCTGCACGACCTGTGG | CCACAGGTCGTGCAGG |
Absorption and fluorescence spectroscopy
Absorption and emission spectra were acquired at room temperature in a 10-mm silica glass cuvette in buffer A at DNA concentrations of 0.1 and 10 µM for measurements of fluorescence and absorption, respectively. Emission spectra were recorded on a Fluorolog spectrofluorometer (SPEX, USA). A Raman emitter RS 664 LP (679.3–1497.7 nm) was used to filter emission spectra from the excitation light. The absorption spectra were recorded on a UV300 spectrophotometer (Spectronic Unicam, UK).
Circular dichroism spectroscopy
Circular dichroism (CD) spectra were acquired on a JASCO J-715 spectropolarimeter (JASCO Corporation, Tokyo, Japan) in 10-mm silica glass cuvettes at room temperature. The CD spectra were acquired in buffer A at a DNA concentration of 10 µM. The melting temperatures of the DNA constructs were determined by the temperature dependence of the molar ellipticity in the global minimum of the CD spectra. A thermostatic water bath combined with a thermoelectric Peltier device was used to control the temperature. The individual points of the melting curve were collected after 15 min of temperature equilibration at each temperature step.
Time-resolved fluorescence anisotropy
Time-resolved fluorescence anisotropy decay measurements were performed to evaluate the rotational mobility of the donor and acceptor fluorophores. The anisotropy decay r(t) was constructed from polarized intensity decays as
| (1) |
where I|| and I⊥ are the intensity decays recorded with the emission polarizer oriented parallel or perpendicular, respectively, relative to the vertically polarized excitations and G is the detector correction factor.
Fluorescence anisotropy measurements were performed in buffer A at a DNA concentration of 0.1 μM. The fluorescence anisotropy measurements were performed with a modified instrumental setup as for lifetime measurements (vide infra) with the addition of a Glan–Taylor prism to separate the two mutually perpendicular polarization channels and an additional avalanche photo diode (SPCM-AQR-16) used to detect the second polarization channel.
The instrument response function was deconvoluted from the lifetime decays of the fluorophores, and the decays were fitted using a Fluofit module (18) of Matlab (Mathworks, USA). In most cases, one short decay component, not related to donor excited-state lifetime, was used to eliminate the direct acceptor excitation and scattered excitation light in the cellular environment. The collected lifetime probability distribution functions were analyzed through the maximum likelihood estimation with the Matlab function ‘gmdistribution.fit’, with the multi-component distribution analysis providing the means, covariance and logarithmic likelihood of the fit.
Time-resolved fluorescence anisotropy decays were fitted to a model consisting of the sum of two exponentials, one accounting for the fast local motion of the fluorophore and one accounting for the slow global tumbling of the entire molecule:
| (2) |
The parameters βF, βS, αF and αS characterize the amplitudes and rotational correlation times of fast (F) and slow (S) motion, respectively.
Time-resolved in vitro FRET measurements and Förster radius determination
The in vitro FRET efficiencies were measured using the time-resolved method for the ensemble of molecules (19). Acquired FRET efficiencies were translated into interprobe distances using Supplementary Equation S1. The Förster radius in Supplementary Equation S1 was calculated from the experimentally acquired medium refractive index, the extinction coefficient of the acceptor, the fluorescence quantum yield of the donor, the orientation factor κ2 and the overlap integral of FRET pair spectra in buffer A (Supplementary Table S3). Quantum yield of ATTO680 was determined relative to rhodamine B according to the procedure of Williams et al. (20). The emission spectra used to determine the quantum yield were corrected for detection efficiency in different spectral regions. The extinction coefficient of the acceptor in buffer A/lysate was estimated from absorbancies acquired for various dilutions of the acceptor in buffer A/lysate relative to the absorbance of the ATTO740 dissolved in water using Lambert–Beer equation (all the measurements were conducted in the linear region of the detector) (Supplementary Table S3). Extinction coefficient of the ATTO740 in water was provided by manufacturer (ATTO-TEC, Germany).
The orientation factor, κ2, was estimated from the steady-state fluorescence anisotropies following the procedure of Dale et al. (21) (Supplementary Figure S1) using a single nano-positioning system according to the approach described by Muschielok et al. (22). Estimation of κ2 from the steady-state anisotropies was preferred to that one based on time-resolved anisotropy although resulting estimate is usually less precise (23). The reasons for this choice were anomalous characteristics of time-resolved anisotropies in crude bacterial lysate lacking exponential decays (Supplementary Figure S3).
FRET data processing
The most likely donor and acceptor positions and their uncertainties in all the terminally labeled DNA constructs were calculated using the Nano Positioning System (NPS) incorporated in FRETnps Tools (22). The probability distribution function (PDF) of acceptor position was derived from FRET measured efficiencies and from the accessible volume (AV) of donor dye simulated according the procedure of Sindbert et al. (23). In agreement with in vitro CD data, the canonical B-DNA model with defined attachment points for acceptor and donor along with the knowledge of the length (20 Å) and width (4.5 Å) of the flexible C6 linker was used in the simulation of the AV for both dyes (23). As the manufacturer has not yet published the structures of the ATTO680 and ATTO740 dyes, each dye was approximated by sphere with a diameter of 5 Å in the AV simulations.
Incorporation of DNA into E. coli cells
The BL21 StarTM(DE3)plysS One Shot (Invitrogen, US) strain of E. coli was used for all in vivo FRET measurements. The DNA constructs were introduced into E. coli cells using heat shock with an optimized protocol allowing the introduction of one to five DNA molecules per cell (for details see Supplementary Data). After DNA incorporation, the cells were washed three times in PBS buffer (137 mM NaCl, 2.7 mM KCl, 10 mM Na2 HPO4, 1.8 mM KH2PO4, pH 7.4) to remove the extracellular, unincorporated DNA constructs. Subsequently, the transfected cells were immobilized in a thin layer of polyvinyl alcohol (MW 145 000, 98% hydrolyzed, Merck) using the spin-coating method (24).
The information on the number of molecules per cell was obtained via visual counting. This approach was made possible by exploiting the element that extends the depth of focus of the microscope objective (EDOF), as described in Fessl et al. (25). When using this element, the setup can also detect molecules that are out of focus using the plane microscope objective. Such defocused molecules increase the fluorescence background and might contaminate measured fluorescence lifetime decays. The EDOF element was therefore used to optimize the procedure for the incorporation of single DNA molecules into the cells.
Single-pair time-resolved in-cell FRET measurements
Escherichia coli cells with incorporated DNA constructs were immobilized in a thin layer of PVA. Intracellular fluorophores were localized and individual molecules were spatially separated by slits in an imaging spectrograph. The lifetimes of single constructs were examined. Lifetime histograms were created from approximately 60 molecules for each oligonucleotide length.
The values of EFRET were measured using the time-resolved method as
| (3) |
where τ ′D and τD are the donor fluorescence lifetimes in the presence and absence of an acceptor, respectively.
The single-pair time-resolved FRET measurements were performed using a setup composed of an inverted Olympus IX70 microscope, Triax 320 imaging spectrograph with back-illuminated liquid nitrogen-cooled CCD camera (Spectrum One, Jobin Yvon, 2048 × 512 pixels, pixel size 13.5 × 13.5 μm) and a picosecond-pulse laser diode module (PicoQuant LDH-D-C-640) that generated pulses at 640 nm with linear polarization as an excitation source. The microscope was equipped with an objective providing 100× magnification (Olympus, NA of 1.35, UplanApo), an Olympus filter cube (Olympus, Japan) containing a Raman emitter RS 664 LP (679.3–1497.7 nm), and a donor bandpass BrightLine HC 720/13 and HC-Laser Clean-up MaxDiode 640/8 (Semrock, Germany). For measurements of the fluorescence lifetimes, a time-correlated single photon counting avalanche photodiode (Perkin Elmer, SPCM-AQR-16) was attached to the side exit of the Triax monochromator (Jobin Yvon Inc., USA). The samples were excited at 640 nm by a total internal reflection prism (25). Acquired in-cell FRET efficiencies were evaluated in terms of the most likely donor and acceptor positions and their uncertainties using the same procedure as described for in vitro data. However, in contrast to in vitro FRET evaluation, the Förster radius constructed from the medium refractive index, the extinction coefficient of the acceptor, the fluorescence quantum yield of the donor, the orientation factor κ2 and the overlap integral of FRET pair spectra all acquired in crude bacterial lysate was employed for EFRET-distance conversion.
Preparation of the bacterial lysate
Bacteria were cultivated following the same procedure as for single molecule experiments in a total volume of 1 l. To wash the cells from the cultivation medium, the cell suspension was pelleted via centrifugation at 4°C for 10 min at 4500g. The supernatant was removed and the cell pellet was resuspended in 200 ml of ice-cold PBS buffer. This procedure was repeated three times with fresh PBS. Following the last step of centrifugation, the PBS buffer was removed and a semi-dry bacterial pellet was obtained. To obtain the bacterial lysate, the bacterial pellet was sonicated on ice. Cell debris from the crude cell homogenate was removed by centrifugation at 4°C for 1 h at 23 500g. The supernatant, herein referred as bacterial lysate, was used for FRET measurements.
RESULTS
ATTO680 and ATTO740 do not affect helical conformation of DNA
In general, the principal disadvantage of the FRET technique is the required presence of donor and acceptor fluorophore tags on a probed NA fragment. In principle, these tags might interact with NA fragments and influence both the structure of the NA fragment under study and affect the mobility of fluorophores, which biases the interpretation of EFRET via modulation of the orientation factor, κ2 (Supplementary Equation S5). To evaluate the potential interference of ATTO680 and ATTO740 fluorophores with conformation of the DNA, the influence of the fluorophores on the conformation of the DNA was investigated using CD spectroscopy, thermo-melting analysis and time-resolved fluorescence anisotropy measurements.
The comparison of CD spectra of unmodified DNA and DNA modified with ATTO680, ATTO740, and both ATTO680 and ATTO740 fluorophores suggested that ATTO680 and ATTO740 did not disturb the global helical geometry of the DNA construct (Supplementary Figure S2). For all the constructs, the CD spectra exhibited characteristic features corresponding to the B-form of DNA. Minor differences among the CD spectra, melting temperatures, and distinct kinetics of anisotropy decays between DNA–ATTO680 and DNA–ATTO740 suggested that while ATTO680 has free mobility and moderately promotes terminal base-pair opening, the mobility of ATTO740 is partially hindered due to stabilizing interaction with the DNA (Supplementary Figure S2–S3 and Supplementary Table S1).
Modulation of EFRET in a DNA duplex series in vitro versus in vivo
To evaluate both the potential of EFRET measurements for characterization of NA structure within a complex cellular environment and impact of the intracellular E. coli environment on helical geometry of our DNA constructs, the variations of EFRET for a series of five DNA duplexes of a length of 8–16 bp terminally labeled with ATTO680/ATTO740 fluorophores in free solution and in living E. coli cells were investigated (Supplementary Figure S4). The resulting EFRET, both in vitro and in vivo, were plotted as a function of the helix length in Figure 2. Overall, in vitro EFRET values decreased with helix length, yet there was an apparent local increase in EFRET at 10 bp. Based on the characteristic pattern of the CD spectrum (Supplementary Figure S2), the observed profile for EFRET dependence on helix length can be associated with helical parameters corresponding to the B-form of DNA.
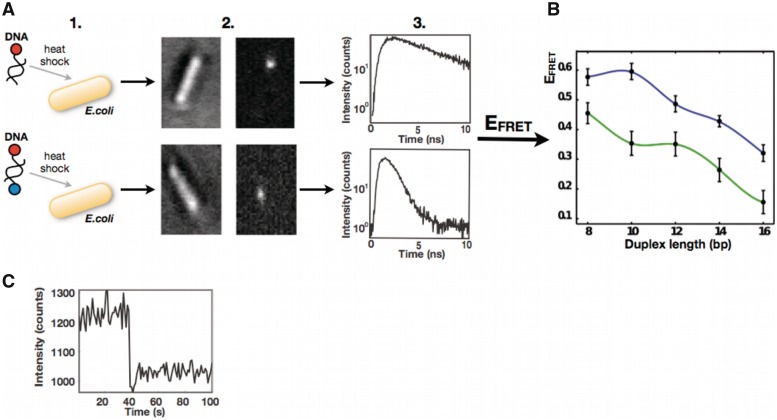
Figure 2.
(A) Simplified workflow of the in-cell spFRET experiment. Step 1—Bacterial cells were separately transformed with DNA constructs labeled with ATTO680 or with the complete FRET pair (ATTO680 and ATTO740). Step 2—Fluorescent molecules inside E. coli cells were localized. Left sub-figure shows transmission image of a bacterial cell with the introduced DNA. The sub-figure on the right is fluorescent image of the same cell. Step 3—Fluorescence lifetimes from the constructs labeled with the donor (ATTO680) and from the constructs labeled with the complete FRET pair (ATTO680 and ATTO740) were acquired and used for calculation of the FRET efficiencies (EFRET). (B) Modulation of EFRET in a duplex DNA series of variable length inside living bacterial cells (green line) versus corresponding modulation in vitro (blue line). The experimental data were fitted with spline. (C) A representative photon-trace from DNA construct labeled with the donor (ATTO680) inside E. coli cell showing single step photobleaching. The trace demonstrates that the fluorescence signal comes from single molecule.
Analogously to the situation in vitro, the in vivo EFRET values decreased with helix length. However, in contrast to in vitro data, the EFRET values displayed a local maximum in EFRET at 12 bp (Figure 2). Comparison of the phase modulation between the in vivo and in vitro EFRET profiles suggested differences in either DNA helical geometry and/or effective relative orientations of the donor and acceptor transition dipole moments. In addition to the differences in phase modulation between in vivo and in vitro EFRET profiles, the in vivo EFRET values appeared systematically reduced in amplitude compared to the in vitro measurements. The reduced EFRET values indicated altered optical properties of environment and fluorophores inside the cells compared to those in the phosphate buffer used for in vitro measurements (Supplementary Equation S5). Indeed, the experimentally determined refractive index of the phosphate buffer, nin vitro = 1.33, was lower than the refractive index of the bacterial lysate, nin vivo = 1.40. However, the difference in refractive indices accounted only for ∼40–50% of reduction in measured EFRET values based on the estimation from Supplementary Equation S2. The fact that the experimentally observed attenuation of EFRET was almost twice as much as it was expected to be based on the corrected refractive index suggests that the cellular environment influences optical properties of fluorophores.
Altogether, the differences in EFRET amplitude and phase modulation between in vitro and in vivo data strongly indicate that for quantitative analysis of an NA structure inside living cells, alterations in both spectral properties of fluorophores and their linker lengths and flexibilities due to the cellular environment need to be considered in the process of the in-cell spFRET data analysis. As for the spectral properties of fluorophores, it is, unfortunately, experimentally impossible to acquire Q0 and J (Supplementary Equation S5) in the context of an intact living cell due to technical difficulties with recording the absorbance spectra and quantum yield of individual molecules. Therefore, to account for altered spectral properties of fluorophores in a cellular environment, we acquired the extinction coefficient of the acceptor, the fluorescence quantum yield of the donor, the orientation factor and the overlap integral of the FRET pair using a bulk amount of DNA mixed with the crude lysate from bacterial cells. Quantitative agreement between the fluorophore emission spectra and lifetimes recorded both in vivo and in the crude bacterial lysate indicated that the fluorophores’ spectral properties in the crude bacterial lysate were similar to those in intact living cells (Supplementary Figure S5 and Supplementary Table S4).
The fluorophores’ spectral properties and orientational factor from fluorescence anisotropies, both acquired in the bacterial lysate along with experimentally acquired refractive index of the bacterial lysate (see ‘Materials and Methods’ section), were subsequently used to calculate Förster distance (R0), whose a priori knowledge is essential for interpretation of in-cell EFRET values in terms of interprobe distances (Supplementary Equations S1 and S5). Supplementary Figure S6 shows the R0 distributions derived from measurements in crude bacterial lysate and diluted buffer. While the distribution of R0 in vitro was characterized by a median distance of R0 = 64.2 Å, and first and third quantiles of 60.1 and 69.5 Å, respectively, the distribution of R0 in the crude bacterial lysate was marked by a median distance of R0 = 47.1 Å, and first and third quantiles of 39.6 and 53.2 Å, respectively. Significantly broader distribution of R0 distances in the bacterial lysate is a consequence of higher fluorescence anisotropies of both dyes in this inherently heterogeneous environment that translates into larger variability of the orientational factor in bacterial lysate compared to that in the diluted solution. The large variability of the orientational factor is reflected in lower precision of distance determination from in vivo EFRET measurements compared to measurements under in vitro conditions. From R0 distributions (Supplementary Figure S6), the precision for the interprobe distance determination from EFRET values was expected to be ∼13.9% and 31.4% for in vitro and in-cell data, respectively (Supplementary Equation S4).
Structural interpretation of in vivo spFRET data
The double-helical DNA fragments of randomized sequences, analogous to those employed in our study, adopt B-DNA conformation in diluted aqueous solutions of monovalent ions (26,27). However, the same DNAs can undergo a B–A transition that is induced by a number of natively occurring agents, such as multivalent ions (28–30), osmolytes mimicking lower water content inside the cells (30–33) and proteins (34–36).
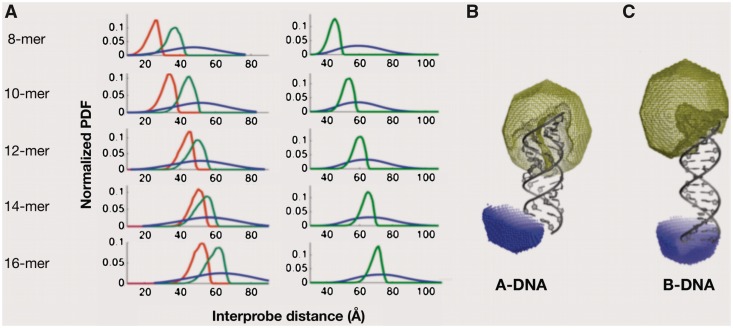
To assess which conformation the DNA adopts inside living prokaryotic cells, and simultaneously assess resolution power of the in-cell spFRET measurements, the derived R0 values from the crude bacterial homogenate were used to express experimentally acquired in-cell FRET efficiencies for a series of five DNA duplexes with a length of 8–16 bp in terms of the donor–acceptor PDF. Resulting interprobe distance PDFs for individual DNA constructs are shown in Figure 3. Experimentally acquired in-cell interprobe distance PDFs for individual constructs were then compared to corresponding PDFs that were simulated based on geometric AVs of fluorophores positioned on the canonical A- and B-form DNA models according to a recently proposed technique by Sindbert et al. (23). Comparison of the experimental interprobe distance PDFs derived from in-cell spFRET efficiencies along with simulated PDFs for A- and B-form of DNA are displayed in Figure 3. Statistical comparison between the experimental and model PDFs clearly indicated better agreement of in-cell spFRET experimental data with B-DNA compared to the A-DNA model (Table 2).
Figure 3.
(A) Left panel: Overlaps between interprobe distance PDFs derived from in-cell FRET measurements (blue line) and ‘in-cell’ PDFs simulated using FRET NPS tools for A- (red line) and B-form (green line) of DNA. Right panel: Overlaps between PDFs derived from in vitro FRET measurements (blue line) and ‘in vitro’ PDFs simulated for B-form of DNA. (B) and (C) AVs of ATTO680 (olive mesh) simulated for A-form and B-form DNA, respectively, and FRET derived and constrained positions of ATTO740 (blue spheres). Both simulations accounted for altered fluorophores properties in cellular environment. For details on the simulation approach, see ‘Materials and Methods’ section.
Table 2.
Probabilistic interpretation of the in-cell FRET data
| bp | A-DNA | B-DNA |
|---|---|---|
| 8 | 0.87 | 0.60 |
| 10 | 0.92 | 0.81 |
| 12 | 0.80 | 0.67 |
| 14 | 0.51 | 0.39 |
| 16 | 0.65 | 0.43 |
Quantification of a distance between the ‘in-cell’ PDFs simulated using FRET NPS tools (22) for A- and B-form of DNA and ‘reference’ interprobe distance PDFs derived from in-cell FRET measurements using the Kolmogorov–Smirnov (KS) statistic. Identical distributions are marked by 0 value in the KS statistics. Note that B-DNA model has lower KS values compared to A-DNA model for all the DNA constructs indicating the B-form of DNA to be a more likely conformation in intact bacterial cells compared to A-DNA.
DISCUSSION
In the present study, we investigated the variation of EFRET for a series of five DNA duplexes with a length of 8–16 bp terminally labeled with ATTO680 and ATTO740 donor–acceptor pair in a solution and in living E. coli cells. Our measurements showed that for the ATTO680/740 donor–acceptor system, the fluorophore properties in the cellular environment differed from those acquired under in vitro conditions. While it is experimentally impossible to acquire all spectral properties of the fluorophores for individual molecules under heterogeneous conditions of a living cell, our measurements indicated that these could be approximated by their measurements using a bulk amount of DNA mixed in a crude cellular homogenate.
Comparison between in vitro R0 value distribution and that in crude bacterial lysate indicated that the precision for the interprobe distance determination from EFRET values acquired in vivo was significantly lower (∼31%) compared to that acquired in diluted buffers (∼13%).
The low level of precision of in vivo-derived distances limits the ab initio structure analysis based on acquired in-cell EFRET data and makes NA structure characterization from in-cell FRET data dependent on probabilistic evaluation. Still, the in-cell EFRET data can be useful to assess the consistency of structural data acquired from EFRET values measured in a complex cellular environment with various structural models. Comparison of interprobe distance PDF derived from experimentally acquired EFRET values in intact bacterial cells and R0 values corresponding to crude bacterial homogenate with the simulated PDFs corresponding to A- and B-forms of DNA indicated the B-form of DNA to be a more likely conformation in intact bacterial cells compared to A-DNA. This finding is in agreement with recent data on conformation of double-helical DNA oligonucleotides inside living eukaryotic cells derived using pulsed electron–electron double-resonance spectroscopy(3,4).
Considering the inherently low precision of in-cell spFRET measurements, we attempted to assess the application potential of the in-cell spFRET for structural analysis of NAs. We simulated several typical situations encountered in DNA structure analysis such as B–A and B–Z transitions, or structural deformations in DNA due to oxidative DNA damage, protein–DNA binding or DNA–drug interactions (for details see Supplementary Figure S7). Using nano-positional models for fluorophore locations derived based on structural information from the PDB database and the derived precision of interprobe distance determination under in-cell-like conditions, we constructed the model interprobe distance PDFs. Comparison of the model interprobe distance PDFs for B- and Z-DNA, as well as for B-DNA and DNA containing α-anomeric adenosine, which are representative of the damaged right-handed double-helical DNA marked by a minor groove bend, revealed that the structural changes corresponding to the B–Z transition or the structural DNA damage couldn’t be readily resolved using in-cell spFRET measurements (Supplementary Figure S7 and Supplementary Table S5). However, comparison between the model PDFs for DNA bound to protein, namely E. coli endonuclease IV, or to base-binding drugs, namely 2-amino-1-methyl-6-phenylimidazo[4,5-2 B]pyridine and cisplatin, and PDF for a naked DNA revealed the same or larger differences than observed for analyzed B–A transition (Supplementary Table S5), which is suggestive that in-cell spFRET could find its applications in structural studies involving DNA–drug and/or protein binding. Nevertheless, the low resolving power of in-cell EFRET implies that for unbiased in vivo structural analysis of NAs, the in-cell FRET should be combined with other in vivo high-resolution techniques such as in-cell NMR or PELDOR (2–4).
Table 3 compares limitations and application potential of the in-cell spFRET with other established techniques for in vivo structural characterization of DNA, namely in-cell NMR and in-cell PELDOR. While both in-cell NMR and PELDOR have higher resolution and precision, respectively, compared to in-cell spFRET, their main drawback lies in the requirement of ultra-high concentrations of exogenous NA that needs to be deposited into the living cells (Figure 1). At the moment, the requirement of ultra-high concentration of exogenous NA for in-cell NMR and PELDOR represents the main source of controversy in the field of cellular structural biology of NAs, as the introduced bulk amount of NA might alter the environmental conditions in the cell and, therefore, affect the structure of the studied molecule. One of the known artifacts of the ultra-high concentration of introduced DNA is the dimerization of double-stranded DNA fragments introduced in X. laevis cells (3). This artifact would be naturally avoided in the in-cell spFRET setup.
Table 3.
A comparison of in-cell spFRET with other techniques for the in vivo structural characterization of DNA
| In-cell NMR | In-cell PELDOR | In-cell spFRET | |
|---|---|---|---|
| Disturbance of native environment | Yes | Yes | No |
| Cell type | X. laevis egg/oocyte | X. laevis egg/oocyte | E. coli, mammalian cellsa |
| Toxicity | Sequence dependentb | Sequence dependentb | No |
| Subcellular localization | Nucleus/cytosolc | Nucleus/cytosolc | Nucleusd |
| Tag requirement | No | Yes | Yes |
| Measurement time span | Hours | < 70 mine | Hours |
| Structural informationf | Short-range | Long-range | Long-range |
aIllustrated here for human epitheloid carcinoma (HeLa) cells (Supplementary Figure S9).
bSee the text for details.
cFor both in-cell NMR and PELDOR, the issue of the intracelllular localization of introduced DNA has not been properly addressed in the literature. In X. laevis, localization experiments using fluorescently-labeled DNA mini-haiprin suggest that ∼90% of introduced DNA is localized in the nucleus and ∼10% in the cytosol (41).
dApplies for mammalian cells (37–40).
fShort-range—typically < 5 Å; long range—typically < 50 Å.
Another consequence of the required ultra-high concentrations of exogenous NA for in-cell NMR and PELDOR is that these techniques are currently limited to X. laevis oocytes and eggs, which are large cells that can be mechanically injected with bulk amount of DNA. In contrast to in-cell NMR and PELDOR, the in-cell spFRET measurements can in principle be performed in both prokaryotic as well as eukaryotic cells. To demonstrate that the in-cell spFRET setup is compatible not only with E. coli cells but also with mammalian cells, we performed in-cell spFRET measurements in human epitheloid carcinoma (HeLa) cells (Supplementary Figure S9). One of the foreseen applications of the in-cell FRET thus could be its use for studies aimed at evaluating the influence of cell-type specific environments on DNA structure. In this respect, however, it needs to be emphasized that not all cell types are amenable to in-cell spFRET measurement. The cell-type choice for in-cell spFRET is generally limited by three factors: the cell’s autofluorescence, light scattering, and size. For example, large cells, such as the X. laevis oocytes/eggs used for in-cell NMR/PELDOR, cannot readily be used for in-cell spFRET measurements because they have high autofluorescence and their size allows focusing only on single molecules close to the cell surface.
One of the important advantages of the in-cell spFRET (in eukaryotic cells) over both in-cell NMR/PELDOR is physiologically relevant localization of the introduced DNA. Under native conditions, i.e. inside living cell, the DNA is localized in the nucleus. Although low concentrations of exogenous DNA introduced into cell’s cytoplasm tend to spontaneously localize and concentrate in the nucleus (37–40), the nuclei of X. laevis oocytes seems unable to accommodate the large quantities of introduced DNA that are required for both in-cell NMR and PELDOR measurements (41). Consequently, the structural information acquired from in-cell NMR/PELDOR is a superposition of information from both the physiologically relevant nucleus and the non-physiologically relevant cytosol (for details see Table 3).
Abstracting from the requirement that an ultra-high concentration of DNA be deposited into the interior of the cells and from the restriction to the X. laevis system, the in-cell PELDOR represents an analogy to the proposed in-cell FRET technique. Both in-cell spFRET and PELDOR provide long-range structural information. Both techniques also require the attachment of reporter tags, namely, fluorophores for FRET and radicals for PELDOR. However, compared with in-cell PELDOR, the in-cell spFRET setup offers one particularly important advantage, namely extended measurement time. Whereas the in-cell spFRET measurement time is primarily limited by the life span of the cell (typically several hours for the ATTO680/ATTO740 donor–acceptor system used in this study), the in-cell PELDOR measurement time is typically limited to ∼70 min due to the rapid reduction of the spin labels by the cellular environment (3,4,42).
Last but not least, the introduction of exogenous NA into cells at the single-molecule level is non-toxic to the cells. While the introduction of single molecules into the cell does not impose any toxicity to the cell regardless of the nature of the nucleic acid fragment, it was observed by our group (unpublished data) and others (43) that the introduction of large quantities of specific DNA motifs might be toxic to the cells. Although reduction of the amount of DNA can diminish its toxicity, the DNA concentrations required to avoid toxicity are far below the detection limits of both in-cell NMR and in-cell PELDOR. In this respect, the in-cell spFRET might be regarded as the only alternative to study DNA molecules that display toxicity at high concentrations.
Taken together, the inherent lack of precision appears to be the main disadvantage of in-cell spFRET. Future development of the in-cell spFRET technique thus should include surveying the effect of the cellular environment on other donor–acceptor systems with particular attention paid to anchoring fluorophores to the NAs to increase precision of interprobe distance determination based on in-cell spFRET data.
Nonetheless, the fact that the FRET measurements in vivo can be achieved on single molecules makes the in-cell spFRET the only technique allowing in vivo quantitative characterization of NA structure without significant disturbance of the native cellular environment. Performing quantitative spFRET measurements compared to bulk in-cell FRET measurements avoids the need to introduce bulk amounts of DNA into the cells and the use of chemical transfectants, which both disturb composition of the intracellular environment. Given the unique features of the in-cell spFRET setup, this technique has the potential to become an important tool to investigate the role of intracellular environment composition variations on the structure of NA through the cell-cycle progression.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–5, Supplementary Figures 1–9, Supplementary Theory and Methods and Supplementary References [44–48].
FUNDING
The Grant Agency of the Academy of Sciences of the Czech Republic [KAN200100801, AV0Z50510513]; Ministry of Education of the Czech Republic [MSM6007665808]. Funding for open access charge: Faculty of Science, University of South Bohemia.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Prof. Petr Smilauer for stimulating discussions.
REFERENCES
- 1.Ito Y, Selenko P. Cellular structural biology. Curr. Opin. Struct. Biol. 2010;20:640–648. doi: 10.1016/j.sbi.2010.07.006. [DOI] [PubMed] [Google Scholar]
- 2.Hansel R, Foldynova-Trantirkova S, Lohr F, Buck J, Bongartz E, Bamberg E, Schwalbe H, Dotsch V, Trantirek L. Evaluation of parameters critical for observing nucleic acids inside living Xenopus laevis oocytes by in-cell NMR spectroscopy. J. Am. Chem. Soc. 2009;131:15761–15768. doi: 10.1021/ja9052027. [DOI] [PubMed] [Google Scholar]
- 3.Krstic I, Hansel R, Romainczyk O, Engels JW, Dotsch V, Prisner TF. Long-range distance measurements on nucleic acids in cells by pulsed EPR spectroscopy. Angew. Chem. Int. Edit. 2011;50:5070–5074. doi: 10.1002/anie.201100886. [DOI] [PubMed] [Google Scholar]
- 4.Azarkh M, Okle O, Singh V, Seemann IT, Hartig JS, Dietrich DR, Drescher M. Long-range distance determination in a DNA model system inside Xenopus laevis oocytes by in-cell spin-label EPR. Chembiochem. 2011;12:1192–1195. doi: 10.1002/cbic.201100281. [DOI] [PubMed] [Google Scholar]
- 5.Selvin PR. The renaissance of fluorescence resonance energy transfer. Nat. Struct. Biol. 2000;7:730–734. doi: 10.1038/78948. [DOI] [PubMed] [Google Scholar]
- 6.Stryer L. Fluorescence energy-transfer as a spectroscopic ruler. Annu. Rev. Biochem. 1978;47:819–846. doi: 10.1146/annurev.bi.47.070178.004131. [DOI] [PubMed] [Google Scholar]
- 7.Stryer L, Haugland RP. Energy transfer - a spectroscopic ruler. Proc. Natl Acad. Sci. USA. 1967;58:719–726. doi: 10.1073/pnas.58.2.719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hurley DJ, Tor Y. Donor/acceptor interactions in systematically modified Ru-II-Os-II oligonucleotides. J. Am. Chem. Soc. 2002;124:13231–13241. doi: 10.1021/ja020172r. [DOI] [PubMed] [Google Scholar]
- 9.Lilley DMJ, Wilson TJ. Fluorescence resonance energy transfer as a structural tool for nucleic acids. Curr. Opin. Chem. Biol. 2000;4:507–517. doi: 10.1016/s1367-5931(00)00124-1. [DOI] [PubMed] [Google Scholar]
- 10.Woodside MT, Anthony PC, Behnke-Parks WM, Larizadeh K, Herschlag D, Block SM. Direct measurement of the full, sequence-dependent folding landscape of a nucleic acid. Science. 2006;314:1001–1004. doi: 10.1126/science.1133601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wozniak AK, Schroder GF, Grubmuller H, Seidel CAM, Oesterhelt F. Single-molecule FRET measures bends and kinks in DNA. Proc. Natl Acad. Sci. USA. 2008;105:18337–18342. doi: 10.1073/pnas.0800977105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ishii M, Ikushima M, Kurachi Y. In vivo interaction between RGS4 and calmodulin visualized with FRET techniques: possible involvement of lipid raft. Biochem. Biophys. Res. Commun. 2005;338:839–846. doi: 10.1016/j.bbrc.2005.10.026. [DOI] [PubMed] [Google Scholar]
- 13.Gaibelet G, Planchenault T, Mazeres S, Dumas F, Arenzana-Seisdedos F, Lopez A, Lagane B, Bachelerie F. CD4 and CCR5 constitutively interact at the plasma membrane of living cells: a confocal fluorescence resonance energy transfer-based approach. J. Biol. Chem. 2006;281:37921–37929. doi: 10.1074/jbc.M607103200. [DOI] [PubMed] [Google Scholar]
- 14.Chilibeck KA, Wu T, Liang C, Schellenberg MJ, Gesner EM, Lynch JM, MacMillan AM. FRET analysis of in vivo dimerization by RNA-editing enzymes. J. Biol. Chem. 2006;281:16530–16535. doi: 10.1074/jbc.M511831200. [DOI] [PubMed] [Google Scholar]
- 15.Schoen I, Krammer H, Braun D. Hybridization kinetics is different inside cells. Proc. Natl Acad. Sci. USA. 2009;106:21649–21654. doi: 10.1073/pnas.0901313106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Uchiyama H, Hirano K, Kashiwasake-Jibu M, Taira K. Detection of undegraded oligonucleotides in vivo by fluorescence resonance energy transfer. J. Biol. Chem. 1996;271:380–384. doi: 10.1074/jbc.271.1.380. [DOI] [PubMed] [Google Scholar]
- 17.Treutlein B, Michaelis J. Direct observation of single RNA polymerase processing through a single endogenous gene in a living yeast cell. Angew. Chem. Int. Ed. Engl. 2011;50:9788–9790. doi: 10.1002/anie.201103809. [DOI] [PubMed] [Google Scholar]
- 18.Enderlein J, Erdmann R. Fast fitting of multi-exponential decay curves. Opt. Commun. 1997;134:371–378. [Google Scholar]
- 19.Huang F, Lerner E, Sato S, Amir D, Haas E, Fersht AR. Time-resolved fluorescence resonance energy transfer study shows a compact denatured state of the B domain of protein A. Biochemistry. 2009;48:3468–3476. doi: 10.1021/bi801890w. [DOI] [PubMed] [Google Scholar]
- 20.Williams ATR, Winfield SA, Miller JN. Relative fluorescence quantum yields using a computer-controlled luminescence spectrometer. Analyst. 1983;108:1067–1071. [Google Scholar]
- 21.Dale RE, Eisinger J, Blumberg WE. Orientational freedom of molecular probes - orientation factor in intra-molecular energy-transfer. Biophys. J. 1979;26:161–193. doi: 10.1016/S0006-3495(79)85243-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Muschielok A, Andrecka J, Jawhari A, Bruckner F, Cramer P, Michaelis J. A nano-positioning system for macromolecular structural analysis. Nat. Methods. 2008;5:965–971. doi: 10.1038/nmeth.1259. [DOI] [PubMed] [Google Scholar]
- 23.Sindbert S, Kalinin S, Nguyen H, Kienzler A, Clima L, Bannwarth W, Appel B, Muller S, Seidel CA. Accurate distance determination of nucleic acids via Forster resonance energy transfer: implications of dye linker length and rigidity. J. Am. Chem. Soc. 2011;133:2463–2480. doi: 10.1021/ja105725e. [DOI] [PubMed] [Google Scholar]
- 24.Clifford JN, Bell TDM, Tinnefeld P, Heilemann M, Melnikov SM, Hotta J, Sliwa M, Dedecker P, Sauer M, Hofkens J, et al. Fluorescence of single molecules in polymer films: Sensitivity of blinking to local environment. J. Phys. Chem. B. 2007;111:6987–6991. doi: 10.1021/jp072864d. [DOI] [PubMed] [Google Scholar]
- 25.Fessl T, Ben-Yaish S, Vacha F, Adamec F, Zalevsky Z. Depth of focus extended microscope configuration for imaging of incorporated groups of molecules, DNA constructs and clusters inside bacterial cells. Opt. Commun. 2009;282:2495–2501. [Google Scholar]
- 26.Wang JC. Helical repeat of DNA in solution. Proc. Natl Acad. Sci. USA. 1979;76:200–203. doi: 10.1073/pnas.76.1.200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kypr J, Chladkova J, Zimulova M, Vorlickova M. Aqueous trifluorethanol solutions simulate the environment of DNA in the crystalline state. Nucleic Acids Res. 1999;27:3466–3473. doi: 10.1093/nar/27.17.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Minyat EE, Ivanov VI, Kritzyn AM, Minchenkova LE, Schyolkina AK. Spermine and spermidine-induced B to A transition of DNA in solution. J. Mol. Biol. 1979;128:397–409. doi: 10.1016/0022-2836(79)90094-9. [DOI] [PubMed] [Google Scholar]
- 29.Xu Q, Shoemaker RK, Braunlin WH. Induction of B-A transitions of deoxyoligonucleotides by multivalent cations in dilute aqueous solution. Biophys. J. 1993;65:1039–1049. doi: 10.1016/S0006-3495(93)81163-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Robinson H, Wang AH. Neomycin, spermine and hexaamminecobalt (III) share common structural motifs in converting B- to A-DNA. Nucleic Acids Res. 1996;24:676–682. doi: 10.1093/nar/24.4.676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Brahms J, Mommaerts WF. A study of conformation of nucleic acids in solution by means of circular dichroism. J. Mol. Biol. 1964;10:73–88. doi: 10.1016/s0022-2836(64)80029-2. [DOI] [PubMed] [Google Scholar]
- 32.Ivanov VI, Minchenkova LE, Minyat EE, Frank-Kamenetskii MD, Schyolkina AK. The B to A transition of DNA in solution. J. Mol. Biol. 1974;87:817–833. doi: 10.1016/0022-2836(74)90086-2. [DOI] [PubMed] [Google Scholar]
- 33.Vorlickova M, Minyat EE, Kypr J. Cooperative changes in the chiroptical properties of DNA induced by methanol. Biopolymers. 1984;23:1–4. doi: 10.1002/bip.360230102. [DOI] [PubMed] [Google Scholar]
- 34.Florentiev VL, Ivanov VI. RNA polymerase: two-step mechanism with overlapping steps. Nature. 1970;228:519–522. doi: 10.1038/228519a0. [DOI] [PubMed] [Google Scholar]
- 35.Beabealashvily RS, Ivanov VI, Minchenkova LE, Savotchkina LP. RNA polymerase-DNA complexes. I. The study of the conformation of nucleic acids at the growing point of RNA in an RNA polymerase-DNA system. Biochim. Biophys. Acta. 1972;259:35–40. doi: 10.1016/0005-2787(72)90471-6. [DOI] [PubMed] [Google Scholar]
- 36.Setlow P. DNA in dormant spores of Bacillus species is in an A-like conformation. Mol. Microbiol. 1992;6:563–567. doi: 10.1111/j.1365-2958.1992.tb01501.x. [DOI] [PubMed] [Google Scholar]
- 37.Chin DJ, Green GA, Zon G, Szoka FC, Jr, Straubinger RM. Rapid nuclear accumulation of injected oligodeoxyribonucleotides. New Biologist. 1990;2:1091–1100. [PubMed] [Google Scholar]
- 38.Leonetti JP, Mechti N, Degols G, Gagnor C, Lebleu B. Intracellular distribution of microinjected antisense oligonucleotides. Proc. Natl Acad. Sci. USA. 1991;88:2702–2706. doi: 10.1073/pnas.88.7.2702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Clarenc JP, Lebleu B, Leonetti JP. Characterization of the nuclear binding sites of oligodeoxyribonucleotides and their analogs. J. Biol. Chem. 1993;268:5600–5604. [PubMed] [Google Scholar]
- 40.Fisher TL, Terhorst T, Cao X, Wagner RW. Intracellular disposition and metabolism of fluorescently-labeled unmodified and modified oligonucleotides microinjected into mammalian cells. Nucleic Acids Res. 1993;21:3857–3865. doi: 10.1093/nar/21.16.3857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hansel R. 2009. Evaluation of parameters critical for observing nucleic acids inside living Xenopus laevis oocytes by in-cell NMR spectroscopy. PhD thesis - Johan Wolfgang Goethe University, Frankfurt am Main, Germany. [DOI] [PubMed] [Google Scholar]
- 42.Azarkh M, Okle O, Eyring P, Dietrich DR, Drescher M. Evaluation of spin labels for in-cell EPR by analysis of nitroxide reduction in cell extract of Xenopus laevis oocytes. J. Magn. Reson. 2011;212:450–454. doi: 10.1016/j.jmr.2011.07.014. [DOI] [PubMed] [Google Scholar]
- 43.Sedoris KC, Thomas SD, Clarkson CR, Muench D, Islam A, Singh R, Miller DM. Genomic c-Myc quadruplex DNA selectively kills leukemia. Mol. Cancer Ther. 2012;11:66–76. doi: 10.1158/1535-7163.MCT-11-0515. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.