Abstract
Gene fusions are common driver events in leukaemias and solid tumours; here we present FusionAnalyser, a tool dedicated to the identification of driver fusion rearrangements in human cancer through the analysis of paired-end high-throughput transcriptome sequencing data. We initially tested FusionAnalyser by using a set of in silico randomly generated sequencing data from 20 known human translocations occurring in cancer and subsequently using transcriptome data from three chronic and three acute myeloid leukaemia samples. in all the cases our tool was invariably able to detect the presence of the correct driver fusion event(s) with high specificity. In one of the acute myeloid leukaemia samples, FusionAnalyser identified a novel, cryptic, in-frame ETS2–ERG fusion. A fully event-driven graphical interface and a flexible filtering system allow complex analyses to be run in the absence of any a priori programming or scripting knowledge. Therefore, we propose FusionAnalyser as an efficient and robust graphical tool for the identification of functional rearrangements in the context of high-throughput transcriptome sequencing data.
INTRODUCTION
Until a few years ago, the importance of gene fusions as driver oncogenic events was considered to be virtually restricted to clonal haematological disorders, such as leukaemias and lymphomas. Recently, oncogenic gene fusions have been identified also in solid tumours (1), indicating that the role of fusions in oncogenesis is broader than previously expected. Fusions are routinely investigated using cytogenetic analyses. These techniques, however, although still largely used, suffer from severe limitations: they require the presence of an adequate number of mitotic cells, which is often a challenging problem in many solid cancers and in some types of leukaemia/lymphoma; they are only able to produce a gross map of the rearrangements, thus requiring further efforts to identify the fusion partners; finally, they are not able to detect cryptic fusions.
The recent development of many selective inhibitors that target proteins abnormally activated in specific types of cancer and, most notably, the successful experience of imatinib for the treatment of chronic myeloid leukaemia (CML), strongly suggest that understanding the biologic, and thus genetic, mechanisms underlying the development of cancer is of primary importance to treat it successfully. In this scenario, the ability to identify the presence of oncogenic fusions even in ‘difficult’ samples, such as many solid cancers, where the oncogenic lesions are still largely unknown, could play a critical role also in clinical research to develop targeted treatment strategies.
Therefore, the availability of user-friendly fusion-detection tools, being able to identify new and known fusions at nucleotide resolution even in the absence of mitotic events and when the availability of cancer cells is limited, can have a profound impact in basic as well as clinical research.
The development of high-throughput short-read sequencing technologies had a dramatic impact in our ability to generate whole-transcriptome data of complex genomes and many pipelines dedicated to digital expression analysis of transcriptome re-sequencing have been developed; however, a limited effort has been yet dedicated to the development of bioinformatics tools focused on the detection of driver gene fusions through transcriptome re-sequencing.
In a pioneeristic paper, Gerstein’s (2) group developed a pipeline for the detection of gene fusions by using paired-end sequences. By using their work as a starting point, we developed FusionAnalyser, a graphical, event-driven tool which makes use of paired-end short-read transcriptome sequences to initially detect and annotate the presence of fusion rearrangements and then to identify the potentially driver event(s) (Supplementary Figure S1). The core of our procedure relies on the concept of using multiple annotation layers: FusionAnalyser initially uses paired reads, mapping to different genes (Bridge reads), to build a data set of candidate fusion events. This data set is then used to generate the first annotation layer (Bridge Annotation Layer, BAL); by taking in account and comparing the strand compatibility among the two fusion partners, the presence of reads mapping to the hypothetical fusion (Junction reads), the frame of the candidate fusions and the presence of a reciprocal event, FusionAnalyser is able to build multiple layers of biological evidence upon the BAL, which allows the user to dynamically filter the biologically relevant events and analyse the results in real-time.
MATERIALS AND METHODS
Algorithms
Our approach to detect fusions in transcriptome sequencing relies on the analysis of short, paired-end reads. These reads are initially aligned to the reference genome: paired reads, mapping to two different genes, are used to generate a first data set of potential intrachromosomal and extrachromosomal fusions candidates (‘Bridge reads’). Subsequently, a second data set, built upon those reads where only one of the two sequences in a pair is successfully mapped to the reference genome (‘Half-mapped Anchor reads’) is generated. The underlying idea is that, in presence of a gene fusion event, a fraction of the unmapped reads of the ‘Anchor’ data set could align to the corresponding fusion region, which is not present in the reference genome. The mapped reads in the latter data set are used as an anchor to tie each Half-mapped event to the corresponding Bridge region. The genomic coordinates of each Bridge event are automatically annotated against an exonic database and the individual Bridge exons are thus identified. Annotated Bridge events mapping to the same two genes are grouped together and reads pertaining to the same group and their associated exons are analysed using a dedicated Junction Prediction Algorithm (JPA, Supplementary Figure S2a) in order to identify the most likely fusion (‘Junction’ region) for each bridge. The Junction candidate is generated by identifying all the exons of each partner being aligned to one or more Bridge reads. If one of the two partner genes is at the 5′ of the fusion (Gene1), according to the Strand prediction algorithm (Supplementary Figure S3), the Gene1-exon contributing to the Junction candidate will be the 3′-most exon among all those receiving the alignment of at least one Bridge read. If the partner gene is at the 3′ of the fusion (Gene2), the Gene2-exon contributing to the Junction candidate will be the 5′-most exon among all those receiving the alignment of at least one Bridge read. Starting from the two candidate breakpoint exons identified by the JPA, the heuristic junction projection module (JPM) algorithm will build all the candidate Junction regions, taking into account the strand mapping of each read pair to the corresponding chromosome and the physical strand occupancy of the associated genes (Supplementary Figure S2b). The depth of the projection can be customized by users, ranging from 0 (i.e. only the two exons deterministically found by the JPA are considered) to the infinity (i.e. all the candidate exons pertaining the two genes are taken into account). These data, together with the corresponding genes and exons, are then stored in a dedicated data set.
All the mapped reads in the Anchor data set are similarly annotated against the RefSeq exonic database to identify the corresponding genes and exons.
Subsequently, the Bridge and Anchor data sets are filtered according to a customizable set of parameters, namely: Phred-scored read quality, frequency of each event, maximum number of undetermined nucleotides (N) in each read, mapping quality, presence of alternative alignments mapping to the paired read gene, quality of the Cigar match, HLA–HLA filtering and alignment homology (Bridge data set only) between the two exons of each Bridge. Optionally, Bridge reads can be further filtered with a user defined list of gene pairs (‘a priori’ filter).
Read quality filter
The Read Quality filter is activated by default. This filter applies to the read quality of each SAM or BAM read. If the read quality of at least n nucleotides in one of two reads of a pair is lower than the threshold, the entire pair is discarded. The read quality threshold is expressed in Phred units.
Hits threshold filter
The Hits Threshold filter is activated by default. This filter is applied to candidate Bridge reads only after the identification of the genes associated to each pair. If the number of events bridging between two genes is lower than the Hits threshold, the corresponding reads are discarded.
N filter
The N filter is activated by default. This filter applies to the sequence of each SAM or BAM read. If the number of undetermined nucleotides within a read is equal or higher than the N threshold filter, the pair is discarded.
Mapping quality filter
The Mapping Quality filter is activated by default. It applies to the mapping quality of each SAM or BAM read. If the mapping quality of one of two reads in a pair is lower than the threshold, the entire pair is discarded.
Alternative alignments filter
During the alignment of paired short reads to the reference human genome, it may occur that a read aligns to multiple regions with an identical alignment score. In this scenario, the aligner may assign that read to the wrong region. The other read of that pair, however, will still align to the correct genomic locus. The overall result is that an artefactual fusion is generated. This is indeed a powerful source of artefacts in mRNAseq fusion analyses. To overcome this problem, the Alternative Alignments filter, which is activated by default, scans the alignment data for the presence of alternative alignments. If present, these data are processed, using the exonic database as reference, to identify the corresponding genes. Then, these data are compared with the alignment(s) and gene(s) of the paired read. If a common gene between the two reads is found, then the data is considered an alternative alignment artefact and thus discarded.
Cigar filter
The alignment of short, paired-end reads to a genome may lead to a perfect match or to a partial match (e.g. a match carrying small insertions, deletions or mismatches). Although the ability to identify suboptimal mapping is critical for single nucleotide or small indel variants identification, the presence of suboptimal matches in fusion discovery is usually detrimental, because it increases the risk of artefacts due to erroneous mapping. This is indeed another important source of artefacts. To overcome this problem, the Cigar filter, which is activated by default, scans the alignment data for the presence of less-than-perfect alignments. If present, these data are discarded.
HLA–HLA filter
The HLA genes typically share an extremely high sequence similarity with one another (e.g. there is a 92% sequence identity between HLA–B and HLA-C) and they are highly polymorphic. The identification of HLA–HLA fusion (or read-through) candidates is most likely the result of errors during the alignment of the sequencing reads to the human genome. This is typically due to the presence of sequencing errors or polymorphisms, which leads to an erroneous mapping of the two paired reads to different HLA genes. Therefore, HLA–HLA events represent sequencing artefacts rather than real fusion events. Although the ‘ex post’ identification of such HLA artefacts is trivial, their presence steals computational power, thus increasing the time required to complete a run. The HLA–HLA filter is active by default.
Alignment Homology filter
The Homology filter tries to filter out gene pairs by comparing the homology of the two corresponding exons. The idea behind this filter is similar to the one of the Alternative Alignments filter, however this approach is more computationally intensive and less potent and should be used only when the Alternative Alignments filter is not applicable (e.g. the XA Tag is not available and a new alignment is not feasible). The Homology filter is inactive by default.
A priori filter
Read-through genes are commonly found in mRNAseq fusion studies. They represent physiological phenomena not related to cancer. However, from an analytical point of view, they mimic intrachromosomal, non-reciprocal fusions. The processing of read-through data may thus steal resources and may therefore slow down the whole process. To overcome this problem, FusionAnalyser allows the user to define a set of custom a priori filtering pairs that can be filtered out in the early phases of the analysis.
After the completion of the filtering step, each filtered Bridge event is scanned against the annotated, filtered Anchor data set. If one of the two genes associated with a Bridge event corresponds to a mapped gene in the Anchor data set, the matched unmapped read is aligned to the candidate Junction regions of the Bridge event, generated by the JPA/JPM, using a dedicated built-in, gapped alignment algorithm. The result of the alignment is then evaluated by a first, computationally fast, scoring algorithm. Alignments passing the first filter are evaluated by a second, more accurate, scoring algorithm. If the alignment succeeds, the Junction is deemed to be valid (Junction read). In this case, FusionAnalyser generates a ‘Junction annotation’ comprising the alignment information and the genomic coordinates, gene names and sequences of the two partner exons involved in the candidate fusion. This annotation is associated with the corresponding Bridge event (BJ data set).
Each Bridge or BJ event will then undergo a series of three further annotation steps:
- –
Strand annotation: by analysing the strand mapping of each read pair to the corresponding chromosome and the physical strand occupancy of the associated genes, the compatibility of the two candidate fusion genes is tested (Supplementary Figure S3). If the two genes/reads are strand compatibles, a ‘Strand annotation’ (S) is associated with the corresponding Bridge event.
- –
Frame annotation: this algorithm will be generated only for the Bridge events associated with a Junction annotation (BJ). The codon frame of each of the two exons in the exon–exon fusion boundary region in each BJ event is retrieved by analysing the frame and length of each exon of the corresponding gene in the exonic database (Supplementary Figure S4). This information is then used to verify whether the frame in the fusion region is conserved. If so, a ‘Frame annotation’ (F) is associated with the corresponding Bridge event (BF).
- –
Reciprocal translocation annotation: FusionAnalyser scans the rearrangement candidates for the presence of reciprocal events before the application of the static filters: if a potential reciprocal translocation is detected, it automatically adapts the filtering strategy by applying the Hits threshold algorithm to the sum of the individual contributions of each of the two reciprocal events. If such an event is found, FusionAnalyser adds a ‘Reciprocal annotation’ to the two corresponding Bridge events (BR).
A multiparameter scoring algorithm, which takes into account the coverage of each candidate and its annotation status is then applied to each Bridge event and its value is associated with the corresponding fusion.
After the completion of the annotation steps for each Bridge event, the corresponding data, together with their associated annotations, are processed for non-volatile storage through a serializing algorithm. Finally, the processed data are loaded in the Visualization and Dynamic Filtering (VIDYF) module. Here, intra and extrachromosomal candidate fusions can be dynamically filtered in line with the following set of parameters: read coverage, overall scoring threshold, presence of Junction reads targeting the fusion breakpoint, strand compatibility of the candidate fusion gene pair, presence of a continuous translation frame in the candidate fusions, presence of a reciprocal translocation, junction alignment score and removal of read duplicates. The fusion data generated through the dynamic filtering process are shown in real-time in a dedicated graphical visualization module (Supplementary Figure S5).
FusionAnalyser
FusionAnalyser is implemented in C# and runs under 64/32 bit Windows (successfully tested under Windows 7, Vista, XP, 2000) and Linux using Mono (successfully tested under Ubuntu and RedHat).
It was designed using streaming and serializing technologies in order to work under a limited memory footprint, so it can be successfully run on standard dual or quad core, 4 GB memory desktop/notebook PC. The typical timing required to complete a run using a 4 Gigabases human transcriptome data set is 6–8 h on a 4 GB, QuadCore Intel i7 X 940 Notebook.
Transcriptome sequencing
All the transcriptome libraries were generated using the Illumina TruSeqTM RNA Sample Preparation Kit. Paired-end 60 base reads were generated using an Illumina Genome Analyzer IIx and the Illumina TruSeqTM SBS kit v5. On average, 4.7 Gigabases per sample were generated.
Alignment to the human genome
All the sequence-processing and alignment steps were performed using a local instance of the Galaxy framework (3). Each 60 bp FastQ sequence was initially split in 2 × 30 bp reads, to maximize the chance of mapping in presence of small exons. Then, transcriptome sequencing data were aligned to the human genome (NCBI36/hg18) using the fast short-reads aligner BWA (4). BWA alignment parameters were set as follows: the fraction of missing alignment, assuming an uniform base error rate of 0.02, was set at 0.04. The maximum number of gaps per sequence was fixed to 1. Given the limited length of the split sequences, seeding was disabled and the mismatch penalty for single nucleotide variants was set at 3. Gap open and gap extension penalties were fixed at 11 and 4, respectively. To improve the efficiency of the alignment, the identification of suboptimal hits was disabled if the best hit was a repeat. The maximum number of alignments to output in the XA tag for discordant read pairs was set to 10. All the reads were treated as paired and the maximum insert size for a properly mapped pair was fixed at 500 bp.
FusionAnalyser settings
The following settings were applied to the analyses of the transcriptome of the CML patients: the mapping quality filter was activated, with a mapping quality filter threshold set to 30 (Phred). The Threshold for the presence of undetermined nucleotides (N) was set to 2. The read quality threshold was set to allow a maximum of 2 nt per read with a read quality of ≤25. The frequency threshold filter was set to 20. The homology filter was disabled. The Cigar filter, the alternative alignment algorithms and the HLA–HLA filter were activated. The intrachromosomal alignment filter threshold, indel malus, mismatch malus, match gain, continuity gain, split threshold and split minimum value were set to 0.9, −2, −2, 1, 1, 0.8 and 5, respectively. The extrachromosomal alignment filter threshold, indel malus, mismatch malus, match gain, continuity gain, split threshold and split minimum value were set to 0.9, −2, −2, 1, 1, 0.8 and 5, respectively. The JPM was activated and set to 3. The a priori filter was disabled. The real-time ‘condense identical reads’ algorithm was activated and the corresponding minimum coverage threshold was set to 5. The following settings were applied to the analyses of the in silico data: the mapping quality filter was activated, with a mapping quality filter threshold set to 30 (Phred). The N-filter was disabled. The read quality threshold was set to allow a maximum of two nucleotides per read with a read quality of ≤25. The frequency threshold filter was disabled for the low coverage analyses and was set to 20 for all the remaining data sets. The homology filter was disabled. The Cigar filter and the alternative alignment algorithms were disabled. The intrachromosomal alignment filter threshold, indel malus, mismatch malus, match gain, continuity gain, split threshold and split minimum value were set to 0.75, −2, −2, 1, 1, 0.8 and 5, respectively. The extrachromosomal alignment filter threshold, indel malus, mismatch malus, match gain, continuity gain, split threshold and split minimum value were set to 0.75, −2, −2, 1, 1, 0.8 and 5, respectively. The a priori filter was disabled.
Patients
Written informed consent was obtained from each subject involved in the study. All the human investigations were performed in accordance with the principles embodied in the declaration of Helsinki.
In silico data
In silico Sequence Alignment/Map (SAM) data were generated by using a dedicated software, which accepts the sequence and coordinates of n 5′ and m 3′ exons from the breakpoint and a RefSeq-based database and the following parameters as input: the simulated read length (Rl), the total amount of in silico generated bases (B), the number of random Bridge events (BrN) and the number of random Junction reads (JnN). The number of non-chimeric random paired reads per run is thus calculated according to the following formula: [B – 2 * Rl * (BrN + JnN)] / (2 * Rl). All the non-chimeric reads are considered to be exonic and generated accordingly. The n and m parameters were set to 4 whenever this was compatible with the exonic structure of the fusion gene.
RESULTS
To assess the ability of FusionAnalyser to identify fusion genes, we generated 1 Gigabase of artificial alignment data (see ‘Materials and Methods’ section for further details) for each of 20 known human translocations (Table 1) occurring in leukaemias (18) and solid cancer (2) and we analysed these data using our tool. In all the cases, FusionAnalyser identified the specific translocation associated with each data set (Supplementary Data S1 and Supplementary Figure S6) and the exact fusion region at exon and nucleotide level for all the translocations under analysis, correctly annotating the presence of a continuous coding frame in each breakpoint junction and predicting the correct orientation of each fusion, through the identification of its 5′ and 3′ partners.
Table 1.
Molecular characteristics of the human fusions analysed using simulated in silico data
| Fusion | Translocation | Exon1 Chr | Exon1 Start | Exon1 End | Exon2 Chr | Exon2 Start | Exon2 End | Disease |
|---|---|---|---|---|---|---|---|---|
| BCR–ABL1 (p210) | t(9;22)(q34;q11) | Chr22 | 21 962 525 | 21 962 600 | Chr9 | 132 719 271 | 132 719 445 | CML |
| BCR–ABL1 (p190) | t(9;22)(q34;q11) | Chr22 | 21 852 551 | 21 854 425 | Chr9 | 132 719 271 | 132 719 445 | ALL |
| CBFB–MYH11 | inv(16)(p13q22) | Chr16 | 65 673 616 | 65 673 712 | Chr16 | 15 728 205 | 15 728 412 | AML |
| CEP110–FGFR1 | t(8;9)(p12;q33) | Chr9 | 12 297 5773 | 12 297 5836 | Chr8 | 3 839 8471 | 38 398 616 | 8p12 MPD |
| ETV6–JAK2 | t(9;12)(p24;p13) | Chr12 | 11 913 624 | 11 914 170 | Chr9 | 5 071 724 | 5 071 861 | ALL |
| NCOA4–RET | inv(10)(q11.2;q11.2) | Chr10 | 51 251 275 | 51 251 384 | Chr10 | 42 932 037 | 42 932 185 | PTC |
| NPM1–ALK | t(2;5)(p23;q35) | Chr5 | 170 751 314 | 170 751 408 | Chr2 | 29 299 711 | 29 299 898 | ALCL |
| NUP98–HOXD13 | t(2;11)(q31;p15) | Chr11 | 3 722 314 | 3 722 455 | Chr2 | 176 667 453 | 176 668 912 | AML |
| PICALM–MLLT10 | t(10;11)(p13–14;q14–21) | Chr11 | 85 365 313 | 85 365 373 | Chr10 | 21 941 282 | 21 941 386 | ALL/AML |
| PML–RARA | t(15;17)(q24;q21) | Chr15 | 72 112 549 | 72 112 808 | Chr17 | 35 758 093 | 35 758 242 | AML |
| ETV6–NTRK3 | t(12;15)(p13;q25) | Chr12 | 11 913 624 | 11 914 170 | Chr15 | 86 284 857 | 86 284 988 | AML |
| ETV6–RUNX1 | t(12;21)(p13;q22) | Chr12 | 11 913 624 | 11 914 170 | Chr21 | 35 187 091 | 35 187 130 | ALL |
| EWSR1–ERG | t(21;22)(q22;q12) | Chr22 | 28 012 911 | 28 013 123 | Chr21 | 38 696 348 | 38 696 429 | ES |
| MLL–MLLT1 | t(11;19)(q23;p13.3) | Chr11 | 117 857 639 | 117 858 017 | Chr19 | 6 213 238 | 6 213 321 | ALL/AML |
| MLL–MLLT3 | t(9;11)(p22;q23) | Chr11 | 117 857 639 | 117 858 017 | Chr9 | 20 353 473 | 20 353 603 | AML |
| RUNX1–RUNX1T1 | t(8;21)(q22;q22) | Chr21 | 35 153 640 | 35 153 745 | Chr8 | 93 098 629 | 93 098 767 | AML |
| SFRS3/BCL6 | t(3;6)(q27;p21) | Chr6 | 36 672 515 | 36 672 723 | Chr3 | 188 932 190 | 188 932 412 | NHL–FL |
| TCF3–PBX1 | t(1;19)(q23;p13) | Chr19 | 1 570 109 | 1 570 233 | Chr1 | 163 028 354 | 163 028 599 | ALL |
| TRIP11–PDGFRB | t(5;14)(q33;q32) | Chr14 | 91 524 380 | 91 524 480 | Chr5 | 149 486 275 | 149 486 370 | AML |
| ZBTB16–RARA | t(11;17)(q23;q21) | Chr11 | 113 532 268 | 113 532 366 | Chr17 | 35 758 093 | 35 758 242 | AML |
The fusion name, translocation, genomic coordinates of the two breakpoint exons and the disorder most commonly associated with each lesion are shown.
CML = Chronic Myeloid Leukaemia, AML = Acute Myeloid Leukaemia, MPD = myeloproliferative disorder, PTC = Papillary thyroid carcinoma, ALCL = Anaplastic Large Cell Lymphoma, ES = Ewing Sarcoma, NHL = Non-Hodgkin Lymphoma, FL = Follicular Lymphoma.
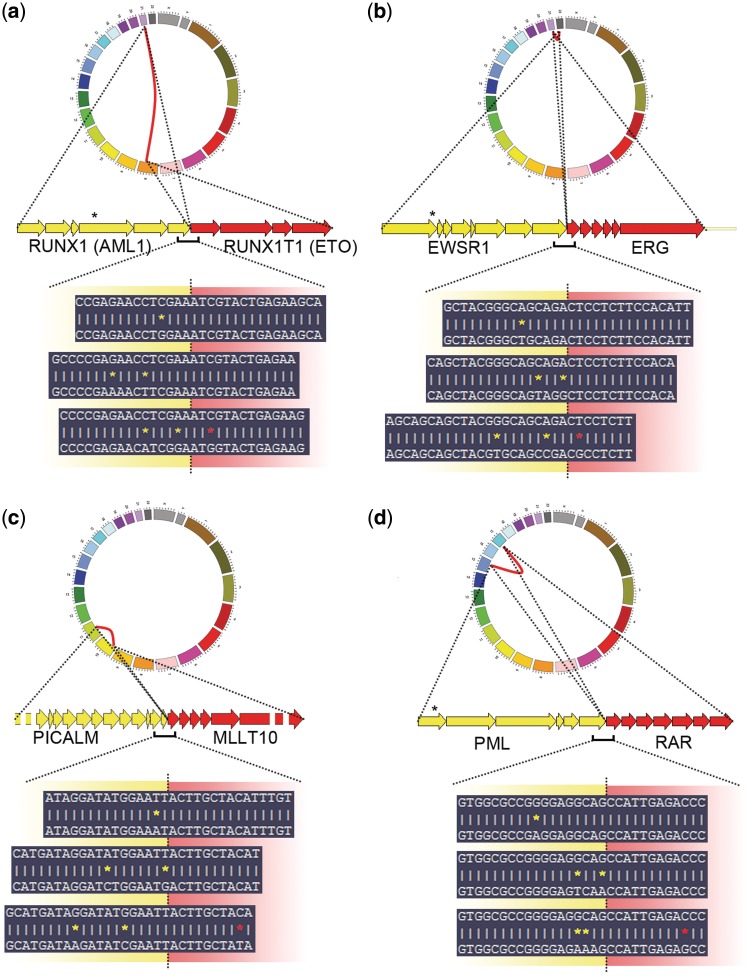
To further test the robustness of our tool, we generated artificial alignment data for four translocations (RUNX1–RUNX1T1, EWSR1–ERG, MLLT10–PICAM and PML–RARA), simulating the presence of 1, 2 or 3 randomly generated single nucleotide variants at a distance of no more than 15 nt from the breakpoint site, to take in account the presence of single nucleotide polymorphisms, somatic variants or sequencing errors in the context of each breakpoint. The analysis of these data sets (Figure 1 and Supplementary Data S2) showed that FusionAnalyser was invariably able to identify all the translocations and to predict the exact breakpoints.
Figure 1.
Analysis of artificial alignment data for four translocations: RUNX1–RUNX1T1 (a), EWSR1–ERG (b), MLLT10–PICAM (c) and PML–RARA (d), simulating the presence of 1, 2 or 3 randomly generated single nucleotide variants within the breakpoint region. In the upper part of each panel, the standard graphical FusionAnalyser output, in the form of a circular diagram reproducing the identified rearrangement, is shown. In the lower part of each panel, three representative junction regions are shown. The upper sequence in each box represents the reference breakpoint sequence, generated by the Junction Prediction/Projection modules; the lower sequence represents part of an anchor read successfully mapped to the breakpoint region despite the presence of 1 (upper box), 2 (middle box) or 3 (lower box) variants. Each variant is highlighted by the presence of a yellow (variant occurring in the first gene of the fusion) or red (variant occurring in the second gene of the fusion) asterisk.
Low coverage fusions
To verify the ability of our tool to detect rearrangements in the context of gene fusions expressed at low levels, we generated eight new data sets (1 Gigabase each) with a progressively decreasing number of reads aligning to the fusion region (RUNX1–RUNX1T1 and PML–RARA; 24, 12, 6 and 2 reads targeting the fusion, with 20, 10, 5 and 1 Bridge and 4, 2, 1 and 1 Junction reads, respectively). Even at the lowest expression level (two fusion reads/16.6 × 106 total reads) FusionAnalyser was consistently able to detect and report the correct translocation, even in presence of a single nucleotide variant within the junction region (Supplementary Figure S7 and Supplementary Data S3). The presence of the translocation was also correctly reported in presence of a single Bridge read targeting the fusion (RUNX1–RUNX1T1 and PML–RARA; Supplementary Data S3), although in this case the absence of any information pertaining to the junction prevented the identification of the breakpoint at nucleotide level.
Heuristic junction prediction
A potential issue when gene fusions are expressed at low levels or with low sequencing coverage is the absence of bridge reads mapping to one or both the breakpoint exons. A similar condition is typically found in presence of small exons, when the exon length is comparable or smaller than the length of the read. In this condition the aligner may fail, leading to a localized coverage drop. In this scenario, the identification of the correct junction is particularly challenging, because limited deterministic information about the fusion exons can be derived. To mimic these situations and put our heuristic algorithm of junction projection under test (see ‘Materials and Methods’ section, Supplementary Figure S2a and b), we generated two new data sets for the RUNX1–RUNX1T1 translocation where we enforced the absence of reads mapping to one of the two breakpoint exons (Supplementary Figure 8a) or to both (Supplementary Figure S8b). Even in complete absence of Bridge reads mapping to the two breakpoint exons, FusionAnalyser identified the presence of the rearrangement, the correct junction at nucleotide level and the corresponding exons (Supplementary Figure S8a, b and Supplementary Data S4).
Complex rearrangements
Several recent reports (5–8) suggest that multiple rearrangements are commonly detected in cancer cells. To test FusionAnalyser in the context of this complex scenario, two new data sets were generated, comprising 6 (5 extra and 1 intrachromosomal events) and 20 (18 extra and 2 intrachromosomal events) rearrangements, respectively. In both cases our tool was able to correctly identify all the translocations at nucleotide level (Supplementary Figure S9 and Supplementary Data S5) and to annotate the coding frame and the orientation of each fusion.
Reciprocal translocations
According to the Mitelman database of chromosomal aberrations in cancer (1), ∼96% of the reported translocations are reciprocal. The ability to identify the presence of these events may thus play an important role in the process of data annotation and validation: the demonstration that a candidate fusion event and its reciprocal coexist in a cancer transcriptome can add a significant layer of evidence to that candidate and may help in discriminating between real translocations and read-through fusions. Transcripts generated through reciprocal translocations are under the control of two different promoters, one for each of the two genes involved in the translocation. If one of the two promoters is weak, an unbalanced expression of the two transcripts may occur, with one of the transcripts being expressed at low levels. Under these circumstances, the information pertaining the latter transcript may be lost during the filtering steps, preventing the detection and annotation of the reciprocal event. To overcome this limitation, we developed a dedicated algorithm to automatically scan the rearrangement candidates for the presence of reciprocal events before the application of the static filters: if a potential reciprocal translocation is detected, FusionAnalyser automatically modifies the Hits threshold algorithm by applying it to the sum of the individual contribution of each reciprocal event, thus raising the overall sensitivity in presence of candidate reciprocal translocations and avoiding the risk of an undesired loss of information.
To test the ability of FusionAnalyser to identify reciprocal translocations, we generated two new data sets where we modelled the presence of reciprocal fusions (PML–RARA+RARA–PML, NPM1–ALK+ALK–NPM1). In all these models, our tool identified each rearrangement and annotated the presence of the corresponding reciprocal translocation (Supplementary Data S6).
Transcriptome analysis of chronic and acute myeloid leukaemia samples
The ideal objective of transcriptome based fusion analyses is the identification of driver rearrangements occurring in patients affected by solid tumours or leukaemias, either to identify new, yet unknown translocations or to diagnose the presence of known ones. However, a critical problem of these studies is the co-detection of a very high number of spurious events generated either during the library preparation or due to misalignments, with no involvement in the pathogenesis of the clonal disorder (9). The presence of such a high background may seriously impair our ability to discriminate the real driver events.
To assess the potential of our approach to the identification of driver rearrangements, we generated paired-end transcriptome sequencing data (4.5, 3.0 and 3.7 Gigabases, respectively) from the peripheral blood of three patients affected by CML in Chronic Phase (CML–CP) at onset of the disease (Table 2). CML patients at onset typically lack the extensive genomic rearrangements that are more typical of the advanced phase of the disease (10) and of many cancer-derived cell lines. Indeed, in all the CML patients under analysis, cytogenetic studies failed to reveal any other genomic alteration besides the presence of the Philadelphia chromosome (data not shown). This approach allowed us to test our tool in a model where only a single ‘real’ rearrangement was bona fide present and thus to assess whether FusionAnalyser was able to filter out the majority of the artefacts and to identify a driver translocation with sufficient specificity. Despite the presence of a relatively high number of BAL events (11, 9 and 75 for transcriptome of patient CML–CP-001, CML–CP-002 and CML–CP-003, respectively), the application of the algorithms of driver fusion identification was sufficient to narrow down our candidates to the single BCR/ABL1 translocation in all the three data sets (Supplementary Figure S10). Moreover, FusionAnalyser correctly reported the absence of the reciprocal ABL1–BCR translocation in CML–CP-003, where loss of the derivative chromosome 9 was known to be present (Table 2).
Table 2.
Summary of clinical details of the three CML patients included in this study
| Patient ID | Age at diagnosis | Sokal Score | WBC at diagnosis (per µl) | Platelets at diagnosis (per µl) | Additional cytogenetic abnormalities | Q-PCR at diagnosis/100 copies of ABL (IS) |
|---|---|---|---|---|---|---|
| CML–CP-001 | 23 | 0.8 | 74.5 × 103 | 748 × 103 | No | 59.5 |
| CML–CP-002 | 52 | 0.66 | 55.7 × 103 | 281 × 103 | No | 60.5 |
| CML–CP-003 | 45 | 0.91 | 34.4 × 103 | 1068 × 103 | Loss of der (9) | 44.2 |
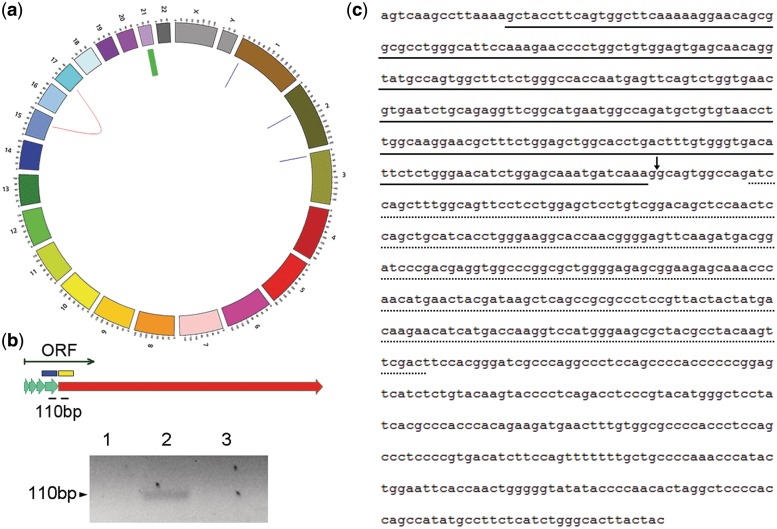
To further put the ability of FusionAnalyser to identify driver events under test, we generated paired-end transcriptome sequencing data (6.4, 6.2 and 4.4 Gigabases, respectively) on three Acute Myeloid Leukaemia (AML, Table 3) specimens in absence of any a priori knowledge about their cytogenetic status. In all the three cases our tool identified a specific fusion event (RUNX1–RUNX1T1 in Patient 1 and PML–RARA in Patients 2 and 3). Subsequent PCR analysis confirmed the correctness of each prediction (data not shown). Interestingly, in patient AML-002, FusionAnalyser identified the presence of a second, in-frame, cryptic, intrachromosomal event localized on chromosome 21, involving two closely related genes: ETS2 and ERG (Figure 2a). The presence of the ETS2–ERG fusion was confirmed by PCR amplification and sequencing (Figure 2b and c). The detailed analysis of the biological and functional role of this fusion will be discussed elsewhere.
Table 3.
Summary of clinical details of the three AML patients included in this study
| Patient ID | Age at diagnosis | Sex | WBC at diagnosis (per µl) | Platelets at diagnosis (per µl) | Haemoglobin at diagnosis (g/dl) |
|---|---|---|---|---|---|
| AML-001 | 34 | Male | 74.5 × 103 | 748 × 103 | 10.9 |
| AML-002 | 18 | Male | 55.7 × 103 | 281 × 103 | 6.3 |
| AML-003 | 64 | Female | 34.4 × 103 | 1068 × 103 | 7.1 |
Figure 2.
Analysis of transcriptome sequencing data of patient AML002 (a): the red curved line highlights the presence of the PML–RARA translocation; the blue lines indicate bona fide read-through events; the thick green line points to the intrachromosomal ETS2–ERG fusion. (b) Schematic model of the ETS2–ERG fusion: the ETS2 exons are shown as thick green arrows; the 3′ ERG exon is shown as a thick red arrow. The thin green arrow shows the open reading frame of the fusion. The blue and yellow boxes indicate the PNT domain of ETS and the ETS domain of ERG, respectively. The two black lines indicate the position of the two primers used for the amplification of the breakpoint region. In the bottom panel, the result of the ETS2–ERG amplification in patients AML001 (1), AML002 (2) and AML003 (3) is shown. (c) Sequence of the ETS2–ERG breakpoint region. The solid black line highlight the PNT domain of ETS, the dotted line the ETS domain of ERG. The black arrow indicates the breakpoint site.
Although the analysis of in silico samples suggests that our tool is able to efficiently manage multiple events (Supplementary Figure S9), it is also conceivable that in silico data are less noisy than real transcriptomes, mostly because library preparation artefacts can be present in the latter case. Therefore, to test FusionAnalyser on real sequencing data in presence of multiple fusions, we conceived a new test, where we combined the alignment data set of one BCR–ABL1 positive patient (CML–CP-002) with patient AML002, in whom the PML–RARA and ETS2–ERG fusions were detected. By using this approach we generated a new, hybrid data set containing three fusions in the context of real transcriptome data. It is important to notice that this approach is even tougher than ‘real’ transcriptome analyses, since in our test the individual contribution of each fusion comes from approximately half of the entire data set and thus the signal-to-noise ratio for each event is halved. In addition, the overall size of the alignment data is doubled, potentially leading to an increase of the background noise against statically defined filters (which were unchanged from previous analyses). Even in presence of these demanding conditions, FusionAnalyser was able to identify all the fusions at exon and nucleotide level (Supplementary Figure S11).
Comparative analysis of three fusion detection tools
A critical step to fully validate a new tool is to compare it with already available packages. Although a direct comparison of different tools in bioinformatics is always challenging, we compared FusionAnalyser with two known fusion detection tools: FusionSeq (2) and FusionHunter (11). The results of the comparison are schematized in Table 4. As an ideal candidate for this test we chose the ‘AML002’ data set because FusionAnalyser was able to identify two fusions, a known (PML–RARA) and a completely new one (ETS2–ERG) and the two fusions were fully validated at exon and nucleotide level using conventional molecular biology techniques. To perform this test we focused on four different criteria:
- 1)
Results: obviously, this is the most important criterion. When the AML002 data set was analysed (Table 4) with FusionHunter under standard settings, the software readily identified the PML–RARA fusion but failed to detect ETS2–ERG. A possible explanation for this behaviour is that FusionHunter implements an homology filtering algorithm in its standard fusion discovery pipeline. The criterion behind this algorithm is that in paired-end sequencing it is possible that the two paired reads are mistakenly aligned to two different genes sharing a high level of homology and this misalignment could be erroneously identified as a fusion by the rearrangement discovery tool. Indeed, ETS2 and ERG are members of the same Ets family of oncogenes and they share a global 34.2% consensus and 25.1% similarity with a peak of 72.6% similarity in the C-terminus. The use of homology filters, albeit potent, is potentially detrimental because it may filter out real fusions involving two homologous genes. Surprisingly, the same analysis failed to identify any fusions under FusionSeq. This could be due to several factors: the first one is that we used the latest FusionSeq version (0.7.0) which is still an alpha and may require some further ‘fine tuning’. The second reason could be that, after the identification of the ‘fusion junction library’, we aligned the whole library against the Anchor reads instead of the entire data set, in order to decrease the challenging computational complexity of this step. Although unlikely, we cannot however exclude that this weakened the power of the analysis.
- 2)
Complexity of installation and configuration. The main criteria used to evaluate the installation and configuration steps are directly linked to the number of dependencies necessary to complete the installation and to the number of ‘hands-on’ steps required to complete the setup (Table 4).
- 3)
Flexibility of the hardware/software configuration required to run the software: the requirements for both FusionSeq and FusionHunter are demanding: a multicore Linux server, possibly not less than eight cores with 32 GB of RAM, was required to efficiently perform the most complex steps of the analysis, while FusionAnalyser was able to smoothly run on a standard dual or quad-core desktop or notebook computer with 4 GB of RAM on a Linux or Windows operative system. These requirements make our tool ideal also for laboratories with no in-site ‘high-throughput sequencing’ infrastructures (where no high-throughput-sized server machines are available) because it allows the analysis of transcriptome data, such as those generated by external companies, with no investment in costly and complex multicore clusters. Another important parameter to assess the flexibility of the three tools is their dependence from other software. FusionHunter is dependent on the Bowtie (12) alignment tool and cannot accept already aligned data sets as input, while FusionSeq and our tool allow the user to choose the preferred aligner. However, while FusionAnalyser accepts either SAM or BAM/BAI alignment files, which are the universally accepted standard alignment formats, FusionSeq requires dedicated ‘mrf’ files. It is worth noticing, however, that our tool is expressly dedicated to the detection of fusions in human transcriptomes while FusionHunter and FusionSeq can ideally be run also on transcriptomes from other species, provided that all the required annotation files are generated (Table 4).
- 4)
Friendliness of use: FusionAnalyser is fully graphical and event-driven: installation, configuration, filtering parameters, input/output files selection and data visualization are entirely managed through graphical windows and point-and-click interfaces thus requiring no background in bioinformatics or scripting knowledge. The output is automatically visualized in a dedicated module, which is able to react to the post-processing filters and selections in real time. FusionHunter requires command-line interaction under a Linux framework and requires manual configuration of initialization files; FusionSeq requires extensive command-line interaction under a Linux framework, the implementation of job parallelization techniques and the development of dedicated scripts.
Table 4.
Comparison of three fusion discovery tools
| CRITERIA | FUSIONANALYSER | FUSIONHUNTER | FUSIONSEQ |
|---|---|---|---|
| FUSIONS DETECTEDa | 2 | 1 | 0 |
| INSTALLATIONb | EASY (0/1 dep.) | EASY (0/1 dep.) | COMPLEX (≥ 4 dep.) |
| CONFIGURATIONc | EASY | NORMAL | COMPLEX |
| MULTIPLE SPECIESd | NO | YES | YES |
| HARDWARE | DUAL/QUAD CORE PC, 4 GBYTES RAM | MULTICORE SERVER | MULTICORE SERVER |
| ALIGNMENT TOOLe | OPEN (SAM/BAM) | CLOSE (Bowtie) | OPEN (.mrf) |
aExpressed as the number of validated fusions identified in the AML002 data set.
bThe complexity of the installation was scored proportionally to the number of dependencies typically required to complete the installation.
cConfiguration scores the complexity and hands-on time required to configure a standard analysis.
dThe ‘Multiple species’ field indicates if the tool is able to analyse transcriptomes from other species besides humans.
eThe ‘Alignment tool’ field indicates if the fusion discovery tool is dependent on a specific aligner. ‘OPEN (SAM/BAM)’ means that any aligner generating correct SAM/BAM alignments can be used to perform the analysis. ‘CLOSE (Bowtie)’ means that only the Bowtie aligner can be used. ‘OPEN (.mrf)’ means that any aligner can be used but the output format must be converted into .mrf files.
DISCUSSION
In this study we described FusionAnalyser, a new graphical tool dedicated to the identification of driver fusion rearrangements through the analysis of short, paired-end transcriptome sequencing data.
To verify the ability of FusionAnalyser to effectively detect rearrangements, we initially tested our tool using an extensive set of in silico generated data characterized by a progressively increasing complexity. In all these models, FusionAnalyser was invariably able to identify and annotate the correct fusion, to annotate the sequence of the fusion region at nucleotide level, to test strand and frame compatibility between the fusion partners and to assess the presence of reciprocal translocations, even in presence of multiple rearrangements, demonstrating the robustness of our approach. Then we generated paired-end transcriptome sequencing data from three patients affected by CML at the onset of the disease. We reasoned that the use of CML patient samples would lead to two major advantages: the first one was the chance to test FusionAnalyser in the contest of patient data, which is the most likely scenario for the application of our tool in the next future; the second was related to the fact that most CML patients at onset present only the t(9;22) translocation, lacking extensive genomic rearrangements: this allowed us to test the ability of FusionAnalyser to identify a single driver translocation with high specificity.
The analysis of these data sets revealed that, in line with previously published data (9), the number of candidate rearrangement events was in the range of 9–75 per patient (Supplementary Figure S10). However, when we dynamically filtered our candidates according to presence of strand compatibility, evidence of junction reads, presence of a coding frame throughout the fusion and reciprocal recombination, we were able to narrow down our driver fusion candidates to the single BCR–ABL1 rearrangement. In a similar analysis done on three AML samples, we were also able to identify a new, cryptic, in-frame ETS2–ERG fusion, which is now under characterization.
Taken globally, these data indicate that FusionAnalyser is a robust discovery software: it is able to identify driver rearrangements from transcriptome paired-end data even in presence of single nucleotide mismatches, such as single nucleotide polymorphisms or sequencing artefacts in the context of the breakpoint region or in presence of extremely low-coverage data. The use of data streaming and serialization, of memory-sparing algorithms and of dynamic parallel programming, allows FusionAnalyser to be run in standard dual or quad-core desktop or notebook machines, saving the precious computational time of servers/workstations to more demanding tasks. The presence of a highly flexible filtering system, comprising read quality filters, frequency of each event, maximum number of undetermined nucleotides in each read, mapping quality, analysis of paired-reads alternative alignments, dynamic removal of read duplicates, quality of the Cigar match, HLA–HLA and alignment homology filtering, together with the use of a fully event-driven graphical interface grants the end-user a significant analytical flexibility even in absence of a priori bioinformatics/scripting knowledge. Therefore we propose FusionAnalyser as a potent and practical tool for the identification of functional rearrangements in the context of high-throughput transcriptome sequencing data.
FusionAnalyser Executable for Windows 32 and 64 bit and for Linux, complete source code, FusionAnalyser manual and a test data set are available at NAR online.
FusionAnalyser is also available for download, together with hg19 and hg18 reference databases, from: http://www.ilte-cml.org/FusionAnalyser.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–11, Supplementary Data 1–6, FusionAnalyser Executable and source code, FusionAnalyser manual, FusionAnalyser test data set.
FUNDING
Italian Ministry of Health [RFPS-2006-333974]; Italian Association for Cancer Research (AIRC) [IG-10092]; Progetti di Ricerca di Interesse Nazionale (PRIN) [20084XBENM_004]; CARIPLO Foundation [2009-2667]; Regione Lombardia [ID-16871 and ID-14546A]. Funding for open access charge: Italian Association for Cancer Research (AIRC) [IG-10092].
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Michela Viltadi for technical assistance.
REFERENCES
- 1.Mitelman F, Johansson B, Mertens F. The impact of translocations and gene fusions on cancer causation. Nat. Rev. Cancer. 2007;7:233–245. doi: 10.1038/nrc2091. [DOI] [PubMed] [Google Scholar]
- 2.Sboner A, Habegger L, Pflueger D, Terry S, Chen DZ, Rozowsky JS, Tewari AK, Kitabayashi N, Moss BJ, Chee MS, et al. FusionSeq: a modular framework for finding gene fusions by analyzing paired-end RNA-sequencing data. Genome Biol. 2010;11:R104. doi: 10.1186/gb-2010-11-10-r104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Goecks J, Nekrutenko A, Taylor J. Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome. 2010;11:R86. doi: 10.1186/gb-2010-11-8-r86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, Sboner A, Esgueva R, Pflueger D, Sougnez C, et al. The genomic complexity of primary human prostate cancer. Nature. 2011;470:214–220. doi: 10.1038/nature09744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pleasance ED, Stephens PJ, O'Meara S, McBride DJ, Meynert A, Jones D, Lin ML, Beare D, Lau KW, Greenman C, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010;463:184–190. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee W, Jiang Z, Liu J, Haverty PM, Guan Y, Stinson J, Yue P, Zhang Y, Pant KP, Bhatt D, et al. The mutation spectrum revealed by paired genome sequences from a lung cancer patient. Nature. 2010;465:473–477. doi: 10.1038/nature09004. [DOI] [PubMed] [Google Scholar]
- 8.Ding L, Ellis MJ, Li S, Larson DE, Chen K, Wallis JW, Harris CC, McLellan MD, Fulton RS, Fulton LL, et al. Genome remodelling in a basal-like breast cancer metastasis and xenograft. Nature. 2010;464:999–1005. doi: 10.1038/nature08989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maher CA, Palanisamy N, Brenner JC, Cao X, Kalyana-Sundaram S, Luo S, Khrebtukova I, Barrette TR, Grasso C, Yu J, et al. Chimeric transcript discovery by paired-end transcriptome sequencing. Proc. Natl Acad. Sci. USA. 2009;106:12353–12358. doi: 10.1073/pnas.0904720106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shteper PJ, Ben-Yehuda D. Molecular evolution of chronic myeloid leukaemia. Semin. Cancer Biol. 2001;11:313–323. doi: 10.1006/scbi.2001.0387. [DOI] [PubMed] [Google Scholar]
- 11.Li Y, Chien J, Smith DI, Ma J. FusionHunter: identifying fusion transcripts in cancer using paired-end RNA-seq. Bioinformatics. 2011;27:1708–1710. doi: 10.1093/bioinformatics/btr265. [DOI] [PubMed] [Google Scholar]
- 12.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.