Abstract
Hfq is an important RNA-binding protein that helps bacteria adapt to stress. Its primary function is to promote pairing between trans-acting small non-coding RNAs (sRNAs) and their target mRNAs. Identification of essential Hfq-binding motifs in up-stream regions of rpoS and fhlA led us to ask the question whether these elements are a common occurrence among other Hfq-dependent mRNAs as well. Here, we confirm the presence of a similar (ARN)x motif in glmS RNA, a gene controlled by two sRNAs (GlmZ and GlmY) in an Hfq-dependent manner. GlmZ represents a canonical sRNA:mRNA pairing system, whereas GlmY is non-canonical, interfacing with the RNA processing protein YhbJ. We show that glmS interacts with both Hfq-binding surfaces in the absence of sRNAs. Even though two (ARN)x motifs are present, using a glmS:gfp fusion system, we determined that only one specific (ARN)x element is essential for regulation. Furthermore, we show that residues 66–72 in the C-terminal extension of Escherichia coli Hfq are essential for activation of GlmS expression by GlmY, but not with GlmZ. This result shows that the C-terminal extension of Hfq may be required for some forms of non-canonical sRNA regulation involving ancillary components such as additional RNAs or proteins.
INTRODUCTION
Small non-coding RNAs (sRNAs) are used to modulate a wide variety of cellular responses in both bacterial and eukaryotic systems (1–4). In eukaryotes microRNAs and siRNAs (∼21 nt) often regulate translation and RNA turnover and defend the genome from invasive nucleic acids (3). Similarly, in bacterial systems, two main classes of sRNAs have been identified based on the origin of the sRNAs relative to their mRNA target. Cis-acting sRNAs base pair extensively with mRNAs as they are transcribed from the opposite strand of their target RNA (4). These systems are often used by mobile genetic elements such as bacteriophages, transposons and plasmids to regulate translation, RNA processing and RNA decay (5,6). Riboswitches represent another ubiquitous class of cis-acting RNAs. Riboswitches respond to cellular stimuli by refolding the mRNAs via their 5′-UTRs to trigger translation regulation or transcription termination (7). In contrast, trans-acting RNAs are synthesized from different genetic loci than their targets and are widely used to modulate stress responses in bacteria. Unlike cis-RNAs, trans-acting RNAs can accommodate imperfect base pairing with their target RNAs and often require the RNA-binding protein Hfq to facilitate complex formation.
Hfq was first identified as a host factor involved in the replication of phage Qβ RNA (8). Hfq belongs to the Sm/Lsm family of proteins and associates into a homo-hexameric structure to form the functional nucleic acid-binding core. In general, the nucleic acid-binding Sm1 and Sm2 domains are highly conserved across bacterial species (7–66 amino acids) and form two distinct RNA-binding surfaces, often called the proximal and distal surfaces, respectively (9). These two RNA-binding surfaces are used to promote sRNA–mRNA base pairing by increasing the local concentration of the two binding partners. Bacterial strains lacking Hfq exhibit increased sensitivity to stress and reduced pathogenesis among other phenotypes (10–14). One of the most intriguing features of Hfq-mediated gene regulation is the network of regulatory responses that allows a single sRNA to trigger a multi-dimensional response to stress (15–17). For instance, the sRNA RyhB regulates multiple genes (including sodB, ftnA, bfR, acnA and sdhC) to help the organism respond to low iron concentration (18). While the use of Hfq in many other stress pathways such as cold shock (DsrA) (19), oxidative stress (OxyS) (20) and sugar stress (SgrS) (21,22) are well established, a common mechanism describing how Hfq controls all these simultaneously is lacking.
The recent discovery of Hfq-binding elements, so-called (ARN)x motifs, found in upstream regions of those mRNAs that are regulated by sRNAs has provided new clues for how Hfq helps mediate gene regulation. In both rpoS and fhlA mRNAs, these Hfq-binding sequences are essential for regulation (23–25) and structural data confirmed that the (ARN)x motif specifically interacts with the distal RNA-binding surfaces of Hfq hexamers (26). If these upstream Hfq-binding motifs are a common feature of regulated messages, this could help explain how Hfq rapidly targets specific mRNAs for regulation during stress. In theory, as described in the fhlA work, Hfq could pre-form complexes with relevant mRNAs and could promote an efficient stress response upon sRNA synthesis if and when it is induced by environmental stimuli (25). Furthermore, recent work on the DsrA-rpoS system has shown that Hfq binding to rpoS refolds the mRNA for a favorable interaction with the sRNA (27). These observations, although currently limited to a few systems, help explain the complex manner by which Hfq facilitates integrated responses to environmental stress.
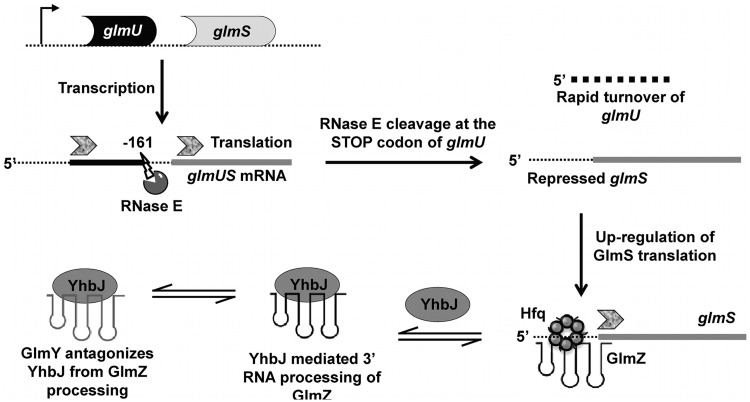
To further understand the importance of upstream (ARN)x motifs in Hfq-dependent gene regulation, the glmS system was studied. glmS encodes an essential enzyme glucosamine-6-phosphate synthase (GlcN-6-P) (28). In many Gram-positive bacteria (such as Bacillus subtilis) the expression of GlmS is tightly regulated by a self-cleaving ribozyme located within glmS that catalyzes cleavage in response to high GlcN-6-P concentrations (29). However, in Gram-negative species, GlmS expression is regulated by two sRNAs GlmZ and GlmY, a dual regulatory system that requires Hfq for activity (30). In addition to Hfq and RNase E, YhbJ, a predicted NTPase, was shown to be involved in this regulatory network in Escherichia coli (31) (Figure 1). In brief, GlmS is encoded as a poly-cistronic message that also includes GlmU, a gene for a bifunctional enzyme that produces UDP-GlcNAc. In order to individually regulate GlmS expression, RNase E cleaves the glmU–glmS transcript at the stop codon of glmU to produce a monocistronic glmS transcript that is translationally repressed by its 5′-UTR structure (30). GlmZ interacts with glmS to unmask the ribosome-binding site (RBS) and promote translation. Additional modulation is provided by YhbJ, a protein involved in GlmZ turnover and thus negatively regulates GlmS expression by decreasing the amount of GlmZ available to activate glmS (30). GlmY is a second sRNA involved in this pathway. GlmY has significant sequence and structural homology to GlmZ such that when it is expressed, GlmY acts as a decoy, recruiting YhbJ away from GlmZ (32). Thus, GlmY prevents GlmZ degradation increasing GlmZ concentration and allowing more glmS-GlmZ adduct formation, with a net result of enhancing GlmS expression.
Figure 1.
Mechanism of Hfq-mediated regulation of GlmS with sRNAs, GlmZ and GlmY [adapted from (31)]. glmS is transcribed as a poly-cistronic message that also includes glmU. Upon transcription, RNase E cleaves at the stop codon of glmU (at −161 nt position with respect to the translation start site of glmS) that separates the translational control of GlmS and GlmU genes. The RNase E cleaved glmS transcript is repressed for translation by its 5′-UTR structure that masks the RBS. Upon synthesis of GlmZ sRNA, GlmS translation is activated in an Hfq-dependent manner. Here the GlmZ–glmS interaction relieves the inhibitory structure of glmS for translation. YhbJ, a predicted NTPase negatively regulates GlmS expression by processing GlmZ sRNA. GlmY, a second sRNA which has structural and sequence homology to GlmZ, antagonizes YhbJ from GlmZ, upregulating GlmS expression. Overall, the GlmS expression is activated by two homologous sRNAs GlmZ and GlmY in a coordinated manner in the presence of Hfq.
In this work, we show that the glmS interaction with Hfq is similar to what was previously reported with fhlA, binding both the proximal and distal surfaces of Hfq. Structural characterization of glmS revealed two potential (ARN)x motifs similar to those found in fhlA and rpoS. Using a GFP fusion system we show that one of the two upstream (ARN)x elements is essential for Hfq-mediated regulation of glmS by both GlmZ and GlmY whereas the other is dispensable. Furthermore, we probed the requirement of the variable C-terminal region of Hfq for regulation—a topic that has been very controversial. We show that the Hfq C-terminus is not required for the classical Hfq regulation involving GlmZ pairing with GlmS. However, at least seven residues of the C-terminal extension (residues 65–72) were essential for GlmY activation of GlmS expression mediated through YbhJ and GlmZ. Thus the C-terminal extension may be involved in systems where ancillary RNA or protein partners (such as YhbJ) must be recruited to the complex during regulation.
MATERIALS AND METHODS
DNA oligonucleotides
The complete list of DNA oligonucleotides used for cloning is provided as Supplementary Table S1.
Bacterial strains, media and growth conditions
The E. coli strain Top10 (Invitrogen) was used for cloning, GFP fusion expression analysis and in all that involved coexpression of GFP fusions with sRNAs and Hfq variants. The Δhfq strains were also derived from Top10 cells and were generated as described previously by replacing hfq with chloramphenicol or kanamycin resistance cassettes using the Quick and Easy Conditional Knockout Kit (Gene Bridges) (33). In order to comply with the usage of antibiotics, the chloramphenicol resistance cassette was removed from the hfq::CamR strain as described in the Quick and Easy Conditional Knockout Kit (Gene Bridges) operational manual. The successful removal of the CamR cassette was verified by antibiotic screening followed by PCR.
Growth in Luria–Bertani (LB) broth or on LB plates at 37°C was used throughout this study. Antibiotics were applied at the following concentrations: 100 µg/ml ampicillin, 100 µg/ml spectinomycin, 34 mg/ml chloramphenicol and 30 mg/ml kanamycin.
Plasmids
Fusion plasmids
All GFP fusion systems were constructed starting from the plasmid pBacEmGH [gift of the Cunningham Lab (34)]. In brief, a glmS mRNA fragment from −162 to +21 relative to the translation start site was cloned behind a pBAD promoter to prepare pNS9006 (Supplementary Figure S2). Primers NS20 and NS21 were used to amplify the glmS fragment from genomic DNA and inserted into pBacEmGH using NotI and NheI restrictions endonucleases. The plasmid was then verified using DNA sequencing. Preparation of ARN mutants in glmS was carried out by synthesizing glmS using primer extension where mutations were introduced by the primers that were used. To construct pNS 9009 that contains the ARN Δ1 mutation in glmS, five cycles of PCR were performed using primers NS23, NS24 and NS27 where the desired mutation was introduced in NS24. The PCR amplification was performed using Taq polymerase according to standard procedure. After the first five cycles, the PCR product was diluted 100-fold into a fresh PCR reaction to amplify the 5′- and 3′-regions of glmS using primers NS22 and NS21. The amplified PCR fragment was then subjected to NotI/NheI restriction digestion before cloning into pBacEmGH. Similarly pNS9010 (derivative of pNS9006::ARNΔ2, prepared using NS23, NS25 and NS27) and pNS9011 (derivative of pNS9006::ARNΔ1Δ2, prepared using NS23, NS26 and NS27) were constructed. Successful construction of the plasmid was validated by DNA sequencing.
sRNA plasmids
sRNA plasmids, pNS9007 (GlmZ) and pNS9008 (GlmY) were based on pNM12 (35). sRNA fragments for GlmZ (NS37/NS38) and GlmY (NS35/NS36) were amplified using DNA from TOP10 E. coli cells. The amplified fragments were then inserted into pNM12 after restriction digestion using MscI/EcoRI. The plasmids were verified by DNA sequencing.
Hfq plasmids
All Hfq constructs were based on the plasmid ptac-KanL94P (Addgene (34) plasmid 15927) (36). Hfq wild-type (pNS9012, NS42/NS43), Hfq65 (pNS9013,NS42/NS44), Hfq72 (pNS9014, NS42/NS45), Hfq87 (pNS9015, NS42/NS46) were amplified using Top10 E. coli genomic DNA. Hfq mutants Y25A Hfq (pNS9017), K56A Hfq (pNS9020) and H57A Hfq (pNS9016) were amplified using a plasmid as template that was prepared previously (9). Clostridium perfringens Hfq (pNS9018, NS47/NS48) and Clostridium difficile Hfq (pNS9019, NS49/NS50) were amplified using C. perfringens and C. difficile genomic DNA. The PCR fragments were then restriction digested with BamHI and HindIII and inserted into ptac-KanL94P. The plasmids were verified by DNA sequencing.
Fluorescence data collection
GFP expression of fusion constructs was measured after lysis of cells grown to early stationary phase. All cultures were grown at 37°C in LB media with appropriate antibiotics at concentrations listed above. Bacterial cultures were generated by diluting overgrown overnight cultures. After 3 h of growth, the cultures were induced with 0.1% of arabinose. Cells were then allowed to grow for another 2.5 h. For Hfq complementation studies using the pTac-plasmid system, Hfq expression was induced using 1 mM IPTG after 1 h of growth of diluted cultures before inducing the expression of GlmS-GFP fusion proteins at 3 h. Cells were grown for an additional 2.5 h, before the absorbance was measured at 600 nm to monitor the growth of cells. 3.0 ml of cells were then harvested by centrifuging at 10 000 rpm for 4 min. The cells were then resuspended in 200 µl of lysis buffer (50 mM Tris–HCl pH 7.5, 25 mM NaCl, 2 mM EDTA). To lyse cells 15 µl of lysozyme (20 mg/ml, Fisher), 30 µl of protease inhibitor solution (complete EDTA-free protease inhibitor tablet dissolved in 8 mL Roche) and 30 µl of 1% TritonX-100 was added followed by an incubation of 30 min at 37°C while shaking. The lysed cells were then centrifuged briefly. An amount of 200 µl of the supernatant was then withdrawn to measure the fluorescence intensities of GFP in a 96-well flat bottom black plate (Corning®). Fluorescence measurements were made at an excitation wavelength of 485 nm and at an emission wavelength of 535 nm using a Tecan GENios Plus multi label plate reader. The detector gain values were consistently set between 95 and 103. All fluorescence measurements were normalized to an identical culture where the expression of GFP was not induced by arabinose to account for cellular fluorescence. All measurements were at least in triplicate and error values for each experiment were based on the standard deviation between independent trials.
RNA preparation for SHAPE and gel shift analysis
A18 RNA was purchased from Thermo Scientific Dharmacon Technologies (Lafayette, CO) and deprotected following the manufacturer’s protocol. RNA quality was assessed using denaturing PAGE and gel purified. For in vitro binding analysis, glmS mRNA was transcribed using a DNA fragment that amplifies the glmS from −161 to +21 in E. coli XL-10 cells using primers NS33 and NS34 and digested with Ssp I prior to transcription. For SHAPE analysis, glmS mRNA was in vitro transcribed using a DNA template that was amplified from XL-10 cells using primers NS31 and NS32, that includes a structure cassette in the 5′- and 3′-regions as previously described (37). In vitro transcription was performed after digesting the amplified product with Ssp I. DsrA was obtained by runoff transcription of pBAU10301 that was digested by Ssp I (38).
Electrophoretic mobility shift assays
All binding reactions were performed in 50 mM Tris–HCl pH 7.5,100 mM KCl and 10 mM MgCl2 at room temperature. Prior to any interactions all RNAs were annealed at 90°C for 3 min in buffer, and cooled to room temperature for 30 min. For all reactions 10 µl aliquots were loaded after diluting with loading buffer (10% (w/v) sucrose, xylene cyanol, bromophenol blue) under a power of 5 W on native 5–7% polyacrylamide (37:1) gels in 1× TBE. Dried gels were visualized by phosphorimaging using a Typhoon 9210 imaging system (GE LifeSciences). Quantification was done using ImageQuant 5.1 (Molecular Dynamics) and Kaleidagraph 3.0 (Synergy). Data were fit using nonlinear least-square analysis to a cooperative binding model shown below [Equation (1)]. Here, L is the ligand concentration and the cooperatively is indicated by n. Typical values for n ranged from 1.5 to 2.1.
| (1) |
In the case of A18, DsrA competition assays, the glmS•Hfq complex was pre-formed and A18 and DsrA was titrated at varying concentrations from 0 to 30 µM.
Chemical SHAPE analysis and footprinting
The method for using SHAPE technology to determine the secondary structure of an mRNA 5′-UTR and the footprinting of Hfq on the folded RNA has been described previously (25,37,39).
Western blot analysis
Total cellular extracts were collected from appropriate strains and were lysed using sonication. Total protein was separated on 15% (w/v) polyacrylamide gels and electroblotted to a PVDF membrane. The membrane was probed with rabbit anti-Hfq polyclonal anti-sera. The antigen–antibody complex was visualized using goat anti-rabbit immunoglobin horseradish peroxidase conjugated antibody (Super Signal West Pico Chemiluminescent Substrate, PIERCE).
RESULTS
glmS interaction with Hfq
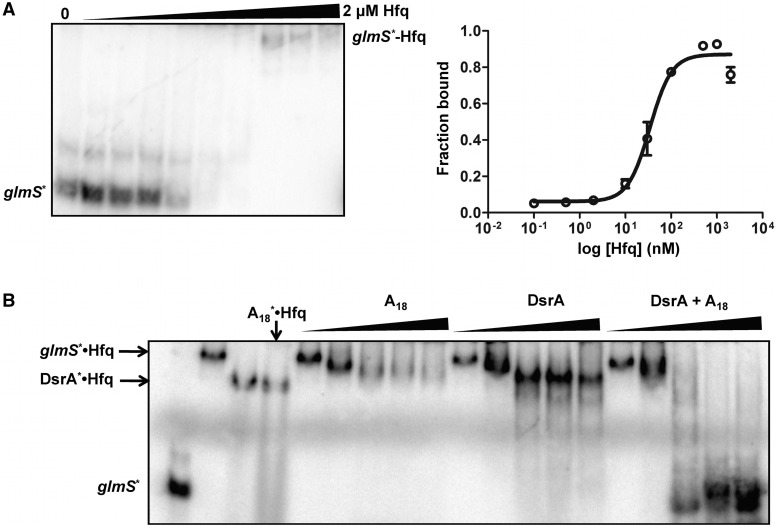
Previous work performed on the glmS regulatory system in E. coli showed the requirement of Hfq for GlmZ- and GlmY-mediated activation of glmS (31,32). While the basic outline of the regulatory network was established, no molecular level understanding was developed. Based on our previous work with OxyS/fhlA (25) and DsrA/rpoS (24) we asked whether Hfq pre-binds the glmS mRNA and how similar the glmS–Hfq interaction is relative to other systems. The glmS mRNA fragment used throughout this study was 182 nt in length. This segment extends from position −161 nt upstream of the GlmS start codon, which is the RNase E processing site that separates the glmU–glmS bicistronic transcript (Figure 1), and extends +21 nt into the coding region. We used this construct to test whether glmS behaves in a manner similar to fhlA (25), and to determine whether one or both RNA-binding surfaces were involved in the interaction with Hfq (Figure 2).
Figure 2.
glmS interaction with Hfq. (A) Analysis of glmS interaction with Hfq (left). Gel shift experiment showing the binary complex between Hfq and glmS mRNA. 32P-labeled glmS* was titrated with varying concentrations of Hfq ranging from 0 to 2 μM. Quantitative analysis of the glmS–Hfq gel (right). As described in materials and methods, thermodynamic constants were determined by non-linear least squares analysis fitted to a cooperative binding model. (B) Competition gel-binding experiment to determine the Hfq-binding surface to glmS. The glmS*–Hfq complex was pre-formed and incubated with increasing amounts of either distal (A18 RNA), proximal (DsrA) or both (A18 and DsrA) binding RNAs (0–30 μM) to compete glmS* away from Hfq.
To measure the binding constant between glmS and Hfq, [5′-32P] glmS mRNA was incubated with varying Hfq concentrations from 0 nM to 2 µM (hexamer). The glmS–Hfq complex was resolved on native gels to provide a KD of ∼30 nM (Figure 2A). This value was comparable to the interaction between fhlA–Hfq and is typical of RNA–Hfq interactions measured previously (9,24,25,40).
To determine which binding surfaces of Hfq interact with Hfq, a competition gel shift assay was carried out where the glmS*–Hfq complex was pre-formed and competed against bona fide distal (A18 RNA) and proximal (DsrA) site binders of Hfq. As observed in Figure 2B, the glmS*–Hfq complex could only be displaced by the simultaneous addition of both DsrA and A18 showing that GlmS interacts with both faces of Hfq similar to fhlA and rpoS (25).
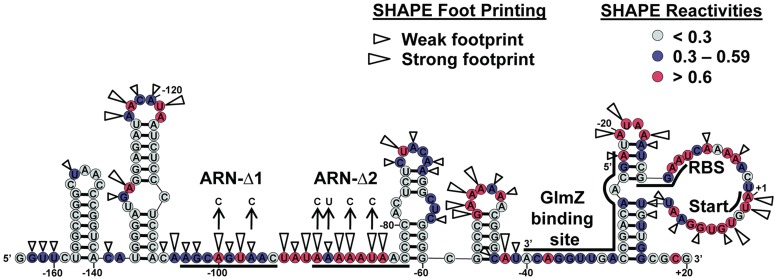
Secondary structure of glmS mRNA and Hfq-binding sites probed using SHAPE
SHAPE (37,41) was carried out to determine the secondary structure of glmS mRNA to identify its functional elements. Figure 3 depicts the secondary structure for glmS mRNA derived from SHAPE constraints, superimposed with the modification intensity data. The functional elements in glmS mRNA such as the potential (ARN)x motifs, GlmZ-binding site, RBS and translation start sites are also depicted in Figure 3. According to the proposed glmS structure, two potential (ARN)x motifs can be identified as candidates that could serve as upstream Hfq-binding sites. To confirm Hfq-binding regions in glmS, SHAPE footprinting was used as described previously (25) and the results are indicated in Figures 3 and Supplementary Figure S1. As expected, both (ARN)x regions footprinted with Hfq among other probable bindings sites indicated in Figure 3.
Figure 3.
SHAPE-derived secondary structure model for glmS mRNA. The NMIA reactivity for each nucleotide corresponds to a given color, nucleotides with reactivities >0.6 are shown in red, from 0.3 to 0.59 in purple and <0.3 in grey. Hfq footprinting data in the presence and absence of 0.5 µM Hfq hexamer are superimposed on the structure. Large wedges indicate where there was a strong protection (>0.6) from wild-type Hfq observed and small wedges where a weak protection (0.3–0.59) was observed. The (ARN)x sites, glmZ-binding site, RBS and the start codon are also shown.
Measuring translational regulation of GlmS using a GFP fusion system
A GFP fusion system was used to measure the Hfq-dependent translational regulation of GlmS by sRNAs GlmZ and GlmY. A schematic diagram of the fusion expression system is shown in Supplementary Figure S2. As described above, a glmS fragment spanning the region of −161 to +21 was N-terminally fused to GFP under the transcriptional control of a pBAD promoter. The fusion system is constructed in a pBAC vector backbone (kindly provided by the Philip Cunningham Lab, Wayne State University, Biological Sciences) which is based on the F-plasmid ori2/RepE replicon, that allows stable maintenance of large genomic fragments as very low or single copy plasmids (42). In addition, the vector also contains an oriV replication origin that can be induced by l-arabinise to produce high copy numbers of the plasmid in an oriV specific manner when expressed in a host strain that can supply the replication initiation protein TrfA in trans (43). However, throughout this study a low copy number of fusion plasmids were maintained since TrfA is not present in these strains.
To monitor expression of GlmS in the presence of sRNAs, GlmZ or GlmY were coexpressed together with the GlmS:GFP reporter. Here vectors pNS9007 and pNS9008, derivatives of the pBAD plasmid series, were used to express GlmZ and GlmY. Therefore coexpression of the GFP fusion and sRNAs were both under the control of a pBAD promoter induced with 0.1% arabinose. The coordinated transcription of mRNAs and their cognate sRNAs is required for efficient Hfq-mediated regulation in vivo (15,16).
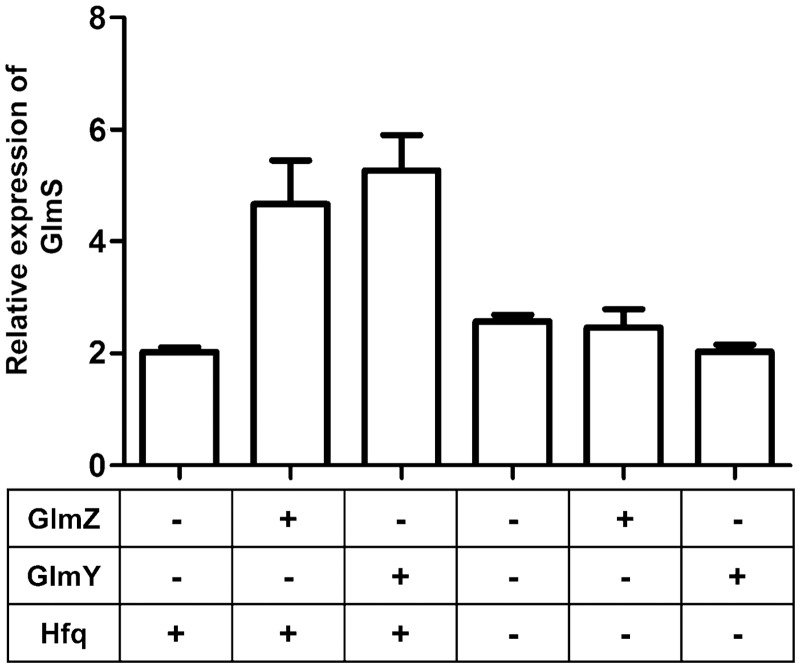
To confirm the proper functioning of the fusion construct and coexpression conditions, GlmS:GFP expression was measured to obtain previously validated observations on the GlmS regulatory system (31,32) and the results are shown in Figure 4. To measure GFP fluorescence, liquid cultures were grown from overgrown overnight cultures. New cultures were then grown at 37°C for 3 h while shaking before inducing with 0.1% l-arabinose. The cultures were left to grow for another 150 min upon induction before the cells were harvested. The growth of each individual culture was monitored by its OD600 absorbance. Cells were consistently collected at their early stationary phase throughout (OD600 ∼1.4 ± 0.2). Lower dilutions from overnight cultures were made for slow growing strains, such as Δhfq cells to reach early stationary phase within the time frame of the assay. Fluorescence was measured upon lysis of 3.0 ml of cell culture (see ‘Materials and Methods’ section). The fluorescence data measured from liquid cultures are shown in Figure 4, where GFP expression is normalized to an identical un-induced culture to correct for cellular fluorescence and used to calculate the fold expression of GlmS.
Figure 4.
Expression of GlmS:GFP measured using GFP fluorescence. Relative expression levels of GlmS are measured in response to sRNAs GlmZ, GlmY and Hfq. Fluorescence was measured from E. coli cells that were grown in liquid media according to the conditions described in materials and methods. The growth of cells was controlled and monitored using the cell density (OD600) where all cultures were harvested upon reaching early stationary phase (OD600 1.4 ± 0.2). Cells were lysed before fluorescence was measured using a Tecan GENios Plus multi label plate reader. All fluorescence measurements were at least triplicated using independent inoculations. To calculate relative expression levels of GlmS, the fluorescence levels of each sample was compared to an identical uninduced culture to correct for cellular fluorescence. GlmS:GFP and sRNA synthesis was controlled using a pBAD promoter inducible by l-arabinose. To measure the Hfq dependence on GlmS expression, experiments were carried out in a Δhfq background.
Basal levels of GlmS expression in the absence of either sRNA are ∼2-fold that of an identical culture that was not induced with arabinose. The presence of either GlmZ or GlmY leads to an additional 2- to 3-fold increase in GlmS:GFP expression. The ability to upregulate GlmS expression is lost in the absence of Hfq, even in the presence of sRNAs. These data correlate nicely with previous observations of this regulatory system (31,32). They show that the GFP fusion construct and the assay conditions are able to capture the effects of sRNAs and Hfq-mediated regulation of GlmS.
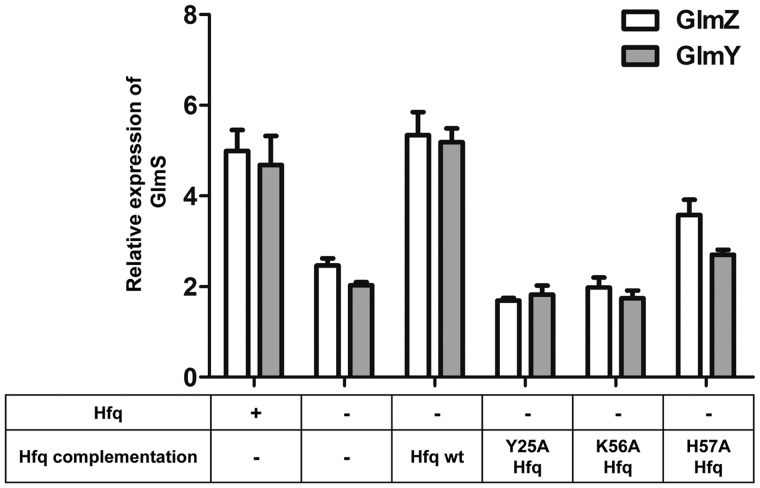
To test whether the translational fusion can recognize defective Hfq species that are incapable of regulating gene expression, proximal and distal mutants of Hfq were used to measure GlmS activation (Figure 5). Y25A Hfq and K56A Hfq mutants have been shown previously to diminish nucleic acid binding at distal and proximal RNA-binding sites, respectively, (9,25) and the H57 proximal residue has been implicated to be crucial for hexamer organization in Pseudomonas aeruginosa (44) and Salmonella typhimurium (45). Data presented in Figure 5, indicates that either proximal (K56A) or distal (Y25A) mutants are incapable of facilitating GlmS upregulation in the presence of sRNAs. However, H57A Hfq shows a weak facilitation towards GlmS expression compared to wild-type Hfq. These data indicate the proper functioning of the GlmS:GFP fusion system, and that it is sensitive to sRNAs (GlmZ/GlmY) and Hfq.
Figure 5.
GlmS:GFP expression levels in the presence of mutant Hfq variants. To further characterize the functionality of the fusion system, GlmS expression levels were monitored in the presence of sRNAs with mutant Hfq variants that were previously shown to deter Hfq function. External Hfq for these experiments was supplemented using a plasmid system where the synthesis of Hfq was under the control of a pTac promoter that is inducible by IPTG. Here Hfq expression was induced prior to GlmS:GFP and sRNA synthesis to allow efficient regulation.
Upstream Hfq-binding (ARN)x elements are essential for regulation
(ARN)x motifs in upstream regions are essential for Hfq-mediated gene regulation of fhlA (25) and rpoS (23,24). Structural evidence also indicates the preferential binding of (ARN)x motifs at the distal site of Hfq (26). Furthermore, a recent study showed the specific enrichment of these sequence elements in genomic SELEX experiments that were performed to identify Hfq binders (46). The above observations lead to a model where, the presence of upstream (ARN)x motifs could be a common occurrence in Hfq-dependent mRNAs. The secondary structure model together with Hfq footprinting data for glmS, indicated the presence of two potential (ARN)x motifs either of which could be required for regulation (Figure 3). The GFP fusion system was used to identify (ARN)x element(s) that are essential for GlmS activation in vivo. Both potential ARN motifs were mutated serially (glmS Δ1 and glmS Δ2) and simultaneously (glmS Δ1Δ2) to identify the role of each in glmS regulation (also see Figure 3).
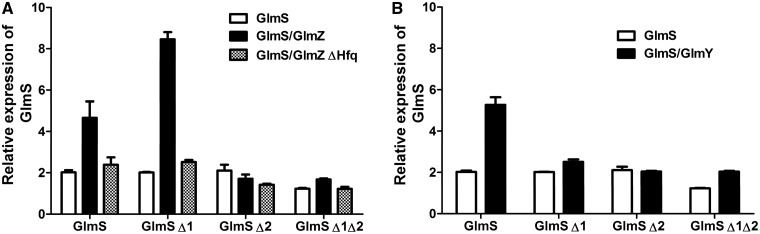
The expression of GlmS Δ1:GFP, GlmS Δ2:GFP and GlmS Δ1Δ2:GFP was monitored in the presence of GlmZ (Figure 6A) and GlmY (Figure 6B). Data in Figure 6A show that, sRNA GlmZ is unable to activate GlmS expression of the glmS Δ2 mutant. However GlmS expression is higher than wild-type levels for the glmS Δ1 mutant. The overactivation caused by glmS Δ1 was absent when both ARN motifs were mutated (Δ1Δ2). To test whether glmS Δ1 activation is Hfq dependent, the regulation was monitored in the absence of Hfq. Data from this experiment reveals that the Δ1 mutation did not change the Hfq-dependence of GlmS activation by GlmZ. However, neither ARN mutant (Δ1 nor Δ2) were able to upregulate GlmS expression in the presence of GlmY (Figure 6B), implying that the Δ1 mutation only affects the GlmZ pathway. These results indicate the requirement of (ARN)x elements for Hfq-mediated regulation of GlmS and that ARN-2 is the most essential Hfq-binding motif for both GlmZ and GlmY pathways.
Figure 6.
Effect of upstream Hfq binding (ARN)x elements in GlmS activation by GlmZ and GlmY. (A) Relative GlmS:GFP expression levels of glmS ARN mutants measured in the presence of GlmZ. Here mutations to glmS ARN elements were made as indicated in above Figure 2. Since two potential ARN motifs were detected they were removed sequentially and simultaneously to produce glmS Δ1, glmS Δ2 and glmS Δ1Δ2 respectively. The expression level of GlmS was measured as indicated in material and methods. The relative upregulation of GlmS was compared to expression levels when in the absence of GlmZ and Hfq. (B) Upregulation of glmS ARN mutants were measured with GlmY and compared to wild-type glmS expression levels.
Requirement of the C-terminal domain of Hfq for regulation of GlmS by sRNAs GlmZ and GlmY
Hfq is present in most Gram-positive and Gram-negative bacteria. In all cases the nucleic acid-binding Sm1 and Sm2 motifs are evolutionary conserved across species which spans positions 7 through 66 based on the E. coli numbering. The greatest variability is observed at the C-terminus of Hfq where E. coli and Salmonella enterica have fairly long tails (∼102 amino acids) while species such as C. difficile and C. perfringens have shorter C-terminal extensions. The functional importance of the C-terminus of Hfq for regulation is unclear and conflicting reports have been presented previously (47–49). Therefore, the necessity of the C-terminal region of Hfq for GlmS activation by sRNAs, GlmZ or GlmY was tested using the GFP fusion system.
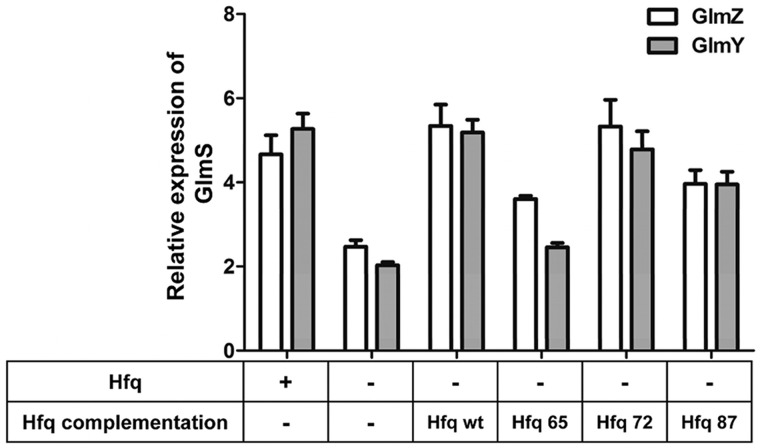
Several C-terminal truncations of E. coli Hfq were coexpressed together with the GlmS:GFP fusion protein and sRNAs in a Δhfq background to identify if the C-terminal amino acids are required for regulation. In addition to wild-type Hfq that consists of 102 amino acids, shortened C-terminal constructs of Hfq87 (87 amino acids), Hfq72 and Hfq65 were prepared. Here the Hfq65 variant can essentially be regarded as the tail-less version of Hfq since it only consists of the core Sm1 and Sm2 motifs. All C-terminal variants of Hfq were constructed using the ptac-KanL94P plasmid as the parent vector (36), that is under the transcriptional control of an IPTG inducible pTac promoter. The assay conditions to measure the GFP expression were similar to those described earlier, although Hfq induction was carried out using 1 mM IPTG, 1 h before arabinose induction to allow synthesis of Hfq prior to sRNA regulation. Relative expression levels of GlmS were monitored as the ability to upregulate GlmS synthesis individually by sRNAs GlmZ and GlmY with varying C-terminal extensions of Hfq (Figure 7).
Figure 7.
Requirement of Hfq C-terminal domain for GlmS:GFP upregulation by GlmZ and GlmY. Activation of GlmS:GFP expression by sRNAs was measured in the presence of Hfq that consists of varying C-terminal extension lengths. In addition to Hfq wild-type (102 amino acids), Hfq65 (65 amino acids), Hfq72 and Hfq87 were tested for its ability to perform regulation. Hfq65 consists only the nucleic acid-binding Sm1 and Sm2 domains while Hfq72 and Hfq87 have 7 aa and 22 aa extensions off the Sm cores. All externally supplemented hfq was under the transcriptional control of a pTac promoter that was induced using 1 mM IPTG. Coexpression and induction conditions of GlmS:GFP, sRNA and Hfq plasmid systems are described in material and methods.
From data shown in Figure 7, it is clear that plasmid borne and native Hfq (Hfq wt) regulate GlmS expression at comparable levels regardless of whether GlmZ or GlmY is used as the activator. GlmS regulation by GlmZ was mostly independent of the length of the Hfq C-terminal extension as Hfq72 and Hfq87 showed identical levels of GlmS expression to wild-type Hfq. Hfq65 on the other hand showed reduced levels of GlmS expression implying a possible effect caused by shortening the Hfq C-terminal tail. Semi-native western blot analysis of cellular Hfq65 showed multiple Hfq conformations relative to wild-type Hfq implying possible destabilization of the Hfq functional core in the absence of C-terminal residues (Supplementary Figure S3). However, all Hfq C-terminal variants (including Hfq65) were better at upregulating GlmS expression than a Δhfq strain in the presence of GlmZ, indicating at least some functional hfq hexamer is present in vivo (Figure 7). Conversely, for GlmY-mediated activation, Hfq65 was unable to upregulate GlmS above basal levels while Hfq72 and Hfq87 were proficient. These data show that the absence of amino acid residues from at least 66–72 affected GlmY’s ability to upregulate GlmS in a manner distinct from GlmZ regulation. However, as described above, GlmZ and GlmY act in a hierarchical manner during regulation. Therefore a loss of GlmY’s ability to recruit YhbJ for self-processing would affect GlmZ’s ability to activate GlmS synthesis. These data provide sufficient evidence to suggest the importance of at least the first seven amino acids of the C-terminal tail of Hfq for some in vivo functions.
Clostridium perfringens and C. difficile Hfq variants are proficient in GlmZ but not GlmY-mediated regulation of GlmS in E. coli
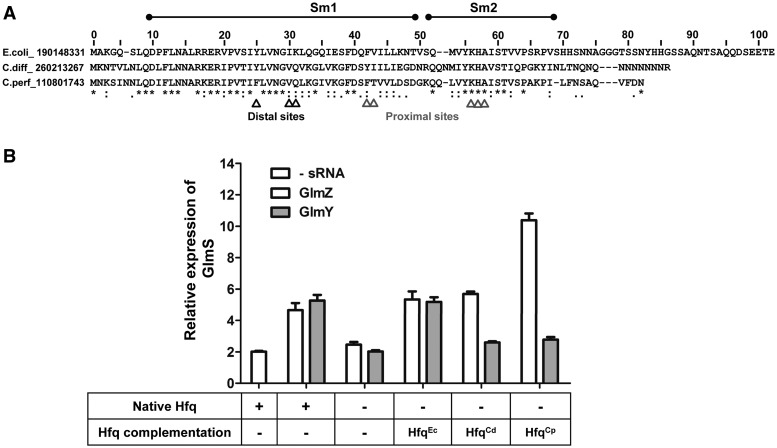
To validate these observations that the first 7-amino acids of the C-terminal tail of Hfq were essential for the GlmY pathway, Hfq variants belonging to Gram-positive bacteria C. difficile and C. perfringens were used to substitute for E. coli Hfq to activate GlmS synthesis. Both HfqCd and HfqCp variants have similar nucleic acid-binding regions (Sm1/Sm2) compared to E. coli. However, not much sequence homology is observed beyond the Sm regions (Figure 8A). Therefore, the nucleic acid-binding properties should be conserved in HfqCd and HfqCp to promote GlmZ–glmS base pairing. However, if there is a need for the C-terminal extension to facilitate regulation, HfqCp and HfqCd may not be able to cross complement GlmY native functions.
Figure 8.
GlmS:GFP upregulation using C. perfringens and C. difficile Hfq variants by GlmZ and GlmY. (A) Sequence homology between E. coli, C. difficile and C. perfringens Hfq variants. Nucleic acid-binding Sm1/Sm2 domains and important amino acid residues in proximal and distal RNA-binding surfaces required for regulation are indicated. (B) Cross-complementation for HfqEc using HfqCp and HfqCd homologues for GlmS upregulation. The ability to activate GlmS expression by GlmZ and GlmY was measured when HfqCd and HfqCp was supplemented in E.coli Δhfq cells.
GlmS activation was monitored in the presence of HfqCd and HfqCp variants with either GlmZ or GlmY sRNAs in a Δhfq background. The results are shown in Figure 8B. HfqCd and HfqCp both successfully promote GlmZ-dependent GlmS activation. In fact, increased upregulation of GlmS is observed for HfqCp in the presence of GlmZ. These data fundamentally confirm that the GlmZ–glmS interaction, which revolves around the ability of Hfq to facilitate base-paring between a sRNA and its target mRNA, can be achieved so long as the core nucleic acid-binding domain is present. However, HfqCd and HfqCp were unable to upregulate GlmS expression via the GlmY pathway. Gel shift assays confirmed that GlmY binds HfqEc and HfqCp, although its affinity for HfqCp is about 10-fold weaker than for HfqEc. (Supplementary Figure S4). In contrast, GlmZ binds HfqCp and HfqEc with only a slight loss of affinity implying some differences in the manner with which GlmZ and GlmY bind Hfq despite their sequence and structural homology. Since the only significant difference between the three Hfq variants are at the C-terminal region, a potential role of the C-terminus is implied for GlmY activation, where additional protein–protein or protein–RNA interactions are likely required.
DISCUSSION
Being able to adapt to environmental stress is vital for the survival and propagation of bacteria. The RNA-binding protein Hfq plays a key role in modulating stress responses in bacteria by regulating gene expression in a sRNA-dependent manner. Hfq functions primarily in promoting sRNA–mRNA base pairing by increasing the local concentrations of sRNAs and target mRNAs through its distal and proximal RNA-binding surfaces. In addition, Hfq acts as a chaperone to trigger structural transitions in RNAs to initiate recognition between cognate sRNA–mRNA pairs (27), or partners directly or indirectly with processing enzymes such as RNase E (50,51), Poly A polymerase (52), PNPase (50), RNA helicases (53,54), RelA (55) and YhbJ (30) to aid stringent gene regulation. These diverse functions enable Hfq to be involved in a cascade of regulatory events that control multiple stress responsive pathways. Since all Hfq networks involve sRNAs to initiate gene regulation at the post-transcriptional level, the sRNA–mRNA interaction can be regarded as the main trigger for downstream regulatory outcomes.
A number of recent reports have indicated that upstream (ARN)x sequence motifs are common among Hfq-dependent mRNAs and help recruit Hfq to these mRNAs (23,25,56). Structural evidence shows that (ARN)x motifs interact with the distal RNA-binding site of Hfq while the proximal site harbors sRNAs during regulation (26). In this work, (ARN)x motifs were identified and characterized within glmS mRNA, a message regulated by sRNAs GlmZ and GlmY in an Hfq-dependent manner. The interaction between Hfq and glmS is similar to what was previously observed for fhlA (25) where the mRNA uses both RNA-binding surfaces to interact with Hfq in the absence of sRNAs. This mode of interaction between mRNAs and Hfq was attributed to a model wherein, the complexes could serve as a precursor RNP complex that enables a rapid response to stress in bacteria.
Examining the proposed secondary structure of glmS and Hfq footprinting data obtained using SHAPE, it was apparent that two potential ARN motifs (ARN-1 and ARN-2) were present upstream of the RBS (Figure 3). To identify which ARN element(s) were essential for regulation, glmS mRNA was fused N-terminally to GFP to measure GlmS expression levels. GlmS translation was upregulated in the presence of either sRNA in an Hfq-dependent manner confirming the proper functioning of the fusion system. Of the two potential ARN motifs, ARN-Δ2 had the most adverse effects on regulation in the presence of either sRNA. However, the loss of ARN-1 caused a hyper-activation of GlmS only in the presence of GlmZ. Nonetheless, upregulation of glmS ARN-Δ1 was Hfq dependent, implying that the regulatory node was still intact despite the changes in glmS functional elements. These results indicate that ARN-2 is the essential (ARN)x motif in glmS required for Hfq-mediated regulation. While we do not completely understand why glmS-ARN-Δ1 led to over-activation, it might result from structural changes in glmS due to the sequence modifications required to ablate the Hfq binding site. Alternatively, it could also be explained by a partitioning model. If Hfq must be present at ARN-2 for glmZ activation, and Hfq can bind at either ARN-1 or ARN-2, destruction of ARN-1 would lead to an increased population of the active adduct (Hfq-binding to ARN-2) triggering a more favorable response than wild-type glmS. The key point however, is that, as observed in previous cases such as in rpoS, rprA (23) and fhlA (25), Hfq-mediated regulation of glmS is absolutely dependent on the presence of an upstream (ARN)x element, adding to the list of networks that rely on such sequence motifs for function and may indicate that this is a global requirement within mRNAs that are destined for Hfq-dependent regulation.
The manner by which Hfq recognizes target mRNAs could be important for understanding the mechanism of stress regulation in bacteria. This interaction could allow Hfq•mRNA complexes to form prior to stress, thus identifying those messages that might be regulated during a time-sensitive response. While we have no definitive data to confirm this hypothesis, this model would explain Hfq’s ability to handle multiple regulatory networks using multiple sRNAs. Recent evidence also suggests that Hfq can be a limiting factor in cells and that a coordinate expression of sRNA and mRNAs is required for the proper functioning of regulatory nodes (15). This implies the intense competition between RNAs to acquire Hfq which in turn explains the need for expressed mRNAs to capture Hfq to facilitate their own regulation. Other functions of this interaction could include initiation of structural rearrangements in the mRNA to facilitate the interaction with sRNA as was observed in the case of DsrA and rpoS (27). However, the structural transitions that are triggered by the mRNA–Hfq interaction alone are insufficient to affect basal translation levels. The glmS–Hfq interaction alone had no effect in GlmS expression, for instance (data not shown). Similar observations were reported for sodB, rpoS, ompC and fhlA, all having only minimal effects on translation upon binding to Hfq (15).
The role of Hfq in stress regulation throughout the bacterial kingdom is well established. Comparison of Hfq variants across species has confirmed the presence of an evolutionary conserved nucleic acid-binding core (Sm1 and Sm2) and a variable C-terminal region that is less well preserved. While the requirement of the Sm core for Hfq function is clear, involvement of the C-terminal region of Hfq in regulation is still controversial (49,57).
To gain more information regarding the function of the C-terminal region of Hfq during regulation, the glmS system was used. A clear advantage of using this system to address this problem stems from the presence of two regulatory layers that are directed by GlmZ and GlmY in an Hfq-dependent manner. While the role of Hfq in facilitating the GlmZ–glmS interaction is well established, the requirement of Hfq by GlmY in YhbJ processing is unclear. Since an interaction between GlmY and glmS is not established, (31) YhbJ could be recruited by a GlmY–Hfq complex potentially through the C-terminal region of Hfq.
When the requirement of the C-terminal region of Hfq for GlmS activation was measured, data indicated that both Hfq72 and Hfq87 were equally proficient at assisting GlmY and GlmZ upregulate GlmS. However, when only the Sm core of Hfq was present (Hfq65), only GlmZ allowed significant upregulation of GlmS expression relative to a Δhfq strain. Although crystallographic data show that C-terminal residues are not essential for hexamer association (58), semi-native western blot analysis of Hfq65 indicates some issues associated with speciation or reduced stability of the functional Hfq hexamer (Supplementary Figure S3) as previously postulated (13). This result explains the reduced activity of GlmZ-mediated upregulation of glmS expression in the presence of Hfq65. However, the inability of GlmY to utilize Hfq65 to any appreciable extent remains puzzling since a significant amount of functional hexamer is still present in these cells based on western blot analysis.
HfqCp and HfqCd were used to cross complement for HfqEc in order to completely rule out the potential differences in Hfq utilizations by sRNAs during regulation. Hfq variants from C. perfringens and C. difficile fully complemented the Δhfq strain when measured based on GlmZ activation of GlmS. GlmY, however, was unable to take advantage of cross-species complementation. Sm cores in HfqCp and HfqCd are highly conserved compared to HfqEc and successfully promote GlmZ–glmS base pairing that represents the classical Hfq mediation process through its nucleic acid-binding domains. The inability of GlmY to use Hfq65, HfqCp and HfqCd to upregulate GlmS suggests that the Sm core alone is insufficient for regulation in this pathway. Since the primary functional role of GlmY is to self-direct the processing complex that utilizes YhbJ and polyA polymerase (PAP I) (31), Hfq C-terminal region may facilitate the assembly of this complex through additional RNA and/or protein interactions with Hfq.
The Hfq C-terminal region has long been predicted as a landing pad for either protein or nucleic acid-binding partners. In fact, recent structural data compares the Hfq C-terminal regions to intrinsically disordered proteins (IDPs) that could potentially facilitate intermolecular interactions (47,57). Pull down experiments carried out to map Hfq-protein complexes indicate that Hfq interacts directly or indirectly with proteins in the bacterial RNA degradosome such as RNase E, polynucleotide phosphorylase (PNPase), poly A polymerase (PAP I) (59,60) and a variety of regulatory proteins in bacteria (T. Lee and A. Feig, unpublished data). Furthermore, a recent study by Prévost et al. proposed a model for the physiological relevance of an RNase E–Hfq interaction during RNA degradation triggered by the sRNA–mRNA recognition (61,62). Since no detectable protein interactions have been reported within the Sm cores, these complexes should be accommodated at the C-terminal region of Hfq. Whether Hfq recruits YhbJ as part of a novel processing complex that affects sRNA turnover or whether it promotes an RNA interaction with GlmY through its C-terminal that is essential for GlmY turnover needs to be tested further.
Overall, this work further illustrates the requirement for Hfq to bind mRNAs through upstream (ARN)x motifs during sRNA-dependent gene regulation. In addition, these studies show that a non-canonical sRNA pathway requires additional Hfq interactions traced to the C-terminal extension that are not essential for the canonical sRNA–mRNA pairing behavior typically carried out by Hfq. C-terminal amino acids 66–72 from E. coli are essential for GlmY-mediated regulation of GlmS, implying a conditional requirement for the C-terminal region depending on the mode of sRNA recruitment and may relate to additional RNA or protein-binding interactions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables 1–2 and Supplementary Figures 1–4.
FUNDING
National Institutes of Health [GM075068 to A.L.F., in part]; Wayne State University, Office of the Vice-President for Research. Funding for open access charge: NIH [GM-075068].
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Repoila F, Darfeuille F. Small regulatory non-coding RNAs in bacteria: physiology and mechanistic aspects. Biol. Cell. 2009;101:117–131. doi: 10.1042/BC20070137. [DOI] [PubMed] [Google Scholar]
- 3.Carthew RW, Sontheimer EJ. Origins and Mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Brantl S. Bacterial chromosome-encoded small regulatory RNAs. Future Microbiol. 2009;4:85–8103. doi: 10.2217/17460913.4.1.85. [DOI] [PubMed] [Google Scholar]
- 5.Brantl S. Regulatory mechanisms employed by cis-encoded antisense RNAs. Curr. Opin. Microbiol. 2007;10:102–109. doi: 10.1016/j.mib.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 6.Wagner EG, Simons RW. Antisense RNA control in bacteria, phages, and plasmids. Annu. Rev. Microbiol. 1994;48:713–742. doi: 10.1146/annurev.mi.48.100194.003433. [DOI] [PubMed] [Google Scholar]
- 7.Breaker RR. Prospects for riboswitch discovery and analysis. Mol. Cell. 2011;43:867–879. doi: 10.1016/j.molcel.2011.08.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Franze de Fernandez MT, Hayward WS, August JT. Bacterial proteins required for replication of phage Q ribonucleic acid. Pruification and properties of host factor I, a ribonucleic acid-binding protein. J. Biol. Chem. 1972;247:824–831. [PubMed] [Google Scholar]
- 9.Mikulecky PJ, Kaw MK, Brescia CC, Takach JC, Sledjeski DD, Feig AL. Escherichia coli Hfq has distinct interaction surfaces for DsrA, rpoS and poly(A) RNAs. Nat. Struct. Mol. Biol. 2004;11:1206–1214. doi: 10.1038/nsmb858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sittka A, Pfeiffer V, Tedin K, Vogel J. The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium. Mol. Microbiol. 2007;63:193–217. doi: 10.1111/j.1365-2958.2006.05489.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ding Y, Davis BM, Waldor MK. Hfq is essential for Vibrio cholerae virulence and downregulates sigma expression. Mol. Microbiol. 2004;53:345–354. doi: 10.1111/j.1365-2958.2004.04142.x. [DOI] [PubMed] [Google Scholar]
- 12.Christiansen JK, Larsen MH, Ingmer H, Sogaard-Andersen L, Kallipolitis BH. The RNA-binding protein Hfq of Listeria monocytogenes: role in stress tolerance and virulence. J. Bacteriol. 2004;186:3355–3362. doi: 10.1128/JB.186.11.3355-3362.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sonnleitner E, Napetschnig J, Afonyushkin T, Ecker K, Vecerek B, Moll I, Kaberdin VR, Blasi U. Functional effects of variants of the RNA chaperone Hfq. Biochem. Biophys. Res. Commun. 2004;323:1017–1023. doi: 10.1016/j.bbrc.2004.08.190. [DOI] [PubMed] [Google Scholar]
- 14.Ansong C, Yoon H, Porwollik S, Mottaz-Brewer H, Petritis BO, Jaitly N, Adkins JN, McClelland M, Heffron F, Smith RD. Global systems-level analysis of Hfq and SmpB deletion mutants in Salmonella: implications for virulence and global protein translation. PLoS One. 2009;4:e4809. doi: 10.1371/journal.pone.0004809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hussein R, Lim HN. Disruption of small RNA signaling caused by competition for Hfq. Proc. Natl Acad. Sci. USA. 2011;108:1110–1115. doi: 10.1073/pnas.1010082108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Adamson DN, Lim HN. Essential requirements for robust signaling in Hfq dependent small RNA networks. PLoS Computat. Biol. 2011;7:e1002138. doi: 10.1371/journal.pcbi.1002138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Storz G, Vogel J, Wassarman KM. Regulation by Small RNAs in Bacteria: Expanding Frontiers. Mol. Cell. 2011;43:880–891. doi: 10.1016/j.molcel.2011.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Papenfort K, Vogel J. Multiple target regulation by small noncoding RNAs rewires gene expression at the post-transcriptional level. Res. Microbiol. 2009;160:278–287. doi: 10.1016/j.resmic.2009.03.004. [DOI] [PubMed] [Google Scholar]
- 19.Majdalani N, Cunning C, Sledjeski D, Elliott T, Gottesman S. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc. Natl Acad. Sci. USA. 1998;95:12462–12467. doi: 10.1073/pnas.95.21.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Altuvia S, Weinstein-Fischer D, Zhang A, Postow L, Storz G. A small, stable RNA induced by oxidative stress: role as a pleiotropic regulator and antimutator. Cell. 1997;90:43–53. doi: 10.1016/s0092-8674(00)80312-8. [DOI] [PubMed] [Google Scholar]
- 21.Kawamoto H, Morita T, Shimizu A, Inada T, Aiba H. Implication of membrane localization of target mRNA in the action of a small RNA: mechanism of post-transcriptional regulation of glucose transporter in Escherichia coli. Genes Dev. 2005;19:328–338. doi: 10.1101/gad.1270605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vanderpool CK. Physiological consequences of small RNA-mediated regulation of glucose-phosphate stress. Curr. Opin. Microbiol. 2007;10:146–151. doi: 10.1016/j.mib.2007.03.011. [DOI] [PubMed] [Google Scholar]
- 23.Soper T, Mandin P, Majdalani N, Gottesman S, Woodson SA. Positive regulation by small RNAs and the role of Hfq. Proc. Natl Acad. Sci. USA. 2010;107:9602–9607. doi: 10.1073/pnas.1004435107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Soper TJ, Woodson SA. The rpoS mRNA leader recruits Hfq to facilitate annealing with DsrA sRNA. RNA. 2008;14:1907–1917. doi: 10.1261/rna.1110608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Salim NN, Feig AL. An upstream Hfq binding site in the fhlA mRNA leader region facilitates the OxyS-fhlA interaction. PLoS One. 2010;5:e13028. doi: 10.1371/journal.pone.0013028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Link TM, Valentin-Hansen P, Brennan RG. Structure of Escherichia coli Hfq bound to polyriboadenylate RNA. Proc. Natl Acad. Sci. USA. 2009;106:19292–19297. doi: 10.1073/pnas.0908744106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Soper TJ, Doxzen K, Woodson SA. Major role for mRNA binding and restructuring in sRNA recruitment by Hfq. RNA. 2011;17:1544–1550. doi: 10.1261/rna.2767211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kobayashi K, Ehrlich SD, Albertini A, Amati G, Andersen KK, Arnaud M, Asai K, Ashikaga S, Aymerich S, Bessieres P, et al. Essential Bacillus subtilis genes. Proc. Natl Acad. Sci. USA. 2003;100:4678–4683. doi: 10.1073/pnas.0730515100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Klein DJ, Ferre-D'Amare AR. Structural basis of glmS ribozyme activation by glucosamine-6-phosphate. Science. 2006;313:1752–1756. doi: 10.1126/science.1129666. [DOI] [PubMed] [Google Scholar]
- 30.Kalamorz F, Reichenbach B, Marz W, Rak B, Gorke B. Feedback control of glucosamine-6-phosphate synthase GlmS expression depends on the small RNA GlmZ and involves the novel protein YhbJ in Escherichia coli. Mol. Microbiol. 2007;65:1518–1533. doi: 10.1111/j.1365-2958.2007.05888.x. [DOI] [PubMed] [Google Scholar]
- 31.Urban JH, Vogel J. Two seemingly homologous noncoding RNAs act hierarchically to activate glmS mRNA translation. PLoS Biol. 2008;6:e64. doi: 10.1371/journal.pbio.0060064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Urban JH, Papenfort K, Thomsen J, Schmitz RA, Vogel J. A conserved small RNA promotes discoordinate expression of the glmUS operon mRNA to activate GlmS synthesis. J. Mol. Biol. 2007;373:521–528. doi: 10.1016/j.jmb.2007.07.035. [DOI] [PubMed] [Google Scholar]
- 33.Lee T, Feig AL. The RNA binding protein Hfq interacts specifically with tRNAs. RNA. 2008;14:514–523. doi: 10.1261/rna.531408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lamichhane TN, Abeydeera ND, Duc AC, Cunningham PR, Chow CS. Selection of peptides targeting helix 31 of bacterial 16S ribosomal RNA by screening M13 phage-display libraries. Molecules. 2011;16:1211–1239. doi: 10.3390/molecules16021211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Majdalani N, Cunning C, Sledjeski D, Elliott T, Gottesman S. DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc. Natl Acad. Sci. USA. 1998;95:12462–12467. doi: 10.1073/pnas.95.21.12462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Besmer E, Market E, Papavasiliou FN. The transcription elongation complex directs activation-induced cytidine deaminase-mediated DNA deamination. Mol. Cell Biol. 2006;26:4378–4385. doi: 10.1128/MCB.02375-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. RNA structure analysis at single nucleotide resolution by selective 2'-hydroxyl acylation and primer extension (SHAPE) J. Am. Chem. Soc. 2005;127:4223–4231. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- 38.Brescia CC, Mikulecky PJ, Feig AL, Sledjeski DD. Identification of the Hfq-binding site on DsrA RNA: Hfq binds without altering DsrA secondary structure. RNA. 2003;9:33–43. doi: 10.1261/rna.2570803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wilkinson KA, Gorelick RJ, Vasa SM, Guex N, Rein A, Mathews DH, Giddings MC, Weeks KM. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol. 2008;6:e96. doi: 10.1371/journal.pbio.0060096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Olejniczak M. Despite similar binding to the Hfq protein regulatory RNAs widely differ in their competition performance. Biochemistry. 2011;50:4427–4440. doi: 10.1021/bi102043f. [DOI] [PubMed] [Google Scholar]
- 41.Wilkinson KA, Merino EJ, Weeks KM. Selective 2'-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 2006;1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- 42.Wild J, Szybalski W. Copy-control pBAC/oriV vectors for genomic cloning. Methods Mol. Biol. 2004;267:145–154. doi: 10.1385/1-59259-774-2:145. [DOI] [PubMed] [Google Scholar]
- 43.Wild J, Hradecna Z, Szybalski W. Conditionally amplifiable BACs: switching from single-copy to high-copy vectors and genomic clones. Genome Res. 2002;12:1434–1444. doi: 10.1101/gr.130502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Moskaleva O, Melnik B, Gabdulkhakov A, Garber M, Nikonov S, Stolboushkina E, Nikulin A. The structures of mutant forms of Hfq from Pseudomonas aeruginosa reveal the importance of the conserved His57 for the protein hexamer organization. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2010;66:760–764. doi: 10.1107/S1744309110017331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Sauer E, Weichenrieder O. Structural basis for RNA 3'-end recognition by Hfq. Proc. Natl Acad. Sci. USA. 2011;108:13065–13070. doi: 10.1073/pnas.1103420108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lorenz C, Gesell T, Zimmermann B, Schoeberl U, Bilusic I, Rajkowitsch L, Waldsich C, von Haeseler A, Schroeder R. Genomic SELEX for Hfq-binding RNAs identifies genomic aptamers predominantly in antisense transcripts. Nucleic Acids Res. 2010;38:3794–3808. doi: 10.1093/nar/gkq032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Beich-Frandsen M, Vecerek B, Sjoblom B, Blasi U, Djinovic-Carugo K. Structural analysis of full-length Hfq from Escherichia coli. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2011;67:536–540. doi: 10.1107/S174430911100786X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Vecerek B, Rajkowitsch L, Sonnleitner E, Schroeder R, Blasi U. The C-terminal domain of Escherichia coli Hfq is required for regulation. Nucleic Acids Res. 2008;36:133–143. doi: 10.1093/nar/gkm985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Olsen AS, Moller-Jensen J, Brennan RG, Valentin-Hansen P. C-terminally truncated derivatives of Escherichia coli Hfq are proficient in riboregulation. J. Mol. Biol. 2010;404:173–182. doi: 10.1016/j.jmb.2010.09.038. [DOI] [PubMed] [Google Scholar]
- 50.Folichon M, Arluison V, Pellegrini O, Huntzinger E, Regnier P, Hajnsdorf E. The poly(A) binding protein Hfq protects RNA from RNase E and exoribonucleolytic degradation. Nucleic Acids Res. 2003;31:7302–7310. doi: 10.1093/nar/gkg915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Moll I, Afonyushkin T, Vytvytska O, Kaberdin VR, Blasi U. Coincident Hfq binding and RNase E cleavage sites on mRNA and small regulatory RNAs. RNA. 2003;9:1308–1314. doi: 10.1261/rna.5850703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mohanty BK, Maples VF, Kushner SR. The Sm-like protein Hfq regulates polyadenylation dependent mRNA decay in Escherichia coli. Mol. Microbiol. 2004;54:905–920. doi: 10.1111/j.1365-2958.2004.04337.x. [DOI] [PubMed] [Google Scholar]
- 53.Resch A, Vecerek B, Palavra K, Blasi U. Requirement of the CsdA DEAD-box helicase for low temperature riboregulation of rpoS mRNA. RNA Biol. 2010;7:796–802. doi: 10.4161/rna.7.6.13768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ikeda Y, Yagi M, Morita T, Aiba H. Hfq binding at RhlB-recognition region of RNase E is crucial for the rapid degradation of target mRNAs mediated by sRNAs in Escherichia coli. Mol. Microbiol. 2011;79:419–432. doi: 10.1111/j.1365-2958.2010.07454.x. [DOI] [PubMed] [Google Scholar]
- 55.Argaman L, Elgrably-Weiss M, Hershko T, Vogel J, Altuvia S. RelA protein stimulates the activity of RyhB small RNA by acting on RNA-binding protein Hfq. Proc. Natl Acad. Sci. USA. 2012;109:4621–4626. doi: 10.1073/pnas.1113113109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Updegrove T, Wilf N, Sun X, Wartell RM. Effect of Hfq on RprA-rpoS mRNA pairing: Hfq-RNA binding and the influence of the 5' rpoS mRNA leader region. Biochemistry. 2008;47:11184–11195. doi: 10.1021/bi800479p. [DOI] [PubMed] [Google Scholar]
- 57.Beich-Frandsen M, Vecerek B, Konarev PV, Sjoblom B, Kloiber K, Hammerle H, Rajkowitsch L, Miles AJ, Kontaxis G, Wallace BA, et al. Structural insights into the dynamics and function of the C-terminus of the E. coli RNA chaperone Hfq. Nucleic Acids Res. 2011;39:4900–4915. doi: 10.1093/nar/gkq1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brennan RG, Link TM. Hfq structure, function and ligand binding. Curr. Opin. Microbiol. 2007;10:125–133. doi: 10.1016/j.mib.2007.03.015. [DOI] [PubMed] [Google Scholar]
- 59.Morita T, Maki K, Aiba H. RNase E-based ribonucleoprotein complexes: Mechanical basis of mRNA destabilization mediated by bacterial noncoding RNAs. Genes Dev. 2005;19:2176–2186. doi: 10.1101/gad.1330405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Mohanty BK, Maples VF, Kushner SR. The Sm-like protein Hfq regulates polyadenylation dependent mRNA decay in Escherichia coli. Mol. Microbiol. 2004;54:905–920. doi: 10.1111/j.1365-2958.2004.04337.x. [DOI] [PubMed] [Google Scholar]
- 61.Prevost K, Desnoyers G, Jacques J-F, Lavoie F, Masse E. Small RNA-induced mRNA degradation achieved through both translation block and activated cleavage. Genes Dev. 2011;25:385–396. doi: 10.1101/gad.2001711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Morita T, Aiba H. RNase E action at a distance: degradation of target mRNAs mediated by an Hfq-binding small RNA in bacteria. Genes Dev. 2011;25:294–298. doi: 10.1101/gad.2030311. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.