Abstract
RNase P processes the 5′-end of tRNAs. An essential catalytic RNA has been demonstrated in Bacteria, Archaea and the nuclei of most eukaryotes; an organism-specific number of proteins complement the holoenzyme. Nuclear RNase P from yeast and humans is well understood and contains an RNA, similar to the sister enzyme RNase MRP. In contrast, no protein subunits have yet been identified in the plant enzymes, and the presence of a nucleic acid in RNase P is still enigmatic. We have thus set out to identify and characterize the subunits of these enzymes in two plant model systems. Expression of the two known Arabidopsis MRP RNA genes in vivo was verified. The first wheat MRP RNA sequences are presented, leading to improved structure models for plant MRP RNAs. A novel mRNA encoding the central RNase P/MRP protein Pop1p was identified in Arabidopsis, suggesting the expression of distinct protein variants from this gene in vivo. Pop1p-specific antibodies precipitate RNase P activity and MRP RNAs from wheat extracts. Our results provide evidence that in plants, Pop1p is associated with MRP RNAs and with the catalytic subunit of RNase P, either separately or in a single large complex.
INTRODUCTION
RNase P is the ubiquitous and essential endonuclease required for tRNA 5′-end maturation; additional functions involve the cleavage of some mRNAs and non-coding RNAs. In most organisms and their organelles, the enzyme consists of one RNA and up to 10 protein subunits (1,2). The catalytic centre resides in the RNA which can function as ribozyme in vitro, although with weak efficiency for more complex organisms (3,4). The composition of nuclear RNase P from yeast and humans is known in detail, but no subunits have hitherto been characterized for the nuclear enzyme from plants. The primitive plastids from the unicellular alga Cyanophora paradoxa contain an RNase P with an essential and catalytically active RNA component (5–8); plastid and mitochondrial genomes from certain algal lineages encode an RNase P RNA (9,10). RNase P from chloroplasts and mitochondria of higher plants however is a protein enzyme (ProRP) similar to the human mitochondrial RNase P (11–13). Previous studies on the nuclear enzyme from carrot and wheat germ suggested the presence of an essential nucleic acid component (14,15). However, the exact molecular composition of the enzyme from higher plants remained unsolved.
In addition to RNase P, the structurally related RNase mitochondrial RNA processing (MRP) is present in eukaryote nuclei and contains a similar RNA subunit. Both enzymes are localized in the nucleolus and form distinct RNP complexes (16,17). Most proteins in the RNase P and MRP complexes are identical, but some are specific for one of the enzymes (1). RNase MRP cleaves the large rRNA precursor at the A3 site and is also involved in mitochondrial DNA replication, cleavage of some cytoplasmic mRNAs and production of siRNAs (18,19). In earlier studies, an RNase MRP RNA gene from Arabidopsis thaliana (AtMRP1) had been identified; the expression of the corresponding RNA and of three tobacco MRP RNAs in vivo was experimentally verified (20). More recently, in silico searches have detected a second, slightly different putative RNase MRP RNA gene in Arabidopsis (AtMRP2), and one in Oryza sativa (21); however, no expression data are available for these RNAs, and the relationship to the enzyme remains unclear.
The largest and one of the central proteins within RNase P and MRP is Pop1p. In yeast and human cells, Pop1p directly binds to several proteins and presumably to the RNA component in these two separate enzyme complexes (1,2,22–24). Its target is possibly the P3 region in RNase P and MRP RNA (25), and binding may be facilitated by the POP6/POP7 heterodimer (26). In all organisms studied, the four highly conserved regions COR1–COR4 are present. Mutational analysis revealed that in yeast, the Pop1 domain consisting of COR1 and COR2 is required for RNP formation. Conserved residues in COR1 and COR4 influence RNase P activity, whereas residues in all four regions contribute to RNase MRP activity (27,28). In A. thaliana, several mRNA splicing variants have been annotated for the single gene encoding this protein (AtPop1p), raising the question of their expression and function.
To get insight into the holoenzyme composition of plant nuclear RNase P and MRP, we have set out to identify the subunits of both Arabidopsis enzymes in silico and to investigate their expression and relation to the enzyme complex. Here, we concentrate on the central protein Pop1p and the RNAs annotated as MRP RNA. These data are complemented by functional studies using the established in vitro tRNA processing system from wheat germ (15). Our expression studies of AtPop1 mRNAs reveal a novel splicing form encoding a hitherto unknown AtPop1p variant. The presence of both Arabidopsis MRP RNAs in vivo was verified. Two novel MRP RNA sequences from wheat are presented, and improved structural models for plant MRP RNAs are suggested. AtPop1p-specific antibodies precipitate RNase P activity and MRP RNAs from wheat, suggesting a close physical association of Pop1p with both enzymes in plants.
MATERIALS AND METHODS
Chemicals were purchased from Applichem, Carl Roth, Merck or Sigma-Aldrich if not stated otherwise, and of the highest purity available. Enzymes were from GE Healthcare, Roche Applied Science, Fermentas, New England Biolabs or Promega and used according to the manufacturer’s instructions. Radionucleotides were purchased from Hartmann Analytic. A. thaliana var. Columbia was grown as described (29); young leaves from 3- to 4-weeks-old plants were used for RNA and protein preparations. Unprocessed wheat germ was obtained from SynPharma (Augsburg, Germany); S23 extract was prepared from defatted germs according to (15) and stored at −20°C. All cloning experiments were performed with gel purified vectors and inserts if not stated otherwise. Sequencing reactions were analysed on ABI Prism sequencers by Institut für Humangenetik (University of Würzburg), IZKF Leipzig or Seqlab, Göttingen. All sequences were obtained from both strands, using universal or internal primers as necessary. Novel nucleotide sequence data are available under the accession numbers: FN673552 (Arabidopsis Pop1 mRNA variant 3), FN689352 (wheat MRP RNA1) and FN689353 (wheat MRP RNA2).
Oligonucleotide primers
Oligonucleotides used in this study were purchased from Eurogentec. For blunt-end cloning of PCR products, phosphorylated amplification primers were used.
Oligo(dT): dT16V; AtPOP1-F: ATGGCTACTACTGCGAATGG; AtPOP1-R: TCAAGAATAAGTGTGAATGCC; AtPOP1-3′ss: AGGAAGGGGATCTAGGG; Anchor2: CGGACGTCACGCGTCGACCTACT20; UAPshort: GTCACGCGTCGACCTAC; Anchor-BRL: GCCACGCGTCGACTAGTACGGGIGGGIGGGIG; AtPOP1-GSP1: GGTCAAGTATCAGGAGGAG; AtPOP1-GSP6R: CAATCGCCTGAGAAACTGG; AtPOP1-GSP7R: GCCAGTGAGAATAGAGTCG; AtPOP1-GSP9R: GCAATGTGATAGCTTGCATC; 3′AtMRP1+2-anti4: AAGCTCCACACACGCGTCTC; 3′AtMRP1+2-anti5: GATTCTCTCGTTACCGAGGC; 3′MRP-anti3: CCCGTTCAGTTAGTCC; 5′MRP: CAGGAAAGTCCCCGG; 5′PlantMRP-CRI: CCAGGAAAGTCCCCGG; 3′PlantMRP-CRVanti-W: TAAGCCCCGTTCWGTTA; 3′PlantMRP-CRVanti-T: TAAGCCCCGTTCTGTTA; WG-MRP1-anti: ACGGACCAAGTGAATAAC; WG-MRP2-anti: GGGCTCCACACTCGTCTC.
Isolation of nucleic acids
Genomic DNA from plants was essentially prepared according to (29) and dissolved in ddH2O.
Total RNA was prepared as described (30) from 5 g Arabidopsis leaves and dissolved in 2.5 ml TE (10 mM Tris–Cl, 1 mM Na2EDTA, pH 7.5). Isolation of mRNA was performed by chromatography on Oligo(dT)-Cellulose. After binding of RNA to the equilibrated matrix in binding buffer [TE (pH 7.5), 0.2 % (w/v) SDS, 0.5 M NaCl], the slurry was transferred into a column and unbound RNAs were removed with washing buffer [0.2 % (w/v) SDS, 0.25 M NaCl in TE]. Poly(A+)-RNAs were eluted with mRNA elution buffer [0.2 % (w/v) SDS in TE] at 65°C and precipitated with EtOH.
Small RNAs were prepared by grinding fresh or frozen plant tissue in ice-cold TE buffer followed by extraction with one volume of phenol/chloroform. Wheat germ was first ground to powder, mixed with two volumes of TE and incubated for 20 min at 4°C before homogenization and phenol extraction. Large nucleic acids were precipitated with 2 M NaCl in an ice bath and removed by centrifugation. Small RNAs were EtOH precipitated from the supernatant, reprecipitated to reduce the salt concentration and dissolved in TE. All RNAs used for RT-PCR analysis were treated with RNase-free DNase I according to the manufacturer’s suggestions, followed by either heat inactivation or phenol extraction, and precipitation.
Cloning of the Arabidopsis Pop1 domain and analysis of AtPop1 mRNAs by Rapid Amplification of cDNA Ends (RACE)
The originally annotated AtPop1p coding sequence (At2g47290.1; AAB63829.1) was cloned by converting poly(A+) RNAs into cDNAs using the anchored oligo(dT) primer dT16V and AMV Superscript Reverse Transcriptase (RT). The open reading frame (ORF) was amplified with Pfu polymerase and the primers AtPOP1-F and AtPOP1-R, and ligated into the SmaI site of pUC19 to give pAtPOP1. Construction of expression vectors for recombinant AtPop1p is described in Supplementary Figure S5. The recombinant Pop1 domain used for functional studies is translated from pRSET-AtPOP1, which contains the unspliced exons 1 and 2 (variant 1 u) attached to the T7 promoter and an N-terminal His-tag. Translation termination within the intron results in a 187 amino acids long recombinant protein consisting of the 151 residues long truncated Pop1 domain attached to the affinity tag.
The 3′-end of AtPop1 mRNAs in vivo was identified by RACE from cDNA prepared as above; consecutive amplifications were performed with the primer pairs AtPOP1-F + Anchor2, AtPOP1-GSP1 + UAPshort and AtPOP1-3'ss + UAPshort. The transcription start site was determined by 5′-RACE using AtPOP1–GSP6R for cDNA synthesis. After tailing the DNA strand with Terminal Transferase and dATP, nested PCR was performed with the primer pairs AtPOP1–GSP7R + Anchor2 and AtPOP1–GSP9R + UAPshort. Cloning of RACE products unbiased by insert size was achieved by electrophoretic fractionation of amplificates and separate ligation of different size classes into pCR-XL-TOPO or pCRII-TOPO, if not stated otherwise.
Preparation of recombinant AtPop1p and antibody production
Recombinant AtPop1p was produced in Rosetta™ (DE3)pLysS (Novagen) transformed with pRSET–AtPOP1. Cultures were grown at 37°C in LB medium containing 100 µg/ml ampicillin, 34 µg/ml chloramphenicol and 2% glucose. Expression was induced at an OD600 of 0.5 by adding 1 mM IPTG and growth was continued at 33°C for 3–4 h. Cell harvest, lysis in 5 ml of denaturing lysis buffer A and affinity chromatography on Ni2+-NTA-Agarose (2 ml column volume) were performed essentially according to the manufacturer’s protocol (Qiagen); all solutions contained 1 mM phenylmethylsulfonyl fluoride (PMSF). Protein renaturation was achieved on-column by a reverse urea gradient (8–2 M). The bound protein was eluted with a dual gradient bringing the pH to 4.5 and concomitantly raising the imidazol concentration to 250 mM. Fractions containing purified protein were identified by SDS–PAGE and protein concentrations were determined according to (31) with BSA as standard. Pooled fractions were stored in elution buffer at 4°C and dialysed against buffer F-GP [20 mM Tris–Cl pH 7.6, 5 mM MgCl2, 20 % (v/v) glycerol, 0.5 mM PMSF] directly before use. For antibody production, the His-tag was removed by digestion with Enterokinase and the reaction products isolated by SDS–PAGE. Approximately 300 µg of protein in gel pieces were used to immunize a rabbit (Eurogentec). Antibodies directed against Escherichia coli proteins were removed from the serum according to (32). For some applications, the IgG fraction was purified by precipitation with 40% ammonium sulfate.
Immunoprecipitation of Pop1p-containing complexes from wheat germ extract
All manipulations were performed at 4°C and under gentle agitation if not stated otherwise. Antibody specificity was determined by western blot analysis of Arabidopsis proteins as described in Supplementary Figure S4.
For native IP, non-specifically binding wheat proteins were first eliminated by overnight incubation of 1 mg wheat S23 protein with 10 mg hydrated protein A-Sepharose beads (Amersham Bioscience) per assay in 1 ml buffer F2 (20 mM Tris–Cl pH 8, 10 mM MgCl2, 20 mM DTT, 20% Glycerol, 1 mM PMSF, 1 µM Pepstatin, 2 µM Leupeptin, 1 U/ml RNasin). The beads were removed by centrifugation and this pretreated extract was used in immunoprecipitations. Coupling of antibodies to protein A-Sepharose and specific binding of antigen were performed essentially as described (33). For immunoprecipitation, 10 mg beads and 25 µg AtPop1p-specific IgG per assay were incubated overnight. After coupling and washing, the beads were collected by centrifugation, equilibrated in buffer F1 (20 mM Tris–Cl pH 8, 5 mM MgCl2) and stored at 4°C. The pretreated S23 extract was bound to the antibody-coupled protein A-Sepharose by overnight incubation. The beads were washed twice with buffer WIP2 [50 mM Tris–Cl pH 7.5, 150 mM NaCl, 1% (v/v) NP40, 0.5% (w/v) sodium deoxycholate] and WIP3 [50 mM Tris–Cl pH 7.5, 500 mM NaCl, 0.1% (v/v) NP40, 0.05% (w/v) sodium deoxycholate], collected by centrifugation, resuspended in 30 µl buffer F1 and assayed for RNase P activity. For analysis of RNAs present in the bound material, the immunoprecipitate was resuspended in 150 µl TE and the nucleic acids were recovered by phenol extraction and EtOH precipitation.
Assay of RNase P activity
Transcription with T7 RNA polymerase and RNA purification by denaturing PAGE was performed as described (34). The radioactive transcripts were visualised by autoradio-graphy or with a Phosphoimager (Molecular Dynamics), eluted and ethanol precipitated. Wheat RNase P activity was assayed in buffer F1 with transcripts obtained from pSu3-wt cut with FokI, yielding E. coli pre-tRNATyr. Reactions contained 5000–10 000 cpm of [32P]-labelled substrate (17–35 fmol) and enzyme preparations as indicated in a total volume of 20 µl. After incubation at 37°C for 30 min, samples were ethanol precipitated and analysed by denaturing PAGE on 8% polyacrylamide (PAA) gels. RNase P assays in immunoprecipitated protein complexes were initiated by addition of [32P]-labelled substrate to the resuspended protein A-Sepharose beads. After incubation at 37°C for 60 min with gentle agitation, the reaction was stopped by addition of 30 µl ddH2O and the slurry extracted with 60 µl TE-saturated phenol. After EtOH precipitation, reaction products were analysed as above. Control reactions with the M1 ribozyme were performed in buffer P2 as described (35).
Identification of AtMRP RNAs in vivo and determination of RNA termini
The cycling protocol for expression analysis by RT-PCR was first optimized by amplification of the two single copy MRP genes from genomic DNA with the primers 5′MRP and 3′MRP-anti3; a Ta of 54°C was used for all further experiments. Simultaneous cDNA synthesis from both AtMRP RNAs was achieved by reverse transcription of total RNA (two independent preparations) with 3′MRP-anti3 and M-MLV RT (Promega) at 50°C. After purification (PCR Purification Kit, Roche), cDNAs were amplified with 5′MRP and 3′MRP-anti3 and analysed by cleavage with EcoRV and electrophoresis on non-denaturing 8% PAA gels in TBE. Plasmids carrying the two MRP genes (pAtMRP1 and 2, respectively) were treated accordingly and used as controls. The 5′-ends of MRP RNAs were determined by RACE as described for Pop1 mRNA, using 3′MRPanti3 for cDNA generation. After dATP tailing and second strand synthesis with Anchor2, nested PCR was performed with UAPshort and the internal primers 3′MRP1+2-anti4 and 3′ATMRP1+2-anti5, and the products were cloned into pCR-XL-TOPO.
Identification and sequence determination of wheat MRP RNA
The core region of wheat MRP RNA genes was amplified from genomic DNA with 5′PlantMRP-CRI and 3′PlantMRP-CRVanti-W. The 5′ region of the RNA was obtained by RACE as described above, using a small RNA preparation from wheat germ and 3′PlantMRP-CRVanti-W as cDNA primer. After 5′ tailing and amplification with anchor2 and the specific primers WG-MRP1anti and WG-MRP2anti, cDNAs were cloned into pCRII-TOPO for sequencing. Identification of MRP RNAs in immunoprecipitates was performed after phenol extraction and DNase I treatment of the bound material. RT-PCR was accomplished with 3′PlantMRP-CRVanti-T as cDNA primer, followed by amplification with the primer pairs 5′PlantMRP-CRI + 3′PlantMRP-CRV-anti-T and 5′PlantMRP-CRI + WG-MRP-anti, cloning and sequencing. Wheat germ RNase P prepared by successive column chromatography (Blue Trisacryl pool) (15) was analysed with the same strategy.
Analysis of RNA populations by non-specific RACE analysis
For identification of unknown RNA species in immunoprecipitates, the RNAs were prepared as above. Following 3′ polyadenylation with poly(A) polymerase, the RNAs were purified with the RNeasy Mini Kit (Qiagen) and reverse transcribed using the d(T16)V primer and AMV RT (Promega). cDNAs were purified through Microcon30 columns (Amicon), and tailed with dCTP and Terminal Transferase. Second strand synthesis was performed with the BRL anchor primer; this primer together with Anchor2 was used for amplification. Reaction products were electrophoretically separated (4% NuSieve GTG Agarose, Cambrex), eluted in different size classes, cloned into pCRII-TOPO and sequenced.
Bioinformatic analysis
The NCBI and EBI tools were used for sequence searches and comparisons by BLAST. Promoter analysis of the AtPOP1 gene was performed with PlantPromDB (36), Athena (37) and Agris (38). RNA structures were constructed with mfold (39), and RNAviz was used for their graphical representation (40).
RESULTS
AtPop1 mRNA is present in multiple splicing isoforms, possibly encoding several distinct proteins
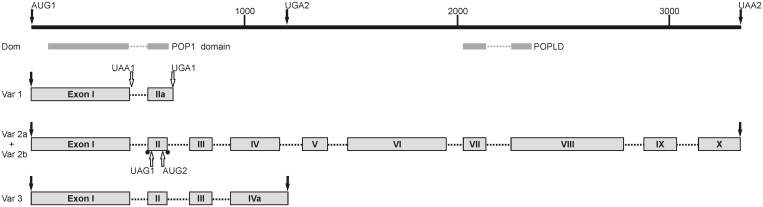
Pop1p has a size of about 100 kDa in most organisms. The original annotation of a Pop1p-related mRNA in A. thaliana however postulated only two spliced exons encoding a 190 residues long protein of 21.5 kDa, starting at AUG1 (TAIR: At2G47290.1; AAB63829.1; Figure 1, variant 1; Supplementary Figure S1). Attempts to clone the corresponding cDNA with AtPOP1-R as cDNA primer exclusively yielded the unspliced sequence (variant 1 u), encoding a 151 amino acids long ORF terminating at an in-frame UAA stop codon at the beginning of intron 1. This cDNA contains the Pop1 domain except for the last eight conserved residues (27,28) and was cloned into pRSET A for heterologous overexpression. The resulting 187 amino acids (21.4 kDa) fusion protein containing an N-terminal affinity tag was used for functional studies.
Figure 1.
Splicing patterns of AtPop1 mRNAs. The 3344 bp long genomic region between AUG1 and UAA2 is shown as a heavy black line; distances in bp are indicated by the ruler above the line. Start and stop codons delimiting the expressed coding regions are drawn as solid black arrows and those with unclear in vivo function as outlined arrows. The position of the conserved regions (Pop1 domain and POPLD) is depicted as heavy grey lines below the genomic region. The exons of mRNA variants 1–3 are drawn as grey boxes identified by roman numerals and connected by dotted lines indicating the length of excised introns. Variant 1 encodes a 190 amino acids protein containing the Pop1 domain; in the unspliced RNA, the ORF terminates at UAA1 after 151 residues. Variants 2a and b are combined into one drawing: 2a encodes an 826 amino acids long ORF extending from AUG1 to UAA2. In variant 2b, the alternative splicing events at the borders of exon 2 are indicated by black dots. The resulting frame shift creates a 659 residues long ORF between AUG2 and UAA2. The novel variant 3 has an extended exon 4 and encodes a 307 amino acids ORF terminating at UGA2. For accession numbers and sequence details, see text and Supplementary Figures S1 and S3.
Two recently annotated longer mRNAs assembled from ESTs differ at the splice site positions of introns 1 and 2 (DQ069804 and DQ069805 on the At2g47290/At2G47300 locus (41); variants 2a and 2b in Figure 1; AT2G47300.2 and AT2G47300.3 in Supplementary Figure S1). Both mRNAs have a shorter exon 2 than variant 1. This splicing pattern explains the exclusive detection of unspliced RNA (variant 1 u) during cloning of the originally annotated mRNA1: the cDNA primer AtPOP1-R is located within the actual intron 2 and thus hybridizes only to unprocessed pre-mRNA. DQ069804 mRNA encodes an 826 amino acids protein of 92.5 kDa, similar to Pop1p from other organisms (Supplementary Figure S3; Ath 2a). DQ069805 mRNA potentially encodes two proteins: the first ORF starts at AUG1; alternative splicing of introns 1 and 2 leads to frameshift and thus to translation termination at UAG1 within exon 2. No cleavage and polyadenylation signals are found downstream of this and the other premature stop codons UAA1 and UGA1. A second ORF starting at AUG2 in the same exon encodes a putative 73.8 kDa protein lacking the Pop1 domain (Ath 2b, 659 amino acids; Supplementary Figure S3). No strong initiation signal (Kozak sequence) is present close to AUG2 and internal initiation is rare in plants, making translation from this start codon unlikely (42,43).
To resolve the discrepancy between the different mRNA annotations, we determined the native RNA termini. Unexpectedly, RACE analysis revealed a novel mRNA of 1214 nt from 5′ untranslated region to polyadenylation site. An ORF of 924 nt extending from AUG1 into the annotated intron 4 encodes a 307 amino acids protein of 34.6 kDa (Figure 1, variant 3; Supplementary Figures S1, FN673552 and S3, Ath 3). The sequence between exons 1 and 4 is identical to DQ069804 mRNA (At2G47300.2) including the rare GC-AG splice sites of intron 2. The 3′-end of the novel mRNA 3 is formed by retaining intron 4, which contains an in-frame stop codon (UGA2), polyadenylation signals and a cleavage site downstream this stop codon. The transcription start site of AtPop1 mRNAs was established by 5′ RACE from cDNA initiated within exon 4 and revealed a 5′ untranslated region of 273 nt, slightly longer than for the annotated mRNAs 2a and 2b. The upstream promoter region contains several transcriptional control elements and factor binding sites at about −235 to +80 nt relative to the postulated transcription start site (Supplementary Figure S1). Examination of global expression databases (e.g. TAIR, SIGnAL) revealed that RNAs derived from this locus are prevalent in fast developing tissue (shoot apex, sperm cells, embryo, flower and seed), and are prone to stage-dependent expression during mature embryo, petal formation and cotyledon stages. The presence of AtPop1 mRNAs in vivo was further confirmed by Northern blot analysis using oligonucleotide probes specific for the two alternative splicing variants of intron 2 (Supplementary Figure S2).
Two MRP RNAs are expressed in A. thaliana and contain signature sequences of RNase P RNA
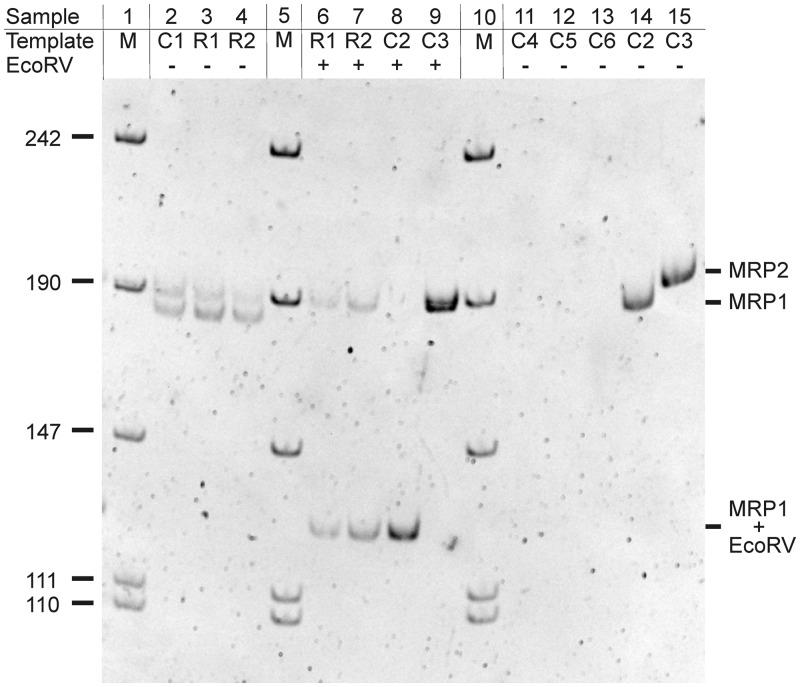
Arabidopsis contains two genes for RNase MRP RNA; data on expression and function are only available for MRP1. To examine whether the AtMRP2 gene is functional, we determined the relative steady-state levels of both AtMRP RNAs in vivo. Due to the high similarity between these sequences, qPCR analysis was not feasible. Instead, an approach was chosen where simultaneous amplification with a single primer pair was followed by restriction cleavage, electrophoresis and quantification of the reaction products. Prior to the analysis, PCR conditions were optimized to amplify the core sequence from both MRP genes with approximately equal efficiency when the two single-copy genes present in genomic DNA were used as template (Figure 2, sample C1). The two amplificates can be distinguished by size and by the unique EcoRV site in MRP2 (Figure 2, C2 and C3). Using two independent Arabidopsis RNA preparations as template, MRP1 was always present in slightly higher amounts than MRP2 (Figure 2, R1 and R2). Examination of PCR products early in linear amplification phase showed that MRP1 was detectable two cycles earlier than MRP2, which correlates to an approximately 4-fold excess of MRP1 over MRP2 (not shown). This is comparable to the ratio between RNase P and MRP RNAs in human cells where approximately twice as many RNase P RNAs than MRP RNA molecules are present (44), but in contrast to yeast, where equal amounts of both RNAs including the RNase P RNA precursor were detected (45).
Figure 2.
Steady-state levels of Arabidopsis MRP1 and MRP2 RNA in vivo. The following templates were used for amplification of MRP sequences: C1: 1ng genomic DNA from A. thaliana; R1, R2: MRP-specific cDNA from DNase-treated total RNA (two independent RNA preparations: 10 ng RNA1 or 14 ng RNA2, respectively); C2, C3: 0.02 ng pAtMRP1 or pAtMRP2; C4: no template; C5, C6: total RNA (83 ng RNA1 or 118 ng RNA2, as used for cDNA synthesis) treated with DNase I. The length of DNA size markers in bp is given to the left. The length of the PCR products indicated to the right is as follows: MRP1, 178 bp; MRP1 + EcoRV, 124 + 54 bp after EcoRV cleavage; MRP2, 184 bp. The ethidium bromide stained gel is shown in inverted colour to enhance contrast.
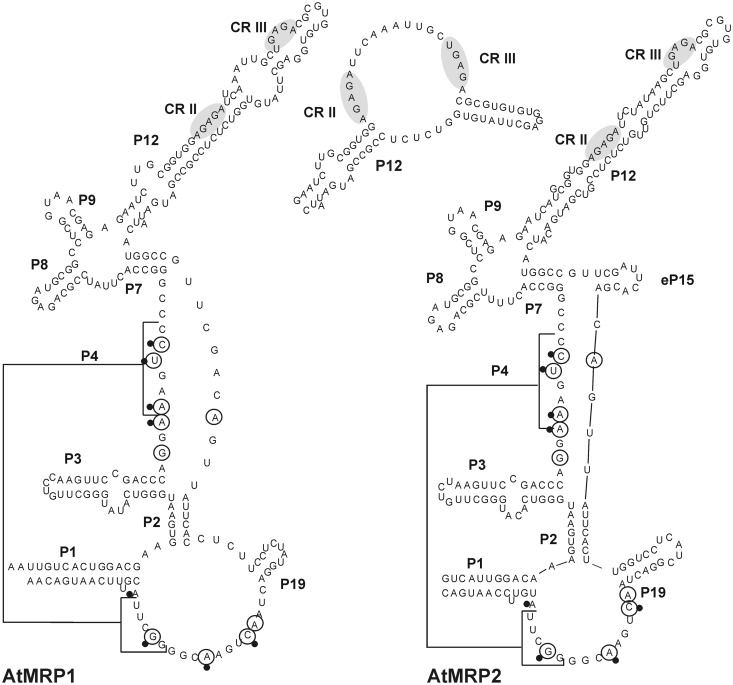
Based on established models for RNase P and MRP RNAs from yeast and humans, and on new sequences from related plant species (our unpublished data), we suggest a novel structure model which differs in several aspects from previous depictions of plant MRP RNAs (Figure 3). A P7 helix connecting the C- and S-domains as in RNase P RNAs is present and in MRP2, a small eP15-like structure can be formed. P3 now accommodates the internal AU-rich loop which is conserved in most known RNase P and MRP RNAs and involved in protein binding (21,25,45). The proximal strand of the Arabidopsis loop resembles the proposed consensus sequence, although it is much smaller than in other eukaryotes (46). Both RNAs conform to the known MRP consensus with the exception that L8 is only four-membered in plants (21,47). The MRP consensus is largely a subset of the eukaryotic RNase P consensus sequence; CR II and CR III however are signature elements specific for RNase P RNA. They have not previously been described in plants, but are present in both AtMRP RNAs. In the published structure representations, both elements were hidden within a paired region of P12. In our alternative model, CR II and CR III are juxtaposed within an internal loop structure in P12, similar to their location in nuclear RNase P RNAs (Figure 3, inset).
Figure 3.
Secondary structure of Arabidopsis RNase MRP RNAs. The RNase P consensus nucleotides found in both RNAs are circled; the MRP consensus is indicated by dots next to the bases. CR II and CR III are indicated by grey shadowing; the alternative P12 structure of AtMRP1 is depicted in the inset (center). MRP RNA1 is shown with the termini as determined by RACE and MRP2 with the ends suggested by (21).
Antibodies specific for AtPop1p recognize two proteins in Arabidopsis and precipitate RNase P activity from wheat germ extracts
Prior to performing immunoprecipitation experiments, the specificity of the antisera created against the recombinant Pop1 domain (AtPop1p, variant 1 u) was examined by western blotting. Using an enriched Arabidopsis protein preparation, a signal at ∼100 kDa and a weak band at 35 kDa were visible (Supplementary Figure S4). This correlates with the ORFs of mRNA variant 2 and the novel variant 3 which encode a 92.5 and 34.6 kDa proteins, respectively. The putative 73.8 kDa ORF encoded by mRNA 2b lacks the Pop1 domain and thus cannot be detected by this antiserum. Functional assessment of Pop1p by in vitro complex formation between the recombinant Pop1 domain and AtMRP RNAs showed only weak binding (Supplementary Figure S5).
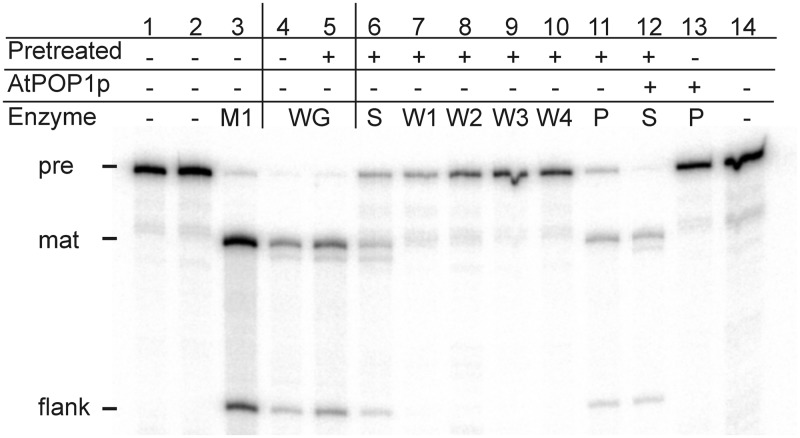
Because Arabidopsis is not amenable to direct biochemical analysis of RNA processing pathways, the well-established tRNA processing system from wheat germ (15,48) was used for coupled immunoprecipitation-activity tests. Wheat germ S23 extract was bound to matrix-coupled AtPop1p-specific antibodies and assayed for RNase P activity (Figure 4). The immunoprecipitate showed significant substrate turnover without degradation (lane 11); no activity was detected in the wash fractions (lanes 7–10). Saturation of the matrix-coupled antibodies with an excess of recombinant antigen prior to incubation with the processing extract completely blocked binding of RNase P activity, showing that recognition by the antisera is highly specific (compare lanes 12 and 13). The residual activity in the supernatant (lane 6) might be derived from mitochondrial RNase P, which is active under these conditions (15); it is a protein-only enzyme and will not be recognized by the antibodies specific for Pop1p (13). This immunoprecipitation experiment shows that AtPop1p and its wheat homologue share antigenic determinants in the Pop1 domain, and that wheat Pop1p is associated with a complex containing RNase P activity.
Figure 4.
Heterologous immunoprecipitation of wheat germ RNase P activity with AtPop1p-specific antibodies. Wheat germ extract was subjected to immunoprecipitation as described and the fractions tested for RNase P activity. S, supernatant; P, Protein A-Sepharose bound precipitate; W, wash fractions. Radiolabelled E. coli pre-tRNATyr was incubated with the following buffers or enzymes: 1, no treatment; 2, incubation in buffer F; 3, E. coli M1 ribozyme in buffer P2; 4, untreated wheat germ extract; 5, extract treated as for immunoprecipitation; 6, supernatant after immunoprecipitation; 7–10, wash fractions; 11, immunoprecipitate; 12 + 13: same as 6 + 11, but the coupled antibody was pre-incubated with AtPop1p antigen prior to binding of the extract; 14: anti-AtPop1p-antibodies as additional control for substrate stability.
Two MRP RNA variants are present in wheat and are associated with purified RNase P preparations
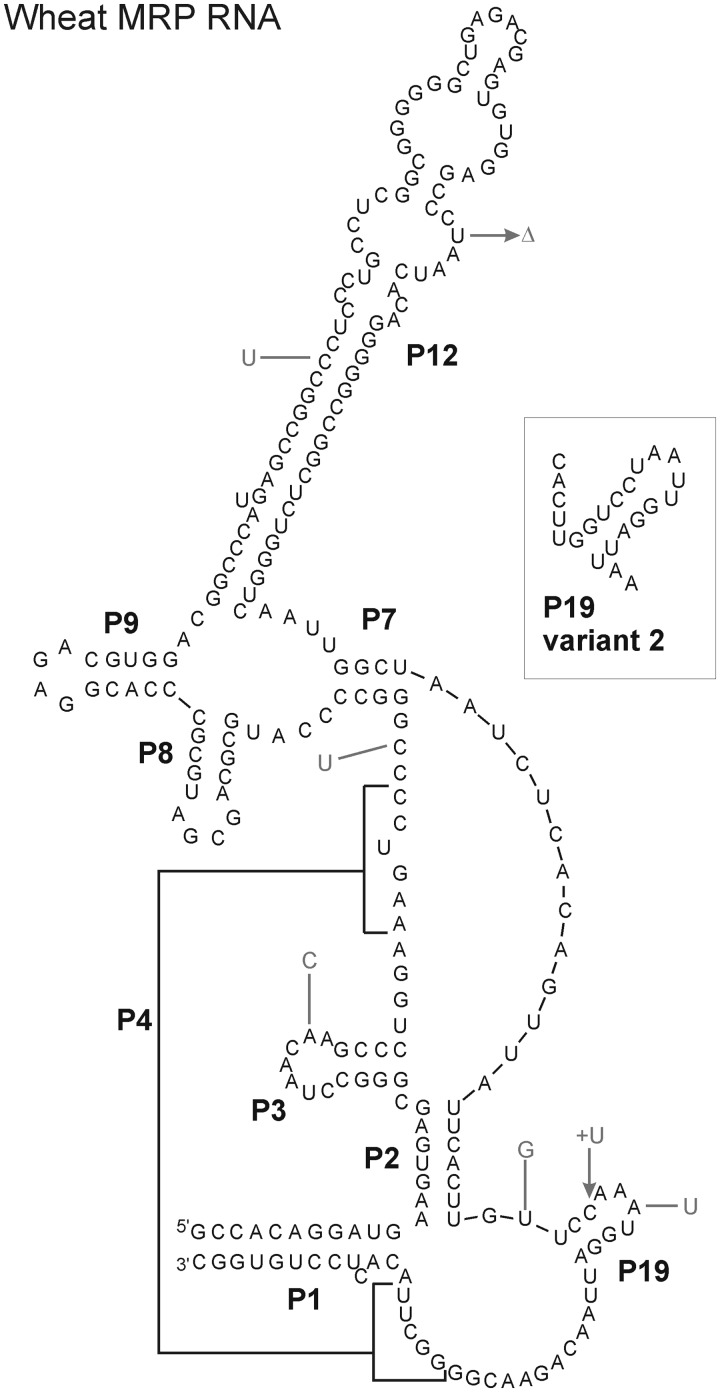
The co-immunoprecipitation of RNase P activity and the RNase P/MRP protein Pop1p in the wheat system raised the question whether plant nuclear RNase P contains an RNA component, as is the case in other eukaryotes. We thus used an RNomics approach to detect unknown RNAs in the immunoprecipitate and in chromatographically purified wheat RNase P fractions. Prior to screening, the RNA quality in these preparations was evaluated by analysis of wheat MRP RNAs. Due to the lack of related sequence data, the core sequence between CR I and CR V was determined by genomic PCR. Completion of the RNA sequences by RACE revealed two slightly different molecules (TaMRP1/2; Figure 5) similar to several short wheat ESTs and to the annotated MRP RNA from Oryza. L8 deviates from the plant consensus GAGA. P3 is shorter than in Arabidopsis and without an internal loop; no CR II–CR III consensus is present in P12.
Figure 5.
Secondary structure of RNase MRP RNAs from wheat. The two wheat RNAs differ at several isolated positions; only the P19 structure is affected by the changes. The sequence of RNA1 is shown; differences in RNA2 are indicated by the corresponding bases positioned outside the structure. The alternative structure of P19 is shown in the insert.
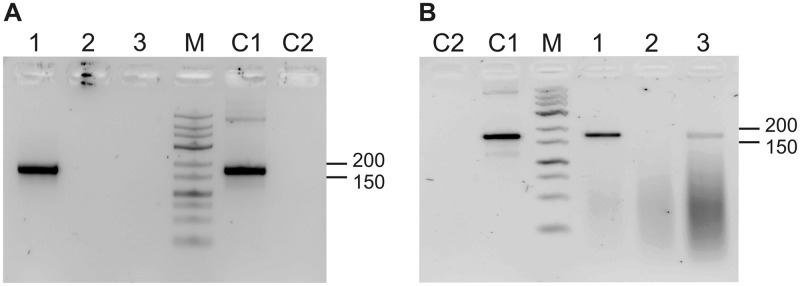
To get insight into the relation of wheat MRP RNAs to the RNase P/MRP complex, the RNA content of partially purified RNase P preparations was examined by RT-PCR and subsequent sequencing. MRP-specific analysis of the immunoprecipitate revealed that only TaMRP2 is associated with the complex (Figure 6); in contrast, both RNA variants were detected in chromatographically purified RNase P (Blue Trisacryl pool) (15). Examination of the total RNA population in these preparations by non-specific RACE (49) showed that in the immunoprecipitate, TaMRP2 and a tRNA were present; cytosolic 18S rRNA, 5.8S rRNA and the 5′ external transcribed sequence prevailed (Table 1). Similarly, the chromatographically purified RNase P preparation contained 18S and 5.8S rRNA in addition to both MRP RNAs. The unequal distribution of the two MRP RNAs in the enzyme preparations can at present only be explained by possible differences in complex stability during the lengthy immunoprecipitation procedure; the presence of rRNAs and tRNA can be attributed to RNase MRP and RNase P activity. The prevalence of rRNAs over tRNA and MRP RNAs may be due to substrate and/or product binding properties of RNase P and MRP. In addition, the variable efficiencies for tailing and cDNA synthesis of the different RNA species during non-specific RACE may bias this qualitative analysis (49). The combined results of RNA analysis and activity-coupled immunoprecipitation experiments suggest that in wheat, Pop1p is associated with MRP RNA and with RNase P activity.
Figure 6.
Detection of RNase MRP RNAs in wheat germ RNase P preparations. The following PCR templates were used: (A) RNAs isolated from AtPop1p-specific wheat germ immunoprecipitate. Sample 1: cDNA prepared from DNase I-treated RNA; 2: DNase I-treated RNA; 3: mock-treated RNA. Nested PCR with MRP-specific primers was performed with templates 1–3, with 10 ng genomic DNA from wheat (C1) or without template (C2). (B) RNAs prepared from purified wheat germ RNase P (Blue Trisacryl pool) were treated as described in A. M: DNA marker (Gene Ruler low range, Fermentas); relevant sizes in bp are indicated to the right. The 170 bp PCR product is specific for wheat RNase MRP RNA. The ethidium bromide stained gel is shown in inverted colour to enhance contrast.
Table 1.
RNA species present in the Pop1p-specific immunoprecipitate
| Gene | Accession number | Cloned fragments | |
|---|---|---|---|
| Nuclear rRNA unit | 5′ ETS | X07841 | 10 |
| 18S | AJ272181 | 13 | |
| 5.8S | FJ229967 | 9 | |
| 25 S | AY049041 | 2 | |
| Others | 5 S rRNA | AY316209 | 2 |
| tRNAAsp | AB005781 | 1 | |
| MRP RNA 2 | FN689353 (this article) | 4 | |
| Mito 18S rRNA | AP008982 | 1 |
All RNA species detected by non-specific RACE of the wheat immunoprecipitate are listed. Only sequences longer than 21 nt were considered; the accession numbers are representatives of multiple hits for each sequence. All listed hits are from wheat with the exception of tRNAAsp (A. thaliana), where no wheat sequence was available. The number of independent clones related to one RNA species is given in the right column. No sequences located within the Internal Transcribed Spacer 1 (ITS1) or ITS2 were detected. 5′ ETS: external transcribed sequence upstream of 18S rRNA.
DISCUSSION
The large central protein Pop1p common to RNase P and MRP was used as a starting point to elucidate the composition of these nuclear enzyme complexes in plants. In Arabidopsis, analysis of Pop1 mRNAs revealed several splicing variants, encoding at least two proteins with considerable size differences (Figure 1). Of the three annotated variants, the original At2G47290 mRNA (variant 1) is not detectable in vivo. The more recently annotated DQ069804 mRNA encodes a 826 amino acids ORF similar to Pop1p from other organisms (Supplementary Figure S3, Ath 2a). DQ069805 mRNA differs by alternative splicing of introns 1 and 2, leading to frameshift and termination of a short ORF within exon 2; a second ORF starting at AUG2 immediately downstream encodes 659 residues and lacks the Pop1 domain, but is congruent with DQ069804 from exon 3 to 10 (Supplementary Figure S3, Ath 2b). Analysis of the differential splicing phenomenon by RACE revealed a novel mRNA of about 1200 nt, encoding a 307 amino acids ORF (Figure 1, variant 3; Supplementary Figure S1, FN673552 and Supplementary Figure S3, Ath 3). Between exons 1 and 4, the mRNA is identical to DQ069804. The 3′-end of this novel mRNA3 however is created by retention of intron 4, which provides an in-frame stop codon (UGA2), polyadenylation signals and a cleavage site downstream of this stop codon (50). The 307 and 826 amino acids long ORFs starting at AUG1 are preceded by a strong initiation signal (43), suggesting translation in vivo. The 659 residues long ORF encoded by mRNA 2b however is lacking a Kozak sequence near AUG2, making efficient translation questionable. Although weak, expression of the 307 and 826 amino acids long proteins in vivo is supported by western analysis using antibodies against the Pop1 domain (Supplementary Figure S4). The 659 residues long ORF does not contain this conserved domain and thus will escape detection.
The four highly conserved regions COR1–COR4 required for complex formation and enzymatic activity of RNase P and MRP are present in the 826 amino acids full-length protein variant 2a from Arabidopsis (27) (Supplementary Figure S3). The 307 residues long protein encoded by the novel mRNA variant 3 contains COR1 and COR2, but lacks COR3 and COR4. In yeast, F89 and Q90 in COR1 are important for the assembly of RNase MRP, but not RNase P. F89 is conserved as F52 in AtPop1p and Q90 is replaced by R53, indicating that this function is at least partially conserved. Mutations of R233 and Q234 in COR2 reduce tRNA processing in yeast; the equivalent positions in AtPop1p are S117 and G118. These differences to conserved amino acids with a known function in yeast suggest a change in the role of Pop1p during plant RNase P evolution. T242, H243 and R249 (COR2) are responsible for RNase P RNA maturation and assembly of the RNase P and MRP complexes in yeast; they are present as T126, H127 and R133 in Arabidopsis. The POP-like domain (POPLD) spanning COR3 may be involved in nucleolar localisation of the protein; COR4 contains glycine-rich RNA binding motifs. Both regions contribute to maturation of yeast RNase P RNA, complex formation and activity of both enzymes. Most of these functionally important residues are present in the AtPop1p variants 2a and 2b; F823 is important for RNase P activity in yeast and is conserved as F750 in Arabidopsis. The adjacent G-rich stretch is replaced by a basic region. The absence of COR3 and COR4 in AtPop1p variant 3 implies that functions related to nucleolar localization, RNase P complex formation and activity are missing in this protein. The annotated hypothetical protein Ath 2b lacks exons 1 and 2 and thus the complete Pop1 domain. The recombinant variant 1 used for functional studies contains this domain and can form complexes with both AtMRP RNAs (Supplementary Figure S5). The weak binding observed in these experiments may be attributed to the lack of the POPLD region in this protein.
Both annotated Arabidopsis MRP RNAs are present in vivo in similar amounts, excluding the presence of a pseudogene. Our new secondary structure models for these RNAs depict all helices conserved in other eukaryotes, as well as the hitherto unassigned CR II and CR III regions unique to RNase P RNAs. We have also established the first complete sequences of two MRP RNAs from wheat, allowing a more reliable structure prediction for plant MRP RNAs (Figure 5). The presence of two MRP RNAs is not unusual: the Arabidopsis genome contains two expressed genes; in Nicotiana, three RNA species have been detected; and several other organisms possess two genes for RNase P or MRP RNA (20,21). Although similar to a previously published genomic sequence from rice, the wheat RNAs deviates from the postulated L8 consensus (21). They have a short P3 without an internal loop, suggesting a different mode of protein binding in Poaceae. CR II and CR III are present in some, but not all known plant MRP sequences, leaving open the question of a possible RNase P-like function in these RNAs.
Antibodies specific for the recombinant Pop1 domain from Arabidopsis precipitate RNase P activity and MRP RNAs from wheat germ extract and purifed enzyme preparations, showing that AtPop1p and its wheat homologue share antigenic determinants in this domain. Likewise, the co-immunoprecipitation of rRNAs and tRNA indicates that Pop1p is associated with RNase P and RNase MRP activity. This may be either in a single large ‘processosome-like’ complex as described for yeast mitochondria (51) or in two similar, but separate complexes as in the nuclei of yeast and human cells (16–18,47). The soluble activity in the wheat immunoprecipitate is probably derived from mitochondrial RNase P, which is a protein-only enzyme unrelated to Pop1p. In Arabidopsis, the same ProRP1 protein also provides RNase P activity in the chloroplasts. Two similar proteins (ProRP2 and 3) localize to the nucleus; their function is still unknown (13). Although these nuclear ProRPs may be associated with the complex and may be responsible for the catalytic activity of nuclear RNase P, we cannot rule out at present that an RNA contributes to RNase P activity in plant nuclei. Further biochemical and genetic characterization of both enzymes will be required to resolve these questions.
ACCESSION NUMBERS
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Figures 1–5, Supplementary Sequence Data and Supplementary Reference [52].
FUNDING
The Deutsche Forschungsgemeinschaft [Scho 515/7-3; Scho 515/7-4] and Fonds der Chemischen Industrie (to A.S.), the FAZIT Foundation (to C.H. and M.K.), and the European Union and Free State of Saxony [ERDF/SMWK, Project 13397] (to P.S.). Funding for open access charge: Deutsche Forschungsgemeinschaft and University of Leipzig Medical School.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The generous scientific and infrastructural support by Prof. H. Beier and Prof. H.J. Gross during the initial phase of this project is greatly acknowledged. We thank Gudrun Grimmer and Birgit Löffler for excellent technical assistance, and Prof. M. Mörl for providing laboratory facilities essential for working with radioactive isotopes.
REFERENCES
- 1.Marvin MC, Engelke DR. RNase P: increased versatility through protein complexity? RNA Biol. 2009;6:40–42. doi: 10.4161/rna.6.1.7566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lai LB, Vioque A, Kirsebom LA, Gopalan V. Unexpected diversity of RNase P, an ancient tRNA processing enzyme: challenges and prospects. FEBS Lett. 2010;584:287–296. doi: 10.1016/j.febslet.2009.11.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kikovska E, Svärd SG, Kirsebom LA. Eukaryotic RNase P RNA mediates cleavage in the absence of protein. Proc. Natl Acad. Sci. USA. 2007;104:2062–2067. doi: 10.1073/pnas.0607326104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pannucci JA, Haas ES, Hall TA, Harris JK, Brown JW. RNase P RNAs from some Archaea are catalytically active. Proc. Natl Acad. Sci. USA. 1999;96:7803–7808. doi: 10.1073/pnas.96.14.7803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baum M, Schön A. Localization and expression of the closely linked cyanelle genes for RNase P RNA and two transfer RNAs. FEBS Lett. 1996;382:60–64. doi: 10.1016/0014-5793(96)00148-2. [DOI] [PubMed] [Google Scholar]
- 6.Baum M, Cordier A, Schön A. RNase P from a photosynthetic organelle contains an RNA homologous to the cyanobacterial counterpart. J. Mol. Biol. 1996;257:43–52. doi: 10.1006/jmbi.1996.0145. [DOI] [PubMed] [Google Scholar]
- 7.Cordier A, Schön A. Cyanelle RNase P: RNA structure analysis and holoenzyme properties of an organellar ribonucleoprotein enzyme. J. Mol. Biol. 1999;289:9–20. doi: 10.1006/jmbi.1999.2762. [DOI] [PubMed] [Google Scholar]
- 8.Li D, Willkomm DK, Schön A, Hartmann RK. RNase P of the Cyanophora paradoxa cyanelle: a plastid ribozyme. Biochimie. 2007;89:1528–1538. doi: 10.1016/j.biochi.2007.08.004. [DOI] [PubMed] [Google Scholar]
- 9.Reith M, Munholland J. Complete nucleotide sequence of the Porphyra purpurea chloroplast genome. Plant Mol. Biol. Rep. 1995;13:333–335. [Google Scholar]
- 10.Turmel M, Otis C, Lemieux C. The complete chloroplast DNA sequence of the green alga Nephroselmis olivacea: insights into the architecture of ancestral chloroplast genomes. Proc. Natl Acad. Sci. USA. 1999;96:10248–10253. doi: 10.1073/pnas.96.18.10248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Thomas BC, Li X, Gegenheimer P. Chloroplast ribonuclease P does not utilize the ribozyme-type pre-tRNA cleavage mechanism. RNA. 2000;6:545–553. doi: 10.1017/s1355838200991465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Holzmann J, Frank P, Löffler E, Bennett KL, Gerner C, Rossmanith W. RNase P without RNA: identification and functional reconstitution of the human mitochondrial tRNA processing enzyme. Cell. 2008;135:462–474. doi: 10.1016/j.cell.2008.09.013. [DOI] [PubMed] [Google Scholar]
- 13.Gobert A, Gutmann B, Taschner A, Gössringer M, Holzmann J, Hartmann RK, Rossmanith W, Giegé P. A single Arabidopsis organellar protein has RNase P activity. Nat. Struct. Mol. Biol. 2010;17:740–744. doi: 10.1038/nsmb.1812. [DOI] [PubMed] [Google Scholar]
- 14.Franklin SE, Zwick MG, Johnson JD. Characterization and partial purification of two pre-tRNA 5′-processing activities from Daucus carrota (carrot) suspension cells. Plant J. 1995;7:553–563. doi: 10.1046/j.1365-313x.1995.7040553.x. [DOI] [PubMed] [Google Scholar]
- 15.Arends S, Schön A. Partial purification and characterization of nuclear ribonuclease P from wheat. Eur. J. Biochem. 1997;244:635–645. doi: 10.1111/j.1432-1033.1997.t01-1-00635.x. [DOI] [PubMed] [Google Scholar]
- 16.Bertrand E, Houser-Scott F, Kendall A, Singer RH, Engelke DR. Nucleolar localization of early tRNA processing. Genes Dev. 1998;12:2463–2468. doi: 10.1101/gad.12.16.2463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.van Eenennaam H, Pruijn GJ, van Venrooij WJ. hPop4: a new protein subunit of the human RNase MRP and RNase P ribonucleoprotein complexes. Nucleic Acids Res. 1999;27:2465–2472. doi: 10.1093/nar/27.12.2465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Esakova O, Krasilnikov AS. Of proteins and RNA: the RNase P/MRP family. RNA. 2010;16:1725–1747. doi: 10.1261/rna.2214510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Maida Y, Yasukawa M, Furuuchi M, Lassmann T, Possemato R, Okamoto N, Kasim V, Hayashizaki Y, Hahn WC, Masutomi K. An RNA-dependent RNA polymerase formed by TERT and the RMRP RNA. Nature. 2009;461:230–235. doi: 10.1038/nature08283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kiss T, Marshallsay C, Filipowicz W. 7-2/MRP RNAs in plant and mammalian cells: association with higher order structures in the nucleolus. EMBO J. 1992;11:3737–3746. doi: 10.1002/j.1460-2075.1992.tb05459.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Piccinelli P, Rosenblad MA, Samuelsson T. Identification and analysis of ribonuclease P and MRP RNA in a broad range of eukaryotes. Nucleic Acids Res. 2005;33:4485–4495. doi: 10.1093/nar/gki756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Houser-Scott F, Xiao S, Millikin CE, Zengel JM, Lindahl L, Engelke DR. Interactions among the protein and RNA subunits of Saccharomyces cerevisiae nuclear RNase P. Proc. Natl Acad. Sci. USA. 2002;99:2684–2689. doi: 10.1073/pnas.052586299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Welting TJ, van Venrooij WJ, Pruijn GJ. Mutual interactions between subunits of the human RNase MRP ribonucleoprotein complex. Nucleic Acids Res. 2004;32:2138–2146. doi: 10.1093/nar/gkh539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Aspinall TV, Gordon JM, Bennett HJ, Karahalios P, Bukowski JP, Walker SC, Engelke DR, Avis JM. Interactions between subunits of Saccharomyces cerevisiae RNase MRP support a conserved eukaryotic RNase P/MRP architecture. Nucleic Acids Res. 2007;35:6439–6450. doi: 10.1093/nar/gkm553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ziehler WA, Morris J, Scott FH, Millikin C, Engelke DR. An essential protein-binding domain of nuclear RNase P RNA. RNA. 2001;7:565–575. doi: 10.1017/s1355838201001996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Perederina A, Esakova O, Koc H, Schmitt ME, Krasilnikov AS. Specific binding of a Pop6/Pop7 heterodimer to the P3 stem of the yeast RNase MRP and RNase P RNAs. RNA. 2007;13:1648–1655. doi: 10.1261/rna.654407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Xiao S, Hsieh J, Nugent RL, Coughlin DJ, Fierke CA, Engelke DR. Functional characterization of the conserved amino acids in Pop1p, the largest common protein subunit of yeast RNases P and MRP. RNA. 2006;12:1023–1037. doi: 10.1261/rna.23206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lygerou Z, Pluk H, van Venrooij WJ, Seraphin B. hPop1: an autoantigenic protein subunit shared by the human RNase P and RNase MRP ribonucleoproteins. EMBO J. 1996;15:5936–5948. [PMC free article] [PubMed] [Google Scholar]
- 29.Beier D, Stange N, Gross HJ, Beier H. Nuclear tRNATyr genes are highly amplified at a single chromosomal site in the genome of Arabidopsis thaliana. Mol. Gen. Genet. 1991;225:72–80. doi: 10.1007/BF00282644. [DOI] [PubMed] [Google Scholar]
- 30.Logemann J, Schell J, Willmitzer L. Improved method for the isolation of RNA from plant tissues. Anal. Biochem. 1987;163:16–20. doi: 10.1016/0003-2697(87)90086-8. [DOI] [PubMed] [Google Scholar]
- 31.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 32.Gruber A, Zingales B. Alternative method to remove antibacterial antibodies from antisera used for screening of expression libraries. Biotechniques. 1995;19:28–30. [PubMed] [Google Scholar]
- 33.Simanis V, Lane DP. An immunoaffinity purification procedure for SV40 large T antigen. Virology. 1985;144:88–100. doi: 10.1016/0042-6822(85)90308-3. [DOI] [PubMed] [Google Scholar]
- 34.Heubeck C, Schön A. Cyanelle ribonuclease P: isolation and structure-function studies of an organellar ribonucleoprotein enzyme. Methods Enzymol. 2001;342:118–134. doi: 10.1016/s0076-6879(01)42540-7. [DOI] [PubMed] [Google Scholar]
- 35.Gimple O, Schön A. In vitro and in vivo processing of cyanelle tmRNA by RNase P. Biol. Chem. 2001;382:1421–1429. doi: 10.1515/BC.2001.175. [DOI] [PubMed] [Google Scholar]
- 36.Shahmuradov IA, Solovyev VV, Gammerman AJ. Plant promoter prediction with confidence estimation. Nucleic Acids Res. 2005;33:1069–1076. doi: 10.1093/nar/gki247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.O'Connor TR, Dyreson C, Wyrick JJ. Athena: a resource for rapid visualization and systematic analysis of Arabidopsis promoter sequences. Bioinformatics. 2005;21:4411–4413. doi: 10.1093/bioinformatics/bti714. [DOI] [PubMed] [Google Scholar]
- 38.Yilmaz A, Mejia-Guerra MK, Kurz K, Liang X, Welch L, Grotewold E. AGRIS: the Arabidopsis Gene Regulatory Information Server, an update. Nucleic Acids Res. 2011;39:D1118–D1122. doi: 10.1093/nar/gkq1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.De Rijk P, Wuyts J, De Wachter R. RnaViz 2: an improved representation of RNA secondary structure. Bioinformatics. 2003;19:299–300. doi: 10.1093/bioinformatics/19.2.299. [DOI] [PubMed] [Google Scholar]
- 41.Xiao YL, Smith SR, Ishmael N, Redman JC, Kumar N, Monaghan EL, Ayele M, Haas BJ, Wu HC, Town CD. Analysis of the cDNAs of hypothetical genes on Arabidopsis chromosome 2 reveals numerous transcript variants. Plant Physiol. 2005;139:1323–1337. doi: 10.1104/pp.105.063479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kozak M. Faulty old ideas about translational regulation paved the way for current confusion about how microRNAs function. Gene. 2008;423:108–115. doi: 10.1016/j.gene.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 43.Sugio T, Matsuura H, Matsui T, Matsunaga M, Nosho T, Kanaya S, Shinmyo A, Kato K. Effect of the sequence context of the AUG initiation codon on the rate of translation in dicotyledonous and monocotyledonous plant cells. J. Biosci. Bioeng. 2010;109:170–173. doi: 10.1016/j.jbiosc.2009.07.009. [DOI] [PubMed] [Google Scholar]
- 44.Puranam RS, Attardi G. The RNase P associated with HeLa cell mitochondria contains an essential RNA component identical in sequence to that of the nuclear RNase P. Mol. Cell. Biol. 2001;21:548–561. doi: 10.1128/MCB.21.2.548-561.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Esakova O, Perederina A, Quan C, Schmitt ME, Krasilnikov AS. Footprinting analysis demonstrates extensive similarity between eukaryotic RNase P and RNase MRP holoenzymes. RNA. 2008;14:1558–1567. doi: 10.1261/rna.1106408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Marquez SM, Harris JK, Kelley ST, Brown JW, Dawson SC, Roberts EC, Pace NR. Structural implications of novel diversity in eucaryal RNase P RNA. RNA. 2005;11:739–751. doi: 10.1261/rna.7211705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Walker SC, Engelke DR. Ribonuclease P: the evolution of an ancient RNA enzyme. Crit. Rev. Biochem. Mol. Biol. 2006;41:77–102. doi: 10.1080/10409230600602634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Stange N, Beier H. A cell-free plant extract for accurate pre-tRNA processing, splicing and modification. EMBO J. 1987;6:2811–2818. doi: 10.1002/j.1460-2075.1987.tb02577.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hüttenhofer A, Vogel J. Experimental approaches to identify non-coding RNAs. Nucleic Acids Res. 2006;34:635–646. doi: 10.1093/nar/gkj469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Loke JC, Stahlberg EA, Strenski DG, Haas BJ, Wood PC, Li QQ. Compilation of mRNA polyadenylation signals in Arabidopsis revealed a new signal element and potential secondary structures. Plant Physiol. 2005;138:1457–1468. doi: 10.1104/pp.105.060541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Daoud R, Forget L, Lang BF. Yeast mitochondrial RNase P, RNase Z and the RNA degradosome are part of a stable supercomplex. Nucleic Acids Res. 2012;40:1728–1736. doi: 10.1093/nar/gkr941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Hasegawa K, Yukawa Y, Sugiura M. In vitro analysis of transcription initiation and termination from the Lhcb1 gene family in Nicotiana sylvestris: detection of transcription termination sites. Plant J. 2003;33:1063–1072. doi: 10.1046/j.1365-313x.2003.01693.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.