Abstract
Phage display has been instrumental in discovery of novel binding peptides and folded domains for the past two decades. We recently reported a novel pIX phagemid display system that is characterized by a strong preference for phagemid packaging combined with low display levels, two key features that support highly efficient affinity selection. However, high diversity in selected repertoires are intimately coupled to high display levels during initial selection rounds. To incorporate this additional feature into the pIX display system, we have developed a novel helper phage termed DeltaPhage that allows for high-valence display on pIX. This was obtained by inserting two amber mutations close to the pIX start codon, but after the pVII translational stop, conditionally inactivating the helper phage encoded pIX. Until now, the general notion has been that display on pIX is dependent on wild-type complementation, making high-valence display unachievable. However, we found that DeltaPhage does facilitate high-valence pIX display when used with a non-suppressor host. Here, we report a side-by-side comparison with pIII display, and we find that this novel helper phage complements existing pIX phagemid display systems to allow both low and high-valence display, making pIX display a complete and efficient alternative to existing pIII phagemid display systems.
INTRODUCTION
Phage display is a powerful technology for the discovery of novel binding peptides and folded domains (1,2). By creating libraries of polypeptides fused to coat proteins on a filamentous phage, one may simultaneously screen >1010 unique sequences for binding to a specific target. To display a polypeptide, the protein of interest (POI) is normally placed in-frame between an N-terminal signal sequence targeting the bacterial periplasm and the mature form of either the minor coat protein pIII, or the major coat protein pVIII (1). The resulting POI coat protein fusions may be encoded either in a complete phage genome usually resulting in multivalent display, or on a phagemid resulting in low-valence display (3). In comparison with phage genome vectors, a phagemid has increased genetic stability and a wider variety of fusion partners may be displayed (1). Due to its ease of handling and smaller molecular size that translate to increased transformation frequencies, the phagemid format is often the display system of choice (3). As the phagemid lacks all phage genes except the capsid protein selected for display, a helper phage is used to provide the genetic information required for phage production (3).
Phage display technology has gained particularly widespread use for the discovery of novel antibody specificities (4,5) and for display and selection of antibody fragments, pIII appears advantageous over pVIII systems (6). Low-valence display is desirable for applications such as high-affinity selection and in vitro affinity maturation where the highest possible monomeric affinity is the goal (7). In comparison, high-valence display facilitates increased retrieval of specific binders during selection (8), it may be used to enrich for open reading frames (ORFs) (9), and it has been used for selection of stabilized antibody fragments (10). High-valence display has also shown beneficial when selecting for internalizing antibodies directed against cell-surface-bound receptors (11). Therefore, an optimal and versatile display system should take advantage of the combined strengths of high and low-valence display. Indeed, systems have been developed to allow for this by the construction of modified helper phages with defective or deleted pIII genes (12–14).
However, pIII is responsible for bacterial host infection, hence fusions to this protein adversely affects infectivity and thereby putative retrieval during selection, and this concern applies especially to high-valence display formats (15). pIII fusions also affect phage propagation translating to bias in selected repertoires (7,16,17). Furthermore, many display systems, and in particular smaller fusions such as antibody scFv display, suffer from heterogeneous oligovalency effects masking true monomeric affinity. The latter effect may result in the preferential retrieval of low affinity binders during selection on polyvalent antigens (18–20).
Alternative display routes have therefore been explored as substitutes to the widely used pIII display. In particular pIX that is found in the same stoichiometry on the phage virion as pIII (about five copies/phage) has been shown to work for affinity selection of both antibody scFv and Fab fragments based on phagemid display (21–24). We recently reported a further modification of pIX display exploiting the N-terminal signal sequence-independent nature of the native pIX for phage integration (25). This phagemid display system is characterized by a low display level in conjunction with robust phagemid packaging, translating onto very efficient affinity selection. As high diversity in selected repertoires is intimately coupled to high display levels during initial selection rounds (8), POI display on all copies of pIX would further broaden the area of application for pIX phagemid display. However, previous reports indicate that folded domain POI display on pIX is strictly dependent on wild-type (wt) complementation, which would make high-valence display unattainable (26–28).
To the contrary, we here demonstrate the use of a novel helper phage termed DeltaPhage that indeed allows for high-valence POI display on pIX. This was achieved by disrupting the pIX ORF in the M13K07 helper phage genome with amber stop codons leading to translational stop in a non-suppressor host, while pIX production, and therefore helper phage production, is allowed in a supE suppressor host that inserts a glutamine residue where non-suppressor hosts terminate translation (29). When used for pIX display phagemid rescue in a non-suppressor host, DeltaPhage therefore results in high-valence POI display as the phagemid is the only source of pIX. No deleterious effects on phagemid titer or phenotype is observed, but a markedly increased antigen reactivity with the DeltaPhage-rescued phages is seen due to avidity effects when tested in both target specific phage capture ELISA and in binding to a cell surface receptor. Phagemid rescue with modified helper phages for high-valence pIII display usually gives 10- to 1000-fold lower end titers than rescue with classical helper phages such as M13K07 (3,30). We do not observe such a decrease in phagemid end titers with our novel helper phage, which makes it easy to obtain highly diverse libraries without the need to upscale production volumes. Neither do we see reduction in infectivity, as no modifications are done to the infection-mediating pIII. In addition, we observe very high phagemid to helper phage packaging ratios and thus less clone dependent bias. Compared in a side-by-side model selection, high-valence scFv display on pIX produced with DeltaPhage led to markedly increased selection efficiency compared to the low-valence counterpart and on par with that seen with high-valence pIII display. In summary, we here report the first pIX display system capable of fully substituting for both low and high-valence pIII display, but devoid of any pIII modifications.
MATERIALS AND METHODS
Phagemids, helper phages, bacterial strains
The pSEX, pFKPDN, pGALD9ΔL and pGALD9ΔLFN phagemids harboring either of the two scFvs specific for the haptens 4-ethoxymethylene-2-phenyl-2-oxazolin-5-one (phOx) or 4-hydroxy-3-iodo-5-nitrophenylacetate (NIP), or the scTCR 4B1A1 has been described previously (25,31). The pFAB-Display phagemid harboring the anti-phOx Fab was a kind gift from Affitech AS (Oslo, Norway). The M13K07, VCSM13 and Hyperphage helper phages were purchased from GE Healthcare (Uppsala, Sweden), New England Biolabs (Ipswich, MA, USA) and Progen Biotechnik GmbH (Heidelberg, Germany), respectively. The Escherichia coli strains XL1-Blue {endA1 gyrA96(nalR) thi-1 recA1 relA1 lac glnV44 F′[::Tn10 proAB+ lacIq Δ(lacZ)M15] hsdR17(rK− mK+)} and TOP10F′ {F′ [lacIq Tn10(tetR)] mcrA Δ(mrr–hsdRMS–mcrBC) φ80lacZΔM15 ΔlacX74 deoR nupG recA1 araD139 Δ(ara–leu)7697 galU galK rpsL(StrR) endA1 λ−} were purchased from Stratagene (LaJolla, CA, USA) and Life Technologies (CA, USA), respectively, whereas MC1061 {F− Δ(ara–leu)7697 [araD139]B/r Δ(codB–lacI)3 galK16 galE15 λ− e14− mcrA0 relA1 rpsL150(strR) spoT1 mcrB1 hsdR2(r-m+)} was obtained from Dr G.P. Smith (Division of Biological Sciences, University of Missouri, USA).
Cell lines, antibodies and materials
The murine T cell clones 4B2A1 and 7A10B2 have been described previously (32). The mAbs GB113 (mIgG2a) specific for the 4B2A1 T cell receptor and AB10 (mIgG2a, unpublished data) were purified from cell supernatant on protein G-sepharose (GE Healtcare, Uppsala, Sweden) and biotinylated according to standard protocols (33). The Absolutely RNA Miniprep Kit was purchased from Stratagene (La Jolla, CA, USA). Sheep-anti M13-HRP and biotinylated rabbit anti-fd antibodies were purchased from GE Healthcare (Uppsala, Sweden) and Sigma-Aldrich (Oslo, Norway), respectively. Streptavidin–PE was obtained from BD Pharmingen (San Diego, CA, USA). Bovine serum albumin (BSA) was purchased from Sigma-Aldrich (Oslo, Norway). Trypsin/EDTA was purchased from BioWhittaker (Lonza Group Ltd., Visp, Switzerland). Restriction Enzymes (RE) and T4 DNA ligase, and RNase H were purchased from New England Biolabs (Ipswich, MA, USA). Pfu Turbo and Phusion DNA polymerases were purchased from Stratagene (LaJolla, CA, USA) and Sigma-Aldrich (Oslo, Norway), respectively. SuperScript II Reverse Transcriptase and rTerminal Transferase were purchased from Roche (Penzberg, Germany) and Invitrogen (Carlsbad, CA, USA), respectively. DNA oligonucleotides were purchased from Eurofins MWG operon (Ebersberg, Germany) and Sigma-Aldrich (Oslo, Norway). The pCR-Blunt II TOPO plasmid was purchased from Invitrogen (Carlsbad, CA, USA). The haptens NIP and phOx conjugated to BSA were prepared essentially as described (34,35). All media and buffers were prepared essentially as described (36).
Mutagenesis of the M13K07 helper phage genome
The amber mutations in the pIX ORF in M13K07 was introduced by QuikChange in vitro mutagenesis according to the manufacturer’s protocol (Stratagene, LaJolla, CA, USA), using the oligonucleotides pIX_amber_F/pIX_amber_R (5′-GCTGGGGGTCAAAGATGAGTTAGAGCTAGGTTTTAGTGTATTCTTTCGC-3′/5′-GCGAAAGAATACACTAAAACCTAGCTCTAACTCATCTTTGACCCCCAGC-3′, inserted bases underlined), and introduced to E. coli MC1061 by electroporation. Successful introduction of the mutation was verified by sequencing. To ensure a clean vector background, the modified pIX fragment was moved into the M13K07 genome using the BsrGI and SnaBI RE sites using standard techniques, followed by heat shock transformation of E. coli XL1-Blue. Both the modified and unmodified M13K07 were introduced into E. coli TOP10F′ cells by transduction with virions produced in the XL1-Blue cells.
Construction of and subcloning of the Fab-pIX display phagemids
The pFAB-Display phagemid is built on the pSEX81 backbone (GenBank accession no: Y14584). The NcoI/HindIII variable heavy (VH) and MluI/NotI variable light (VL) gene cassettes are preserved and these segments are fused to hIgG1 CH1 (HindIII/SpeI cassette) and hCκ (MluI/EcoRV cassette), respectively, as two consecutive separate ORFs of which the latter is in-frame to pIII (31). The pIX encoding sequence was PCR amplified from M13K07 using the primers pIX_EcoRV/pIX_NheI (5′-ATATGATATCAGAATGAGTGTTTTAGTGTATTCTTTCGCC-3′/5′-ATATGCTAGCTTATCATGAGGAAGTTTCCATTAAACGGG-3′) and moved into the pFAB-Display phagemid on compatible EcoRV/NheI RE sites, thereby exchanging the pIII encoding region, resulting in a N-terminal in-frame fusion to pIX of the upstream MluI/NotI-defined VLCκ-cassette. The pelB leader upstream of the VL segment was removed and the original MluI site moved 5′-prime of the ORF by exchanging a SpeI/BmgBI segment with a synthetic fragment harboring the changes (Genscript, CA, USA) using standard techniques. The resulting phagemid was transformed into E. coli XL1-Blue and confirmed by sequencing.
Helper phage and phagemid packaging and titration
Small-scale packaging of E. coli XL1-Blue or TOP10F′ harboring the DeltaPhage genome was done in 10 ml 2× YT containing 50 µg/ml kanamycin using sterile 50-ml tubes, and incubation at 37°C overnight with agitation. The cells were pelleted by centrifugation and the supernatant was sterile filtered into fresh 50 ml tubes using 0.2 µm filters followed by infectious titration. Up-scaled helper phage packaging was done in 1–2 l baffled shaker flasks using 200 ml volumes and rigorous agitation. The virions were purified and concentrated by PEG/NaCl precipitation as described (37). Virion concentration was determined by optical density (OD) by the formula: virions/ml = [(A269nm − A320nm) × 6.083 × 1016]/genome size (38). Phagemid rescue from E. coli XL1-Blue and TOP10F′ was done essentially as described (39) using a helper phage MOI20. Where applicable, virions were purified and concentrated by PEG/NaCl precipitation from 50 ml sterile filtered supernatant and resuspended in 1 ml PBS, pH 7.4. Virion assembly was monitored by infectious spot titration as described (40) with the exception of Hyperphage-rescued samples that required a modified titration protocol (41).
Agarose gel electrophoresis of intact virions
Samples of equal volume normalized to 1 × 1012/ml PEG precipitated phages (determined by OD as described above) of M13K07 and DeltaPhage, respectively, were separated for 3 h at 60 V at RT on a 1% agarose gel in Tris–Acetate–EDTA (TAE) buffer. The gel was then incubated with 0.2 M NaOH for 1 h, rinsed with dH2O and neutralized with 1 M Tris–HCl, pH 7.0 for 15 min. The ssDNA bands from denatured phage virions were visualized by 30 min incubation with SYBR Safe DNA gel stain (Invitrogen, Carlsbad, CA, USA) solution (1:10 000 v/v in dH2O) and imaged on a Bio-Rad Gel Doc 2000 work station (Bio-Rad, Hercules, CA, USA).
Electron microscopy
Samples normalized to 1 × 1010/ml were agitated for 10 s on a vortex shaker. An amount of 5 µl drops of each sample were applied to a clean surface and 100 mesh copper grids placed on top of the drops in order to absorb the particles for 5 min. The grids were then washed for 5 min on a total of three drops of PBS pH 7.4 followed by a 10 min wash on six drops of triple distilled water. The samples were subsequently stained with a 4% aqueous uranyl–acetate solution for 30 s and air-dried before observation in a Philips CM200 transmission electron microscope at 80 kV. Images were recorded at a magnification of 25 000× and a measuring grid was overlayed. Intersections with the grid were counted and the length of each particle was calculated using π/4 * I * d (I being the number of intersections and d being the distance between the grid lines) (42).
Phage capture ELISA
The two antigens NIP-BSA and phOx-BSA were adsorbed to MaxiSorpTM microtiter plate wells (Nunc, Roskilde, Denmark) in concentrations of 5–10 µg/ml in PBS, pH 7.4 ON at 4°C. The wells were blocked with PBSTM (PBS supplemented with 0.05% v/v Tween 20 and 4% w/v skim milk) for 1 h at room temperature (RT). Phage preparations were then added and allowed to react for 1.5 h at RT before captured phages were detected with anti M13-HRP (1:5000) for 1 h at RT. A 3× washing step with PBST (PBS supplemented with 0.05% v/v Tween 20) was applied between each incubation step. The wells were developed with TMB soluble (Merck KGaA, Darmstadt, Germany), and the reaction terminated by addition of 1 M HCl, equilibrated, and the absorbance read at A450nm.
RACE cloning and construction of a mAb GB113 derived scFv for phage display
GB113 cells frozen on nitrogen were thawed quickly before being pelleted and subjected to total RNA isolation using the Absolutely RNA Miniprep Kit. First strand cDNA was synthesized with SuperScript II Reverse Transcriptase using 500 ng total RNA as template and 2 pmol of primer (for light chain: primer IgκC cDNA (5′-TGCTGTCTTTGCTGTCCTGAT-3′) and for heavy chain: primer IgG2a cDNA (5′-TAG AGG TCA GAC TGC AGG ACA-3′). The RNA was removed by RNase H treatment before the cDNA was subjected to poly dCTP 3′-tailing with rTerminal Transferase. The VL and VH genes were then retrieved by PCR using the primers PolyG_NotI_fwrd: 5′-ATATGCGGCCGCGGGGGGGGGGGGGGGG-3′ (both VL and VH), 3′IgκC_rev: 5′-CAAGAAGCACACGACTGAGGC (VL only) and 3′Ig2a_rev: 5′-CTTGACCAGGCATCCTAGAGTCA-3′ (VH only). The amplified fragments were then blunt cloned into the vector pCR-Blunt II TOPO and sequenced. New primers were designed for amplification of the VL and VH genes to exclude the leader sequences and constant regions. The VL and VH genes were amplified with the following primer pairs: GB113_IGKV_scFv_fw (5′-ATCCATGGCCCAAATTGTTCTCACCCAGTCTCCA-3′) and GB113_IGKV_scFv_rv (5′-ATATATAAGCTTTTTCAGCTCCAGCTTGGTCC-3′), and GB113_IGHV_scFv_fw (5′-ATATACGCGTACAGATCCAGTTGGTACAGTCTGG-3′) and GB113_IGHV_scFv_rv (5′-ATATGCGGCCGCTGAGGAGACTGTGAGAGTGGT-3′). After PCR amplification the scFv construct was assembled by sequential cloning of the GB113 VL and VH genes into the both the two phagemids pFKPDN and pGALD9ΔL using the NcoI/HindIII and MluI/NotI sites, respectively.
Flow cytometry
4B2A1 and 7A10B2 T cells frozen on nitrogen were thawed on ice, washed once in PBS and aliquots of 2 × 105 cells were distributed into a V-shaped 96-well dish (NUNC, Roskilde, Denmark). The total volumes were adjusted to 250 µl/well with 5% w/v BSA/PBS (pH 7.4). The plate was centrifuged at 300g/5 min at RT and the supernatants discarded. Aliquots of 50 µl/well of phages with normalized titers of 5 × 1011 cfuampR/ml pre-blocked in 5% BSA/PBS were added (corresponding to 830 pM phages/well) and the plate incubated for 1 h at 4°C with gentle agitation. Control wells received 5% BSA/PBS only. The cells were washed by adding 200 µl/well with 5% BSA/PBS, the cells pelleted by centrifuged at 300g/5 min/RT and the supernatants discarded. All phage-containing wells received 50 µl biotinylated anti-fd (10 µg/ml in 5% BSA/PBS), whereas the control wells received either 50 µl mAb biotinylated GB113 or AB10 (isotype control), both at 10 µg/ml in 5% BSA/PBS (corresponding to 67 nM/well assuming a MW of 150 kDa), followed by a 30 min incubation at 4°C with gentle agitation. In addition, some wells received mAb GB113 equimolar to the phage samples (830 pM). The wells were washed as above and all wells received 50 µl streptavidin-PE (2 µg/ml) followed by a 15 min incubation at 4°C with gentle agitation. The wells were washed as above, fixed with 200 µl/well 2% PFA and kept in the dark until analysis on a FACScalibur (BD Biosciences). Data analysis was done using the CellQuest Pro (v5.2.1) software (BD Biosciences).
Spiked phage library selection
Fresh phage samples were prepared, PEG precipitated and titrated as described. Ag specific virions were then spiked into an unspecific virion background at a 1:107 level, giving a known diversity of 107, corresponding to a medium sized combinatorial library. For phOx–BSA selection, the scFv anti-phOx was spiked into a scFv anti-NIP background, and vice versa. The initial input was 1010 cfuampR resulting in a complexity level of 103 for all the libraries. For selection, antigen was immobilized in MaxisorpTM microtiter plate wells (NUNC, Roskilde, Denmark) using 100 µl volumes of 1 µg/ml per well. Prior to panning, wells were blocked with PBSTM for 2 h at RT, before 100 µl of the respective pre-blocked (in PBSTM) phage preparations were added and allowed to react for 1.5 h at RT with agitation. The wells were washed 9× with PBST followed by 5× with dH2O using a microtiter plate washer before antigen bound phages were eluted by adding 100 µl/well Trypsin/EDTA for 10 min with agitation. The eluates were then used to infect log-phase E. coli XL1-Blue (M13K07 and Hyperphage-rescued samples) or TOP10F′ (DeltaPhage-rescued samples) cultures in 9 ml of YT-TAG (2×YT supplemented with 30 µg/ml tetracycline, 100 µg/ml ampicillin and 0.1 M glucose), incubated for 15 min at 37°C with low agitation before 1 ml of YT-TAG supplemented with the appropriate helper phage at MOI 10 was added. The incubation was continued for 15 min at 37°C with low agitation followed by 30 min at 37°C with rigorous agitation, followed by centrifugation at 3220g/10 min/RT. The supernatants were discarded and the pellets gently resuspended in 2× YT containing 100 µg/ml ampicillin and 50 µg/ml kanamycin, and all samples were incubated at 30°C with rigorous agitation until A600nm > 1. The samples were then centrifuged at 3220g/10 min/RT and the supernatants sterile filtered into fresh 15 ml tubes using 0.2 µm filters. These supernatants were then pre-blocked as before, and subjected to a second round of panning, identical to the first.
Single clone screening from the spiked selection
Single colonies were obtained by transduction of panning output supernatants into fresh E. coli cells. These were then picked, and grown ON in 400 μl YT-TAG in a MegaBlock 96-deep well plate (Sarstedt, Nümbrecht, Germany) using a Titramax 1000 (Heidolph Instruments GmbH & Co. KG, Schwabach, Germany) at 37°C/900-rpm. The colonies were re-inoculated by transferring 10 μl of culture to fresh deep-well plates, each well containing 400 µl YT-TAG, and incubated 3 h/37°C/900-rpm. The cultures were super infected with the appropriate helper phages and incubated for 30 min/37°C without agitation followed by 30 min/37°C/900-rpm. The cultures were pelleted by centrifugation, the supernatant discarded and the pellets re-suspended in 2× YT supplemented with 100 μg/ml ampicillin and 50 μg/ml kanamycin, and incubated ON/30°C/900-rpm. The bacteria were pelleted by centrifugation and the supernatant used in an antigen specific phage capture ELISA as described above.
RESULTS
Design of a helper phage for high-valence pIX display
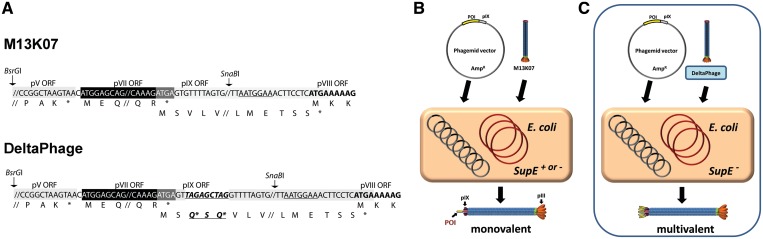
To create a helper phage encoding conditionally mutant pIX, named DeltaPhage, two amber codons were introduced into the M13K07 genome (43) by the addition of 9 nt (5′-TAGAGCTAG-3′) close to the pIX start codon, but after the pVII translational stop (Figure 1A). Introduction of two consecutive amber codons were chosen based on previous observations that a single amber codon may be insufficient to achieve complete knock down in the non-suppressor host (13). As superior suppression has been observed if the amber codon is immediately followed by a purine (29), the two amber codons were separated by 5′-AGC-3′ encoding a small hydrophilic serine residue. As a consequence of this insertion, two glutamines and a serine anticipated to be solvent exposed in the intact phage were added in the N-terminal portion of the full-length protein (26). In contrast to larger N-terminal pIX fusions that completely abrogated phage production in the absence of wt complementation (26), we hypothesized that this small modification would have minimal, or no effect on pIX function when complementing a pIX display phagemid. While phagemid rescue with a standard helper phage should lead to low-valence phagemid display (Figure 1B), phagemid rescue in a non-suppressor host with DeltaPhage should result in phages carrying the POI on all copies of pIX (Figure 1C).
Figure 1.
(A) Schematic illustration of the pV, pVII, pIX and pVIII genomic region of M13 important for the DeltaPhage design. The modification (bold, italic, underlined) was inserted between residue two and three in the pIX ORF downstream of the pVII translational stop overlapping with the pIX translational start. The region is framed by the unique BsrGI/SnaBI restriction sites shared by the fd and M13 genomes. (B) Basic outline of phagemid rescue using the standard helper phage M13K07, which renders low-valence display independent of supE genotype of the producing host. The phagemid is selectively propagated as a plasmid in E. coli, as it carries an ampicillin resistance marker. For phage assembly to occur, the host is super-infected with a helper phage containing all elements necessary for new virion assembly. As the helper phage also produces the wt pIX found as a POI-pIX fusion protein on the phagemid, the resulting phages carries a blend of both rendering low-valence display. (C) Basic outline of phagemid rescue using the DeltaPhage helper phage, which renders high-valence pIX display in a supE producing host due to complete lack of wt pIX production.
Phage production characteristics of DeltaPhage
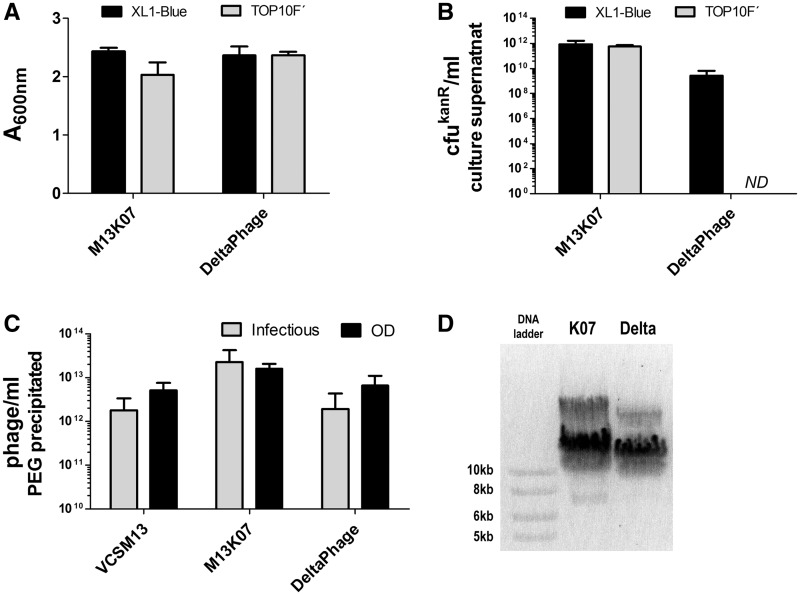
To study how the amber mutations in DeltaPhage affected helper phage propagation and assembly in a suppressor and a non-suppressor host, we cultured E. coli XL1-Blue and TOP10F′ with M13K07 or DeltaPhage. Both host strains showed roughly equal cell density after overnight culturing in selective medium (kanamycin at 50 µg/ml) regardless of helper phage (Figure 2A). Thus, the pIX modification had no pronounced phenotypic effect on host cell viability beyond that observed for M13K07.
Figure 2.
Host-cell propagation and phage assembly characteristics of DeltaPhage. (A) Single colonies of either E. coli XL1-Blue or TOP10F′ harboring either DeltaPhage, or M13K07 as control, were grown ON in selective medium before the cell density was measured at A600nm (values shown are extrapolated from measurements in one-fourth dilutions). The results are given as mean ± SD of three independent colonies of each sample assessed in parallel. (B) The M13K07 and DeltaPhage phage content in the supernatants from small-scale ON cultures were determined by infectious titration and the results given as the average kanamycin resistant colony forming units/ml (cfukanR/ml). The results are given as mean ± SD of three independent colonies of each sample assessed in parallel. ND = not detected. (C) The VCSM13, DeltaPhage and M13K07 phage content in PEG concentrated samples derived from up-scaled parallel cultures determined by either infectious titer, or total virion content determined by OD using the formula (((A269nm – A320nm) × 6.083 × 1016)/genome size = virions/ml) (38). Genome sizes: VCSM13, 8669 bp; M13K07, 8669 bp; DeltaPhage, 8678 bp. The results are given as mean ± SD of four to six independent experiments. (D) Agarose gel analysis of intact helper phage virions as described in ‘Materials and Methods’ section. The picture is representative of three experiments employing three independently packaged phage samples of both M13K07 (K07) and DeltaPhage (Delta).
In infectious titration of the culture supernatants, we observed equal phage end titers for the two host strains when producing M13K07 (Figure 2B). In contrast, a >100-fold reduction in titers of DeltaPhage was seen when compared with the M13K07 control in the suppressor strain XL1-Blue. In the non-suppressor strain TOP10F′, DeltaPhage production was completely abolished, as expected (Figure 2B). To date, repeated testing has yet to detect a single, infectious DeltaPhage particle produced from TOP10F′ (data not shown).
The reduction in titer seen with DeltaPhage indicated that the pIX mutations had a direct and negative influence on the phage production capacity. These initial experiments were performed in small volumes and disposable tubes. When up-scaled to shaker flasks allowing more rigorous agitation no significant difference in titers between DeltaPhage and both of the classical helper phages M13K07 and VCSM13 was seen (Figure 2C). Thus, the intended conditional pIX expression phenotype designed for DeltaPhage had been achieved, in that helper phage stocks for use in subsequent phagemid rescue could be readily produced in the suppressor strain XL1-Blue, whereas pIX expression, and hence, helper phage production, was completely abolished in the non-suppressor strain TOP10F′.
Amber mutations in pIX have previously been reported to yield infectious polyphage (44). Notably, we observed no difference between infectious phage counts (measured as cfukanR) and total phage counts (measured by OD) with DeltaPhage (Figure 2C). Depending on the titration method used, a slightly lower infectious than total virion count is expected even in wt systems (45). A significant discrepancy between the infectious and total virion count would have indicated the presence of polyphages. The absence of polyphage beyond that normally occurring was confirmed by gel electrophoresis and electron microscopy (Figure 2D and Supplementary Table S1).
DeltaPhage performance in phagemid rescue
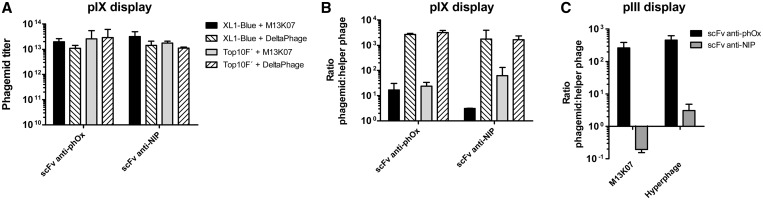
To characterize how DeltaPhage performed as a helper phage, we rescued a pIX display phagemid encoding either of two different scFv fusions with anti-phOx or anti-NIP specificity and determined the phage content in the culture supernatant by infectious titration. These two scFvs were selected due to their properties in the pIII display system where anti-phOx shows high display levels and is well tolerated, whereas the anti-NIP shows an intermediate display level and is poorly expressed (25). The phagemids were rescued with M13K07 or DeltaPhage in either E. coli XL1-Blue (suppressor), or TOP10F′ (non-suppressor) using a standard phagemid rescue protocol followed by PEG precipitation and concentration. The results show that phagemid particles were produced and that similar and very high titers were reached with both host strains and helper phages (Figure 3A). We also determined the phagemid (ampR) to helper phage genome (kanR) ratio of the rescued samples based on titers determined by selective growth on the appropriate antibiotics. The phagemids rescued with DeltaPhage exhibited a larger phagemid to helper phage ratio than the corresponding samples rescued with M13K07, as the ratio with DeltaPhage was consistently >103, while it varied from 3 to 60 with M13K07 (Figure 3B). This effect was independent of host strain and also contrasts the large variation in helper phage packaging seen after M13K07 rescue of the pIII display phagemids (Figure 3C). Thus, we confirm that the heterogeneous packaging of pIII display phagemids is not observed using pIX display (25), and the effect is further enhanced with DeltaPhage. The heterogeneity in phagemid packing seen with the pIII system was also alleviated when Hyperphage was used for phagemid rescued, though to a lesser extent than seen with the pIX phagemids and DeltaPhage (Figure 3C).
Figure 3.
Assessment of DeltaPhage in pIX display phagemid rescue. (A) Two pIX display phagemids (encoding either an anti-phOx or anti-NIP scFv) were propagated in 50 ml cultures of E. coli XL1-Blue and TOP10F′ followed by rescue by either M13K07 or DeltaPhage helper phages. Phages were precipitated and concentrated by PEG, followed by infectious titration and the results given as cfuampR/ml. (B) Phagemid to helper phage ratios were determined by cfuampR/cfukanR from parallel infectious titration of the same samples. The results are given as mean ± SD of three independent colonies of each sample assessed in parallel. (C) Two pIII display phagemids (encoding either the anti-phOx or anti-NIP scFv) were propagated in E. coli XL1-Blue and rescued with either M13K07, or Hyperphage. Phagemid to helper phage ratios were determined as above. The results are given as mean ± SD of three to four independent colonies of each sample assessed in parallel.
Determination of POI-pIX display levels by antigen specific phage capture ELISA
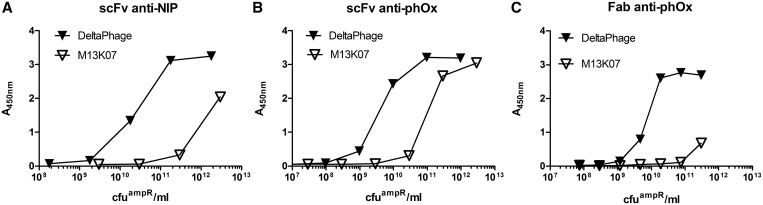
We expected the phenotype of phages rescued with DeltaPhage in a non-suppressor host to be high valence with respect to the POI fusion. This should directly translate into an increase in antigen reactivity as a result of the avidity effect compared to rescue with a standard helper phage, such as M13K07. To assess this, serial dilutions of the two different scFv specificities anti-phOx and anti-NIP rescued with M13K07 in XL1-Blue or DeltaPhage in TOP10F′ were tested for binding to a constant amount of antigen in a phage capture ELISA. Indeed, we observed a pronounced increase in antigen reactivity from the DeltaPhage-rescued samples (Figure 4A and B). The scFv anti-phOx (Kd ∼ 1 nM) and anti-NIP (Kd ∼ 370 nM) differ in their monomeric affinities (46,47). The effect of high-valence display was most pronounced for the low affinity anti-NIP specificity, where reactivity was increased by almost two orders of magnitude, when rescued with DeltaPhage compared to the standard system. In contrast, the difference between M13K07 and DeltaPhage rescued scFv anti-phOx was about one order of magnitude
Figure 4.
Functional analysis of pIX display levels at low and high-valence display. Serial dilutions of phagemid-rescued samples displaying either scFv anti-phOx (A), scFv anti-NIP (B) or (C) Fab anti-phOx were applied in a phage capture ELISA as described in ‘Material and Methods’ section. Antigen-bound phages were detected by anti-M13HRP and the data shown as function of number of phagemids (cfuampR)/ml solution. The data is representative of three to four independent experiments.
We also generated a Fab-pIX fusion utilizing the same variable regions as in the anti-phOx scFv and tested the performance of DeltaPhage in phagemid rescue. Again, antigen reactivity was tested using serial dilutions of phage in antigen specific phage capture ELISA (Figure 4C). As was seen with the scFv counterpart, we observed a marked increase in antigen reactivity of the DeltaPhage rescued phages compared with those rescued with M13K07, and conclude that DeltaPhage permits high-valence pIX display of both two domain scFvs and four domain heterodimeric Fabs.
Cell surface receptor binding of phage with multivalent pIX display
In the phage capture ELISA the antigen was small haptens presented as a dense array in microtiter plate wells. A more challenging antigen would be a transmembrane cell surface bound receptor presented in its native form. We therefore constructed a scFv version of the monoclonal antibody GB113 (see ‘Materials and Methods’ section) that exclusively reacts with the murine T cell receptor 4B2A1 (33). The GB113 scFv was displayed at low and high valence both from a pIII and pIX display phagemid, and tested for binding to 4B2A1 T cells in flow cytometry (Figure 5). Due to the large size of the phage virion, there is a solubility threshold of ∼1013 phages/ml, which corresponds to 16.6 nM. To allow for a direct comparison of all phage versions, we set the sample concentration to a normalized titer of 5 × 1011 cfuampR /ml, which corresponds to a phage input of 830 pM, the threshold set by the low phage numbers in the Hyperphage rescued samples.
Figure 5.
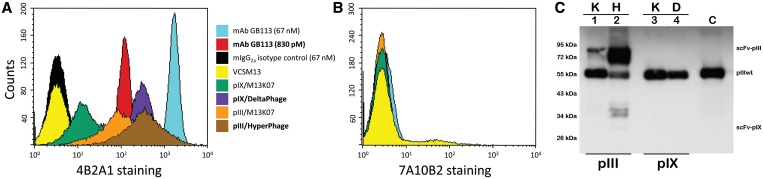
Phage staining of 4B2A1 T cells by flow cytometry. A normalized phage input of 5 × 1011 cfu/ml, which corresponds to a concentration of 830 pM was used throughout. A total of 2 × 105 T cells were incubated with the indicated phages followed by detection using anti-fdbio and SA–PE as described in ‘Materials and Methods’ section. Cells were also stained with the 4B2A1 clonotype-specific mAb GB113. The samples were analyzed on a FACScalibur flow cytometer and the histograms showing staining on the cognate 4B2A1 T cells (A), or the negative control T cells 7A10B2 (B) were generated using CellQuest Pro (BD Biosciences). (C) SDS–PAGE/western blot analysis of the phage samples tested in flow. Normalized phage samples of 2 × 109 cfuampR/lane were used and detected using an anti-pIII antibody as described in ‘Materials and Methods’ section. Key to figure: K, M13K07-rescued phagemid; H, Hyperphage-rescued phagemid; D, DeltaPhage-rescued phagemid; C, M13K07.
We found all GB113 scFv-displaying phages to specifically stain the 4B2A1 T cells (Figure 5A). The high-valence phages (pIX/DeltaPhage and pIII/Hyperphage) stained with comparable intensities, while the low-valence pIII (rescued with M13K07) exhibited ∼4-fold stronger staining than the low-valence pIX phages [mean fluorescent intensity (MFI), data not shown]. The naked phage (VCSM13) and the isotype control mAb did not stain 4B2A1, and all samples were negative on the 7A10B2 T cells expressing an unrelated T cell receptor with the same pMHC specificity as 4B2A1 (Figure 5B). There was a >8-fold difference in target reactivity between pIX low and high-valence display (MFI, data not shown), while on pIII this difference was no more than ∼3-fold (MFI, data not shown). Thus, the results confirm that at low valence, the pIX display level is lower than that of pIII (25). Due to the much smaller molecular size of an antibody than a phage particle, which permits a higher molar input, a significantly stronger staining was seen with the parent antibody than with the phages using standard antibody conditions corresponding to 67 nM (10 µg/ml). However, when phage and antibody inputs were equal, the phages exhibited the strongest staining (Figure 5A). Although the latter observation may in part be attributed to differences in the detection sandwich, it may well reflect the difference in binding between the bivalent antibody and the putative pentavalent display on the phage.
True differences in display levels and adequate normalization of the samples were confirmed by SDS–PAGE/anti-pIII western blotting (Figure 5C). The anti-pIII detection was chosen due to its proven robustness and the lack of any corresponding anti-pIX specificity available. Here, the difference in pIII display levels were readily visible (lane 1 and 2), confirming previous observations (12), as well as the uniform amount of wt pIII showing equal phage input. As expected, the pIX fusions are not visible in this analysis, which was pIII specific (lanes 3 and 4).
Comparison of low and high-valence pIII and pIX display in antigen selection
The primary application of phage display technology is to isolate specific binders from large and diverse libraries. We therefore made mock libraries and did parallel affinity selections with phages displaying a specific scFv in low or high valence on either pIII or pIX. The scFvs were expressed from phagemids and the variation in display valence was achieved by using either standard M13K07 rescue, or Hyperphage for the pIII unit and DeltaPhage for the pIX unit, respectively. A total of 1000 antigen specific phages were spiked into 1010 phages with the alternative specificity using the scFv anti-phOx and anti-NIP phages creating a mock library with a diversity of 107 (corresponding to a library of medium diversity) and an input complexity of 1000. Importantly, the two scFvs are not cross-reactive (Supplementary Figure S1). We then enriched for either phOx or NIP positive clones. Selection was done by adding the libraries to wells coated with either phOx or NIP conjugated to BSA. The wells were stringently washed, and bound phages were eluted by cleavage of the fusion by incubation with trypsin. The eluates were then used to infect E. coli cells to amplify the phages, and the selection was repeated using the phage containing supernatants as input. The unselected libraries as well as amplified supernatants from the first and second round of selection were used for transduction to provide single clones for screening. In each case, 40 randomly picked clones were tested for binding to both antigens in an antigen specific phage capture ELISA (Supplementary Figure S1). The results are summarized in Table 1.
Table 1.
Single phage colony screening after spiked phOx/NIP selectiona
| Display capsid | Specificity | Helper phage | Round of selection |
||
|---|---|---|---|---|---|
| 0 | 1 | 2 | |||
| pIII | phOx | M13K07 | 0/40 | 3/40 | 20/40 |
| Hyperphage | 0/40 | 8/40 | 38/40 | ||
| NIP | M13K07 | 0/40 | 0/40 | 4/40 | |
| Hyperphage | 0/40 | 9/40 | 34/40 | ||
| pIX | phOx | M13K07 | 0/40 | 3/40 | 13/40 |
| DeltaPhage | 0/40 | 8/40 | 37/40 | ||
| NIP | M13K07 | 0/40 | 1/40 | 8/40 | |
| DeltaPhage | 0/40 | 7/40 | 38/40 | ||
aThe target-specific scFv was selected from 1:107 spiked mock libraries. Forty randomly chosen single colonies were packaged from each library before (round 0) and from each of two consecutive selection rounds. Target reactivity was tested using an antigen-specific phage capture ELISA. Clones were regarded as positive if they exhibited at least 3-fold higher response than the background signal. The results are given as: number of positive clones/total number of clones tested.
The screening procedure showed low efficiency of retrieval for low-valence display after the first round of selection, yielding between no anti-NIP (pIII) to 7.5% positive clones for anti-phOx (both pIII and pIX). As anticipated, high-valence display proved more efficient in retrieval of binders, yielding ∼20% positive clones with no prominent differences between the two systems. After the second round, the fraction of positive clones were overall increased independently of capsid and valence. Notably, the high-valence versions of both pIII and pIX display now had reached almost complete retrieval of the target specific scFv at similar levels of enrichment between 85% and 95% positive clones. Three of four low-valence libraries exhibited a retrieval efficiency of only 10–32%. The exception was the scFv anti-phOx displayed on pIII, showing 50% positive clones. In summary, high-valence display on pIX performed at least as well as the pIII counterpart after two rounds of selection, showing that DeltaPhage is indeed well suited for use in improved selection of specific binders as compared to low-valence display.
DISCUSSION
pIX is strictly required for phage assembly (26). In principle, one would therefore predict high-valence pIX display of phagemid encoded POI fusions after selective inactivation of the helper phage encoded pIX. A conditional pIX inactivation would allow for normal helper phage packaging without the need for an exogenous capsid source (12,48), and this approach was therefore chosen. We disrupted translation of the protein in the E. coli host through the introduction of suppressible amber stop codons in the pIX ORF. When expressed in amber suppressor host strains with the supE genotype, such as E. coli XL1-Blue, translational read-through by insertion of a glutamine residue occurs, hence resulting in production of a full-length protein (29). In non-suppressor strains, such as E. coli TOP10F′, the amber codon leads to translational stop, and therefore neither pIX protein nor phage production.
Until now, the consensus has been that display on pIX is dependent on wt complementation, making high-valence display unachievable (26,27). A very recent report does however show that genomic multivalent pIX display is permitted using small peptides, but argues that larger fusions such as folded structures are prohibited, which is in line with previous reports (26,28). Here, we clearly demonstrate that this is not the case. To the contrary, we report multivalent display of large molecules such as antibody scFv and even Fab. This was achieved using the novel helper phage DeltaPhage and phagemid technology. The helper phage itself is easily produced in the suppressor host and, in contrast to some of the other engineered helper phages for high-valence pIII display, we do not observe any pronounced reduction in end titers, making high titer stocks easily obtainable (12,49).
The phage genome is sensitive to genetic manipulations (44). When compared to the M13K07 backbone from which DeltaPhage was constructed, we did observe a very mild reduction in end titer dependent on the growth conditions of the bacterial host during phage packaging. We did however, not observe a large increase in polyphage formation due to pIX amber manipulation (44).
Although functional affinity effects are case specific and may vary with the experimental conditions applied, it is usually associated with a marked increase in target reactivity (50,51). Indeed this is also what we observed when comparing low and high-valence pIX display in ELISA. A previous report suggests putative restrictions in target binding of a fusion displayed from the narrow pVII/pIX tip due to small and closely packed capsids as compared to display on the larger and highly flexible three domain pIII capsid (52). However, our results clearly show that even a large and complex fusion such as a heterodimeric Fab was readily displayed on pIX and strongly benefited from multivalent display. Moreover, the T cell receptor specific scFv fusion exhibited a very strong effect of multivalent display and showed comparable binding to multivalent pIII display. Thus, our results show that multivalent display on pIX may be allowed for even large fusions of multidomain topology.
When used in affinity selection of binders from mock libraries, we observed a significant improvement in retrieval of the target specific unit at high-valence display when compared to the low-valence display counterparts, both with respect to rounds of selection needed and in the absolute amount of positive hits obtained. In these model selections, there was no difference in efficiency between multivalent pIII and pIX display. Moreover, the human scFv anti-phOx unit used herein is particularly well adopted for both pIII display and prokaryotic expression as it has been expressed and matured through two independent pIII phage display selections (46). This is not the case for the anti-NIP scFv used that originates from a hybridoma, and which exhibits toxicity effects in the pIII system (25). It is therefore noteworthy that the largest discrepancy between the pIII and pIX systems was seen for low-valence scFv anti-NIP selection where pIX appeared the most effective selector. This discrepancy was only apparent at low-valence display when wt capsid complementation was operational, and not at high-valence display. However, when larger and more diverse libraries are used, introducing stronger clone competition during amplification, this may well change, as high-valence pIII display is well known to unpredictably affect both functional display, phage titer and the infectivity of the phage (15,53,54). Here, pIX display may offer several advantages over high-valence pIII display. (i) High end titers are easily obtained, which provides the opportunity to screen libraries of higher diversity. (ii) The phagemid to phage vector packaging ratios are uniformly very high and high-valence pIX display does not affect infectivity in any way, both of which prevents that selected clones are lost (25). (iii) pIX display seems to a large extent to overcome the clone dependent bias often observed for pIII display. This prevents loss of binders due to unfavorable growth conditions in addition to heterogeneous phagemid packaging (25). (iv) The pIX system apparently exhibits better discrimination between low and high-valence display than is observed using pIII. This may prove to be a significant advantage in primary selection, as well as in affinity maturation, as it suggests that there are less avidity effects when using low-valence pIX display than with its pIII counterpart. Indeed, effective high affinity selection has been shown to benefit strongly from low display levels (7,20,55). Interestingly, a previous report also point out an apparent stronger enrichment in low-valence affinity selection by use of pIX display as compared to pIII display (22).
In summary, the combination of high and low-valence pIX display reported herein offers the possibility to now tap into both the favorable phenotypes of multivalent display by initially selecting for specificity and increased retrieval whereby switching to low-valence display and efficient affinity selection in later rounds (3,8). In effect, this system may therefore out-compete current pIII based systems as pIX display is devoid of any negative pIII effects.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1 and Supplementary Figure 1.
FUNDING
The Norwegian Research Council [203373/I30]. Funding for open access charge: Nextera AS.
Conflict of interest statement. A patent application covering the helper phage described in the article has been submitted by GÅL and IS. All other authors have no financial conflicts of interest.
Supplementary Material
ACKNOWLEDGEMENTS
The authors would like to thank Sathiyaruby Sivaganesh for excellent technical assistance.
REFERENCES
- 1.Bratkovic T. Progress in phage display: evolution of the technique and its applications. Cell. Mol. Life Sci. 2010;67:749–767. doi: 10.1007/s00018-009-0192-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rakonjac J, Bennett NJ, Spagnuolo J, Gagic D, Russel M. Filamentous bacteriophage: biology, phage display and nanotechnology applications. Curr. Issues Mol. Biol. 2011;13:51–76. [PubMed] [Google Scholar]
- 3.Bradbury AR, Marks JD. Antibodies from phage antibody libraries. J. Immunol. Methods. 2004;290:29–49. doi: 10.1016/j.jim.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 4.Dubel S, Stoevesandt O, Taussig MJ, Hust M. Generating recombinant antibodies to the complete human proteome. Trends Biotechnol. 2010;28:333–339. doi: 10.1016/j.tibtech.2010.05.001. [DOI] [PubMed] [Google Scholar]
- 5.Bradbury AR, Sidhu S, Dubel S, McCafferty J. Beyond natural antibodies: the power of in vitro display technologies. Nat. Biotechnol. 2011;29:245–254. doi: 10.1038/nbt.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kretzschmar T, Geiser M. Evaluation of antibodies fused to minor coat protein III and major coat protein VIII of bacteriophage M13. Gene. 1995;155:61–65. doi: 10.1016/0378-1119(94)00897-2. [DOI] [PubMed] [Google Scholar]
- 7.Pavoni E, Monteriu G, Cianfriglia M, Minenkova O. New display vector reduces biological bias for expression of antibodies in E. coli. Gene. 2007;391:120–129. doi: 10.1016/j.gene.2006.12.009. [DOI] [PubMed] [Google Scholar]
- 8.O'Connell D, Becerril B, Roy-Burman A, Daws M, Marks JD. Phage versus phagemid libraries for generation of human monoclonal antibodies. J. Mol. Biol. 2002;321:49–56. doi: 10.1016/s0022-2836(02)00561-2. [DOI] [PubMed] [Google Scholar]
- 9.Hust M, Meysing M, Schirrmann T, Selke M, Meens J, Gerlach GF, Dubel S. Enrichment of open reading frames presented on bacteriophage M13 using hyperphage. BioTechniques. 2006;41:335–342. doi: 10.2144/000112225. [DOI] [PubMed] [Google Scholar]
- 10.Jespers L, Schon O, Famm K, Winter G. Aggregation-resistant domain antibodies selected on phage by heat denaturation. Nat. Biotechnol. 2004;22:1161–1165. doi: 10.1038/nbt1000. [DOI] [PubMed] [Google Scholar]
- 11.Becerril B, Poul MA, Marks JD. Toward selection of internalizing antibodies from phage libraries. Biochem. Biophys. Res. Commun. 1999;255:386–393. doi: 10.1006/bbrc.1999.0177. [DOI] [PubMed] [Google Scholar]
- 12.Rondot S, Koch J, Breitling F, Dubel S. A helper phage to improve single-chain antibody presentation in phage display. Nat. Biotechnol. 2001;19:75–78. doi: 10.1038/83567. [DOI] [PubMed] [Google Scholar]
- 13.Baek H, Suk KH, Kim YH, Cha S. An improved helper phage system for efficient isolation of specific antibody molecules in phage display. Nucleic Acids Res. 2002;30:e18. doi: 10.1093/nar/30.5.e18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Soltes G, Barker H, Marmai K, Pun E, Yuen A, Wiersma EJ. A new helper phage and phagemid vector system improves viral display of antibody Fab fragments and avoids propagation of insert-less virions. J. Immunol. Methods. 2003;274:233–244. doi: 10.1016/s0022-1759(02)00294-6. [DOI] [PubMed] [Google Scholar]
- 15.Løset GÅ, Kristinsson SG, Sandlie I. Reliable titration of filamentous bacteriophages independent of pIII fusion moiety and genome size by using trypsin to restore wild-type pIII phenotype. BioTechniques. 2008;44:551–554. doi: 10.2144/000112724. [DOI] [PubMed] [Google Scholar]
- 16.Shi B, Wang H, Guo S, Xu Y, Li Z, Gu J. Protein III-based single-chain antibody phage display using bacterial cells bearing an additional genome of a gene-III-lacking helper phage. BioTechniques. 2007;42:760–765. doi: 10.2144/000112461. [DOI] [PubMed] [Google Scholar]
- 17.Zanconato S, Minervini G, Poli I, Lucrezia DD. Selection dynamic of Escherichia coli host in M13 combinatorial peptide phage display libraries. Biosci. Biotechnol. Biochem. 2011;75:812–815. doi: 10.1271/bbb.110099. [DOI] [PubMed] [Google Scholar]
- 18.Cwirla SE, Peters EA, Barrett RW, Dower WJ. Peptides on phage: a vast library of peptides for identifying ligands. PNAS. 1990;87:6378–6382. doi: 10.1073/pnas.87.16.6378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lowman HB, Bass SH, Simpson N, Wells JA. Selecting high-affinity binding proteins by monovalent phage display. Biochemistry. 1991;30:10832–10838. doi: 10.1021/bi00109a004. [DOI] [PubMed] [Google Scholar]
- 20.de Haard HJ, van Neer N, Reurs A, Hufton SE, Roovers RC, Henderikx P, de Bruine AP, Arends JW, Hoogenboom HR. A large non-immunized human Fab fragment phage library that permits rapid isolation and kinetic analysis of high affinity antibodies. J. Biol. Chem. 1999;274:18218–18230. doi: 10.1074/jbc.274.26.18218. [DOI] [PubMed] [Google Scholar]
- 21.Simons GF, Konings RN, Schoenmakers JG. Genes VI, VII, and IX of phage M13 code for minor capsid proteins of the virion. PNAS. 1981;78:4194–4198. doi: 10.1073/pnas.78.7.4194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gao C, Mao S, Kaufmann G, Wirsching P, Lerner RA, Janda KD. A method for the generation of combinatorial antibody libraries using pIX phage display. PNAS. 2002;99:12612–12616. doi: 10.1073/pnas.192467999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Shi L, Wheeler JC, Sweet RW, Lu J, Luo J, Tornetta M, Whitaker B, Reddy R, Brittingham R, Borozdina L, et al. De novo selection of high-affinity antibodies from synthetic fab libraries displayed on phage as pIX fusion proteins. J. Mol. Biol. 2010;397:385–396. doi: 10.1016/j.jmb.2010.01.034. [DOI] [PubMed] [Google Scholar]
- 24.Tornetta M, Baker S, Whitaker B, Lu J, Chen Q, Pisors E, Shi L, Luo J, Sweet R, Tsui P. Antibody Fab display and selection through fusion to the pIX coat protein of filamentous phage. J. Immunol. Methods. 2010;360:39–46. doi: 10.1016/j.jim.2010.06.001. [DOI] [PubMed] [Google Scholar]
- 25.Løset GÅ, Roos N, Bogen B, Sandlie I. Expanding the versatility of phage display II: improved affinity selection of folded domains on protein VII and IX of the filamentous phage. PLoS One. 2011;6:e17433. doi: 10.1371/journal.pone.0017433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Endemann H, Model P. Location of filamentous phage minor coat proteins in phage and in infected cells. J. Mol. Biol. 1995;250:496–506. doi: 10.1006/jmbi.1995.0393. [DOI] [PubMed] [Google Scholar]
- 27.Gao C, Mao S, Lo CH, Wirsching P, Lerner RA, Janda KD. Making artificial antibodies: a format for phage display of combinatorial heterodimeric arrays. PNAS. 1999;96:6025–6030. doi: 10.1073/pnas.96.11.6025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ploss M, Kuhn A. Membrane insertion and assembly of epitope-tagged gp9 at the tip of the M13 phage. BMC Microbiol. 2011;11:211. doi: 10.1186/1471-2180-11-211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bossi L. Context effects: translation of UAG codon by suppressor tRNA is affected by the sequence following UAG in the message. J. Mol. Biol. 1983;164:73–87. doi: 10.1016/0022-2836(83)90088-8. [DOI] [PubMed] [Google Scholar]
- 30.Soltes G, Hust M, Ng KK, Bansal A, Field J, Stewart DI, Dubel S, Cha S, Wiersma EJ. On the influence of vector design on antibody phage display. J. Biotechnol. 2007;127:626–637. doi: 10.1016/j.jbiotec.2006.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Løset GÅ, Lunde E, Bogen B, Brekke OH, Sandlie I. Functional phage display of two murine a/b T-cell receptors is strongly dependent on fusion format, mode and periplasmic folding assistance. Protein Eng. Des, Sel. 2007:20, 461–472. doi: 10.1093/protein/gzm044. [DOI] [PubMed] [Google Scholar]
- 32.Bogen B, Snodgrass R, Briand JP, Hannestad K. Synthetic peptides and beta-chain gene rearrangements reveal a diversified T cell repertoire for a lambda light chain third hypervariable region. Eur. J. Immunol. 1986;16:1379–1384. doi: 10.1002/eji.1830161111. [DOI] [PubMed] [Google Scholar]
- 33.Bogen B, Lauritzsen GF, Weiss S. A stimulatory monoclonal antibody detecting T cell receptor diversity among idiotype-specific, major histocompatibility complex-restricted T cell clones. Eur. J. Immunol. 1990;20:2359–2362. doi: 10.1002/eji.1830201030. [DOI] [PubMed] [Google Scholar]
- 34.Michaelsen TE, Aase A, Westby C, Sandlie I. Enhancement of complement activation and cytolysis of human IgG3 by deletion of hinge exons. Scand. J. Immunol. 1990;32:517–528. doi: 10.1111/j.1365-3083.1990.tb03192.x. [DOI] [PubMed] [Google Scholar]
- 35.Nakela O, Kaartinen M, Pelkonen JL, Karjalainen K. Inheritance of antibody specificity V. Anti-2-phenyloxazolone in the mouse. J. Exp. Med. 1978;148:1644–1660. doi: 10.1084/jem.148.6.1644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sambrook J, Russell DW. Molecular Cloning: A Laboratory Manual. 3 edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 37.Marks JD, Hoogenboom HR, Bonnert TP, McCafferty J, Griffiths AD, Winter G. By-passing immunization. Human antibodies from V-gene libraries displayed on phage. J. Mol. Biol. 1991;222:581–597. doi: 10.1016/0022-2836(91)90498-u. [DOI] [PubMed] [Google Scholar]
- 38.Mount JD, Samoylova TI, Morrison NE, Cox NR, Baker HJ, Petrenko VA. Cell targeted phagemid rescued by preselected landscape phage. Gene. 2004;341:59–65. doi: 10.1016/j.gene.2004.07.006. [DOI] [PubMed] [Google Scholar]
- 39.Welschof M, Terness P, Kipriyanov SM, Stanescu D, Breitling F, Dorsam H, Dubel S, Little M, Opelz G. The antigen-binding domain of a human IgG-anti-F(ab')2 autoantibody. PNAS. 1997;94:1902–1907. doi: 10.1073/pnas.94.5.1902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Koch J, Breitling F, Dubel S. Rapid titration of multiple samples of filamentous bacteriophage (M13) on nitrocellulose filters. BioTechniques. 2000;29:1196–1198. doi: 10.2144/00296bm08. [DOI] [PubMed] [Google Scholar]
- 41.Løset GÂ, Kristinsson SG, Sandlie I. Reliable titration of filamentous bacteriophages independent of pIII fusion moiety and genome size by using trypsin to restore wild-type pIII phenotype. Biotechniques. 2008;44:551–552, 554. doi: 10.2144/000112724. [DOI] [PubMed] [Google Scholar]
- 42.Weibel ER. Practical Methods for Biological Morphometry. London: Academic Press; 1979. [Google Scholar]
- 43.Vieira J, Messing J. Production of single-stranded plasmid DNA. Methods Enzymol. 1987;153:3–11. doi: 10.1016/0076-6879(87)53044-0. [DOI] [PubMed] [Google Scholar]
- 44.Lopez J, Webster RE. Morphogenesis of filamentous bacteriophage f1: orientation of extrusion and production of polyphage. Virology. 1983;127:177–193. doi: 10.1016/0042-6822(83)90382-3. [DOI] [PubMed] [Google Scholar]
- 45.Barbas CF. Phage display: A Laboratory Manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2001. [Google Scholar]
- 46.Marks JD, Griffiths AD, Malmqvist M, Clackson TP, Bye JM, Winter G. By-passing immunization: building high affinity human antibodies by chain shuffling. Biotechnology. 1992;10:779–783. doi: 10.1038/nbt0792-779. [DOI] [PubMed] [Google Scholar]
- 47.Løset GÅ, Roux KH, Zhu P, Michaelsen TE, Sandlie I. Differential segmental flexibility and reach dictate the antigen binding mode of chimeric IgD and IgM: implications for the function of the B cell receptor. J. Immunol. 2004;172:2925–2934. doi: 10.4049/jimmunol.172.5.2925. [DOI] [PubMed] [Google Scholar]
- 48.Kramer RA, Cox F, van der Horst M, van der Oudenrijn S, Res PC, Bia J, Logtenberg T, de Kruif J. A novel helper phage that improves phage display selection efficiency by preventing the amplification of phages without recombinant protein. Nucleic Acids Res. 2003;31:e59. doi: 10.1093/nar/gng058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Soltes G, Hust M, Ng KKY, Bansal A, Field J, Stewart DIH, Dubel S, Cha S, Wiersma EJ. On the influence of vector design on antibody phage display. J. Biotechol. 2007;127:626–637. doi: 10.1016/j.jbiotec.2006.08.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rheinnecker M, Hardt C, Ilag LL, Kufer P, Gruber R, Hoess A, Lupas A, Rottenberger C, Pluckthun A, Pack P. Multivalent antibody fragments with high functional affinity for a tumor-associated carbohydrate antigen. J. Immunol. 1996;157:2989–2997. [PubMed] [Google Scholar]
- 51.Pack P, Muller K, Zahn R, Pluckthun A. Tetravalent miniantibodies with high avidity assembling in Escherichia coli. J. Mol. Biol. 1995;246:28–34. doi: 10.1006/jmbi.1994.0062. [DOI] [PubMed] [Google Scholar]
- 52.Kwasnikowski P, Kristensen P, Markiewicz WT. Multivalent display system on filamentous bacteriophage pVII minor coat protein. J. Immunol. Methods. 2005;307:135–143. doi: 10.1016/j.jim.2005.10.002. [DOI] [PubMed] [Google Scholar]
- 53.Huovinen T, Sanmark H, Yla-Pelto J, Vehniainen M, Lamminmaki U. Oligovalent Fab display on M13 phage improved by directed evolution. Mol. Biotechnol. 2010;44:221–231. doi: 10.1007/s12033-009-9231-3. [DOI] [PubMed] [Google Scholar]
- 54.Kuba H, Furukawa K. An optimized procedure for efficient phage display of antibody fragments with a low folding efficiency. Protein Expr. Purif. 2009;65:148–153. doi: 10.1016/j.pep.2009.01.016. [DOI] [PubMed] [Google Scholar]
- 55.Rothe C, Urlinger S, Lohning C, Prassler J, Stark Y, Jager U, Hubner B, Bardroff M, Pradel I, Boss M, et al. The human combinatorial antibody library HuCAL GOLD combines diversification of all six CDRs according to the natural immune system with a novel display method for efficient selection of high-affinity antibodies. J. Mol. Biol. 2008;376:1182–1200. doi: 10.1016/j.jmb.2007.12.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.