Abstract
Streptococcal surface enolase (SEN) is a major plasminogen-binding protein of group A streptococci. Our earlier biochemical studies have suggested that the region responsible for this property is likely located at the C-terminal end of the SEN molecule. In the present study, the gene encoding SEN was cloned from group A streptococci M6 isolate D471. A series of mutations in the sen gene corresponding to the C-terminal region (428KSFYNLKK435) of the SEN molecule were created by either deleting one or more terminal lysine residues or replacing them with leucine. All purified recombinant SEN proteins with altered C-terminal ends were found to be enzymatically active and were analyzed for their Glu- and Lys-plasminogen-binding activities. Wild-type SEN bound to Lys-plasminogen with almost three times more affinity than to Glu-plasminogen. However, the recombinant mutant SEN proteins with a deletion of Lys434-435 or with K435L and K434-435L replacements showed a significant decrease in Glu- and Lys-plasminogen-binding activities. Accordingly, a streptococcal mutant expressing SEN-K434-435L showed a significant decrease in Glu- and Lys-plasminogen-binding activities. Biochemical and functional analyses of the isogenic mutant strain revealed a significant decrease in its abilities to cleave a chromogenic tripeptide substrate, acquire plasminogen from human plasma, and penetrate the extracellular matrix. Together, these data indicate that the last two C-terminal lysine residues of surface-exposed SEN contribute significantly to the plasminogen-binding activity of intact group A streptococci and hence to their ability to exploit host properties to their own advantage in tissue invasion.
Group A Streptococcus (Streptococcus pyogenes or GAS) expresses a variety of surface proteins that bind to several mammalian proteins to adhere to, invade, and proliferate in host cells (20, 21, 28-30). Since most of these proteins are multifunctional in nature and their expression is controlled by the surrounding environment, understanding of the mechanisms involved in GAS pathogenesis has become more complex than previously believed (9, 17, 26, 27, 36). The plasmin(ogen)-binding property of GAS is viewed as an important virulence characteristic that may contribute to tissue invasion, even in the immunocompetent host (8, 25). GAS may interact with plasminogen either by directly involving its specific surface proteins in the binding process or indirectly by first binding to fibrinogen with its fibrinogen-binding surface proteins, followed by plasminogen (8). Plasminogen (∼90 kDa) is a single-chain plasma proenzyme that comprises a preactivation peptide (67 or 77 residues, ∼8 kDa), followed by five disulfide-bonded triple-loop kringle structures (65 kDa) and a 25-kDa light-chain serine proteinase domain (11, 35). The native form of plasminogen is called Glu-plasminogen because it has an amino-terminal glutamic acid residue. When Glu-plasminogen is cleaved by plasmin, an 8-kDa preactivation peptide is released, leaving behind a truncated form of zymogen called Lys-plasminogen (∼83 kDa). The majority of GAS strains that cause human diseases secrete a plasminogen activator, streptokinase (Ska). The latter forms a 1:1 stoichiometric complex with plasminogen, converts other plasminogen molecules into active plasmin, and is not regulated by host fluid phase α2-antiplasmin (Apl) (10).
Since Lys-plasminogen is readily activated to plasmin, conversion of Glu-plasminogen to Lys-plasminogen is essential and a key mechanism by which optimal stimulation of plasminogen on the eukaryotic cell surface occurs (8, 19). This may be true even for the GAS, which may preferentially bind Glu-plasminogen (2, 45). Furthermore, plasmin(ogen) has domains that are known to bind exposed C-terminal lysine residues of streptococcal surface proteins and since the affinity of Lys-plasminogen for such residues is known to be substantially higher than that of Glu-plasminogen, studying the mechanisms used by GAS to preferentially bind to Lys-plasminogen is essential to understand the role of this property in GAS virulence.
In addition to the plasminogen-binding streptococcal secreted protein Ska, three more distinct surface-bound proteins that directly bind plasmin(ogen) have been characterized in GAS. Two of them belong to a family of glycolytic enzymes, glyceraldehyde-3-phosphate dehydrogenase (termed streptococcal surface dehydrogenase or plasmin-binding receptor [Plr]) and α-enolase (termed streptococcal surface enolase [SEN]). These proteins are found on the surface of most of the M types and bind to Lys-plasminogen (24, 33, 34). However, the third surface protein, PAM (plasminogen-binding GAS M-like protein), binds to Glu-plasminogen and is found only in selected M types, such as M33, M41, M52, M53, and M56, most of which have shown the skin as a preferred tissue site for infection (2, 45). Recently, enolase has also been shown to be a Lys-plasminogen-binding protein on the surface of Streptococcus pneumoniae (3). Similarly, eukaryotic cells also express several receptors that bind Glu- and Lys-plasminogen (37). SEN is also expressed on the surface of hematopoietic cells and may serve as a plasminogen receptor (15, 18, 23, 28).
Our earlier studies and those of others have indicated that Plr, in comparison with SEN, is a weak Lys-plasmin(ogen)-binding protein and hence Plr may not play a major role in the strong Lys-plasminogen-binding activity of intact GAS (12, 24, 33, 34, 49, 50). On the basis of several biochemical parameters, we reported that the carboxy-terminal lysine residues of SEN might play an important role in the strong Lys-plasminogen-binding activity of GAS (34). The ubiquitous presence of SEN on the surface of almost all M types suggests that SEN may play a major role in the strong direct Lys-plasminogen-binding activity of GAS, including those serotypes that have been shown to bind Glu-plasminogen (2, 34, 45).
In the present study, by using a site-directed mutagenesis approach, we have investigated the contribution of the C-terminal lysine residues of the SEN molecule to the Lys- and Glu-plasminogen-binding ability of GAS. Further, we have evaluated their possible roles in the tissue invasion process by studying the ability of GAS to acquire exogenous plasminogen from human plasma and digest extracellular matrix (ECM) proteins.
MATERIALS AND METHODS
Bacterial strains, plasmids, and growth conditions.
S. pyogenes type M6 strain D471 (D471-M6) was from the Rockefeller University Streptococcal Culture Collection. Escherichia coli strains XL1-Blue and BL-21 were used as hosts for cloning and expression experiments, respectively. E. coli strains were grown in Luria-Bertani broth or on Luria-Bertani agar plates. Ampicillin (50 μg/ml), spectinomycin (100 μg/ml), and chloramphenicol (10 μg/ml) were used in growth media as and when required. GAS strains were grown in Todd-Hewitt broth supplemented with 0.5% yeast extract (THY) or in Proteose Peptone 3 blood agar plates supplemented with spectinomycin (up to 500 μg/ml) when required. E. coli vector pET14b (Novagen) was used to make different His-tagged mutant SEN proteins. Vector pFW5 containing the spectinomycin resistance gene (aad9) was used to transform strain D471-M6 and was obtained from Andreas Podbielski, University Hospital Rostock, Rostock, Germany (38).
DNA techniques.
Genomic DNA was prepared from the protoplasts of wild-type strain D471-M6 and mutant strains as previously described (33, 41). Restriction endonuclease digestions, DNA ligations, and agarose gel electrophoresis were performed in accordance with standard techniques (41). Nucleotide sequences of both the forward and reverse primers used in the present study are described in Table 1 and were custom synthesized by Gene Link Inc., Hawthorne, N.Y. Southern hybridization was performed on the AlwNI digest of genomic DNA of wild-type and mutant GAS strains with a sen gene-specific, alkaline phosphatase-labeled probe (Amersham Pharmacia, Piscataway, N.J.) in accordance with standard techniques (41). DNA sequencing was performed at the Protein/DNA Technology Center at The Rockefeller University, New York, N.Y.
TABLE 1.
Primers used in this study
| Primer | No. | Sequencea |
|---|---|---|
| sen-F-NdeI | 1 | 5′ CTACGGCCATATGTCAATAATAACTGATGTGTATGCTCG 3′ |
| sen-R-BamHI | 2 | 5′ CGCGGATCCTTTTTTTAAGTTATAGAATGATTTGATACC 3′ |
| sen-ΔK435 | 3 | 5′ CGCGGATCCTACTATTTTAAGTTATAGAATGATTTGATACC 3′ |
| sen-ΔK434-435 | 4 | 5′ CGCGGATCCTACTATAAGTTATAGAATGATTTGATACC 3′ |
| sen-K435L | 5 | 5′ CGCGGATCCTACTATAATTTTAAGTTATAGAATGATTTGATACC 3′ |
| sen-K434-435L | 6 | 5′ CGCGGATCCTACTATAATAATAAGTTATAGAATGATTTGATACCTTTG 3′ |
| sen-K428-434-435L | 7 | 5′ CGCGGATCCTATAATAATAAGTTATAGAATGATAAGAVACCTTTGTA 3′ |
| sen-DST-F | 8 | 5′ CTGCAGATTAGTCTAGUGGACTCATTTATCC 3′ |
| sen-DST-R | 9 | 5′ CATATGTGTTGAGCAACTAGCTGCTTTGACG 3′ |
| sen-F-SalI | 10 | 5′ ACGCGTCGACATG TCAATAATAACTGATGTGTATGCTCG 3′ |
| spc-1-F | 11 | 5′ GGTACTTACATGTTTGGATCAGG 3′ |
| spc-2-R | 12 | 5′ CCATTCAATATTCTCTCCAAG 3′ |
| sen-C-F | 13 | 5′ GGTGAAGTTGCUCAATACAAAGG 3′ |
| sen-DST-N-R | 14 | 5′ CACATTCTACGAATGACC 3′ |
| sen-DST-C-R | 15 | 5′ CGACAGAGCAGTTGATATCTTGG 3′ |
Restriction sites introduced into the primer sequences are underlined.
Site-directed mutagenesis of the sen gene and production of His-tagged SEN proteins.
On the basis of the known sequence of GAS M1 enolase (http://www.genome.ou.edu), the open reading frame encoding D471-M6 enolase (sen) was amplified by PCR with a pair of primers, sen-F-NdeI (primer 1, forward) and sen-R-BamHI (primer 2, reverse) (Fig. 1A and Table 1) (16). The PCR-amplified DNA product was digested with NdeI and BamHI, ligated into vector pET-14b, and transformed into E. coli XL1 Blue. To construct mutant SEN proteins with altered C-terminal ends, forward primer 1 was kept common to all mutagenesis experiments (Fig. 1A). Point mutations, as shown in Fig. 1A, were introduced at the 3′ end of the sen gene, with a series of reverse primers (Table 1). Primer sen-ΔK435 (primer 3 in Table 1) was used for deletion of the last AAA codon encoding a lysine (K435), primer sen-ΔK434-435 (primer 4) was used for deletion of the last two codons encoding K434 and K435, primer sen-K435L (primer 5) was used for replacement of the last AAA codon with a TTA codon encoding L435, primer sen-K434-435L (primer 6) was used for replacement of the last two AAA codons with two TTA codons encoding two leucine residues, L434-435, and primer sen-K428-434-435L (primer 7) was used for replacement of lysines at positions 428, 434, and 435 with leucines (Fig. 1A). The PCR-amplified DNA fragments obtained from each pairs of primers were digested with NdeI and BamHI and then ligated to vector pET-14b, yielding a series of recombinant plasmids, pET-senΔK435, pET-senΔK434-435, pET-senK435L, pET-senK434-435L, and pET-senK428-434-435L. All of the point mutations described above were verified by DNA sequencing of the mutated sen gene. Wild-type and mutated SEN proteins were expressed in E. coli BL21(DE3)/pLysS under the inducible phage T7 promoter. The expression of these proteins was induced for 3 h with 1 mM isopropyl-β-d-thiogalactopyranoside (IPTG). Histidine-tagged proteins were purified by Ni+-nitrilotriacetic acid affinity chromatography. Protein concentration was determined with a bicinchoninic acid protein assay kit (Pierce) with bovine serum albumin as the standard protein.
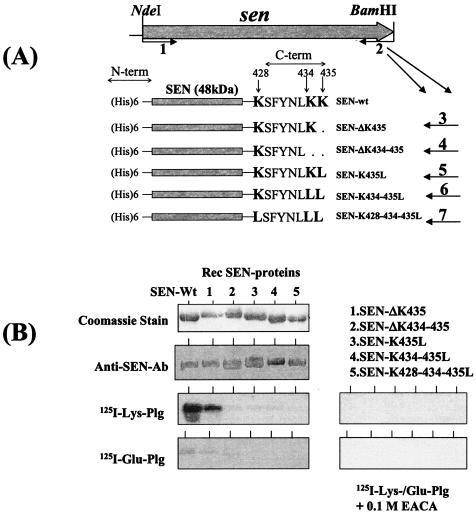
FIG. 1.
(A) Strategy used to delete or replace the C-terminal (C-term) lysine residues (K428, K434, and K435) by site-directed mutagenesis and the resultant His-tagged recombinant SEN proteins. Primer 1 (sen-F-NdeI) is a common forward primer and contains an NdeI site. Primers 2 to 7 are reverse primers each with a restriction site for BamHI used to construct wild-type SEN (SEN-wt) and mutated SEN proteins with their C-terminal and penultimate lysine residues either deleted or replaced with leucines (see Table 1). N-term, N terminal. (B) Western blot analysis showing the antibody reactivities of different purified recombinant (Rec) SEN proteins with affinity-purified rabbit polyclonal anti-SEN antibodies (Ab) and their plasminogen (Plg)-binding activities with 125I-labeled Glu-plasminogen and 125I-labeled Lys-plasminogen in the presence or absence of 0.1 M EACA.
Construction of strains D471-M6-SEN-K434-435L and D471-M6-aad.
To construct plasmid pFWsenK434-435L, the primer 1 sequence was modified at its 5′ end, where the NdeI site was replaced with a SalI site (primer 10). Wild-type sen and mutated sen (sen K434-435L) were amplified by PCR with primers 10 and 2 and primers 8 and 6, respectively (Table 1). The 1.3-kb DNA fragment, corresponding to the downstream region of sen in the D471-M6 chromosome (sen DST), was amplified by PCR containing a forward primer containing a PstI restriction site, sen DST-F (primer 8), and a reverse primer with an NdeI site, sen DST-R (primer 9). After digestion with the appropriate restriction enzymes, the PCR-amplified DNA products corresponding to the wild-type or mutated sen gene and the sen DST region were, respectively, cloned into the multiple cloning sites located upstream (MCS-I) and downstream (MCS-II) of the aad9 gene in pFW5 (38). The orientations of all DNA inserts and mutations in the specified places were confirmed by DNA sequencing. Plasmids pFWsenK434-435L and pFWsen were introduced into strain D471-M6 by electroporation with a Gene Pulser II electroporator (Bio-Rad) as previously described (43). Presumed mutants were identified and confirmed by PCR, Southern blotting, and DNA sequencing with primers 11, 12, 13, and 15 (Table 1).
α-Enolase activity.
To determine α-enalase activity, the enzyme reaction was performed at room temperature in 100 mM HEPES buffer, pH 7.0, containing 10 mM MgSO4, 7.7 mM KCl, and 3 mM 2-phosphoglycerate in a final volume of 1.0 ml as previously described (34). The change in absorbance as a result of the conversion of 2-phosphoglycerate to phosphoenolpyruvate was monitored spectrophotometrically at 240 nm. The rate of phosphoenolpyruvate release was recorded as absorbance per minute.
Labeling of plasminogen.
Purified human Lys-plasminogen and Glu-plasminogen (Enzyme Research Labs, Indianapolis, Ind.) were radiolabeled with Na125I (100 mCi/ml; ICN Radiochemicals) by the chloramine T method with IODO-Beads (Pierce) as previously described (34). The labeled plasminogen was separated from free iodine with a G-25 column (PD-10; Amersham Pharmacia) and stored at −70°C. Typically, the measured specific radioactivity of 125I-labeled plasminogen ranged between 1.6 × 106 and 2.2 × 106 cpm/μg of protein (Glu-plasminogen, 63.2 μCi/nmol; Lys-plasminogen, 83.5 μCi/nmol).
Blot overlay plasminogen-binding assay.
Cell wall extracts from the wild-type and mutant GAS strains were prepared with mutanolysin in 30% raffinose buffer as previously described (33). Proteins in the cell wall extracts or purified recombinant SEN proteins were resolved by SDS-15% PAGE and blotted electrophoretically onto polyvinylidene difluoride membranes as previously described (33, 34). Blots were incubated at room temperature for 3 h in HBST gel blocking buffer (50 mM HEPES-NaOH, pH 7.4, containing 0.15 M NaCl, 1% acidified bovine serum albumin, 0.5% gelatin, 0.5% Tween 20, and 0.04% NaN3) and probed for 3 h at room temperature in blocking buffer containing 125I-labeled human Lys- or Glu-plasminogen (3 × 105 cpm/ml, i.e., 15 to 20 ng/ml) in the presence or absence of 0.1 M EACA (ɛ-aminocaproic acid). The blots were then washed five times with half-strength HBST gel buffer containing 0.35 M NaCl, dried, and autoradiographed as previously described (34).
Plasminogen binding to immobilized SEN.
The ligand-binding analysis was carried out with 96-well microtiter break-apart plates (Nunc) as previously described (34). The plates were coated with purified, His-tagged SEN proteins (1 μg per well) in 0.05 M sodium carbonate buffer, pH 9.6, for 1 h at 37°C, followed by overnight incubation at 4°C. The plates were washed and blocked with HBST gel buffer for 1 h at 37°C. A range of concentrations of aprotinin-treated, 125I-labeled Glu- or Lys-plasminogen (serial twofold dilutions of the starting concentration, 5.5 or 11.32 μg/ml, i.e., 632 or 1,364 nM, respectively) was added to the SEN-coated wells with or without 0.1 M EACA in a final volume of 100 μl of HBST gel buffer and incubated for 2 h at room temperature. The plates were then washed with HBST gel buffer and dried at room temperature. Each well was then subjected to radioactivity counting (counts per minute) with a Beckman mini-γ-counter. The binding of different concentrations of plasminogen to bound mutant SEN proteins was determined in duplicate wells. The specific plasminogen-binding activity of bound SEN was determined after subtracting values of nonspecific plasminogen-binding activity obtained in the presence of 0.1 M EACA from those of the test samples. The results obtained from three separate experiments were plotted and analyzed statistically, assuming the one-site-binding and two-site-binding nonlinear-curve models. The extracted goodness-of-fit values of these two models were then tested by the F test. The affinity dissociation constant (Kd) of each mutant SEN protein and the maximum amount of bound ligand (Bmax) were then obtained with GraphPad Prism version 3.03 software.
Plasminogen-binding activity of intact streptococci.
The wild-type and mutant GAS M6 strains were grown to stationary phase in Todd-Hewitt broth and washed twice with phosphate-buffered saline. The optical density at 600 nm of all samples was adjusted to 1.0, which is equivalent to ∼5 ×108 CFU/ml. The individual bacterial suspensions were then centrifuged, and the resulting pellets were resuspended in 1/20 of the original volume of HBST blocking buffer for 1 h. A 100-μl volume of each concentrated bacterial suspension (∼109 CFU) was used in the plasminogen-binding assay with a 96-well deep-well plate (400-μl capacity). Serial twofold dilutions of aprotinin-treated, 125I-labeled Lys-plasminogen and 125I-labeled Glu-plasminogen (starting at 15 and 13.2 μg/well, respectively) were then added separately to the bacterial suspension with or without 0.1 M EACA in a final volume of 150 μl of HBST gel buffer. Samples were incubated at room temperature for 2 h with constant mixing. Bacteria were then washed three times in 200 μl of HBST gel buffer and pelleted by centrifugation at 800 × g for 5 min, and the supernatant from each well was aspirated. The pellets were resuspended in the original reaction volume (150 μl) and transferred to a separate break-apart 96-well plate to determine the radioactivity of the bound plasminogen. The specific binding of plasminogen to GAS strains was determined and evaluated as described above by subtracting the background values obtained in the presence of 0.1 M EACA. The results were analyzed statistically with a t test.
Acquisition of plasminogen from human plasma by wild-type and mutant GAS strains.
Overnight cultures of the wild-type, mutant, and control strains were washed, concentrated, and prepared for subsequent experiments as described above. A 100-μl volume of each bacterial suspension was suspended in 100 μl of normal human plasma and incubated at room temperature for different time periods (1 and 2 h) under constant rotation. At each interval, bacteria were pelleted, washed (three times), resuspended in 200 μl of sample buffer, and finally boiled for 2 min. Bacteria were then pelleted, and the supernatant containing the released plasminogen was subjected to SDS-PAGE and Western blot analyses with affinity-purified anti-plasminogen antibody as described below. After treatment with plasma, a duplicate set of samples obtained at two different time points was similarly washed and pelleted. To activate streptococcal surface-bound plasminogen, samples were treated with Ska (40 U, 2 μl; Calbiochem). To determine the proteolytic activity of activated plasminogen, 12.5 μg of the chromogenic substrate Val-Leu-Lys-paranitroanilide (2.5 μg/μl; Sigma) was added to Ska-treated samples and incubated for 1 h at 37°C with constant mixing. At the end of the incubation period, bacteria were pelleted and the resulting supernatants were subjected to spectrophotometric analysis at 405 nm with a 96-well plate reader (PolarGalaxy; BMG Lab Technologies, Offenberg, Germany). Specific proteolytic activity was measured after subtracting the background proteolytic activity obtained with bacteria alone without plasma-Ska treatment.
Proteolytic activity of Ska-activated bound plasminogen on streptococcal strains.
As described above, 100 μl of a concentrated bacterial suspension (∼109 bacteria) was incubated with 10.0 μg of Lys-plasminogen for 1 h at room temperature and the unbound free plasminogen was then removed by repeated washing. The presence of bound plasminogen was estimated by determining its proteolytic activity on the chromogenic substrate Val-Leu-Lys-paranitroanilide after Ska treatment (40 U, 2 μl) in the absence or presence of 2 μg of human Apl (Enzyme Research Laboratories Inc.) as described before (34). The background proteolytic activity on the substrate, obtained for each strain in the absence of Lys-plasminogen-Ska treatment, was subtracted from those of corresponding test samples. Each analysis was carried out with three or four samples.
Proteolytic activity of tissue plasminogen activator (tPA)- and Ska-activated streptococcus-bound plasminogen on the ECM and streptococcal penetration of the ECM.
The assay of the proteolytic activity of tPA- and Ska-activated, streptococcus-bound plasminogen on the ECM and streptococcal penetration of the ECM was performed essentially as described before, with some modifications (47). Briefly, 50 μl of diluted ECM gel (1:4 in minimum essential medium [MEM]; Sigma) was dispensed on PET track-etched membranes in Transwell culture chamber inserts (3 μm pore size; Becton Dickinson Labware, Franklin Lakes, N.J.) to be used with 24-well tissue culture plates. The latter containing ECM gel were initially kept at 4°C, gelled at 37°C for 1 h, and air dried in a laminar-flow hood overnight at room temperature. Before the start of the experiment, ECM-containing gel in polyethylene terapthalate membrane inserts was reconstituted with 100 μl of MEM at 37°C for 60 min and then 600 μl of MEM was added to the bottom wells. Wild-type, mutant, and control GAS strains (50 μl, 5 × 108 CFU), as described above, were added to the upper wells either alone or after preincubation with Lys-plasminogen as described above. One microgram of tPA (1 μl/well) or 40 U of Ska (2 μl/well), with or without 2 μg of Apl (2 μl), was added to wells containing plasminogen-coated streptococci. After 6 h of initial incubation at 37°C, 50-μl samples were taken from the lower wells and cultured on Proteose Peptone sheep blood agar plates to determine the number of streptococci that penetrated the ECM. Each set of experiments was carried out with two inserts. Statistical evaluations of the result were based on three independent experiments. The increase in the total bacterial counts in the original inoculum in MEM after 6 h was less than 2% of the initial inoculum.
Western immunoblotting.
All protein samples were resolved by SDS-14% PAGE and transferred onto polyvinylidene difluoride membranes (Bio-Rad) as previously described (33, 34). Membranes were blocked overnight at room temperature, and the desired protein bands were visualized with protein-specific antibodies and corresponding appropriate alkaline phosphatase-conjugated anti-immunoglobulin G at a dilution 1:2,000 (Bio-Rad) as described before (34).
RESULTS
Identification of the sen gene and plasminogen-binding activities of recombinant SEN proteins.
The complete gene encoding SEN was amplified by PCR with the D471-M6 genomic DNA template and a pair of primers (Table 1) designed from the nucleotide sequence of the enolase gene available from the M1 genome sequence database (16, 34). The PCR-amplified DNA fragment corresponding to the sen gene from strain D471-M6 was sequenced. The comparison of the deduced amino acid sequence obtained from the sen gene of D471-M6 with that from a similar gene of strain SF370-M1 (accession no. AE004092) (16) revealed their identity throughout the sequence except one substitution at residue 76 (E[GAG]76 to K[AAG]76). The C-terminal sequence of SEN ends with two lysine residues (Fig. 1A). In the present study, different recombinant SEN proteins with mutations in their C-terminal ends (428KSFYNLKK435) and a His tag at their N-terminal ends were expressed in E. coli and further purified (Fig. 1A). All His-tagged recombinant SEN molecules showed enzymatic (enolase) activities similar to that of SEN purified from the cell wall extracts of strain D471-M6 (34). The enzymatic activities (which varied between 0.325 and 0.473 AU/min/5 μg) were found to be essentially the same, with no significant difference between the two (data not shown).
Plasminogen-binding activities of different SEN constructs were first qualitatively analyzed by Western blot ligand overlay assay with 125I-labeled Lys-plasminogen and 125I-labeled Glu-plasminogen as probes. As shown in Fig. 1B, SEN bound to both forms of plasminogen; however, the binding of SEN to Lys-plasminogen was found to be much stronger than its binding to Glu-plasminogen (Fig. 1B). Removal of the last lysine residue (ΔK435) (retaining the penultimate K434 residue of SEN) did not change the plasminogen-binding activity of the mutated SEN protein (SEN-ΔK435). However, deletion of the last two lysine residues of SEN (SEN-ΔK434-435) or their replacement with leucine (SEN-K435L, SEN-K434-435L, SEN-K428-434-435L) significantly decreased the Glu-plasminogen- and Lys-plasminogen-binding activities of mutant SEN proteins. In the same assay, the Lys-plasminogen- and Glu-plasminogen-binding activities of wild-type SEN and SEN-ΔK435 were found to be completely abolished in the presence of a lysine analog, EACA (0.1 M). This finding suggested that the C-terminal lysine residues of SEN play a crucial role in its plasminogen-binding activity.
Specific plasminogen-binding activity of SEN and its mutant forms.
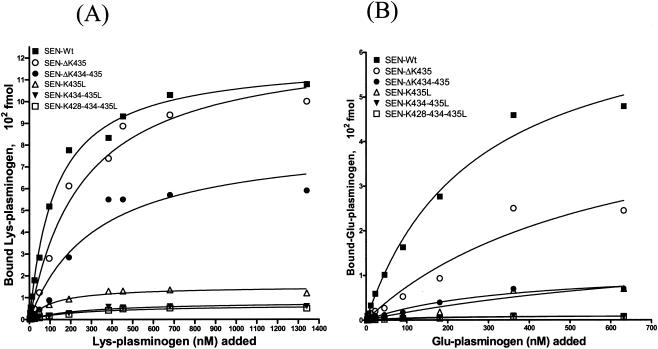
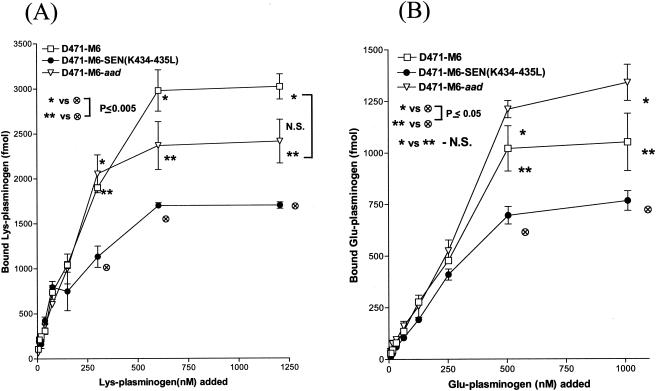
To determine the effect of site-specific mutagenesis at the C-terminal end of SEN on specific Lys-plasminogen- and Glu-plasminogen-binding activities, a quantitative solid-phase assay was carried out with 96-well microtiter plates coated with wild-type or mutated SEN protein. After reacting the 96-well microtiter plates containing different SEN proteins with a range of concentrations of 125I-labeled Lys-plasminogen (3 to 1,340 nM) and 125I-labeled Glu-plasminogen (2.8 to 632 nM), the specific plasminogen-binding activities of wild-type and mutated SEN proteins were determined (Fig. 2). The results were analyzed with the one-site- and two-site-binding nonlinear-curve models. The goodness-of-fit values of the two models were compared with the F test, which indicated that the one-site-binding model fit the data better than the two-site-binding model. The extracted Kd values and corresponding Bmax (maximum amount of bound radioligand) values for Lys- and Glu-plasminogen are described in Table 2. All of the binding sites on wild-type SEN were approaching saturation with 1,000 to 1,340 nM Lys-plasminogen and 400 to 632 nM Glu-plasminogen. Deletion of the C-terminal lysine residues (K434 and/or K435) of wild-type SEN caused a decrease in the plasminogen-binding affinity of the different resultant mutant SEN proteins (SEN-ΔK435 and SEN-ΔK434-435) and thus an increase in their dissociation constants (Fig. 2 and Table 2). The changes in Kd values were even more pronounced when the C-terminal lysine residues of wild-type SEN were replaced with leucines. The mutant SEN proteins (SEN-K435L, SEN-K434-435L, and SEN-K428-434-435L) showed no ability to bind plasminogen (Bmax, 50 to 130 fmol). The results shown in Fig. 2B and Table 2 also revealed similar findings with Glu-plasminogen, which, in comparison with Lys-plasminogen (Fig. 2A and Table 2), bound SEN with significantly lower affinity, as also revealed by Western blot analysis (Fig. 1). Together, these data (Fig. 1 and 2 and Table 2) clearly indicate that the C-terminal lysine residues of the SEN molecule play a determining role in maintaining the proper conformation and integrity of the high-affinity Lys-plasminogen-binding site and thus play an important role in the Glu-plasminogen- and Lys-plasminogen-binding activity of SEN. On the basis of these results, we considered the replacement of the last two lysines with leucines (SEN-K434-435L) in the SEN sequence in vivo, as an ideal strategy to create a mutant GAS strain that may lack or possess altered plasminogen-binding activity.
FIG. 2.
Solid-phase ligand-binding assay showing specific Lys-plasminogen-binding (A) and Glu-plasminogen-binding (B) activities with different His-tagged recombinant SEN proteins. The assay was performed in 96-well microtiter plates. The results were analyzed by the one-site- and two-site-binding nonlinear-curve models with GraphPad Prism version 3.03 software. The best-fit curve shown is the one-site-binding curve, as validated by the F test. The extracted affinity binding values (Kd and Bmax) for each curve are in Table 2. Each data point represents an average value of three experiments, each done with duplicate samples. SEN-Wt, wild-type SEN.
TABLE 2.
Affinity constants of different His-tagged SEN proteins for Lys- and Glu-plasminogena
| His-tagged sen protein | Lys-plasminogen
|
Glu-plasminogen
|
||
|---|---|---|---|---|
| Kd (nM) | Bmax (102 fmol) | Kd (nM) | Bmax (102 fmol) | |
| SEN-Wtb | 127 | 10.70 | 267 | 4.79 |
| SEN-ΔK435 | 260 | 11.0 | 600 | 2.45 |
| SEN-ΔK434-435 | 327 | 5.9 | 995 | 0.7 |
| SEN-K435L | 1,119 | 1.3 | 1,029 | 0.7 |
| SEN-K434-435L | ND | 0.6 | ND | 0.13 |
| SEN-K428-434-435L | ND | 0.5 | ND | 0.08 |
Kd, equilibrium dissociation constant; Bmax, maximal amount of radioligand bound per well containing the specific SEN recombinant protein; ND, undeterminable.
SEN-Wt, wild-type SEN.
Introduction of the sen K434-435L mutation into wild-type streptococcal strain D471-M6 and characterization of mutant strain D471-M6-SEN-K434-435L.
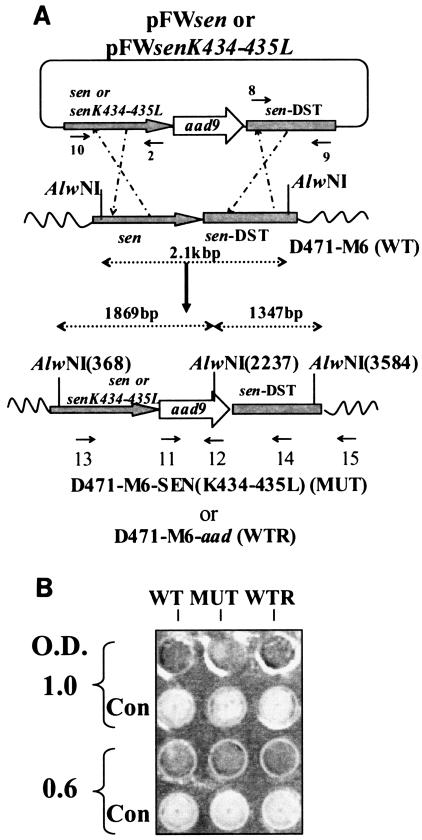
The mutated sen gene (sen K434-435L, 1.32 kb) and the downstream region of the sen gene (sen DST, 1.317 kb) were cloned, respectively, into the upstream and downstream regions (multiple cloning sites 1 and 2, respectively) of the aad9 gene harbored by suicide vector pFW5. The newly constructed plasmid pFWsenK434-435L was then introduced into wild-type GAS strain D471-M6 by electroporation, resulting in successful replacement of the lysine residue with leucine at positions 434 and 435 of the sen gene (Fig. 3A). The structure of the insertion was confirmed by Southern blotting and by DNA sequencing of appropriate PCR-amplified products. To ascertain that the insertion of the spectinomycin resistance gene (aad9) did not affect the plasminogen-binding property of the wild-type strain, aad9 was also introduced immediately downstream of the wild-type sen gene in strain D471 to create a control GAS strain, D471-M6-aad (Fig. 3A).
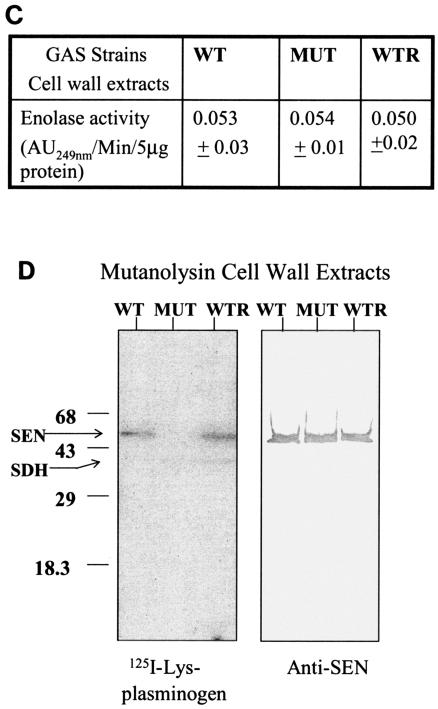
FIG. 3.
(A). Plasmid construction for replacement of the wild-type (WT) sen gene with a sen K434-435L mutation in strain D471-M6. As described in Materials and Methods, the PCR-amplified DNA fragments corresponding to the mutated sen gene and the flanking downstream regions of sen (sen DST) were digested with SalI/BamHI and PstI/NdeI, respectively, and ligated to the appropriate sites upstream and downstream of aad9 (spectinomycin resistance marker gene), respectively, into pFW5 to yield plasmid pFWsenK434-435L. The latter was then introduced into strain D471-M6 and integrated into the streptococcal chromosome via homologous recombination, resulting in the creation of a mutant (MUT) strain, D471-M6-SEN-K434-435L. A similar introduction of pFWsen created a control strain (D471-M6-aad [WTR]) that contained the aad9 gene between the wild-type sen gene and the sen DST region. Primers 10 and 15, primers 11 and 12 (derived from the aad9 gene), primers 13 and 12, and primers 11 and 15 were used to verify the chromosomal insertion of aad9 downstream of sen. The sen K435-434L mutation was verified by DNA sequence analyses of the PCR fragment obtained with primers 13 and 12. (B) Surface expression of SEN on the D471-M6, D471-M6-SEN-K434-435L, and D471-M6-aad strains. Surface expression of SEN was measured by colony blot analysis with a MultiScreen 96-well filter plate (0.22-μm-pore-size low-protein-binding Durapore membrane; Millipore) and a 96-well plate vacuum manifold. A 100-μl volume of different growth phase cultures (mid log [optical density at 600 nm = 0.6] and late log [optical density at 600 nm = 1.0]), normalized at an optical density (O.D.) at 600 nm of 1.0, were dispensed into the wells and blotted. They were then stained with anti-SEN polyclonal antibody and corresponding alkaline phosphatase conjugate and developed as shown. The control (Con) represents bacteria stained with only conjugate antibody. (C) Enolase activity of mutanolysin extracts (5 μg) of the D471-M6, D471-M6-SEN-K434-435L, and D471-M6-aad GAS strains (adjusted to an optical density at 600 nm of 1). Enolase activity was measured as described in Materials and Methods. The values shown are averages of three different experiments ± the standard deviation. (D) Plasminogen-binding activities of SEN found in the mutanolysin-extracted cell walls of streptococcal strains D471-M6, D471-M6-SEN-K434-435L, and D471-M6-aad. The reactivity of these bands with rabbit anti-SEN affinity-purified antibody is shown. SDH, streptococcal surface dehydrogenase; AU249, units of absorbance at 249 nm. The values on the left are molecular sizes in kilodaltons.
The wild-type and isogenic mutant GAS strains showed similar patterns in their growth curves and reactivities with affinity-purified anti-SEN antibodies, as determined by colony Western blot assay, indicating that there was no defect in their surface expression of SEN (Fig. 3B). Similarly, the site-specific mutation and the introduction of the aad9 gene immediately downstream of the altered sen gene did not affect the expression of the Ska-encoding gene (ska). This was revealed by the fact that the supernatants of overnight cultures of the D471-M6, D471-M6-SEN-K434-435L, and D471-M6-aad strains showed the presence of equal amount of Ska and no further release of it into the supernatant over a period of 3 h (data not shown). Since there was no further release of Ska from the washed pellets, we concluded that Glu-plasminogen would not be converted to Lys-plasminogen or to plasmin and thus would not affect the integrity of other streptococcal surface proteins and generate de novo lysine-binding sites.
The amounts of SEN present in the mutanolysin cell wall extracts of wild-type (D471-M6), control (D471-M6-aad), and mutant (D471-M6-SEN-K434-435L) GAS strains were initially analyzed by determining enolase activity. As shown in Fig. 3C, mutanolysin extracts of all of the strains showed similar amounts of enolase activity per 5 μg of total protein. This was further confirmed by Western blotting with anti-SEN antibody (Fig. 3D). The plasminogen-binding ability of enolase in each extract was then analyzed by measuring 125I-labeled Lys-plasminogen (Fig. 3D). The results showed that despite the presence of the same amount of SEN in the cell wall extracts of all of the GAS strains, the SEN protein present in the mutant GAS strains (D471-M6-SEN-K434-435L and D471-M6-SEN-K434-435L) did not bind 125I-labeled Lys-plasminogen. Similar results were obtained when 125I-labeled Glu-plasminogen was used as a probe, except for the fact that the intensity of the plasminogen binding to SEN from D471-M6 and D471-M6-aad was significantly weaker, as expected (data not shown). As also shown in Fig. 3D, a second weak plasminogen-binding protein of ∼39 kDa corresponds to SDH and is equivalently expressed in the wild-type and mutated strains.
Together, these results indicated that the mutation (sen K434-435L) was introduced at an appropriate site, which resulted in a desired functional change in the targeted protein (SEN) without affecting the enolase activity and expression profiles of other plasminogen-binding secreted (Ska) and nonsecreted (Plr) proteins.
Specific plasminogen-binding activity of the D471-M6, D471-M6-SEN-K434-435L, and D471-M4-aad strains.
The Lys-plasminogen- and Glu-plasminogen-binding activities of mutant GAS strain [D471-M6-SEN-K434-435L] were compared with those of the wild-type (D471-M6) and control (D471-M6-aad) GAS strains to understand the contribution of the C-terminal lysine residues of SEN in the streptococcal ability to bind plasminogen. The D471-M6-aad strain, which served as an internal control, was used to ensure that insertion of the aad9 gene did not change its plasminogen-binding activity nonspecifically. Since it was possible to discriminate the difference between the plasminogen-binding activities of wild-type and mutant GAS strains at a concentration of 109 CFU, all subsequent plasminogen-binding assays and functional analyses were carried out with this bacterial concentration (Fig. 4). Plasminogen-binding activity was inhibited by EACA, a lysine analog, in a dose-dependent manner (the concentrations inhibiting Glu-plasminogen binding by 50% were 2 mM EACA [D471-M6] and 1 mM EACA [D471-M6-SEN-K434-435L]; the concentrations inhibiting Lys-plasminogen binding by 50% were 0.5 mM EACA [D471-M6] and 1.5 mM EACA [D471-M6-SEN-K434-435L]) (data not shown). At a concentration of 4 mM EACA, ≥80% of the plasminogen-binding activity was inhibited. All of the plasminogen-binding sites on strain D471-M6 were saturated with 600 to 1,200 nM Lys-plasminogen and 500 to 1,000 nM Glu-plasminogen. Both Lys- and Glu-plasminogen bound to GAS strains in a dose-dependent manner; however, the Lys-plasminogen-binding activities of D471-M6 and D471-M6-SEN-K434-435L were found to be almost three times their Glu-plasminogen-binding activities (Fig. 4). Since there was no significant difference between the Lys-plasminogen-binding activities of D471-M6 and D471-M6-aad (2,400 to 3,000 fmol; P > 0.05), our results confirmed that introduction of the add9 gene did not affect the plasminogen-binding ability of strain D471-M6 (Fig. 4A). On the other hand, strain D471-M6-SEN-K434-435L showed a statistically significant reduction (1,700 fmol, ∼43%; P = 0.003) in Lys-plasminogen-binding activity compared to that of the wild-type strain (Fig. 4A). A similar difference was also observed for the Glu-plasminogen-binding activity of the mutant strains compared with that of wild-type strain D471-M6 (Fig. 4B). Together, these data confirmed that SEN, in particular its C-terminal lysine residues, K434 and K435, contributes significantly to the Lys-plasminogen-binding activity of GAS.
FIG. 4.
Specific dose-dependent 125I-labeled Lys-plasminogen-binding (A) and 125I-labeled Glu-plasminogen-binding (B) activity of streptococcal wild-type strain D471-M6, isogenic mutant strain D471-M6-SEN-K434-435L, and the wild-type strain containing the spectinomycin resistance gene, D471-M6-aad. Each point represents the mean of three independent experiments, each carried out with duplicate samples, ± the standard error of the mean. The specific amount of plasminogen bound was determined on the basis of the specific activity of the 125I-labeled plasminogen added. N.S., no statistically significant difference (P > 0.05).
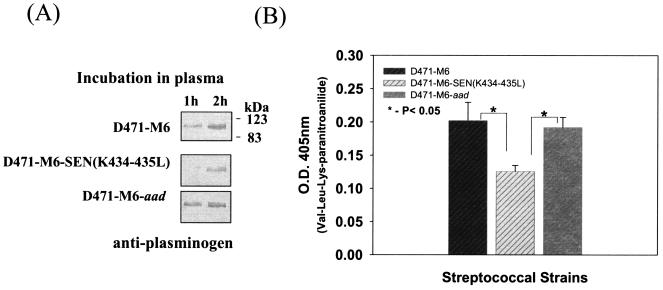
Streptococcal ability to acquire plasminogen from human plasma.
The D471-M6, D471-M6-SEN-K434-435L, and D471-M6-aad strains were separately incubated with human plasma to determine the impact of the mutations in the sen gene on the ability to acquire plasminogen from human plasma. At different time intervals, the amount of GAS-bound plasminogen was determined initially by Western blot analysis with specific polyclonal rabbit anti-human plasminogen antibody. The results, as shown in Fig. 5A, revealed weaker acquisition of human plasma plasminogen by mutant GAS strain D471-M6-SEN-K434-435L after 60 min, compared to those of strains D471-M6 and D471-M6-aad. These results were then confirmed by determining the Ska-mediated proteolytic activity of bound plasminogen on the chromogenic substrate Val-Leu-Lys-paranitroanilide (Fig. 5B). The results revealed a significant decrease (P < 0.05) in the surface-bound proteolytic activity of the mutant GAS strain, compared to those of the wild-type and control GAS strains. These results are in agreement with the findings described in Fig. 4.
FIG. 5.
Acquisition of plasminogen from human plasma by streptococcal strains D471-M6, D471-M6-SEN-K434-435L, and D471-M6-aad. (A) Immunochemical detection of plasminogen acquired from human plasma by streptococcal strains at different time points by Western blot analysis. (B) Proteolytic activity of streptococcus-bound plasminogen acquired from human plasma after reaction with Ska on the chromogenic substrate Val-Leu-Lys-paranitroanilide. Vertical bars represent the average of results from four or five samples ± the standard error of the mean. O.D., optical density.
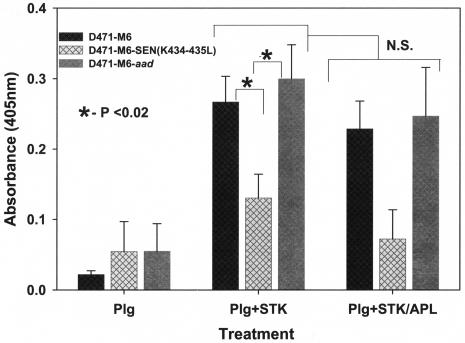
Proteolytic activity of Ska-activated streptococcus-bound plasminogen.
In support of previous findings, the Ska-activated proteolytic activity of Lys-plasminogen bound to mutant GAS strain D471-M6-SEN-K434-435L was compared with those of the D471-M6 and D471-M6-aad GAS strains by using the chromogenic substrate described above. The results of this assay also revealed a significant decrease in the ability of the mutant GAS strain to bind Lys-plasminogen and hence a corresponding decrease (P < 0.05) in Ska-mediated proteolytic activity compared to similar activities displayed by the strains expressing wild-type SEN (Fig. 6). As expected, Ska-mediated surface-bound plasminogen activation was not inhibited by Apl (Fig. 6). These findings, along with those described in Fig. 5, confirmed that the C-terminal lysine residues of SEN play a significant role in the in vitro and in vivo plasminogen-binding ability and the corresponding surface proteolytic activity of GAS.
FIG. 6.
Proteolytic activity of Ska (STK)-activated, streptococcus-bound Lys-plasminogen (Plg) in the presence or absence of Apl. Proteolytic activities were measured by determining cleavage of the Val-Leu-Lys-paranitroanilide chromogenic substrate. Changes in optical density (O.D.) at 405 nm were measured spectrophotometrically. The mean results from three independent experiments ± the standard error of the mean are shown. N.S., no statistically significant difference (P > 0.05).
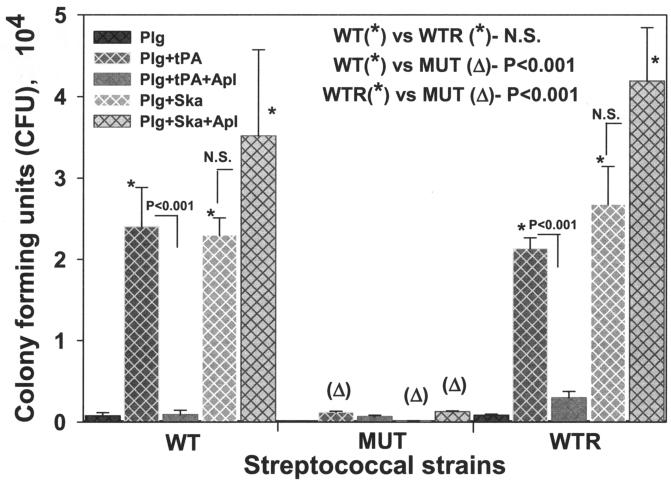
Penetration of ECM by GAS with surface-bound, tPA- and Ska-activated plasminogen.
SEN-mediated plasminogen-binding activity and the corresponding enhancement of the tPA- and Ska-catalyzed surface proteolytic activity of GAS suggested that the C-terminal mutation in SEN might also influence their in vivo pathological functions (Fig. 5 and 6) (34). The D471-M6, D471-M6-SEN-K434-435L, and D471-M6-aad GAS strains, preincubated with plasminogen and activated with tPA and Ska, were subjected to an ECM degradation-penetration assay with Transwell membrane inserts coated with the ECM proteins. In the absence of any treatment, none of these strains showed an ability to penetrate the ECM (data not shown). As shown in Fig. 7, in the presence of plasminogen alone, none of the GAS strains showed a significant ability to digest and penetrate the ECM, indicating the absence of streptococcal surface-associated plasmin-proteolytic activity. However, the outer wells of the Transwell system containing GAS strains that were pretreated with Lys-plasminogen, followed by tPA and Ska, showed a significant increase (P < 0.001) in the number of CFU of only the wild-type D471-M6 and control D471-M6-aad strains and not in the number of CFU of mutant strain D471-M6-SEN-K434-435L. These results clearly indicated that the wild-type and control GAS strains bound plasminogen with high affinity, and its conversion to proteolytically active plasmin allowed them to degrade and then penetrate the ECM efficiently. As expected, tPA-mediated ECM degradation was significantly inhibited by Apl, and hence, treatment of GAS-bound plasminogen with tPA in the presence of Apl significantly decreased (P < 0.001) the CFU counts of all the GAS strains in the outer wells. In contrast to this, Ska-mediated plasminogen activation and subsequent ECM degradation were unaffected by Apl.
FIG. 7.
Streptococcal penetration of ECM protein-coated Trans-well membranes (3-μm pore size). GAS strains D471-M6 (WT), D471-M6-SEN-K434-435L (MUT), and D471-M6-aad (WTR) were added to the upper wells either alone or after preincubation with Lys-plasminogen (Plg). Streptococci preincubated with Lys-plasminogen were treated separately with tPA and Ska or in combination with Apl. The presence in the lower wells of streptococci that penetrated the membrane was determined by counting CFU on blood agar plates. Mean values obtained with three or four individual Trans-wells ± the standard error of the mean are shown. The inset showing the statistically significant difference between the CFU counts of two strains highlights the comparison of the effects of different treatments on the ability of GAS strains to penetrate the membrane. N.S., no statistically significant difference (P > 0.05).
In concurrence with the results shown in Fig. 5 and 6, we conclude from these data (Fig. 7) that the C-terminally located lysine residues play an important role in the streptococcal ability to bind exogenous plasminogen. Moreover, the surface-acquired plasmin-mediated proteolytic activity of GAS may play an essential role in tissue invasion by allowing their penetration of the ECM barrier.
DISCUSSION
The ability of GAS to capture unregulatable forms of plasmin on the cell surface after interacting with human plasmin(ogen) has been postulated as an important virulence factor since plasmin can turn bacteria into proteolytic organisms and confer a tissue-invasive phenotype (8, 25). Among the GAS surface proteins that directly or indirectly bind to plasminogen, genetic proof of the role of their C-terminal or internal lysine residues in the plasminogen-binding activity of GAS is available for Plr and PAM (2, 8, 13, 24, 39, 40, 48-50). On the basis of the reported affinity constants of plasminogen for these proteins, SEN may be considered the strong plasminogen-binding protein (3, 34). Although Plr was the first well-characterized protein showing direct binding activity with plasmin(ogen), reports on its mutational analyses and the comparison of its plasminogen-binding ability with that of SEN have revealed that Plr is a weak plasminogen-binding protein (24, 33, 34, 48, 50). Further, studies with GAS transformed with the plr gene lacking the C-terminal lysine residue clearly indicated that Plr does not play a major role as a streptococcal plasminogen-binding receptor (49). A recent report, however, indicated that Plr remains associated with M or M-like protein and this association may allow Plr to indirectly acquire plasmin generated by the M-protein-fibrinogen-mediated fibrinolytic complex (13, 14). In the present study, we have shown for the first time that SEN, a ubiquitously found surface protein of GAS, binds to both Lys- and Glu-plasminogen.
The enolase activity of SEN is essential for streptococcal survival, and this protein is expressed by a single gene, as revealed by our unpublished data and the genome sequence data of three GAS M types (M1, M3, and M18) (1, 16, 32, 44). Hence, it was not feasible to use the gene knockout approach as used for PAM to understand the role of SEN as a plasminogen receptor (39, 40, 46). Since the C-terminal lysine residues of SEN do not contribute to its innate enolase activity, and the ɛ-amino group of the lysine side chains may be essential for binding to a kringle structure of plasminogen, we replaced the C-terminal lysines with leucine without affecting their enzymatic activity (Fig. 1 and 2) (32). The qualitative and quantitative data presented in Fig. 1 and 2 clearly indicate that the presence of the C-terminal lysine residues is crucial for the plasminogen-binding ability of SEN. The fact that the mutant strain showed a decrease in its plasminogen-binding ability to ∼43% of the total plasminogen-binding ability of the wild-type strain is not surprising, since most of the identified plasminogen-binding receptors are multifunctional and a single bacterial species may express multiple plasminogen-binding proteins with varied affinity for plasminogen (2, 24, 32-34). The residual plasminogen-binding activity of the mutant strain could be due to a combination of the following factors: (i) weak plasminogen-binding activity of Plr, (ii) possible creation of a de novo plasminogen-binding C-terminal lysine site as a result of innate proteolytic activity of GAS, and/or (iii) the existence of an additional conformational Lys-plasminogen- or Glu-plasminogen-binding site on SEN in its natural form, when expressed on the streptococcal surface in association with other surface proteins as recently reported for S. pneumoniae ENO (4, 8, 33, 34). Despite the presence of the residual plasminogen-binding activity displayed by the mutant strain, it also showed a substantial decrease in its surface-bound plasmin(ogen)-derived Ska- or tPA-mediated proteolytic activity and a significantly reduced ability to penetrate the ECM barrier. These results thus highlight the essential role of the SEN-mediated plasminogen-binding ability of GAS in epithelial cell and tissue damage during pharyngitis and soft-tissue invasion (34a).
By using pam and ska knockout bacterial mutants, the pathogenic roles of the corresponding proteins in GAS pathogenicity have been studied in in vivo infection models (45, 46). While this approach is ideal for a protein having a single defined function, it is not possible for essential proteins such as SEN and Plr (17, 31, 32-34). In the host, the activation of plasminogen and its proteolytic activity are also under the tight homeostatic regulation of several components that constitute the complex intravascular and pericellular fibrinolytic systems (6-8). In addition, it is known that in the presence of ATP, the complex between plasminogen and Ska develops an autophosphorylating activity on the N-terminal portion of Ska, which is essential for plasminogen-activating activity (42). The amidolytic activity of plasminogen-Ska is shown to be greatly diminished by micromolar concentrations of ATP and heparin oligosaccharides (5, 42) Considering these factors, it is not surprising that previous animal model studies with pam and ska gene knockout streptococcal strains and the plasminogen knockout (Plg−/−) transgenic mouse infection model with an M type 49 GAS strain could not conclusively establish an epidemiological correlation between GAS M types and plasminogen-mediated disease association or the loss of virulence of GAS (22, 45, 46).
With an appropriate animal infection model or an in vitro tissue culture model, further studies on the plasminogen-binding ability of GAS under different environmental conditions with the streptococcal mutant deficient in Glu- and/or Lys-plasminogen binding may help us to understand the complex dynamics of intermolecular interactions between virulence factors and host-pathogen interactions that decide the final outcomes of the disease. The streptococcal mutant described in the present study may also serve as an important tool to understand the role of SEN in modulating the intravascular and pericellular fibrinolytic systems during systemic and pharyngeal infections, respectively.
Acknowledgments
V.P. is an Established Investigator of the American Heart Association. This work was supported by American Heart Association Established Investigator grant AHA-9940228N (V.P.) and U.S. Public Health Service grants AI-42827 (V.P) and AI-11822 (V.A.F).
We thank Andreas Podbielski (University Hospital Rostock, Rostock, Germany) for the pFW5 vector. We are grateful to David Dubnau and Ines Chen for helpful advice and critically reading the manuscript.
Editor: B. B. Finlay
REFERENCES
- 1.Beres, S. B., G. L. Sylva, K. D. Barbian, B. Lei, J. S. Hoff, N. D. Mammarella, M.-Y. Liu, J. C. Smoot, S. F. Porcella, L. D. Parkins, D. S. Campbell, T. M. Smith, J. K. McCormick, D. Y. M. Leung, P. Schlievert, and J. M. Musser. 2002. Genome sequence of a serotype M3 strain of group A Streptococcus: phage-encoded toxins, the high-virulence phenotype, and clone emergence. Proc. Natl. Acad. Sci. USA 99:10078-10083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berge, A., and U. Sjobring. 1993. PAM, a novel plasminogen-binding protein from Streptococcus pyogenes. J. Biol. Chem. 268:25417-25424. [PubMed] [Google Scholar]
- 3.Bergmann, S., M. Rohde, G. S. Chhatwal, and S. Hammerschmidt. 2001. α-Enolase of Streptococcus pneumoniae is a plasmin(ogen)-binding protein displayed on the bacterial cell surface. Mol. Microbiol. 40:1273-1287. [DOI] [PubMed] [Google Scholar]
- 4.Bergmann, S., D. Wild, O. Diekmann, R. Frank, D. Bracht, G. S. Chhatwal, and S. Hammerschmidt. 2003. Identification of a novel plasmin(ogen)-binding motif in surface displayed α-enolase of Streptococcus pneumoniae. Mol. Microbiol. 49:411-423. [DOI] [PubMed] [Google Scholar]
- 5.Bishop, C., D. M. Rankine, and J. H. Talbott. 1959. The nucleotide in normal human blood. J. Biol. Chem. 234:1233-1237. [PubMed] [Google Scholar]
- 6.Blasi, F. 1993. Urokinase and urokinase receptor: a paracrine/autocrine system regulating cell migration and invasiveness. Bioessays 15:105-111. [DOI] [PubMed] [Google Scholar]
- 7.Blasi, F. 1997. uPA, uPAR, PAI-I: key intersection of proteolytic, adhesive and chemotactic highways. Immunol. Today 18:415-417. [DOI] [PubMed] [Google Scholar]
- 8.Boyle, M. D. P., and R. Lottenberg. 1997. Plasminogen activation by invasive human pathogens. Thromb. Haemostasis 77:1-10. [PubMed] [Google Scholar]
- 9.Caparon, M. G., R. T. Geist, J. Perez-Casal, and J. R. Scott. 1992. Environmental regulation of virulence in group A streptococci: transcription of the gene encoding M protein is stimulated by carbon dioxide. J. Bacteriol. 174:5693-5701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Castellino, F. J., J. M. Sodetz, W. J. Brockway, and G. E. Siefring. 1996. Streptokinase. Methods Enzymol. 45:244-257. [DOI] [PubMed] [Google Scholar]
- 11.Castellino, F. J., and J. R. Powell. 1981. Human plasminogen. Methods Enzymol. 80:365-378. [DOI] [PubMed] [Google Scholar]
- 12.Christner, R., Z. Li, R. Raeder, A. Podbielski, and M. D. P. Boyle. 1997. Identification of key gene products required for acquisition of plasmin-like activity by group A streptococci. J. Infect. Dis. 175:1115-1120. [DOI] [PubMed] [Google Scholar]
- 13.D'Costa, S. S., and M. D. P. Boyle. 1998. Interaction of a group A streptococcus within human plasma results in assembly of a surface plasminogen activator that contributes to occupancy of surface plasmin-binding structures. Microb. Pathog. 24:341-349. [DOI] [PubMed] [Google Scholar]
- 14.D'Costa, S. S., T. G. Romer, and M. D. P. Boyle. 2000. Analysis of expression of a cytosolic enzyme on the surface of Streptococcus pyogenes. Biochem. Biophys. Res. Commun. 278:826-832. [DOI] [PubMed] [Google Scholar]
- 15.Dudani, A. K., C. Cummings, S. Hashemi, and P. R. Ganz. 1993. Isolation of a novel 45 kDa plasminogen receptor from human endothelial cells. Thromb. Res. 69:185-296. [DOI] [PubMed]
- 16.Ferretti, J. J., W. M. McShan, D. Ajdic, D. J. Savic, G. Savic, K. Lyon, C. Primeaux, S. Sezate, A. N. Suvorov, S. Kenton, H. S. Lai, S. P. Lin, Y. Qian, H. G. Jia, F. Z. Najar, Q. Ren, H. Zhu, L. Song, J. White, X. Yuan, S. W. Clifton, B. A. Roe, and R. McLaughlin. 2001. Complete genome sequence of an M1 strain of Streptococcus pyogenes. Proc. Natl. Acad. Sci. USA 98:4658-4663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Fischetti, V. A. 2000. Surface proteins on gram-positive bacteria, p. 11-26. In V. A. Fischetti, R. P. Novick, J. J. Ferretti, D. A. Portnoy, and J. I. Rood (ed.), Gram-positive pathogens. ASM Press, Washington, D.C.
- 18.Fontan, P. A., V. Pancholi, M. M. Nociari, and V. A. Fischetti. 2000. Antibodies to streptococcal surface enolase react with human α-enolase: implications in poststreptococcal sequelae. J. Infect. Dis. 182:1712-1721. [DOI] [PubMed] [Google Scholar]
- 19.Gong, Y., S.-K. Kim, J. Felez, D. K. Grella, F. J. Castellino, and L. A. Miles. 2001. Conversion of Glu-plasminogen to Lys-plasminogen is necessary for optimal stimulation of plasminogen activation on the endothelial cell surface. J. Biol. Chem. 276:19078-19083. [DOI] [PubMed] [Google Scholar]
- 20.Hollingshead, S. K., V. A. Fischetti, and J. R. Scott. 1986. Complete nucleotide sequence of type 6 M protein of the group A Streptococcus: repetitive structure and membrane anchor. J. Biol. Chem. 261:1677-1686. [PubMed] [Google Scholar]
- 21.Kehoe, M. A. 1994. Cell-wall-associated proteins in gram-positive bacteria, p. 217-261. In J.-M. Ghuysen and R. Hakenbeck (ed.), Bacterial cell wall. Elsevier Science, New York, N.Y.
- 22.Li, Z., V. A. Ploplis, E. L. French, and M. D. Boyle. 1999. Interaction between group A streptococci and the plasmin(ogen) system promotes virulence in a mouse skin infection model. J. Infect. Dis. 179:907-914. [DOI] [PubMed] [Google Scholar]
- 23.Lopez-Alemany, R., P. Correc, L. Camoin, and P. Burtin. 1994. Purification of the plasmin receptor from human carcinoma cells and comparison to alpha-enolase. Thromb. Res. 75:371-381. [DOI] [PubMed] [Google Scholar]
- 24.Lottenberg, R., C. C. Broder, M. D. P. Boyle, S. J. Kain, B. L. Schroeder, and R. Curtiss III. 1992. Cloning, sequence analysis, and expression in Escherichia coli of a streptococcal plasmin receptor. J. Bacteriol. 174:5204-5210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lottenberg, R., D. Minning-Wenz, and M. D. P. Boyle. 1994. Capturing host plasmin(ogen): a common mechanism for invasive pathogens? Trends Microbiol. 2:20-24. [DOI] [PubMed] [Google Scholar]
- 26.McIver, K. S., A. S. Heath, and J. R. Scott. 1995. Regulation of virulence by environmental signals in group A streptococci: influence of osmolarity, temperature, gas exchange, and iron limitation of emm transcription. Infect. Immun. 63:4540-4542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McIver, K. S., and J. R. Scott. 1997. Role of mga in growth phase regulation of virulence genes of the group A streptococcus. J. Bacteriol. 179:5178-5187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Miles, L. A., C. M. Dahlberg, J. Plescia, J. Felez, K. Kato, and E. F. Plow. 1991. Role of cell-surface lysines in plasminogen binding to cells: identification of alpha-enolase as a candidate plasminogen receptor. Biochemistry 30:1682-1691. [DOI] [PubMed] [Google Scholar]
- 29.Molinari, G., and G. S. Chhatwal. 1999. Streptococcal invasion. Curr. Opin. Microbiol. 2:56-61. [DOI] [PubMed] [Google Scholar]
- 30.Molinari, G., M. Rohde, C. A. Guzman, and G. S. Chhatwal. 2000. Two distinct pathways for the invasion of Streptococcus pyogenes in non-phagocytic cells. Cell. Microbiol. 2:145-154. [DOI] [PubMed] [Google Scholar]
- 31.Navarre, W. W., and O. Schneewind. 1999. Surface proteins of gram-positive bacteria and mechanisms of their targeting to the cell wall envelope. Microbiol. Mol. Biol. Rev. 63:174-229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pancholi, V. 2001. Multifunctional α-enolase: its role in diseases. Cell. Mol. Life Sci. 58:902-920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pancholi, V., and V. A. Fischetti. 1992. A major surface protein on group A streptococci is a glyceraldehyde-3-phosphate dehydrogenase with multiple binding activity. J. Exp. Med. 176:415-426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pancholi, V., and V. A. Fischetti. 1998. α-Enolase, a novel strong plasmin(ogen) binding protein on the surface of pathogenic streptococci. J. Biol. Chem. 273:14503-14515. [DOI] [PubMed] [Google Scholar]
- 34a.Pancholi, V., P. Fontan, and H. Jin. 2003. Plasminogen-mediated group A streptococcal adherence to and pericellular invasion of human pharyngeal cells. Microb. Pathog. 35:293-303. [DOI] [PubMed]
- 35.Parry, M. A. A., X. C. Zhang, and W. Bode. 2000. Molecular mechanisms of plasminogen activation: bacterial co-factors provide clues. Trends Biochem. Sci. 25:53-59. [DOI] [PubMed] [Google Scholar]
- 36.Perez-Casal, J., M. G. Caparon, and J. R. Scott. 1991. Mry, a trans-acting positive regulator of the M protein gene of Streptococcus pyogenes with similarity to the receptor proteins of two-component regulatory systems. J. Bacteriol. 173:2617-2624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Plow, E. F., T. Herren, A. Redlitz, L. A. Miles, and J. L. Hoover-Plow. 1995. The cell biology of the plasminogen system. FASEB J. 9:939-945. [DOI] [PubMed] [Google Scholar]
- 38.Podbielski, A., B. Spellerberg, M. Woischnik, B. Pohl, and R. Lutticken. 1996. Novel series of plasmid vectors for gene inactivation and expression analysis in group A streptococci (GAS). Gene 177:137-147. [DOI] [PubMed] [Google Scholar]
- 39.Ringdahl, U., and U. Sjobring. 2000. Analysis of plasminogen-binding M proteins of Streptococcus pyogenes. Methods 21:143-150. [DOI] [PubMed] [Google Scholar]
- 40.Ringdahl, U., M. Svensson, A. C. Wistedt, T. Rennes, R. Kellner, W. Muller-Esterl, and U. Sjobring. 1998. Molecular co-operation between protein PAM and streptokinase for plasmin acquisition by Streptococcus pyogenes. J. Biol. Chem. 273:6424-6430. [DOI] [PubMed] [Google Scholar]
- 41.Sambrook, J., and D. W. Russell. 2001. Molecular cloning: a laboratory manual, 3rd ed. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, N.Y.
- 42.Serrano, R. L., P. Rodriguez, S. V. Pizzo, and M. Gonzalez-Gronw. 1996. ATP-regulated activity of the plasmin-streptokinase complex: a novel mechanism involving phosphorylation of streptokinase. Biochem. J. 313:171-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Simon, D., and J. J. Ferretti. 1991. Electrotransformation of Streptococcus pyogenes with plasmid and linear DNA. FEMS Microbiol. Lett. 82:219-224. [DOI] [PubMed] [Google Scholar]
- 44.Smoot, J. C., K. D. Barbian, J. J. Van Gompel, L. M. Smoot, M. S. Chaussee, G. L. Sylva, D. E. Sturdevant, S. M. Ricklefs, S. F. Porcella, L. D. Parkins, S. B. Beres, D. S. Campbell, T. M. Smith, Q. Zhang, V. Kapur, J. A. Daly, L. G. Veasey, and J. M. Musser. 2002. Genome sequence and comparative microarray analysis of serotype M18 group A Streptococcus strains associated with acute rheumatic fever outbreaks. Proc. Natl. Acad. Sci. USA 99:4668-4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Svensson, M. D., U. Sjobring, and D. E. Bessen. 1999. Selective distribution of a high-affinity plasminogen-binding site among group A streptococci associated with impetigo. Infect. Immun. 67:3915-3920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Svensson, M. D., U. Sjobring, F. Luo, and D. Bessen. 2002. Roles of the plasminogen activator streptokinase and the plasminogen-associated M protein in an experimental model for streptococcal impetigo. Microbiology 148:3933-3945. [DOI] [PubMed] [Google Scholar]
- 47.Virkola, R., K. Lahteenmaki, T. Eberhard, P. Kuusela, L. van Alphen, M. Ullberg, and T. K. Korhonen. 1996. Interaction of Haemophilus influenzae with the mammalian extracellular matrix. J. Infect. Dis. 173:1137-1147. [DOI] [PubMed] [Google Scholar]
- 48.Winram, S. B., and R. Lottenberg. 1996. The plasmin-binding protein Plr of group A streptococci is identified as glyceraldehyde-3-phosphate dehydrogenase. Microbiology 142:2311-2320. [DOI] [PubMed] [Google Scholar]
- 49.Winram, S. B., and R. Lottenberg. 1998. Site-directed mutagenesis of streptococcal plasmin receptor protein (Plr) identifies the C-terminal Lys334 as essential for plasmin binding, but mutation of the plr gene does not reduce plasmin binding to group A streptococci. Microbiology 144:2025-2035. [DOI] [PubMed] [Google Scholar]
- 50.Winram, S. B., L. C. Richardson, and R. Lottenberg. 1995. Mutational analysis of a plasmin receptor protein expressed by group A streptococci, p. 199-202. In J. J. Ferretti, M. S. Gilmore, T. R. Klaenhammer, and F. Brown (ed.), Genetics of streptococci, enterococci and lactococci. Karger, Basel, Switzerland. [PubMed]