Abstract
A new strategy to create site-specific, homogeneous, and bright silver nanoclusters (AgNCs) with high-stability was demonstrated by triplex DNA as template. By reasonable design of DNA sequence, homogeneous Ag2 cluster was obtained in the predefined position of CG.C+ site of triplex DNA. This strategy was also explored for controlled alignment of AgNCs on the DNA nanoscaffold. To the best of our knowledge, this was the first example to simultaneously answer the challenges of excellent site-specific nucleation and growth, homogeneity and stability against salt of DNA-templated AgNCs.
INTRODUCTION
As promising substitutes to organic dyes and quantum dots, few-atom fluorescent noble metal nanoclusters of <2 nm in diameter have attracted a great deal of attention in recent years because of their potential applications in nanotechnology and nanomedicine (1–7). In particular, much effort has been dedicated to the synthesis of fluorescent silver nanoclusters (AgNCs) using DNA as a template due to the low toxicity, good biocompatibility and unique optical properties of the AgNCs obtained (8–12). For instance, a series of DNA-based AgNCs with emission from visible to near-infrared region have been produced, which have showed great promise for biological labelling, biosensing and information storage (9). However, up to this stage, site specificity and homogeneity still remain a big challenge, and these features are critical for the precise organization of metallic nanoparticles into sophisticated nanostructures and their further applications in electronics, sensors, nanomedicine and many other fields. For instance, mismatched double-stranded DNA (dsDNA) or hairpin DNA have been used to create AgNCs by us and others (13,14), in which the mismatched or single-stranded loop region of hairpin DNA could serve as the template. However, the exact binding site remains unknown and the as-prepared AgNCs usually suffer from highly heterogeneity and lack stability against Cl− (15). Very recently, Liu et al. (16) reported a method for the synthesis of water-soluble fluorescent AgNCs with a narrow size distribution by use of the well-known Tollens reaction. Through covalently incorporation a serious number of sugar moieties into a DNA sequence at adjacent positions, Ag(I) ions can be reduced to Ag02 by Tollens reaction (17–19). These Ag clusters could then act as nucleation sites for further Ag depositions. However, this approach is limited to sugar-modified DNA sequence, which would compromise their further applications. Herein, we reported a new strategy for the fabrication of site-specific, homogenous and highly stable fluorescent AgNCs by use of triplex DNA as an efficient template.
Recently, it has been demonstrated that the properties of AgNCs are highly dependent on the nature of the template molecules used for nanocluster synthesis: the formation of AgNCs relies on the base sequence of the DNA template; the stabilities and fluorescence properties of AgNCs are dependent on the microenvironments provided by DNA templates (8–12,14,20–22). Therefore, the fact that the properties of AgNCs are highly dependent on the nature of the DNA template opens the opportunities to modulate the in situ site-specific fabrication of AgNCs by choosing some appropriate DNA templates.
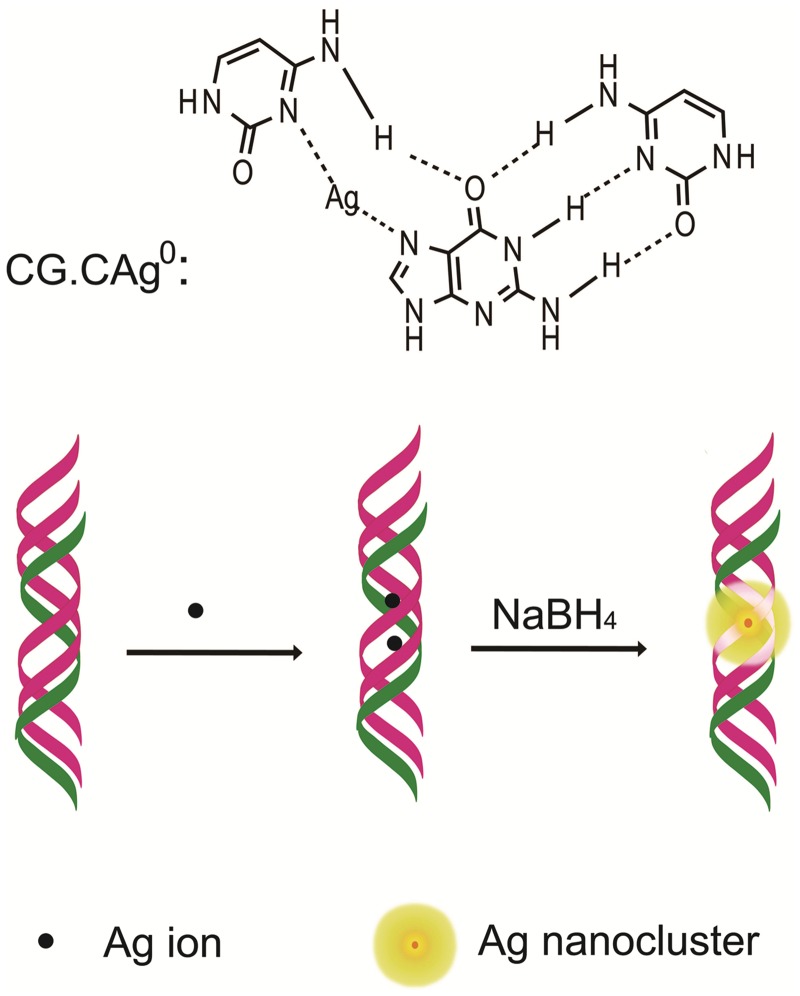
The DNA triplex has become one of the most useful recognition motifs in the design of new molecular biology tools, diagnostic/therapeutic agents and sophisticated DNA-based nanomaterials (23–26). In these strategy, a triplex-forming oligonucleotide binds to the major groove of the target duplex in a sequence-specific manner through Hoogsteen or reverse Hoogsteen base pairing (24–26). Recently, in the efforts to improve the stability of triplex, Ihara et al. (30) reported a method for stabilization of the triplex DNA using Ag(I) ions. They found that Ag(I) ion could specifically displace an N3 proton of a cytosine in the CG.C+ base site of triplex DNA to form a new triplet (CG.CAg+), which would stabilize the parallel-motif triplex even at neutral pH (30). The specific interaction of Ag(I) ion with CG.C+ triplet ensured that Ag(I) ion could only work on Hoogsteen hydrogen bonding in the CG.C+ triplet. Inspired by this unique feature, we expect that site-specific formation of fluorescent AgNCs could be realized by reducing these Ag(I) ions in situ in the triplet CG.CAg+ base sites (Scheme 1). Meanwhile, we speculated that the species of AgNCs could be controlled by the number and position of CG.C+ base site in triplex DNA, which offers the possibility to produce homogeneous AgNCs in DNA template. Indeed, our results described below showed that homogeneous Ag2 clusters were obtained by exploring the Ag-stabilized triplex DNA as template. The as-prepared AgNCs are indicated to be at the very position of CG.C+ site and have unusual stability against Cl−. To the best of our knowledge, this is the first time that site-addressable, homogeneous and ultra-stable AgNCs are acquired by using unmodified DNA as a template. Moreover, by taking advantages of this triplex DNA-directed fabrication, we realized the precisely controlled alignment of fluorescent AgNCs on DNA nanoscaffold.
Scheme 1.
Schematic representation of the triplex DNA-templated preparation of fluorescent AgNCs.
MATERIALS AND METHODS
Materials
Silver Nitrate (AgNO3, 99.9995%) and sodium borohydride (NaBH4, 98%) were purchased from Alfa Aesar and used without further purification. All other chemicals were perchased from Sigma–Aldrich and used as supplied. DNA oligonucleotides were purchased from Sangon (Shanghai, China). Phosphate buffers (10 mM and 100 mM NaNO3) with different pH value were used for all the analysis except that NH4Ac/HAc buffer (pH = 6.5) was used for MS measurement. Nanopure water (18.2 MΩ; Millpore Co., USA) was used in all experiments.
Measurements
Fluorescence spectra were acquired using a Jasco FP-6500 spectrofluorometer (Jasco International Co., Japan). All fluorescence measurements were carried out under ambient temperature using a quartz cell of 1-cm path length. UV melting curves were determined using a Cary 300 UV/Vis spectrophotometer (Varian Inc., CA, USA), equipped with a thermoelectric temperature controller. All melting studies were carried out using 1-cm path length cells with Teflon stoppers. Absorbance changes at 260 nm versus temperature were collected at a heating rate of 1.0°C min−1, over the temperature range of 10–90°C. Circular dichroism (CD) spectra were measured on a Jasco J-810 spectropolarimeter with a computer-controlled water bath. The optical chamber of CD spectrometer was deoxygenated with dry purified nitrogen (99.99%) for 45 min before use and kept the nitrogen atmosphere during experiments. Three scans were accumulated and automatrically averaged. The TEM images of the AgNCs were acquired on a TECNAI G2 transmission electron microscope (Philips Electronic Instruments Co., NL, USA) at 200 kV. Samples were prepared by pipetting 5 μl of sample solution onto copper grids coated with an ultrathin carbon film (400 mesh; Ted Pella, Redding, CA, USA). The grid was dried overnight under vacuum before measurement.
All the mass spectra were collected on a Thermo LTQ XL ion trap mass spectrometer (Thermo, San Jose, CA, USA) in negative-ion mode. The sample solutions were infused into the mass spectrometer via a Harvard syringe pump (Holliston, MA, USA) at a flow rate of 5 ml/min. The spectra presented represent the average of 200 scans. The needle voltage was set to 3.5 kV. The capillary voltage was set to −20 V and the tube lens offset to −190 V. The heated capillary temperature was set at 250°C and nitrogen sheath and auxiliary gas flows of 45 and 5 arbitrary units.
Preparation of fluorescent silver nanoclusters in the triplex DNA templates
Triplex DNAs were prepared by mixing equimolar amounts of appropriate strands in phosphate buffer, heating to 90°C and slowly cooling to room temperature. The AgNCs were synthesized by cooling the DNA solution and AgNO3 to 0°C and then adding NaBH4 followed by vigorous shaking for 2 min. The solutions were kept in the dark prior to measurements. Unless otherwise specified, final concentrations were 2 µM for each triplex DNA template, 12 µM for AgNO3 and 48 µM for NaBH4. The triplex strands for MS spectra are 10 µM.
Control assembly of fluorescent AgNCs on DNA nanoscaffold
A 59-mer dsDNA (D3, 2 µM) was first prepared by mixing equimolar amounts of appropriate strands in phosphate buffer (DNA sequences were shown in Table 1), heating the mixture to 90°C, and slowly cooling it to room temperature. After that, ssDNA S3 (the molar ratio of S3 to D3 was 2:1) was added to D3 solution, heated to 90°C, and slowly cooled to room temperature to form the triplex–duplex–triplex (T–D–T) structure. The synthesis procedures were the same as those for triplex DNA-templated AgNCs.
Table 1.
The sequences and a labeling scheme of DNA used in this study
| Name | Sequences (5′–3′) | Base length |
|---|---|---|
| Triplex 1 (T1) | 5′-GAG AGG AGA GAG AAG AGG AAG-3′ | 21 |
| 3′- CTC TCC TCT CTC TTC TCC TTC -5′ | ||
| 5′-CTC TCC TCT CTC TTC TCC TTC-3′ | ||
| Duplex 1 (D1) | 5′-GAG AGG AGA GAG AAG AGG AAG-3′ | 21 |
| 3′- CTC TCC TCT CTC TTC TCC TTC -5′ | ||
| S1 | 5′-CTC TCC TCT CTC TTC TCC TTC-3′ | 21 |
| Triplex 2 (T2) | 5′-TTT TTT TTT TTT TTT TTT TTT-3′ | 21 |
| 3′-AAA AAA AAA AAA AAA AAA AAA-5′ | ||
| 5′-TTT TTT TTT TTT TTT TTT TTT-3′ | ||
| Duplex 2 (D2) | 5′-TTT TTT TTT TTT TTT TTT TTT-3′ | 21 |
| 3′-AAA AAA AAA AAA AAA AAA AAA-5′ | ||
| S2 | 5′-TTT TTT TTT TTT TTT TTT TTT-3′ | 21 |
| T1C | 5′-TCTCTCTCTCTCTT-3′ | 14 |
| 3′-AAGAGAGAGAGAGA-5′ | ||
| 5′-TTCTCTCTCTCTCT-3′ | ||
| T2C | 5′-TTCCTTCCTTCCTT-3′ | 14 |
| 3′-AAGGAAGGAAGGAA-5′ | ||
| 5′-TTCCTTCCTTCCTT-3′ | ||
| T3C | 5′-TTCCCTTTTCCCTT-3′ | 14 |
| 3′-AAGGGAAAAGGGAA-5′ | ||
| 5′-TTCCCTTTTCCCTT-3′ | ||
| T2C-2 | 5′-TTCCTTTTTTCCTT-3′ | 14 |
| 3′-AAGGAAAAAAGGAA-5′ | ||
| 5′-TTCCTTTTTTCCTT-3′ | ||
| T2C-4 | 5′-TTCCTTCCTTCCTTCCTT-3′ | 18 |
| 3′-AAGGAAGGAAGGAAGGAA-5′ | ||
| 5′-TTCCTTCCTTCCTTCCTT-3′ | ||
| D3 | 5′-TT CCC TTTT CCC TTT ATA TAT ATA TAT ATA TAT ATA TAT ATA TAT TT CCC TTTT CCC TT-3′ | 59 |
| 3′-AAGGGAAAAGGGAAA TAT ATA TAT ATA TAT ATA TAT ATA TAT ATA AAGGGAAAAGGGAA-5′ | ||
| S3 | 5′-TT CCC TTTT CCC TT-3′ | 14 |
RESULTS AND DISCUSSION
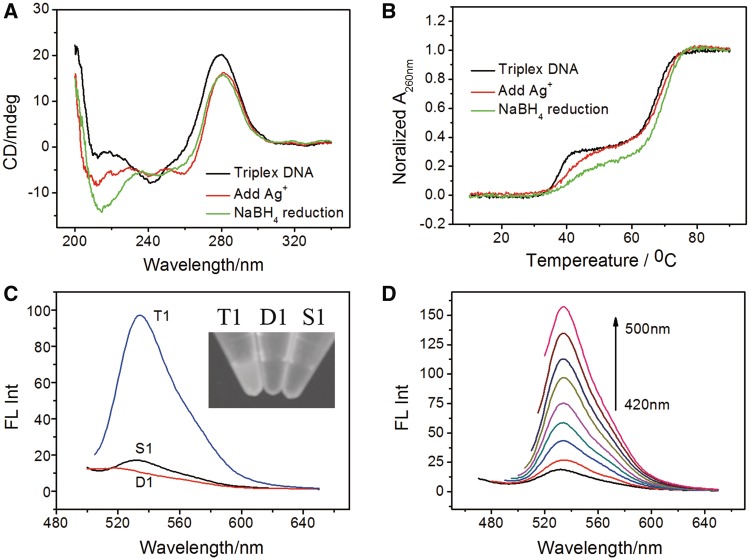
A 21-mer triplex DNA (T1) containing CG.C+ triplets was first employed as template for synthesis of AgNCs (DNA sequences were shown in Table 1). T1 was formed by binding an oligopyrimidine single strand (S1) in the major groove of the duplex strand (D1). The formation of T1 was confirmed by its characteristic UV melting curve and CD spectra of triplex DNA (Figure 1A and B, black line). The AgNCs were obtained by mixing T1 solution with AgNO3, followed by reduction with NaBH4. The presence of negative bands around 210 nm, known as an indicator of the DNA triplex, in both T1-Ag(I) ion and T1-AgNCs indicated that the formation of AgNCs did not destroy the triplex structure of T1 (Figure 1A) (30,31). Moreover, the more negative ellipticity at 210 nm of T1 in the presence of Ag(I) ion and AgNCs, which was due to the coordination distance in N-Ag(I)/Ag(0)-N was longer than that of the Hoogsteen hydrogen bonds in CG.G+, revealing that the Ag(I) ions/AgNCs were involved in the T1 DNA (30,31). In addition, the UV melting studies showed that the T1, T1-Ag(I) ion and T1-AgNCs were all contained two typical triplex melting transitions, in which the triplex strands first melted to single strand and duplex (Tm1), then the duplex in turn melted to single strands (Tm2) (27–29,32–34) (Figure 1B) and these melting temperatures were increased in the presence of Ag(I) ions/AgNCs. These results further demonstrated that the T1 retained its integrity during the synthesis of AgNCs.
Figure 1.
(A) CD spectra of (a) T1, (b) T1-Ag(I) ions and (c) T1-AgNCs. (B) UV melting analysis of T1, T1-Ag(I) ion and T1-AgNCs. (C) Fluorescence emission spectra acquired for T1, D1, S1 solutions, upon excitation at 480 nm. Inset: Fluorescence images of T1, D1, S1-templated AgNCs samples (from left to right). (D) Fluorescence emission spectra of T1-AgNCs with excitation wavelength from 420 nm to 500 nm. [DNA] = 2 µM, [AgNO3] = 12 µM, [NaBH4] = 48 µM.
The fluorescence properties of T1-AgNCs were then studied. As expected, strong emission was observed for the T1-AgNCs solution (Figure 1C). Supplementary Figure S1 shown the variation in the fluorescence intensities of AgNCs as a function of time (λex = 480 nm/λem = 534 nm). The fluorescence reached a maximum at about 3 days after adding of NaBH4 to Ag(I) ions/DNA solutions. In contrast, very weak fluorescence was observed when D1 or S1 was used as template (Figure 1C and Supplementary Figure S1), which ruled out the possibility that the fluorescence was originated from D1/S1-templated AgNCs. In additions, a triplex DNA containing only TA.T triplets (T2) was also used for the synthesis of AgNCs. In Supplementary Figure S2, little emission was observed when T2 was acted as template, which indicated that the CG.C+ triplet was indispensible for the formation of fluorescent AgNCs. The fluorescence quantum yield of the T1-templated AgNCs was 33.2% (relative to rhodamine B in ethanol), which was comparable with previous reports (9). Besides, the initial Ag(I) ions: triplex strand concentration stoichiometry had a strong influence on the formation of the AgNCs. The fluorescence intensity reached a maximum at 6 Ag(I) ions: DNA strand (Supplementary Figure S3). Moreover, pH effect was also studied. The AgNCs emission intensity was higher in acidic solutions, which was consistent with the well-known fact that DNA triplex containing CG.C+ triplets was more stable in acidic conditions (Supplementary Figure S4 and Supplementary Table S1) (27–33). In addition, we also measured the fluorescence spectra at the same pH by diluting the AgNCs samples prepared at different pHs in neural solutions. In Supplementary Figure S4C, the fluorescent intensities of these AgNCs sample prepared at different pHs became almost identical to each other after diluted in same neural solutions. This result showed further evidence that the pH-dependent stability of DNA triplex have significant influence on the fluorescent properies of AgNCs.
The above result confirmed that triplex DNA with CG.C+ triplets could serve as a novel efficient template for the synthesis of fluorescent AgNCs. More importantly, we observed an interesting fluorescence phenomenon in which the emission peak of T1-AgNCs was not change over a range of excitation wavelengths (Figure 1D). This unique feature was distinct from any other DNA-templated AgNCs system, where the emission peak was red-shifted with increasing excitation wavelength. Previous studies indicated that the appearance of the multiple emissions was due to the presence of multiple species of fluorescent AgNCs containing different number of Ag atoms in DNA-AgNCs solution (13,14,34–36). Therefore, the unchanged emission peak of T1-AgNCs in our system suggested that there might be just one single species of fluorescent AgNCs in the T1-AgNCs solution.
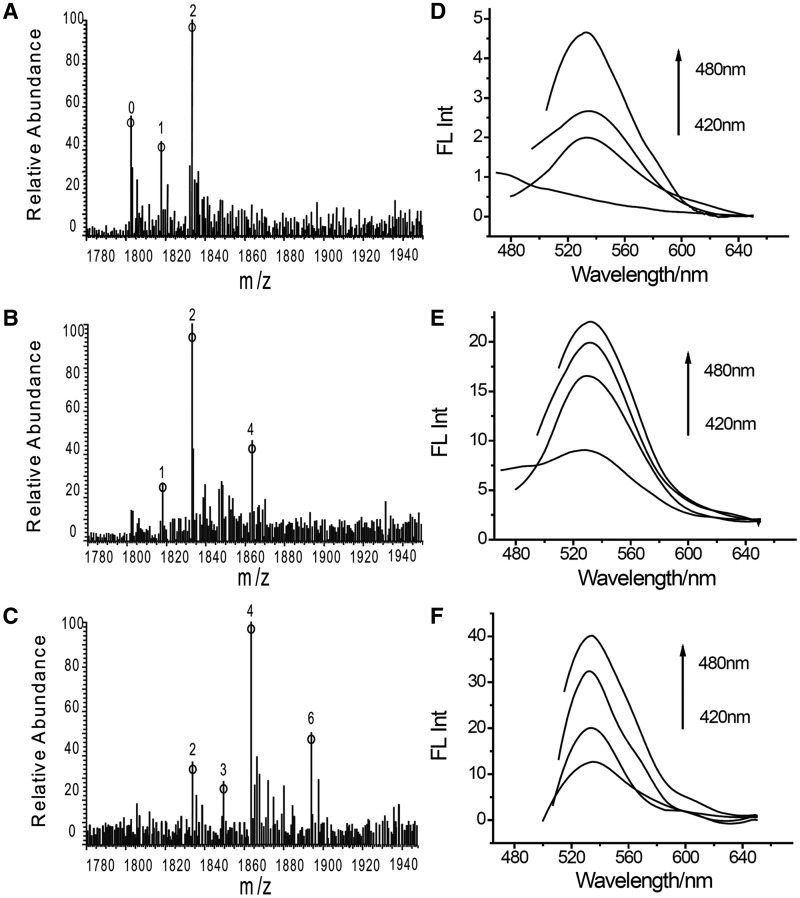
Mass spectra analysis has been proved to be a useful tool for characterization of the AgNCs species formed in DNA template (8,13,14). In order to demonstrate the homogeneity of triplex DNA-templated AgNCs, a 14-mer triplex DNA T2C, which were more suitable for mass spectra study than T1, was used for synthesis and characterization of AgNCs. T2C (Sequence was shown in Table 1) was designed to contain three sets of two successive CG.C+ triplets, separated by eight TA.T triplets. At lower ratio of Ag(Ι) ions/DNA (1:1), MS studies (Figure 2A) showed the presence of dominant T2C-2Ag species and small amount of T2C-1Ag species at lower ratio of Ag(Ι) ions/DNA (1:1). The fluorescent properties of T2C-AgNCs are similar to that of T1-AgNCs: changing the excitation wavelengths had nearly no effect on the emission peak of T2C-AgNCs (Figure 2D). Considering that single Ag atom or Ag(Ι) ion is non-fluorescent, the only one fluorescent species in T2C-AgNCs solution should be the Ag2NCs. This is different from any other current DNA-templated AgNCs, where multiple species of AgNCs are coexisted (13,14,34–36). By increasing the ratio of Ag(Ι) ions:DNA to 3:1 and 6:1, MS studies showed that dominant T2C-4Ag species and T2C-6Ag species were obteined, respectively (Figure 2B and C). Meanwhile, the fluorescent intensity of T2C-AgNCs was increased as the ratio of Ag(Ι) ions/DNA increased while their emission peaks were identical to that of T2C-AgNCs at lower ratio of Ag(Ι) ions/DNA. Furthermore, their emission peaks were unchanged with the excitation wavelength (Figure 2E and F). Thus, the observed T2C-4Ag and T2C-6Ag species at higher ratio of Ag(Ι) ions/DNA should be attributed to the presence of two Ag2 clusters and three Ag2 clusters in the triplex template, which further indicated the homogeneity of T2C-AgNCs. We also performed the absorption and excitation spectrum studies of T2C-AgNCs. In Supplementary Figure S6, the single peaks in the absorption spectra and unchanged excitation peak of triplex DNA-AgNCs at different Ag(Ι) ions/DNA ratio further suggested the presence of only one species of AgNCs in the as-prepared sample. Moreover, considering that T2C contains three sets of two successive CG.C+ triplets and Ag(I) ions could specifically bind to the CG.C+ base site of triplex DNA, (30) homogenous Ag2 in T2C was implied to formed by the in situ reduction of bound Ag(I) ion in the CG.C+ triplets site of T2C. We further investigated the influence of the number of the set of two successive CG.C+ triplets on AgNCs synthesis. The triplex DNA T2C-2 and T2C-4 containing two and four sets of two successive CG.C+ triplets, respectively, were used as AgNCs template to compare with T2C containing three sets of two successive CG.C+ triplets. In Supplementary Figures S7 and S8, emission intensity of AgNCs increased with the increasing of number of the set of two successive CG.C+ triplets while the corresponding emission wavelengths unchanged. These results further supported our conclusion that fluorescent Ag2 clusters were formed specifically on the CG.C+ sites.
Figure 2.
ESI-TOF mass spectra of the T2C-AgNCs in different ratio of Ag(I) ions /DNA: (A) 1:1, (B) 3:1 and (C) 6:1, [DNA] = 10µM. A series of fluorescence emission spectra for AgNCs formed in T2C at corresponding ratio of (D) 1:1, (E) 3:1 and (F) 6:1, [DNA] = 5 µM.
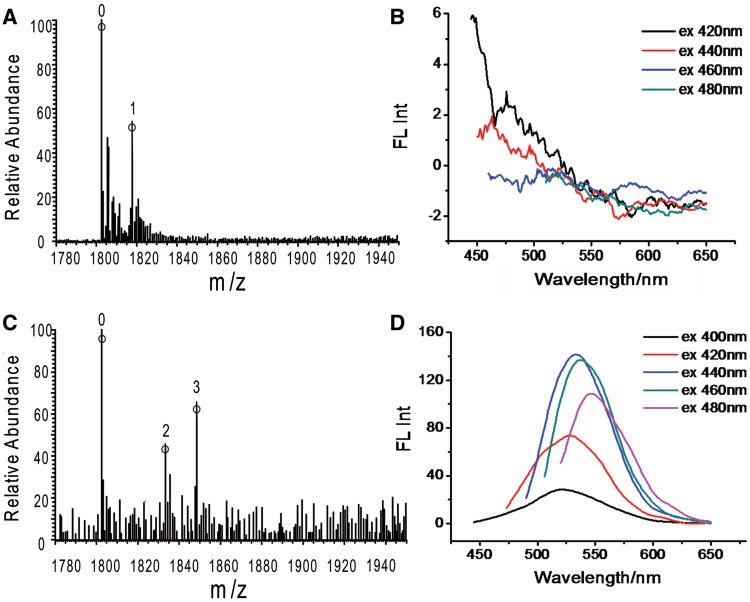
To show further evidence of the in situ reduction of bound Ag(Ι) ion, another two 14-mer triplex DNA, i.e. T1C and T3C were used for AgNCs synthesis (Supplementary Figure S5). Like T2C, both T1C and T3C were designed to contain six CG.C+ triplets. However, the relative position of CG.C+ in T1C and T3C were different (Figure 3). In T1C triplex, six CG.C+ triplets were separated by eight TA.T triplets, created six discrete CG.C+ triplets. In Figure 3B, no emission was observed when T1C was used as template. MS spectra showed that only one Ag atom was presented in T1C at lower ratio of Ag(Ι) ions /DNA (1:1; Figure 3A). MS results combining with fluorescent spectra research inferred that discrete Ag(0) atom rather than fluorescent AgNCs were formed in T1C, which was corresponding to the discrete position of CG.C+ in T1C. For T3C triplex with two sets of three successive CG.C+ triplets, the emission peak of T3C-AgNCs (Figure 3D) was found to be red-shifted with increasing excitation wavelength, revealing the presence of multiple species of AgNCs in T3C-AgNCs. MS studies (Figure 3C) showed the presence of dominant T3C-3Ag and small amount of T3C-2Ag in T3C-AgNCs solution at lower ratio of Ag(Ι) ions /DNA(1:1). Combining the fluorescence and the MS studies, the T3C-2Ag and T3C-3Ag species might be the T3C-Ag2 and T3C-Ag3 nanoclusters. Overall, the presence of Ag atom, Ag2 clusters and Ag3 clusters as dominant species in T1C (with discrete CG.C+ triplet site), T2C (with two successive CG.C+ triplet sites) and T3C (with three successive CG.C+ triplet sites), respectively, further demonstrated the in situ reduction of bound Ag(I) ion and the formation of AgNCs in the very position of CG.C+ triplet site in triplex DNA. In particular, to the best of our knowledge, the formation of Ag2 cluster in CG.C+ triplet site of T2C triplex was the first report of using DNA as template to synthesis homogeneous and site-addressable AgNCs.
Figure 3.
(A and C) ESI-TOF mass spectra of T1C and T3C. [DNA] = 10 µM. (B and D) Fluorescence emission spectra of T1C and T3C solutions. The ratio of Ag(I) ions to DNA is 1:1, [DNA] = 5 µM.
In addition, the as-prepared triplex DNA-AgNCs were very stable against salt (Supplementary Figure S9). No obvious change of the fluorescence properties was observed for the T1-AgNCs in aqueous solution containing 200 mM NaCl for several hours (Supplementary Figure S9B). That was inaccessible to most of other DNA-templated AgNCs (15). The homogeneity and ultra-stability against salt of triplex-AgNCs implied that, compared with single-stranded DNA and duplex DNA, triplex DNA could provide better protection for the AgNCs against further growth to larger nanostructures and quenching in solution. Importantly, this ultrastability under high salt condition would be highly beneficial its application in bioimaging and nanoassembly (8–12,37).
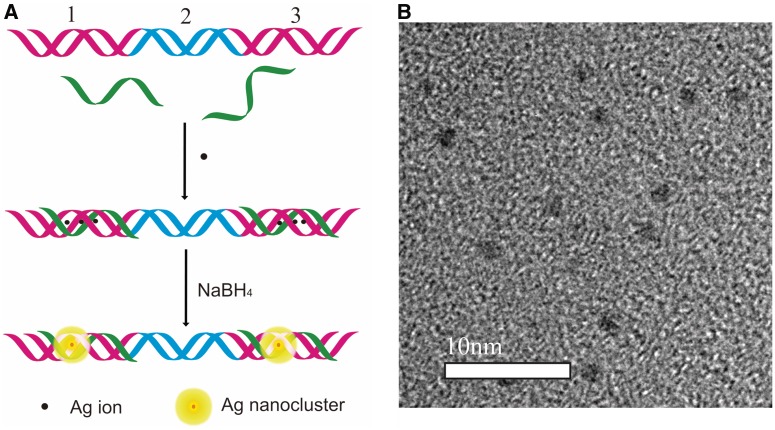
Recently, DNA-mediated assembly of noble metal nanomaterial has received much attention due to their potential application in nanoelectronics, biosensing, clinical diagnostic and dynamic materials (37–44). Using mismatched dsDNA as template, we have reported the controlled alignment of fluorescent AgNCs and use it as an efficient scaffold for SNP detection (13). In situ immobilization of fluorescent AgNCs on DNA origami has also been reported by Liu’s group using sugar-modified DNA as template (16). As demonstrated here, by taking advantage of the site-specific, homogeneous and highly stable feature of our system, we could carry out a convenient and efficient platform for control assembly of fluorescent AgNCs on DNA nanoscaffold. For this purpose, a 59-mer dsDNA (D3, Table 1) was used as a model DNA scaffold. In order to realize the control assembly of fluorescent AgNCs on D3 scaffold, ssDNA (S3) are introduced to D3 solution. S3 could specifically bind to the region 1 and region 3 of D3, resulting in a triplex–duplex–triplex (T–D–T) structure (Figure 4A). As the DNA duplex could not host the formation of AgNCs (13,14), they could act as a rigid linker to create nanostructures with alternating metalized and non-metalized parts. Importantly, fluorescent AgNCs could be selectively formed at the desired sites of triplex region 1 and region 3 through triplex addressable strategy. In that case, the metalized DNA scaffold would appear in the form of an AgNCs dimer separated by a duplex DNA linker with a fixed distance. Indeed, the TEM image (Figure 4B and Supplementary Figure S10) and the corresponding distance distribution histogram (Supplementary Figure S11) showed evidence that the nanocluster dimers with a separation distance of 5.96 ± 1.85 nm were generated by the triplex addressable strategy. The length of the duplex between the nanoclusters was calculated to be 10.54 nm. The shorter distance might be primarily due to a decrease in the persistence length of DNA strand by silver ions and silver nanostructures. The flexibility of the DNA and the process of depositing the DNA nanoclusters onto copper grids for TEM imaging might also affect the distance (45). The fluorescence spectrum of 59-mer DNA-templated AgNCs was shown in Supplementary Figure S12. These result indicated that fluorescent AgNCs dimers were obtained in triplex DNA.
Figure 4.
(A) Schematic representation of the controlled alignment of AgNCs on DNA template. (B) TEM image of AgNCs dimers formed on 59-mer DNA, [DNA] = 2 µM, [AgNO3] = 12 µM, [NaBH4] = 48 µM.
Compared with other methods for control assembly of fluorescent AgNCs, the triplex addressable strategy presented here is more attractive for several reasons. First, the strategy is easy to operate, avoiding any covalent labelling or modification of DNA. Secondly, DNA triplex is a very useful building block for sequence-specific labelling and construction of DNA nanomaterial since the specific binding of the a third single strand in the major groove of the duplex strand in the formation of triplex DNA is an elegant and simple way to specifically target a defined sequence in complex DNA structure. Moreover, triplex DNA formation is easily controlled by a change in pH and temperature. This indicates that the triplex DNA-AgNCs system could be easily applied to more complicated DNA nanomaterial and could be steered by temperature and pH. Moreover, the ultra-stability and homogeneity of triplex DNA-AgNCs offered great potential for further use of AgNCs assembly in practical application.
CONCLUSION
In conclusion, we have successfully demonstrated a new strategy to create site-specific, homogeneous and bright AgNCs with high-stability against salt using triplex DNA as templates. We found that the formation of AgNCs is highly dependent the presence and position of CG.C+ triplets in triplex DNA. By reasonable design of DNA sequence, homogeneous Ag2 cluster could be obtained in the very position of CG.C+ site of triplex DNA. The site-specific strategy was further explored for controllable alignment of Ag clusters at the pre-defined positions on the DNA nanoscaffold. To the best of our knowledge, this is the first example to simultaneously answer the challenges of excellent site-specific control of NC nucleation and growth, homogeneity and stability against salt of DNA-templated AgNCs. By combining the strategy described herein with the inherent biodiversity and molecular self-assembly properties of DNA, we anticipate that this metalized DNA-based structures might be important for the construction of novel DNA-based nanomaterials and nanomechanical devices with more sophisticated functions and would be highly beneficial in future nanomechanical, DNA origami and electronic applications.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Table 1 and Supplementary Figures 1–12.
FUNDING
The National Basis Research Program of China [2012CB720602, 2011CB936004]; the National Natural Science Foundation of China [20831003, 90813001, 20831006, 90913007, 20903086]. Funding for open access charge: National Natural Science Foundation of China and Chinese Academy of Sciences.
Conflict of interest statement. None declared.
Supplementary Material
REFRENCES
- 1.Lee TH, Dickson RM. Discrete two-terminal single nanocluster quantum optoelectronic logic operations at room temperature. Proc. Natl Acad. Sci. USA. 2003;100:3043–3046. doi: 10.1073/pnas.0635474100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wilcoxon JP, Abrams BL. Synthesis, structure and properties of metal nanoclusters. Chem. Soc. Rev. 2006;35:1162–1194. doi: 10.1039/b517312b. [DOI] [PubMed] [Google Scholar]
- 3.Templeton AC, Wuelfing WP, Murray RW. Monlayer-protected cluster molecules. Acc. Chem. Res. 2000;33:27–36. doi: 10.1021/ar9602664. [DOI] [PubMed] [Google Scholar]
- 4.Yu J, Patel SA, Dickson RM. In vitro and intracellular production of peptide-encapsulated fluorescent silver nanoclusters. Angew. Chem., Int. Ed. 2007;46:2028–2030. doi: 10.1002/anie.200123456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Guo C, Irudayaraj J. Fluorescent Ag clusters via a protein-directed approach as a Hg(II) ion sensor. Anal. Chem. 2011;83:2883–2889. doi: 10.1021/ac1032403. [DOI] [PubMed] [Google Scholar]
- 6.Zheng J, Dickson RM. Individual water-soluble dendrimer-encapsulated silver nanodots fluorescence. J. Am. Chem. Soc. 2002;124:13982–13983. doi: 10.1021/ja028282l. [DOI] [PubMed] [Google Scholar]
- 7.Xu H, Suslick KS. Sonochemical synthesis of highly fluorescent Ag nanoclusters. ACS Nano. 2010;4:3209–3214. doi: 10.1021/nn100987k. [DOI] [PubMed] [Google Scholar]
- 8.Petty JT, Zheng J, Hud NV, Dickson RM. DNA-templated Ag nanocluster formation. J. Am. Chem. Soc. 2004;126:5207–5212. doi: 10.1021/ja031931o. [DOI] [PubMed] [Google Scholar]
- 9.Richards CI, Sungmoon C, Hsiang JC, Antoku Y, Vosch T, Bongiorno A, Tzeng YL, Dickson RM. Oligonucleotide-stabilized Ag nanocluster fluorophores. J. Am. Chem. Soc. 2008;130:5038–5039. doi: 10.1021/ja8005644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yeh H, Sharma J, Han J, Martinez JS, Werner JH. A DNA-silver nanocluster probe that fluoresces upon hybridization. Nano Lett. 2010;10:3106–3110. doi: 10.1021/nl101773c. [DOI] [PubMed] [Google Scholar]
- 11.Sengupta B, Ritchie CM, Buckman JG, Johnsen KR, Goodwin PM, Petty JT. Base-directed formation of fluorescent silver clusters. J. Phys. Chem. C. 2008;112:18776–11782. doi: 10.1021/jp804031v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Neidig ML, Sharma J, Yeh H, Martinez JS, Conradson SD, Shreve AP. Ag K-edge EXAFS analysis of DNA-templated fluorescent silver nanoclusters: insight into the structural origins of emission tuning by DNA sequence variations. J. Am. Chem. Soc. 2011;133:11837–11839. doi: 10.1021/ja202797w. [DOI] [PubMed] [Google Scholar]
- 13.Huang Z, Pu F, Hu D, Wang C, Ren J, Qu X. Site-specific DNA-programmed growth of fluorescent and functional silver nanoclusters. Chem. Eur. J. 2011;17:3774–3780. doi: 10.1002/chem.201001795. [DOI] [PubMed] [Google Scholar]
- 14.Gwinn EG, O’Neill P, Guerrero AJ, Bouwmeester D, Fygenson DK. Sequence-dependent fluorescent of DNA-hosted silver nanoclusters. Adv. Mater. 2008;20:279–283. [Google Scholar]
- 15.Sharma J, Yeh HC, Yoo H, Werner JH, Martinez JS. A complementary palette of fluorescent silver nanoclusters. Chem. Commun. 2010;46:3280–3282. doi: 10.1039/b927268b. [DOI] [PubMed] [Google Scholar]
- 16.Pal S, Varghese R, Deng Z, Zhao Z, Kumar A, Yan H, Liu Y. Site-specific synthesis and in situ immobilization of fluorescent silver nanoclusters on DNA nanoscaffolds by use of the Tollens reaction. Angew. Chem., Int. Ed. 2011;50:4176–4179. doi: 10.1002/anie.201007529. [DOI] [PubMed] [Google Scholar]
- 17.Keren K, Berman RS, Braun E. Patterned DNA metallization by sequence-specific localization of a reducing agent. Nano Lett. 2004;4:323–326. [Google Scholar]
- 18.Burley GA, Gierlich J, Mofid MR, Nir H, Tal S, Eichen Y, Carell T. Directed DNA metallization. J. Am. Chem. Soc. 2006;128:1398–1399. doi: 10.1021/ja055517v. [DOI] [PubMed] [Google Scholar]
- 19.Wirges CT, Timper J, Fischler M, Sologubenko AS, Mayer J, Simon U, Carell T. Controlled nucleation of DNA metallization. Angew. Chem., Int. Ed. 2008;48:219–223. doi: 10.1002/anie.200803123. [DOI] [PubMed] [Google Scholar]
- 20.Xu H, Suslick KS. Water-soluble fluorescent silver nanoclusters. Adv. Mater. 2010;22:1078–1082. doi: 10.1002/adma.200904199. [DOI] [PubMed] [Google Scholar]
- 21.Shukla S, Sastry M. Probing differential Ag+-nucleobase interactions with isothermal titration calorimetry (ITC): towards patterned DNA metallization. Nanoscale. 2009;1:122–127. doi: 10.1039/b9nr00004f. [DOI] [PubMed] [Google Scholar]
- 22.Lan GY, Huang CC, Chang HT. Silver nanoclusters as fluorescent probes for selective and sensitive detection of copper ions. Chem. Commun. 2010;46:1257–1259. doi: 10.1039/b920783j. [DOI] [PubMed] [Google Scholar]
- 23.Blommers MJ, Natt F, Jahnke W, Cuenoud B. Dual recognition of double-stranded DNA by 2’-aminoethoxy-modifiedoligonucleotides: the solution structures of an intramolecular triplex obtained by NMR spectroscopy. Biochemistry. 1998;37:17714–11725. doi: 10.1021/bi9816352. [DOI] [PubMed] [Google Scholar]
- 24.Chen Y, Lee SH, Mao C. A DNA nanomachine based on a duplex-triplex transition. Angew. Chem., Int. Ed. 2004;43:5335–5338. doi: 10.1002/anie.200460789. [DOI] [PubMed] [Google Scholar]
- 25.Tumpane J, Kumar R, Lundberg EP, Sandin P, Gale N, Nandhakumar IS, Albinsson B, Lincoln P, Wilhelmsson LM, Brown T, et al. Triplex addressability as a basis for functional DNA nanostructures. Nano Lett. 2007;7:3832–3839. doi: 10.1021/nl072512i. [DOI] [PubMed] [Google Scholar]
- 26.Takezawa Y, Maeda W, Tanaka K, Shionoya M. Discrete self-assembly of iron ions inside triple-stranded artifical DNA. Angew. Chem., Int. Ed. 2009;48:1081–1084. doi: 10.1002/anie.200804654. [DOI] [PubMed] [Google Scholar]
- 27.Htun H, Dahlberg JE. Topology and formation of triple-stranded H-DNA. Science. 1989;243:1571–1576. doi: 10.1126/science.2648571. [DOI] [PubMed] [Google Scholar]
- 28.Plum GE, Park YW, Singleton SF, Dervan PB, Breslauer KJ. Thermodynamic characterization of the stability and the melting behavior of a DNA triplex-a spectroscopic and calorimetric study. Proc. Natl Acad. Sci. USA. 1990;87:9436–9440. doi: 10.1073/pnas.87.23.9436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xing F, Song G, Ren J, Chaires JB, Qu X. Molecular recognition of nucleic acids: coralyne binds strongly to poly(A) FEBS Lett. 2005;579:5035–5039. doi: 10.1016/j.febslet.2005.07.091. [DOI] [PubMed] [Google Scholar]
- 30.Ihara T, Ishii T, Araki N, Wilson AW, Jyo A. Silver ion unusally stabilizes the structure of a paralled-motif DNA triplex. J. Am. Chem. Soc. 2009;131:3826–3287. doi: 10.1021/ja809702n. [DOI] [PubMed] [Google Scholar]
- 31.Arakawa H, Neault JF, Tajmir-Riahl HA. Silver(I) complexes with DNA and RNA studied by fourier tranform infrared spectroscopy and capillary electrophoresis. Biophys. J. 2001;81:1580–1587. doi: 10.1016/S0006-3495(01)75812-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Song G, Xing F, Qu X, Chaires JB, Ren J. Oxazine 170 induces DNA:RNA:DNA triplex formation. J. Med. Chem. 2005;48:3471–3473. doi: 10.1021/jm050131g. [DOI] [PubMed] [Google Scholar]
- 33.Feng L, Li X, Peng Y, Geng J, Ren J, Qu X. Spectral and electrochemical detection of protonated triplex formation by a small-molecule anticancer agent. Chem. Phys. Lett. 2009;480:9–12. [Google Scholar]
- 34.Lesniak W, Bielinska AU, Sun K, Janczak KW, Shi X, Baker JR, Balogh LP. Silver/dendrimer nanocomposites as biomarkers: fabrication, characterization, in vitro toxicity, and intracellular detection. Nano Lett. 2005;11:2123–2130. doi: 10.1021/nl051077u. [DOI] [PubMed] [Google Scholar]
- 35.Shen Z, Duan HW, Frey H. Water-soluble fluorescent Ag nanoclusters obtained from multiarm star poly (acrylic acid) as “molecular hydrogel” templates. Adv. Mater. 2007;19:349–352. [Google Scholar]
- 36.Zhang J, Xu S, Kumacheva E. Photogeneration of fluorescent silver nanoclusters in polymer microgels. Adv. Mater. 2005;17:2336–2340. [Google Scholar]
- 37.Pal S, Deng Z, Ding B, Yan H, Liu Y. DNA-origami-directed self-assembly of discrete silver-nanoparticle architectures. Angew. Chem. Int. Ed. 2010;49:2700–2704. doi: 10.1002/anie.201000330. [DOI] [PubMed] [Google Scholar]
- 38.Rothemund PWK. Folding DNA to create nanoscale shapes and pattens. Nature. 2006;440:297–302. doi: 10.1038/nature04586. [DOI] [PubMed] [Google Scholar]
- 39.Béccrril HA, Wooley AT. DNA-templated nanofabrication. Chem. Soc. Rev. 2009;38:329–337. doi: 10.1039/b718440a. [DOI] [PubMed] [Google Scholar]
- 40.Keren K, Krueger M, Gilad R, Ben-Yoseph G, Sivan U, Braun E. Sequence-specific molecular lithography on single DNA molecules. Sceince. 2002;297:72–76. doi: 10.1126/science.1071247. [DOI] [PubMed] [Google Scholar]
- 41.Zhao W, Gao Y, Kandadai SA, Brook MA, Li Y. DNA polymerizattion on gold nanoparticles through rolling circle amplification: towards novel scaffolds for three-dimensional periodic nanoassemblies. Angew. Chem. Int. Ed. 2006;45:2409–2413. doi: 10.1002/anie.200600061. [DOI] [PubMed] [Google Scholar]
- 42.Beyer S, Nickels P, Simmel F. Periodic DNA nanotemplates synthesized by rolling circle amplification. Nano Lett. 2005;5:719–722. doi: 10.1021/nl050155a. [DOI] [PubMed] [Google Scholar]
- 43.Sharma J, Ke Y, Lin C, Chhabra R, Wang Q, Nangreave J, Liu Y, Yan H. DNA-tile-directed self-assembly of quantumm dots into two-dimensional nannopatterns. Angew. Chem., Int. Ed. 2008;47:5157–5159. doi: 10.1002/anie.200801485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rotaru A, Dutta S, Jentzsch E, Gothelf K, Mokhir A. Selective dsDNA-templated formation of copper nanoparticles in solution. Angew. Chem., Int. Ed. 2010;49:5665–5667. doi: 10.1002/anie.200907256. [DOI] [PubMed] [Google Scholar]
- 45.Zinchenko AA, Baigl D, Chen N, Pyshkina O, Endo K, Sergeyev VG, Yoshikawa K. Conformational behavior of giant DNA through binding with Ag+ and metallization. Biomacromolecules. 2008;9:1981–1987. doi: 10.1021/bm800235j. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.